Abstract
Nuclear hormone receptors (NRs) are evolutionarily conserved ligand-dependent transcription factors. They are essential for human life, mediating the actions of lipophilic molecules, such as steroid hormones and metabolites of fatty acid, cholesterol, and external toxic compounds. The C2H2-type zinc finger proteins (ZNFs) form the largest family of the transcription factors in humans and are characterized by multiple, tandemly arranged zinc fingers. Many of the C2H2-type ZNFs are conserved throughout evolution, suggesting their involvement in preserved biological activities, such as general transcriptional regulation and development/differentiation of organs/tissues observed in the early embryonic phase. However, some C2H2-type ZNFs, such as those with the Krüppel-associated box (KRAB) domain, appeared relatively late in evolution and have significantly increased family members in mammals including humans, possibly modulating their complicated transcriptional network and/or supporting the morphological development/functions specific to them. Such evolutional characteristics of the C2H2-type ZNFs indicate that these molecules influence the NR functions conserved through evolution, whereas some also adjust them to meet with specific needs of higher organisms. We review the interaction between NRs and C2H2-type ZNFs by focusing on some of the latter molecules.
Keywords: Broad-Complex, Tramtrack, and Bric-a-brac (BTB)/poxvirus and zinc finger (POZ), coregulator, evolution, Krüppel-associated box (KRAB), noncoding RNA, SCAN
Abbreviations
AF, activation function; APL, acute promyelocytic leukemia; AR, androgen receptor; Bcl2, B-cell lymphoma 2; BTB, Broad-Complex, Tramtrack, and Bric-a-brac; CAR, constitutive androstane receptor; CBP, CREB-binding protein; CHD8, chromodomain helicase DNA-binding protein 8; CLAMP, chromatin-linked adaptor for MSL protein; CNS, central nervous system; CoCoA, coiled-coil coactivator; COUP-TF, ovalbumin upstream promoter-transcription factor; CtBP, C-terminal tail-binding protein; CTCF, CCCTC-binding protein; DAX-1, dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1; DBD, DNA-binding domain; DRIP, VDR-interacting protein; EAR2, V-erbA-related protein 2; E2F1, E2F transcription factor 1; ER, estrogen receptor; FXR, farnesoid X receptor; Gas5, growth arrest-specific 5; GCNF, germ cell nuclear factor; GIOT-1, gonadotropin-inducible ovarian transcription factor-1; GR, glucocorticoid receptor; HAT, histone acetyltransferase; HDAC, histone deacetylase; HNF4, hepatocyte nuclear factor 4; HP1, heterochromatin protein 1; HTLV-1, human T-cell leukemia virus type-1; HUB1, HTLV-1 U5RE-binding protein 1; KAP-1, KRAB-associated protein-1; KLF, Krüppel-like factor; KRAB, Krüppel-associated box; KRIP1, KRAB-A-interacting protein 1; LBD, ligand-binding domain; lincRNA, long intergenic ncRNA; lncRNA, long ncRNA; LXR, liver X receptor; miRNA, microRNA; MR, mineralocorticoid receptor; MSL, male-specific lethal; nc, protein noncoding; NCoA, nuclear receptor coactivator; NCoR, nuclear receptor corepressor; NOR1, neuron-derived orphan receptor 1; NRs, nuclear hormone receptors; NTD, N-terminal domain; NUR77, nerve growth factor 1B; NURR1, nuclear receptor related 1; Paris, parkin-interacting substrate; p/CAF, p300/CBP-associated factor; PCG-1α, PPARγ coactivator-1α; PCGEM1, prostate-specific transcript 1; PLZF, promyelocytic leukemia zinc finger protein; PNR, photo-specific nuclear receptor; POZ, poxvirus and zinc finger; PPAR, peroxisome proliferator-activated receptor; PR, progesterone receptor; PRC, polycomb-repressive complex; PRDM, PR domain zinc finger protein; PRNCR1, prostate cancer associated noncoding RNA 1p; PXR, pregnane X receptor; RAR, retinoic acid receptor; ROR, RAR-related orphan receptor; RXR, retinoid X receptor; SETDB1, histone H3K9 methyltransferase SET domain bifurcated 1; SF-1, steroidogenic factor-1; SHP, small heterodimer partner; SMRT, silencing mediator for retinoid and thyroid hormone receptors; SP, specificity protein; SR, steroid hormone receptor; SRA, steroid receptor RNA activator; STAT5, signal transducer and activator of transcription 5; TAD, topologically associated domain; TFIIIA, transcription factor IIIA; TFIIIC, transcription factor IIIC; TFIID, transcription factor IID; TIF1, transcription intermediary factor 1; TLX or TLL, timeless receptor; TR, thyroid hormone receptor; TRAP, TR-associated protein; TRIM, tripartite motif protein; TRE, TR response element; U5RE, U5 repressive element; VDR, vitamin D receptor; WT1, Wilms tumor 1; YY1, Yin-Yang 1; ZER6, zinc finger-estrogen receptor interaction, clone 6; ZF, zinc finger; ZNF, zinc finger protein.
Introduction
Gene transcription is essential for maintaining life of organisms through expression of the effector component proteins and influences every aspect of their activities. However, skewed gene expression leads to the development of various disorders in humans, such as metabolic abnormalities, infection, inflammation, and cancer. Thus, higher organisms have developed a sophisticated regulatory system of gene transcription to control the expression of encoding proteins in appropriate places (organs/tissues) at the right timing, to create their complex body components, and to maintain their appropriate functionality.1 This is also evident by the fact that higher organisms have expanded their genome content, particularly the nonprotein-coding genome area, which harbors plenty of regulatory elements, such as promoters, enhancers, and insulators (98.5% of the entire genome in humans), in contrast to the area and the number of the protein-coding genes, which remained unchanged from lower to higher organisms (~20 000-30 000 genes).2 Thus, expansion of the noncoding genomic area provides a more complex and sophisticated regulation on the expression of a limited number of coding genes. Interestingly, a substantial part of the nonprotein-coding genome area expresses numerous numbers and different forms of nonprotein-coding (nc) RNAs, which provides an additional layer of the regulatory mechanisms on gene transcription and protein function.3,4
In addition to genome regulatory elements and abundant ncRNAs for organizing gene transcription, transcription factors are the major protein components that translate cellular internal/external changes to gene transcription.5 Indeed, small changes in their expression/activation can result in dramatic yet concordant alteration in the entire functional network in organisms.5 They usually contain the structural domain(s) for binding to DNA and the effector components attracting the transcriptional regulatory machinery.6 Transcription factors include 2 large families, nuclear hormone receptors (NRs) and C2H2-type zinc finger proteins (ZNFs).7 The former proteins are evolutionarily conserved and function as ligand-dependent transcription factors by mediating the biological actions of small lipophilic compounds.7 On the contrary, the latter molecules demonstrate characteristic expansion of their member proteins in higher organisms and act as transcription factors and/or transcriptional regulators/cofactors through their multiple zinc fingers (ZFs) and other distinct functional modules.8 Involved in diverse and important biological functions as well as exposed to the strong evolutional pressure, NRs and C2H2-type ZNFs have developed mutual and multilayered interactions to coordinate each other’s activities. In this review, we will discuss these 2 families and highlight their roles and mutual interaction from an evolutional point of view.
Nuclear Hormone Receptors
NRs are transcription factors, the activity of which is dependent on their association with their cognate ligands.7 NRs are essential for organisms to support their numerous activities, including development, reproduction, energy metabolism, neural, musculoskeletal, gastrointestinal, and immune functions.7,9 Upon binding to ligand, NRs, either by translocating into the nucleus or by constitutively residing in this subcellular component, interact physically with their specific DNA sequences called response elements.10 DNA-bound NRs then change the transcription rates of their responsive genes by communicating with many co-regulatory molecules.11
The NR family consists of more than 200 members in general and more than 40 in mammals (48 in humans) currently cloned and characterized across species.7,12 Their encoding genes are categorized into 7 major subfamilies from NR0 to NR6, depending on their phylogenetic proximity13 (Table 1). NRs have common structural features: the N-terminal domain (NTD), a middle DNA-binding domain (DBD), and the C-terminal ligand-binding domain (LBD)12,14 (Figure 1). There is a short sequence called hinge region (HR) between DBD and LBD. The NTD of NRs is the most variable domain, presumably mediating receptor-specific transcriptional activity through the transactivation domain(s) (activation function-1 [AF-1]) residing in this domain.15 In contrast, the DBD is highly conserved among NRs and consists of 2 C4-type ZFs through which it directly binds receptor-specific response elements.12 These sequences are mostly present in tandem, as many of NRs bind these sequences as a homo- or a heterodimer.16 The LBD has a ligand-binding pocket and a transactivation domain (AF-2), which is created through ligand-dependent conformational changes inside this domain.17 This second transactivation domain interacts with the histone acetyltransferase (HAT) coactivators, including the CREB-binding protein (CBP)/p300 and the nuclear receptor coactivators (NCoAs), through the latter’s LxxLL coactivator motifs, and stimulates NR’s ligand-dependent transcriptional activity by cooperating with the first transactivation domain located in NTD. In addition to HAT coactivators, NRs interact with numerous cofactor molecules, including mediator/TR-associated protein (TRAP)/VDR-interacting protein (DRIP) and switch/ sucrose non-fermenting (SWI/SNF) protein complexes, and each of their component proteins appears to influence the transcriptional activity of a specific fraction of the NR-responsive genes.18
Table 1.
NR Family Proteins.
| Protein name | Gene name | Typea | Ligands | Biological actions | Reference |
|---|---|---|---|---|---|
| TRα | NR1A1 | 2 | Thyroid hormones | Energy metabolism, CNS development and function | Cheng et al19 |
| TRβ | NR1A2 | 2 | Thyroid hormones | Energy metabolism, CNS development and function | Cheng et al19 |
| RARα | NR1B1 | 2 | All-trans retinoic acid, 9-cis retinoic acid | Brain and other organ development | Giguere20 |
| RARβ | NR1B2 | 2 | All-trans retinoic acid, 9-cis retinoic acid | Brain and other organ development | Giguere20 |
| RARγ | NR1B3 | 2 | All-trans retinoic acid, 9-cis retinoic acid | Brain and other organ development | Giguere20 |
| PPARα | NR1C1 | 1 | Fatty acids, leukotriene B4, fibrates | Lipid and energy metabolism | Berger and Moller21 |
| PPARβ/δ | NR1C2 | 1 | Fatty acids | Lipid and energy metabolism | Berger and Moller21 |
| PPARγ | NR1C3 | 1 | Fatty acids, prostaglandin J2 | Lipid and energy metabolism | Knouff and Auwerx22 |
| Rev-erbα | NR1D1 | 3 | Heme (structural) | Circadian rhythm | Yin et al23 |
| Rev-erbβ | NR1D2 | 3 | Heme (structural) | Lipid and energy metabolism | Yin et al23 |
| RORα | NR1F1 | 1 | Cholesterol, cholesteryl sulfate | Brain and immune development, lipid and bone metabolism | Zhang et al24 |
| RORβ | NR1F2 | 1 | Retinoic acid | Brain function? | Zhang et al24 |
| RORγ | NR1F3 | 1 | Desmosterol, zymosterol | Immune response (T-helper 17) | Zhang et al24 |
| LXRα | NR1H3 | 1 | Oxysterol | Cholesterol and fatty acid metabolism | Jakobsson et al25 |
| LXRβ | NR1H2 | 1 | Oxysterol | Cholesterol and fatty acid metabolism | Jakobsson et al25 |
| FXRα | NR1H4 | 1 | Bile acids | Bile acid synthesis and cholesterol metabolism | Chiang26 |
| FXRβb | NR1H5 | 1 | Lanosterol | Cholesterol metabolism | Chiang26 |
| VDR | NR1I1 | 2 | 1.25-dihydroxyvitamin D | Bone and calcium metabolism | Jurutka27 |
| PXR | NR1I2 | 1 | Xenobiotics | Xenobitoics | Kliewer28 |
| CAR | NR1I3 | 1 | Xenobiotics | Xenobiotics | Willson and Kliewer29 |
| HNF4α | NR2A1 | 3 | Fatty acids (Linoleic acid?) | Glucose and lipid metabolism | Sladek and Giguere30 |
| HNF4γ | NR2A2 | 3 | Fatty acids (Linoleic acid?) | Glucose and lipid metabolism | Sladek and Giguere30 |
| RXRα | NR2B1 | 1 | 9cis-retinoic acid | Hetro-dimerize with other NRs | Giguere31 |
| RXRβ | NR2B2 | 1 | 9cis-retinoic acid | Hetro-dimerize with other NRs | Giguere31 |
| RXRγ | NR2B3 | 1 | 9cis-retinoic acid | Hetro-dimerize with other NRs | Giguere31 |
| TR2 | NR2C1 | 3 | Regulation of sex steroid receptor and PPAR activity | Lee et al32 | |
| TR4 | NR2C2 | 3 | Cellular differentiation/homeostasis, oxidative stress | Lee et al32 | |
| TLX | NR2E1 | 3 | Neural and retinal development, brain function | Benod et al,33 Wang et al34 | |
| PNR | NR2E3 | 3 | Embryonic/retinal development | Kobayashi et al35 | |
| COUP-TFI | NR2F1 | 3 | CNS development and function | Tsai and Tsai36 | |
| COUP-TFII | NR2F2 | 3 | CNS development and function | Tsai and Tsai36 | |
| EAR2 | NR2F6 | 3 | Brain function and hematopoiesis | Zhu et al37 | |
| ERα | NR3A1 | 2 | Estradiol | Reproduction and female body composition | Couse and Korach38 |
| ERβ | NR3A2 | 2 | Estradiol | Reproduction and female body composition | Koehler et al39 |
| ERRα | NR3B1 | 3 | Regulation of estrogen actions | Horard and Vanacker40 | |
| ERRβ | NR3B2 | 2 | Diethylstilbestrol | Regulation of estrogen actions | Horard and Vanacker40 |
| ERRγ | NR3B3 | 2 | Diethylstilbestrol | Regulation of estrogen actions | Horard and Vanacker40 |
| GR | NR3C1 | 2 | Glucocorticoids | Stress response and immune regulation | Kino12 |
| MR | NR3C2 | 2 | Mineralocorticoids | Water and electrolyte homeostasis | Funder41 |
| PR | NR3C3 | 2 | Progestins | Reproduction and female body composition | Graham and Clarke42 |
| AR | NR3C4 | 2 | Androgens | Reproduction and male body composition | Heinlein and Chang,43 Quigley et al44 |
| NUR77 | NR4A1 | 3 | Brain function, steroidogenesis, and immune activity | Bassett and White,45 Hsu et al,46 Ranhotra47 | |
| NURR1 | NR4A2 | 3 | Brain function (dopaminergic system) and immune activity | McMorrow and Murphy,48 Perlmann et al49 | |
| NOR1 | NR4A3 | 3 | Immune regulation and hematopoiesis | McMorrow and Murphy,48 Perlmann et al,49 Mullican et al50 | |
| SF-1 | NR5A1 | 3 | Phosphoinositol (structural) | Sexual development, reproduction, and steroidogenesis | Ozisik et al51 |
| LRH-1 | NR5A2 | 3 | Cell proliferation and immune regulation | Stein and Schoonjans52 | |
| GCNF | NR6A1 | 3 | Embryogenesis and germ cell differentiation | Zechel53 | |
| DAX-1 | NR0B1 | 3 | Development of steroid-producing/regulating organs | El-Khairi et al54 | |
| SHP | NR0B2 | 3 | Inhibition of other NR activity, intermediary metabolism | Zhang et al55 |
Note. NR = nuclear hormone receptor; TR = thyroid hormone receptor; CNS = central nervous system; RAR = retinoic acid receptor; PPAR = peroxisome proliferator-activated receptor; ROR = RAR-related orphan receptor; LXR = liver X receptor; FXR = farnesoid X receptor; VDR = vitamin D receptor; PXR = pregnane X receptor; CAR = constitutive androstane receptor; HNF = hepatocyte nuclear factor; RXR = retinoid X receptor; TLX = Timeless receptor; PNR: photo-specific nuclear receptor; COUP-TF = chicken ovalbumin upstream promoter-transcription factors; EAR = V-erbA-related protein; GR = glucocorticoid receptor; MR = mineralocorticoid receptor; PR = progesterone receptor; NUR77 = nerve growth factor 1B; NURR1 = Nuclear receptor related1; NOR1 = neuron-derived orphan receptor1; SF-1 = steroidogenic factor -1; GCNF = germ cell nuclear factor; DAX-1 = dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1; LRH-1 = liver receptor homolog-1; SHP = small heterodimer partner.
1: metabolic sensor (adopted orphan) receptor, 2: endocrine receptor, 3: orphan receptor.
Figure 1.
Linearized structure of the representative NRs and C2H2-type ZNFs, and their functional domains.
Note. Linearized molecules of the human NRs (RARα, RXRα, GRα, NOR1, SF-1, GCNF, and DAX-1 with gene names in parentheses), C2H2-type ZNFs (poly-ZNFs: KLF9 and CTCF, BTB/POZ-ZNF: PLZF and KRAB-ZNF: ZNF764), and the PLZF-RARα fusion protein are shown. Their functional domains are indicated with different colors. C2H2-type ZFs are shown in pink. DAX-1 does not have NTD and DBD, but is composed of a repetitive domain, which contains 3 LxxLL-like motifs used for interacting with selected NRs.57 PLZF-RARα is a fusion protein found in the acute promyelocytic leukemia with t(11;17)(q23;q21) translocation. This fusion protein consists of the N-terminal part of PLZF containing BTB-POZ domain and 2 ZFs, and the C-terminal part of RARα harboring DBD and LBD.58 Black triangle indicates the junction between PLZF and RARα protein sequences. Distribution of functional domains is based on Ensembl (http://asia.ensembl.org/index.html). NRs = nuclear hormone receptors; ZNFs = zinc finger proteins; RAR = retinoic acid receptor; NTD = N-terminal domain; DBD = DNA-binding domain; HR = hinge region; LBD = ligand-binding domain; KLF = Krüppel-like factor; RXR = retinoid X receptor; CTCF = CCCTC-binding protein; GR = glucocorticoid receptor; PLZF = promyelocytic leukemia zinc finger protein; BTB/POZ = Broad-Complex, Tramtrack, and Bric-a-brac/poxvirus and zinc finger; NOR = neuron-derived orphan receptor; KRAB = Krüppel-associated box; SF -1 = steroidogenic factor -1; GCNF = germ cell nuclear factor; DAX-1 = dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1.
NRs are empirically and functionally categorized into 3 major groups: (1) metabolic sensor (or adopted orphan) receptors, (2) endocrine receptors, and (3) orphan receptors.7 NRs of the first group, including peroxisome proliferator-activated receptors (PPARs), liver X receptors (LXRs), pregnane X receptor (PXR), and constitutive androstane receptor (CAR), act as sensors for lipid metabolites, such as fatty acids, prostanoids, oxysterols, bile acids, dietary lipids, and xenobiotics, by binding to these compounds as ligands with relatively low affinity.7 NRs in the second group (endocrine receptors), including steroid hormone receptors (SRs) (glucocorticoid receptor [GR], mineralocorticoid receptor [MR], androgen receptor [AR], progesterone receptor [PR] and estrogen receptors [ERs]), thyroid hormone receptors (TRs), retinoic acid receptors (RARs), and the vitamin D receptor (VDR), act as receptors for steroid hormones (glucocorticoids, mineralocorticoids, androgens, progestins, and estrogens), retinoid acids, thyroid hormones, and vitamin D, with high affinity interaction to these compounds.7 The last group, orphan receptors, including Nerve growth factor IB (NUR77), chicken ovalbumin upstream promoter-transcription factors (COUP-TFs), hepatocyte nuclear factor 4s (HNF4s), and dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 (DAX-1), do not regularly have known ligands.7 However, some of these receptors (eg, NHF4s and steroidogenic factor-1 [SF-1]) can constitutively bind “structural” ligands, which function as components of their LBDs.59-61
NRs, particularly SRs, are among the molecules most well examined phylogenetically.56,62 Some NRs are present even in simple metazoans with multiple cellular components, whereas substantial numbers of NRs have been reported for metazoans like Caenorhabditis elegans.56 The ancestral SR first appeared in primitive vertebrate lamprey, and all SRs can be found in ray-finned fish.62 Therefore, NRs are observed from the early evolutional period particularly before and around the time of vertebrate emergence and, thus, their functionality is relatively conserved.
NRs have a major impact on diverse regulatory activities in humans (Figure 2 and Table 1). For example, a substantial number of NRs (TRα and β, timeless receptor [TLX or TLL], COUP-TFs, RARs, RAR-related orphan receptor [RORs], V-erbA-related protein 2 [EAR2], NUR77, nuclear receptor related 1 [NURR1], neuron-derived orphan receptor 1 [NOR1], and germ cell nuclear factor [GCNF]) play critical roles in the embryonic development, especially in the development/differentiation of the central nervous system (CNS).63-67 Another biological actions of NRs (such as by TRs, COUP-TFs, PPARs, Rev-erbβ, RORα, LXRs, farnesoid X receptor [FXR], VDR, HNF4s, GR, MR, and small heterodimer partner [SHP]) lie in the regulation of energy, intermediary and electrolyte metabolism, such as for glucose, cholesterol, fatty acids, sodium/potassium, and calcium, consistent with the fact that many of the NRs employ metabolites of these biological pathways as their ligands.68-70 Reproduction and development/maintenance of reproductive organs are also important biological activities regulated by NRs, such as ERs, estrogen-related receptors (ERRs), PR, AR, NUR77, SF-1, and DAX-1.71,72 Furthermore, most of the NRs, including PPARs, RORs, LXRs, VDR, ERs, GR, PR, NUR77, NURR, NOR1, and RORs, have immune regulatory activities, whereas some others play roles in hematopoiesis.73,74 Rev-erbs and RORs are important components of the circadian CLOCK system that synchronizes the body’s activities to diurnal day-night changes caused by rotation of the earth.75 PXR and CAR play a central role in the xenobiotic metabolism by binding toxic metabolites/exogenous compounds as their ligands and by regulating the expression of their metabolizing enzymes including cytochrome P450-related reductases.29
Figure 2.
NRs have diverse regulatory actions on human activities.
Note. NRs virtually influence every aspect of human activities, including embryonic development, energy metabolism, immunity, reproduction, electrolyte/bone/skeletal muscle maintenance, circadian rhythm, and xenobiotics through their various members. Examples of the NRs that have regulatory roles in the indicated biological activities are shown. NRs = nuclear hormone receptors; TRs = thyroid hormone receptors; TLX = timeless receptor; COUP-TFs = ovalbumin upstream promoter-transcription factors; RARs = retinoic acid receptors; RORs = RAR-related orphan receptors; EAR2 = V-erbA-related protein 2; NURR1 = nuclear receptor related 1; NOR 1 = neuron-derived orphan receptor 1; GCNF = germ cell nuclear factor; CNS = central nervous system; GR = glucocorticoid receptor; PPAR = peroxisome proliferator-activated receptor; LXR = liver X receptor; FXR = farnesoid x receptor; HNF4 = hepatocyte nuclear factors 4; NUR77 = nerve growth factor 1B; SHP = small heterodimer partner; MR = mineralocorticoid receptor; VDR = vitamin D receptor; ER = estrogen receptor; ERR = estrogen-related receptors; AR = androgen receptor; PXR = pregnane X receptor; CAR = constitutive androstane receptor; PR = progesterone receptor; SF-1 = steroidogenic factor-1; DAX-1 = dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1.
C2H2-Type ZNFs
C2H2-type ZNFs are another major group of transcription factors. Zinc fingers are small peptide domains with a secondary structure stabilized by a zinc ion bound to the cysteine and/or histidine residues of the finger.76 Different combinations of these 2 residues lead to the development of different classes of ZFs, such as C2H2, C2HC, C2C2 (C4: eg, NRs), and C2C2C2C2 fingers. Among the combinations of ZFs, the C2H2-type ZF motif is known as the “classic ZF.”76 In humans, more than 700 proteins have this motif, constituting the largest class of putative human transcription factors.77 The C2H2-type ZF comprises up to 30 amino acids in the consensus sequence CX2-4CX12HX2-8H (X refers to any amino acid), which forms 1 α-helix and 2 β-sheets, respectively, in the C- and N-terminal portion.78-80 These secondary structures of the peptide fold into a stable assembly through hydrophobic interactions and enclosure of a zinc ion by 2 conserved cysteine and 2 conserved histidine residues.81,82 Multiple ZFs are usually present in tandem and are connected by linkers with conserved amino acid sequences.83 C2H2-type ZNFs function primarily as DNA-binding proteins by forming various modes of contacts to target DNA double helices with their ZFs, as shown in the interaction of the transcription factor IIIA (TFIIIA) with DNA (Figure 3). In addition to the primary role as DNA-binding factors, some C2H2-type ZNFs use their ZFs for interacting with other proteins or double-stranded RNAs, which may be important for their communication with other proteins/RNAs also attracted to the multimolecule transcriptional complex formed on DNA.83-85
Figure 3.
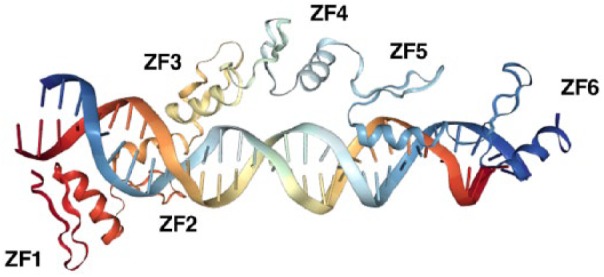
Binding of C2H2-type ZNF to DNA.
Note. Crystallographic structure of the xenopus transcription factor IIIA (TFIIIA) ZFs and the target DNA (5S ribosomal RNA gene internal control region) (PDB: 1SF6) is shown.86 TFIIIIA has 9 C2H2-type ZFs and its N-terminally located 6 ZFs (ZF1 to ZF6) are demonstrated. ZF1 to ZF3 are positioned relatively tightly to the major groove of DNA, whereas ZF4 to ZF6 face only one side of the DNA double helix and form a loose extended association to DNA in which only ZF5 makes a direct contact. Thus, ZFs of TFIIIA differentially contribute to the TFIIIA-DNA interaction by wrapping around its target sequences. ZNF = zinc finger protein; ZF = zinc finger.
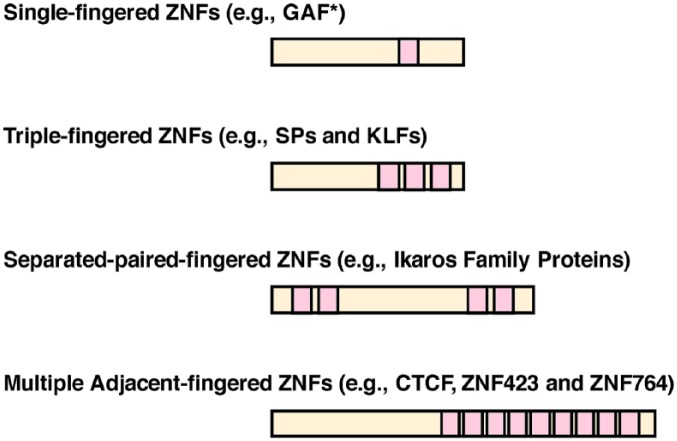
Based on the number, localization, and arrangement of ZFs within a molecule, C2H2-type ZNFs can be divided into 4 classes: (1) single-fingered, (2) triple-fingered, (3) separated-paired-fingered, and (4) multiple adjacent-fingered, and this classification may explain to some extent the functions of the proteins in the respective class87 (Figure 4). C2H2-ZNFs that interact with NRs are found among triple-fingered or multiple adjacent-fingered ZNFs. Besides ZFs, C2H2-type ZNFs harbor additional structural modules, such as the Broad-Complex, Tramtrack, and Bric-a-brac (BTB)/poxvirus and zinc finger (POZ), Krüppel-associated box (KRAB), and SCAN domains.8 These domains are usually located in the N-terminal portion and function as platforms for protein-protein interactions, whereas ZFs are positioned in the C-terminal area8,88 (Figure 4). The BTB/POZ and KRAB domains have transcriptional regulatory activity (mostly repressive), while the SCAN domain does not.8 Among human C2H2-type ZNFs, about 7% have a BTB/POZ domain (BTB/POZ-ZNFs), 43% harbor a KRAB domain (KRAB-ZNFs), and 7% contain a SCAN domain (SCAN-ZNFs).77 Sixty-seven percent of the human C2H2-type ZNFs have only ZFs without any of these domains (thus, they are “poly-ZNFs”).77 Some C2H2-type ZNFs contain multiple of the same or of different domains, whereas the SCAN domain appears as a sole domain in one molecule.88
Figure 4.
Subtypes of C2H2-type ZNFs based on the number and arrangement of their ZFs.
Note. C2H2-type ZNFs can be categorized into 4 subtypes (single-fingered, triple-fingered, separated-paired-fingered, and multiple adjacent-fingered ZNFs) based on the number and arrangement of their ZFs. Schematic protein organization of these C2H2-type ZNF subtypes are shown. C2H2-type ZFs are shown in pink. *: GAF (GAGA factor) is a drosophila protein.89 ZNF = zinc finger protein; ZFs = zinc fingers; SPs = specificity proteins; KLFs = Krüppel-like factors; CTCF = CCCTC-binding protein.
Evolution of C2H2-Type ZNFs
The C2H2-type ZNFs are present throughout the organisms from yeasts to humans. They have expanded their member proteins following evolution, becoming the largest transcription factor family in humans, in contrast to the C4-type ZNFs (mostly NRs), the numbers of which are similar in vertebrates but show a characteristic increase in Nematodes77,90 (Table 2). The number of C2H2-type ZNFs particularly increases in vertebrates, eg, becoming almost double in humans compared with Takifugu fish or to Zebrafish77 (Table 2). BTB/POZ-ZNFs are seen from yeasts to humans, while KRAB-ZNFs and SCAN-ZNFs appear in vertebrates and mammals, respectively.77 The number of KRAB-ZNFs and SCAN-ZNFs increase significantly in higher organisms with the former demonstrating this tendency more obviously77 (Table 3). Thus, the addition of these domains to C2H2-type ZNFs, particularly the KRAB domain, appears to be required for the functions specific to higher organisms, such as the complex transcriptional regulation of genes that enables development, differentiation, and/or function of their complicated organs and tissues. As poly-ZNFs and BTB/POZ-ZNFs are observed early in evolution, they may support more fundamental and/or conserved functions of the organisms, such as general transcriptional regulation and early embryonic development shared among different organisms. In contrast, KRAB-ZNFs and SCAN-ZNFs are likely to play roles in the activities specific to higher organisms. For example, the characteristic expression of the KRAB-ZNFs in human brain appears to be required for the transcriptional network that drives human-specific brain functions, such as enhanced cognitive activities and significantly larger brain size, compared with other primates like chimpanzees.91
Table 2.
Number of the C2H2-Type and C4-Type Genes in the Genome of Various Species.
| Organism | Total number of genes | Number of C2H2-type genes | Number of C4-type genes |
|---|---|---|---|
| Human | 23 299 | 712 (3.0%) | 48 (0.21%) |
| Mouse | 24 948 | 573 (2.3%) | 47 (0.19%) |
| Rat | 21 278 | 466 (2.2%) | 47 (0.22%) |
| Zebrafish | 20 062 | 344 (1.7%) | 53 (0.26%) |
| Drosophila | 13 525 | 298 (2.2%) | 21 (0.16%) |
| Anopheles | 14 653 | 296 (2.0%) | 20 (0.14%) |
| Caenorhabditis elegans | 19 564 | 173 (0.9%) | 270 (1.3%) |
| Caenorhabditis briggsae | 11 884 | 115 (0.9%) | 167 (1.4%) |
Note. Modified from Klug and obtained permission for Table use from the Journal.
Table 3.
Number of C2H2-Type ZNFs Through Evolution.
| Species | C2H2-type ZNF | BTB/POZ-ZNF | KRAB-ZNF | SCAN-ZNF |
|---|---|---|---|---|
| Human | 712 | 50 | 304 | 53 |
| Mouse | 583 | 44 | 219 | 38 |
| Cow | 482 | 41 | 106 | 28 |
| Dog | 329 | 41 | 61 | 17 |
| Chicken | 224 | 26 | 33 | 0 |
| Xenopus | 347 | 30 | 21 | 0 |
| Zebrafish | 405 | 46 | 0 | 0 |
| Takifugu fish | 364 | 41 | 0 | 0 |
| Drosophila | 251 | 11 | 0 | 0 |
| Anopheles | 263 | 9 | 0 | 0 |
| Ciona | 103 | 7 | 0 | 0 |
| Caenorhabditis elegans | 108 | 1 | 0 | 0 |
Note. Modified from Emerson and Thomas and obtained permission for Table use from the Journal. ZNF = zinc finger protein; BTB/POZ = Broad-Complex, Tramtrack, and Bric-a-brac/poxvirus and zinc finger; KRAB = Krüppel-associated box.
Regulation of NR Activity by C2H2-Type ZNFs
NRs regulate and influence numerous critical activities of organisms as we explained above,9 and C2H2-type ZNFs are among their important partners. Based on the previous observation for other proteins,92 it appears that C2H2-type ZNFs appeared from lower organisms, such as poly-ZNFs and BTB/POZ-ZNFs, may regulate the actions of NRs, which are fundamental and/or commonly observed in many organisms. On the contrary, the C2H2-type ZNFs found only in higher organisms like KRAB-ZNFs seem to modulate the actions of NRs involved in complex regulatory mechanisms or in adjusting the NR actions for species-specific needs. Although overall actions of C2H2-type ZNFs on NRs are still largely unknown at this moment, the above-indicated tendency can be seen in some of these molecules91,93 (Figure 5). Below, we review the functional interactions between C2H2-type ZNFs and NRs (listed in Table 4), focusing on their physiologic and pathologic importance. NRs functionally and/or physically interact with poly-ZNFs, BTB/POZ-ZNFs, and KRAB-ZNFs, but none of them have been found to cooperate with SCAN-ZNFs to date.
Figure 5.
Gene network formed between NRs and evolutionarily old or new C2H2-type ZNFs.
Note. NR genes are considered as evolutionarily old genes, as most of the family genes appear before and around the time of vertebrate emergence.56 Thus, NRs are well incorporated in the gene network formed between other old genes including those encoding C2H2-type ZNFs and support fundamental functions shared by several organisms.94 On the contrary, newly appeared genes found only in higher organisms, such as KRAB-ZNFs, have less gene communication, but support the NR-related functions important and specific to respective species (eg, brain function in humans). Solid and dotted lines indicate well-established and newly developing gene networks, respectively. NRs = nuclear hormone receptors; ZNFs = zinc finger proteins.
Table 4.
C2H2-Type ZNFs That Interact With NRs.
| Name | Synonym | Zinc fingers |
Interacting NRs | Domain |
References | ||
|---|---|---|---|---|---|---|---|
| Type | Number | BTB/POZ | KRAB | ||||
| SP1 | Triple-fingered | 3 | GR, MR, AR, PPARγ | Kolla and Litwack,95 Kolla et al,96 Suehiro et al97 | |||
| KLFs | Triple-fingered | 3 | ER, PR, GR, SHP, PPARγ | McConnell and Yang,98 Oishi et al,99 Simmen et al100 | |||
| YY1 | Multiple adjacent-fingered | 4 | PXR, VDR, GR | Bookout et al,9 Deng et al,101 Nem et al,102 Raval-Pandya et al103 | |||
| WT1 | Multiple adjacent-fingered | 4 | SF-1, DAX-1, AR, RARα, VDR, PPARβ | Goodyer et al,104 Kim et al,105 Nachtigal et al,106 Reizner et al,107 Zaia et al108 | |||
| Zip67 | ZNF653 | Multiple adjacent-fingered | 5 | SF-1, LXRα, NUR77, GR, ER | Borud et al109 | ||
| Zac1 | LOT1, Zac | Multiple adjacent-fingered | 7 | TRβ, AR, ERα, PPARγ | Barz et al,110 Huang and Stallcup111 | ||
| ZNF217 | ZABC1 | Multiple adjacent-fingered | 7 | ERα | Frietze et al85 | ||
| ZNF536 | Multiple adjacent-fingered | 9 | RARα | Qin et al112 | |||
| CTCF | Multiple adjacent-fingered | 11 | NRs, ERα, TR | Carroll et al,113 Kim et al,114 Ross-Innes et al,115 Awad et al,116 Lutz et al117 | |||
| ZNF366 | Multiple adjacent-fingered | 11 | ERα | Lopez-Garcia118 | |||
| ZNF423 | Roaz, OAZ, Zfp104 | Multiple adjacent-fingered | 30 | RARα, RXRα, PPARγ | Gupta et al,119 Huang et al,120 Ingle et al121 | ||
| Kaiso | ZNF348, ZBTB33 | Triple-fingered | 3 | NRs associating with NCoR | + | Klose and Bird122 | |
| PLZF | ZBTB16 | Multiple adjacent-fingered | 9 | RARs | + | Martin et al,123 Ward et al,124 Wasim et al125 | |
| ZNF746 | PARIS | Multiple adjacent-fingered | 4 | PPARs, NRs associating with PCG-1α | + | Shin et al126 | |
| ZNF282 | HUB1 | Multiple adjacent-fingered | 5 | ERα, GR, THR, AR | + | Wu et al,18 Yu et al127 | |
| ZNF764 | Multiple adjacent-fingered | 7 | AR, GR, MR, TRs | + | Kino et al128 | ||
| ZNF398 | ZER6 | Multiple adjacent-fingered | 9 | ERα | + | Conroy et al130 | |
| ZNF461 | GIOT-1 | Multiple adjacent-fingered | 12 | SF-1, NUR77 | + | Song et al131 | |
Note. ZNF = zinc finger protein; NR = nuclear hormone receptor; BTB/POZ = Broad-Complex, Tramtrack, and Bric-a-brac/poxvirus and zinc finger; KRAB = Krüppel-associated box; SP = specificity protein; GR = glucocorticoid receptor; MR = mineralocorticoid receptor; AR = androgen receptor; PPAR = peroxisome proliferator-activated receptor; KLF = Krüppel-like factor; ER = estrogen receptor; PR = progesterone receptor; SHP = small heterodimer partner; YY 1 = Yin-Yang 1; PXR = pregnane X receptor; VDR = vitamin D receptor; WT1 = Wilms tumor 1; SF -1 = steroidogenic factor -1; DAX-1 = dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1; LOT1 = lost on transformation 1; ZABC = zinc finger amplified in breast cancer; RAR = retinoic acid receptor; LXR = liver X receptor; TR = thyroid hormone receptor; CTCF = CCCTC-binding protein; ZBTB = zinc finger and BTB; NCoR = nuclear receptor corepressor; PLZF = promyelocytic leukemia zinc finger protein; PARIS = parkin-interacting substrate; PCG-1α = PPARγ coactivator-1α; HUB = HTLV-1 U5RE-binding protein; ZER = zinc finger-estrogen receptor interaction; GIOT = gonadotropin-inducible ovarian transcription factor.
Poly-ZNFs
Poly-ZNFs that do not contain BTB/POZ, KRAB, or SCAN domain are the oldest members of the C2H2-type ZNF family.77 Most of these proteins are evolutionarily conserved.98,132,133 Many of the triple-fingered and some of the multiple adjacent-fingered poly-ZNFs function as classic transcription factors, while others act as transcriptional modulators incorporated in the transcriptional regulatory complex formed on other DNA-binding factors. Representatives of the triple-fingered poly-ZNFs interacting with NRs are the specificity protein (SP)/Krüppel-like factor (KLF) family transcription factors, while those of the multiple adjacent-fingered poly-ZNFs also influencing the NR activities, include the Yin-Yang 1 (YY1) and Wilms tumor 1 (WT1) transcription factors, and the CCCTC-binding protein (CTCF) transcriptional regulator.
SP/KLF Transcription Factors
This family consists of 2 subgroups, SP1- and KLF-related proteins.133 The former group is composed of 9 members (SP1 to SP9) in humans, while the latter has 17 (KLF1 to KLF17).98 Their orthologs are present across species from drosophila to humans.98 These ZNFs have 3 C2H2-type ZFs in their C-terminal portion98 (Figure 1). SP proteins contain the Bottomhead box (BTD) just N-terminally to their ZFs, whereas KLFs do not have this motif.133,134 Both SP- and KLF-related proteins bind the DNA sequences called GC and GR boxes (5’-GGGGCGGGG-3’ and 5’-GGTGTGGGG-3’, respectively) frequently found in CpG islands associated with the promoter of downstream coding sequences.133 SP1, a representative of the SP family proteins, is essential for the transcription of TATA-less genes, facilitating the formation of the transcription initiation complex on their promoters through interaction with TFIID, which is a multiprotein complex including the TATA-binding protein and plays a central role in the RNA polymerase II–mediated transcription.135 Due to its interaction with TFIID and binding to CpG islands, SP1 is required for the transcription of a substantial number of protein-coding genes, obviously functioning as a component of the general transcriptional complex. In contrast, KLF family proteins function as “regular” transcription factors, by binding to the response elements located in the promoter or enhancer regions and by stimulating the transcription of downstream protein-coding genes.98 Knockout mice for the SP/KLF family member genes generally demonstrate early embryonic/perinatal death or severe functional defects in various organs.98 Thus, these poly-ZNFs play important roles in the general transcriptional regulation and/or in the organ development/differentiation observed in the early embryonic phase shared by many organisms. Characteristically, SP/KLF transcription factors have 3 major modes of action on NRs136 (Figure 6). First, SPs/KLFs and NRs mutually influence each other’s transcriptional activity through either direct or indirect interaction at their regulating genes. Second, SPs/KLFs modulate the expression of NRs, and augment/repress the transcriptional activity of NRs. Third, NRs alter the expression of certain SP/KLF family proteins, employing them as mediators of NR activities.
Figure 6.
Regulatory mechanisms of the SP/KLF family transcription factors on NRs.
Note. SP/KLF transcription factors have 3 major modes of actions on NRs: (1) SPs/KLFs influence NR-induced transcriptional activity through either direct or indirect interaction on their responsive genes. SPs/KLFs and NRs may also bind DNA directly or indirectly. (2) SPs/KLFs alter the expression of NRs, and subsequently modulate their transcriptional activity. (3) NRs alter the expression of certain SP/KLF family proteins, employing as mediators of their activities. Some SP/KLFs, the expression of which is regulated by particular NRs, influence the transcriptional activity of such NRs through the mechanism (1), forming a feed-forward transcriptional regulatory loop between them. SP = specificity protein; KLF = Krüppel-like factor; NRs = nuclear hormone receptors.
SP1 is the most well-studied member of the SP subfamily. SP1 can modulate the transcriptional activity of ERα/β, PR, AR, PPARγ, COUP-TFs, and SF-1, through either direct or indirect interaction with these receptors.137 For example, SP1 regulates estrogen-induced cell growth by influencing the transcriptional activity of ERs on the genes encoding c-fos, cyclin D1, B-cell lymphoma 2 (bcl2) and E2F transcription factor 1 (E2F1) and the hormonal activation of the mitogen-activated kinase pathway.137 SP1 is required for AR to repress gonadotropin-stimulated luteinizing hormone receptor expression.138 SP1 enhances AR-induced expression of the p21 cell cycle component protein.139 Furthermore, SP1 is required for PPARγ-mediated repression of the thromboxan gene possibly by tethering this receptor to the promoter site where SP1 binds.140 Finally, SP1 facilitates SF-1-mediated expression of the steroidogenic acute regulatory protein (StAR), a key molecule for cholesterol trafficking in mitochondria and thus for steroidogenesis.141 SP1 and some of these NRs seem to interact physically through the former’s C-terminal domain and latter’s DBD. In addition, SP1 appears to influence the expression of a substantial number of NRs through its ubiquitous DNA-binding sites found in the promoter region of the NR genes.97 This indicates that SP1 regulates the activity of these NRs by changing their expression levels.
KLF family proteins are important transcription factors involved in cell proliferation, differentiation, and induction of apoptosis by regulating the expression of numerous effector molecules, including cyclin D1, cyclin B1, and c-Myc, and are essential for cardiac and vascular development and hematopoiesis in the embryonic phase. 98They are also required for adipogenesis, lymphocyte and monocyte/macrophage differentiation, and maintenance of intestinal epithelial cells after birth.98 KLF proteins influence the transcriptional activity of key NRs involved in these biological pathways, such as PPARγ (KLF5, 2 and 7) for adipogenesis and ERs (KLF9) for breast tissue differentiation and breast cancer proliferation.98,99 KLF4, KLF9, KLF11, and KLF13 regulate hormone-dependent proliferation and differentiation of uterine endometrium and myometrium by influencing directly or indirectly the transcriptional activity of ERs and PR on the E2F transcription factors and the Wnt/β-catenin signaling pathway.98,99 KLF9 and KLF13, respectively, modulate ER- and PR-mediated signaling pathway by stimulating the expression of these receptors.100 KLF11 cooperates with GR for the expression of the monoamine oxidase-A gene, the protein product of which degrades monoamine-related neurotransmitters important for the actions of CNS.142 KLF4 also cooperates with GR for the expression of genes involved in the formation of the skin barrier function in prenatal fetus.98,143 The orphan receptor SHP inhibits the transcriptional activity of KLF6 on the matrix metalloproteinase-9 gene to regulate endothelial cell motility.144
In addition to acting as regulators of the NRs’ transcriptional activity, KLFs function as their downstream mediators, being their encoding genes responsive to NRs. For example, the KLF9 gene promoter contains glucocorticoid response elements (GREs) and induction of KLF9 by GR is important for the survival of the newly differentiated hippocampal granule cells originating from adult neural stem cells (these neuronal cells play important roles in memory consolidation) as well as for the regulation of the inflammatory response in macrophages.136,145,146 Glucocorticoids/GR are known to have strong effects on these activities.147,148 Induction of KLF15 by glucocorticoids is important for the expression of enzymes that catalyze branched-chain amino acids and for the proteasomal degradation of cellular proteins, which ultimately contribute to the development of muscle wasting observed with chronic glucocorticoid excess.136 In colon cancer, PPARγ stimulates the expression of KLF10, which underlies the anti-cancer effect of this receptor.136 As many of the above-indicated KLFs, the expression of which is regulated by NRs, also function as regulators for the transcriptional activity of corresponding NRs, they form a feed-forward transcriptional regulatory loop in the signaling pathways functional for NRs (Figure 6).
YY1 and WT1
YY1 is an ubiquitously expressed transcription factor containing 4 C2H2-type ZFs in its C-terminal portion.149 As the name “Yin-Yang” indicates, YY1 regulates the transcription of its responsive genes positively or negatively.149 YY1 influences cell proliferation and differentiation, and is essential for the embryonic development.149 In addition, YY1 contributes to cancer biology. Its aberrant expression is found in various cancers, and it modulates the expression of genes involved in cancer development and progression.149 YY1 physically interacts with AR and regulates the latter’s transcriptional activity on the prostate-specific antigen gene, which is an indicator of prostate cancer progression.101 YY1 cooperates with PXR for the regulation of the cytochrome P450 enzymes involved in xenobiotic metabolism, such as CYP3A4 and CYP3A5.102 YY1 represses the transcriptional activity of VDR on vitamin D-responsive genes in osteoblastic cells.149 In renal cells, it represses VDR-mediated transcription of the 24(OH) gene whose encoding enzyme inactivates vitamin D (25-dihydroxyvitamin D3).103 YY1, GR and the signal transducer and activator of transcription 5 (STAT5) cooperate with each other in stimulating the transcription of the growth hormone gene by forming a transcription factor complex on its promoter.151
WT1 is also a transcription factor containing 4 C2H2-type ZFs in its C-terminal portion. WT1 regulates cell proliferation and differentiation, and is essential for controlling the transition between the mesenchymal and epithelial state of cells.152 It plays critical roles in the development of urogenital organs, including kidneys, gonadal organs, and external genitalia.153 WT1 is also involved in hematopoiesis, blood vessel formation, and development of various benign/malignant tumors, including Wilms tumor, acute myeloid leukemia, uterine sarcoma, and breast cancer.153,154 WT1 facilitates SF-1-induced transcriptional activity on the genes associated with sexual differentiation, whereas DAX-1 opposes to this WT1/SF-1 cooperation.106 Furthermore, WT1 contributes to the sexual differentiation by regulating the expression of DAX-1.105 WT1 interacts with ERα and suppresses estrogen-stimulated expression of the insulin-like growth factor-1 receptor, antagonizing to the estrogen-dependent growth of breast cancer cells.107 WT1 also represses the transcriptional activity of AR, functioning as a negative regulator for prostate cancer development and progression.108 In renal cells, WT1 stimulates the expression of VDR and augments vitamin D-induced modulation of their cell fate.155,156 WT1 suppresses RARα expression, while PPARβ inhibits WT1 expression and suppresses melanoma cell proliferation.104,157
CCCTC-Binding Protein
CTCF consists of 11 C2H2-type ZFs (Figure 1) and acts as an architectural protein for maintaining/regulating 3-dimensional chromatin interaction by cooperating with the cohesin protein complex and other accessory factors, including TFIIIC, ZNF143, PR domain zinc finger protein 5 (PRDM5), and chromodomain helicase DNA-binding protein 8 (CHD8).93 CTCF is evolutionarily conserved and widely present in bilaterian phyla, but absent in other eukaryotes.132 CTCF forms chromatin loops through which it organizes long-range DNA interactions between 2 and more genomic areas.93 Although it is called as “CCCTC-binding” factor, it can bind sequences with extensive variations by using selected sets of its ZFs.93 The human genome contains ~15 000 CTCF-binding sites and some of them are distributed in insulators or borders of the topologically associated domains (TADs), but substantial CTCF-binding sites are found inside TADs and intragenomic regions.93,158 This distribution pattern of the CTCF-binding sites indicates that CTCF acts not only as an organizer of TADs by blocking the action of the regulatory gene sequences/propagation of specific histone marks but also as a regulator of the enhancer/promoter switch (Figure 7). Thus, CTCF regulates the transcriptional activity of the associated protein-coding genes positively or negatively by changing their interaction to specific enhancers/promoters, ultimately contributing to the tissue/phase-specific expression of these genes. In addition to acting as an organizer for chromatin interactions, CTCF plays roles in the recombination of immunoglobulin genes and transcriptional pausing and alternative splicing.159 As CTCF acts as a general architectural protein, it virtually influences the transcriptional activity of all NRs. The effect of CTCF was examined in details for the transcriptional regulation by ERα.113,114,160 In a genome-wide analysis, CTCF-binding sites are enriched in ERα-binding regions, suggesting that CTCF acts as a cofactor for ERα-mediated transcription by facilitating the association of DNA-bound ERα and the promoter regions of ERα-regulating genes in a 3-dimensional fashion.161 Furthermore, CTCF-binding sites are more likely to overlap with cell line–specific ERα-binding sites, indicating that CTCF determines tissue-specific actions of estrogens by organizing enhancer/promoter interaction of the ERα-regulating genes through formation of the chromatin loops.115 CTCF also cooperates with TR. Some enhancers/promoters responding to TR through their TR response elements (TREs) also have CTCF-binding sites.116 Indeed, about 18% of CTCF-binding sites harbor TREs.162 CTCF- and TR-binding sites sometimes form composite elements having their binding sites closely, and CTCF-mediated enhancer-blocking activity is dependent on the presence of the TR bound on these composite elements,162 indicating that TR modulates the genome regulatory activity of CTCF.
Figure 7.
Organization of TADs and chromatin looping by CTCF for differential expression of NR-responsive genes.
Note. CTCF organizes 3-dimensional chromatin interaction for the formation of TADs and chromatin looping, in cooperating with the cohesin protein complex and other component proteins. Chromatin loop-forming activity of CTCF is essential for differential use of enhancers/promoters by NR-responding genes, and underlies NR-associated organ/tissue-specific functions. TADs = topologically associated domains; CTCF = CCCTC-binding protein; NR = nuclear hormone receptor; NRE = NR-response element.
Other Poly-ZNFs
Zip67, Zac1, ZNF217, and ZNF536 are multiple adjacent-fingered poly-ZNFs. Their biological/physiological actions are not well elucidated, but they act as cofactors for the transcriptional activity of some NRs. Similar to other multiple adjacent-fingered poly-ZNFs, such as ZNF592 and ZNF687, these ZNFs are likely to be incorporated in the transcriptional complex formed on NRs and modulate their transcriptional activity through communication with other regulatory molecules inside the complex.11
Zip67 or ZNF653 contains 5 ZFs and was identified as a regulator of SF-1-induced transcriptional activity.109 Zip67 directly interacts with SF-1 LBD and represses SF-1-induced transcriptional activity.109 Zip67 also represses the transcriptional activity of LXRα, NUR77, GR, and ER, suggesting that Zip67 functions as a general cofactor for some NRs.109
Zac1 or PLAG1 (PLAG-like zinc finger 1) has 7 ZFs and was first identified as a putative transcriptional activator involved in the regulation of apoptosis and cell cycle.163,164 Zac1 physically interacts with the HAT coactivators, p300 and p300/CBP-associated factor (p/CAF) through its ZFs, and regulates the latter’s HAT activity.165,166 Zac1 can coactivate or corepress the hormone-dependent transcriptional activity of AR, ERα, and TRβ1.111 Zac1 also stimulates PPARγ expression by binding to the proximal portion of the PPARγ promoter, suggesting that Zac1 exerts its anti-proliferative effects on various cancer cells through modulating the expression of this receptor.110
ZNF217 possesses 8 ZFs. ZNF217 acts as a coactivator of ERα-induced transcriptional activity in various cell lines.85 In the chromatin immunoprecipitation (ChIP) followed by high throughput sequence analysis, ZNF217- and ERα-binding sites are clustered in the distal enhancer region, and ZNF217 interacts physically with ERα through the former’s C-terminal portion harboring ZFs and the latter’s hinge region.85 Interestingly, expression of ZNF217 was correlated with that of ERα in breast cancer tissues, and its higher expression is linked to worse prognosis of the patients with breast cancer, possibly by enhancing the proliferation-stimulating activity of ERα.85 Indeed, ZNF217 is found to be overexpressed in more than 20% of the breast cancers, and its elevated expression is associated with aggressive tumor behavior.167
ZNF536 was identified as a novel protein interacting with the C-terminal tail-binding protein (CtBP) in the yeast two-hybrid screening.112 ZNF536 has 10 ZFs, is highly conserved in vertebrates, and is most abundantly expressed in the neural cells especially of developing brain.112 ZNF536 regulates neuronal differentiation by repressing retinoic acid/RAR-induced transcriptional activity, presumably through competing with RARα for binding to the latter’s response elements, as ZNF536 can bind these elements similar to RARα.112
ZNF366 is an 11 ZFs-containing protein identified as a molecule interacting with ERα in the yeast two-hybrid screening using an ERα mutant lacking LBD as bait.118 ZNF366 acts as a corepressor for ERα-induced transcriptional activity by interacting with receptor-interacting protein 140 (RIP140) and CtBP. ZNF366 represses estrogen-responsive genes in breast cancer cells,118 suggesting that it may function as a negative factor for estrogen-dependent proliferation of breast cancer cells.
ZNF423 or Roaz contains 30 ZFs and was first described as a binding partner of the EBF1/OLF1 transcription factor.168,169 ZNF423 is important for the differentiation of adipocytes and olfactory neurons and for the development of B-cell lymphoma.170 ZNF423 is a critical factor required for the retinoic acid–induced differentiation of the neuroblastoma cell lines, acting as a cofactor for RAR/RXR-induced transcriptional activity by binding directly to RARα and RXRα.120 High expression of ZNF423 in neuroblastoma tissues is associated with good prognosis of the patients harboring this tumor,120 suggesting that ZNF423 is a promising clinical marker and/or a treatment target.171
BTB/POZ-ZNFs
BTB/POZ-ZNFs are also evolutionarily conserved proteins observed from C elegans and insects in the evolutionary tree.8,77 The BTB/POZ domain spans ~120 amino acids and is also found in some actin-binding proteins containing a kelch motif.8 BTB/POZ domain has multiple activities, including oligomerization between family members and transcriptional repression through interaction with repressive cofactors.8 There are 2 BTB/POZ-ZNFs that interact with NRs, Kaiso, and the promyelocytic leukemia zinc finger protein (PLZF).
Kaiso
Kaiso, or ZNF348 or ZBTB33 (zinc finger and BTB-containing protein 33), is a triple-fingered ZNF having a BTB/POZ domain in its N-terminus and 3 ZFs in the C-terminus. Kaiso physically interacts with the p120-catenin signaling molecule and binds methylated CpG dinucleotides in the consensus sequence CGCG.122 In addition, Kaiso is physically associated with the nuclear receptor corepressor (NCoR) through its BTB/POZ domain and facilitates deacetylation of histone tails; thus, it acts as a methylation-dependent transcriptional repressor by binding to methylated CpG islands of the target gene promoters.122 As many NRs attract NCoR for their repression of responsive genes, Kaiso may be a key molecule for the interplay between gene silencing by DNA methylation and NR-mediated transcriptional repression through histone deacetylation. Kaiso represses GR expression by binding to the proximal promoter region of the GR gene in a methylation-dependent fashion, influencing the anti-apoptotic activity of this receptor in breast cancer cells.172 Kaiso also interacts with the unmethylated consensus sequence, CTGCNA, and represses expression of the molecules involved in the Wnt signaling pathway.122
Promyelocytic Leukemia Zinc Finger Protein
PLZF or ZBTB16 (zinc finger and BTB-containing protein 16) is a BTB/POZ-ZNF with 9 ZFs (Figure 1). PLZF was first identified as the gene for chromosomal translocation t(11;17)(q23;q21) in acute promyelocytic leukemia (APL), which produces the PLZF-RARα fusion protein172 (Figure 1). PLZF plays a role in the control of cell cycle progression and is involved in the forebrain organization, hindbrain segmentation and musculoskeletal/limb development, differentiation of myeloid cells, and spermatogenesis.174-176 PLZF binds through its BTB/POZ domain some repressive cofactors, such as the silencing mediator for retinoid and thyroid hormone receptors (SMRT), NCoR, Sin3 and histone deacetylase 1 (HDAC1), and forms a transcription-repressive complex.177 The PLZF-RARα fusion protein disrupts normal transcriptional regulation organized by retinoic acids, leading to the development of APL by reciprocally attracting the polycomb-repressive complex 1 (PRC1) and PRC2 to RARα-binding sites.178 Progesterone and glucocorticoids strongly stimulate PLZF expression in human endometrial stromal cells and myometrial smooth muscle cells, suggesting that PLZF plays a role in the proliferation and/or differentiation of these cells during the menstrual cycle influenced by these steroid hormones.
KRAB-ZNFs
KRAB-ZNFs form the largest subfamily in C2H2-type ZNFs, comprising nearly half of these proteins in humans77 (Table 3). KRAB-ZNFs appeared in the tetrapod vertebrates and they demonstrate a significant lineage-specific expansion in higher organisms.77 This expansion is caused by segmental gene duplication, as evidenced by the fact that the genes encoding KRAB-ZNFs are found as clusters in particular portions of chromosomes, such as chromosome 19q13, 6p22, 16p13, and 16p11 in humans.179 Rapid expansion of KRAB-ZNFs is particularly observed in the primate lineage, and substantial proportion of the human KRAB-ZNFs has no mouse orthologs.77,180 These pieces of evidence indicate rapid diversification of KRAB-ZNFs and their potential importance in the regulation of the biological functions specific to primates including humans. Indeed, they appear to be responsible for some of the major transcriptional differences found between humans and other primates.91 Furthermore, some human KRAB-ZNF genes (eg, PRDM9) demonstrate diversity in the number of their encoding ZFs among human populations, indicating that they may underlie the variability observed between different populations/individuals.181
KRAB-ZNFs are well known to act as transcriptional repressors, but some of them also function as transcriptional enhancers.8,88 KRAB-ZNFs have the KRAB domain in their N-terminal portion and multiple ZFs in the C-terminal area, which function respectively as a transcriptional regulatory unit and an anchoring component to DNA, RNA, and/or other protein molecules.8 Most of the KRAB-ZNFs have more than 4 ZFs and some contain more than 30 of these motifs (average number of ZFs: 7.5 and 8.5 in mouse and human, respectively), and they selectively use some of their multiple ZFs for interacting with specific DNA sequences and RNA/protein molecules, indicating a potential of having many different partner/target molecules/genes.8,77 The KRAB domain spans approximately 75 amino acids, and has one or both of the KRAB-A and KRAB-B boxes.8 KRAB-A box consists of 45 amino acids and acts as a strong transcriptional repressor domain by recruiting several cofactor molecules including the KRAB-associated protein-1 (KAP-1),182,183 which is also known as the KRAB-A-interacting protein 1 (KRIP1), tripartite motif protein 28 (TRIM28), or transcription intermediary factor 1β (TIF1β).184 In contrast, functions of KRAB-B box are not well known, but it enhances the activity of KRAB-A box.185 KAP-1 binds KRAB domain as an oligomer and further recruits the heterochromatin protein 1 (HP1), HDACs, and the histone H3K9 methyltransferase SET domain bifurcated 1 (SETDB1) through which KRAB-ZNFs silence their associating genes by changing the chromatin status/structure.184,186 Importantly, KAP-1 acts as a coactivator for some NRs, similar to its homologous molecule TIF1α (TRIM24).187-189 TIF1s physically interact with NRs and participate in their transcriptional regulation188,190; thus, they are a link for the functional interaction between KRAB-ZNFs and NRs. Several KRAB-ZNFs, such as ZNF746 (Paris), ZNF282, ZNF764, ZNF398, and ZNF461, influence the transcriptional activity of NRs.
ZNF746
ZNF746 also known as the parkin-interacting substrate (Paris) is a KRAB-ZNF containing 4 ZFs. ZNF746 functions as a transcription repressor by binding to the TATTTT[T/G] consensus motif.126 ZNF746 is a substrate of the ubiquitin E3 ligase Parkin and represses the transcription of the PPARγ coactivator-1α (PGC-1α) gene in neurons.126 Thus, the reduction of Parkin expression or the presence of its inactivating gene mutations results in increase of ZNF746 protein levels in these cells. Elevated ZNF746 in turn reduces PCG-1α expression and contributes to the development of neurodegeneration observed in Parkinson disease through down-regulating the expression of PCG-1α-dependent genes.126 PGC-1α is a coactivator for several NRs, including PPARs, ERs, RXRα, HNF4α, and GR, by interacting physically with these receptors through its LxxLL coactivator motif, and regulates intermediary metabolism, adaptive thermogenesis, mitochondrial biogenesis, and adipocyte differentiation/function in part through these receptors.191 Thus, ZNF746 potentially influences the activity of these receptors indirectly by stimulating the expression of PGC-1α.
ZNF282
ZNF282 is a KRAB-ZNF containing 5 ZFs and was originally identified as a protein that binds the U5 repressive element (U5RE) of the human T-cell leukemia virus type-I (HTLV-I) (thus, it is also called the HTLV-I U5RE-binding protein 1: HUB1).192 ZNF282 physically interacts and synergistically cooperates with the coiled-coil coactivator (CoCoA), a cofactor interacting with NCoA HAT coactivators and contributing to NR-induced transcriptional activity.193,194 ZNF282 enhances ERα-mediated transcriptional activity in the presence of CoCoA and stimulates estrogen-dependent proliferation of breast cancer cells.127 ZNF282 also enhances the transcriptional activity of GR, TR, and AR.127 However, in vivo importance of such ZNF282-mediated regulation of NR activity is not known yet.
ZNF764
ZNF764 consists of 7 ZFs in its C-terminal portion and one KRAB-A domain in its N-terminal area (Figure 1). ZNF764 was identified as a potential gene causing the multiple steroid hormone resistance observed in a patient with 16p11.2 microdeletion.128 The 16p11.2 gene segment is highly susceptible to segmental duplications/deletions in the great ape lineage.195 The gene rearrangement in the 16p11.2 segment is still ongoing in modern humans. For example, length of this segment has almost doubled in humans and chimpanzees compared with their ancestor orangutans.195 This activity has also generated in the segment human-specific genes with beneficial effects, whereas it causes various congenital disorders by impacting appropriate copy numbers of residing genes, such as in the case of ZNF764.128,195 In human cells, ZNF764 enhances the transcriptional activity of GR, AR, MR, and TRs in cooperation with KAP-1.128 In a genome-wide binding study using ChIP-sequencing in human HeLa cells, ZNF764- and GR-binding sites are found in close proximity, indicating that ZNF764 modulates GR transcriptional activity by incorporated in the transcriptional complex formed on DNA-bound GR.129 The exact physiologic functions of ZNF764 in humans are not known, but it appears to regulate the GR activity involved in the stress response organized by the hypothalamic-pituitary-adrenal axis and the AR activity required for fetal development of male-type external genitalia.128
ZNF398
ZNF398 also known as ZER6 (zinc finger-estrogen receptor interaction, clone 6) is a KRAB-ZNF containing 6 ZFs and was identified as an ERα-interacting protein in a yeast two-hybrid screening using full-length ERα as bait.130 Alternative splicing generates 2 ZER6 isoforms, p52-ZER6 and p71-ZER6. ERα physically interacts with p52-ZER6 in a ligand-dependent fashion but not with p71-ZER6, due to the presence of the HUB1 domain in the latter isoform.130 ERα strongly represses the transcriptional activity of ZER6 on the gene containing its response elements.130 As ZER6 is specifically expressed in breast tissues, ZER6 may mediate some actions of estrogens in this organ.196 ZER6 is also expressed in ERα-positive, but not ERα-negative breast cancers, suggesting its potential role in the regulation of estrogen-dependent proliferation in this malignancy.196
ZNF461
ZNF461, also called as the gonadotropin-inducible ovarian transcription factor-1 (GIOT-1), was first identified as a molecule induced by the follicular stimulating hormone.197 ZNF461 is predominantly expressed in steroidogenic tissues, including testicular Leydig cells, ovarian granulosa and theca cells and adrenocortical cells, indicating its primary role in the regulation of steroidogenesis in mammals.198 ZNF461 contains one KRAB-A domain and 12 ZFs, and acts as a transcriptional repressor.197 Expression of ZNF461 is regulated by SF-1 and NUR77, respectively, in ovarian granulose cells and testicular K28 Leydig cells.198 On the contrary, ZNF461 represses SF-1-mediated transcriptional activity by physically interacting with SF-1 and by attracting HDAC2, indicating a presence of auto-regulatory loop between ZNF461 and SF-1 in the steroid hormone production by gonads and adrenal glands.131
Conclusions and Future Perspectives
NRs are evolutionarily conserved ligand-dependent transcription factors that mediate the actions of lipophilic hormones and metabolites and/or intermediates of several important biological pathways.7 NRs are essential for human life, by affecting every aspect of human activities.7,9 On the contrary, C2H2-type ZNFs form an evolutionarily expanding family of transcription factors, characterized by multiple, tandemly arranged ZFs and distinct functional modules, such as BTB/POZ, KRAB, and SCAN domains.8 Among them, KRAB-ZNFs have expanded their members significantly in higher organisms, possibly organizing the species-specific biological actions through the transcriptional regulatory network that has also increased its complexity following evolution.2,77,90 Substantial numbers of C2H2-type ZNFs function as DNA-binding transcription factors, whereas others may act as transcriptional cofactors incorporated in the transcription complex formed on NRs. As both NRs and C2H2-type ZNFs have diverse regulatory actions on human biology, these 2 groups mutually interact with each other through their evolution/diversification, ultimately contributing not only to the functions conserved through evolution but also to the complex gene regulation and/or species-specific activities of higher organisms. Research on the former mainly mediated by the interaction between NRs and older C2H2-ZNFs has demonstrated major progress, as it plays roles in more fundamental biological systems shared by many organisms. In contrast, importance of the latter, eg, for the human-specific actions of KRAB-ZNFs, has just been recognized recently. Indeed, these younger genes tend to have less functional communication with other genes, in contrast to older genes that were well incorporated in the gene interaction network built through long evolutional history94 (Figure 5). Nevertheless, the new genes including KRAB- or SCAN-ZNFs appear to be emerged to support the activities important for respective species, such as superb CNS functions observed in humans. Thus, revealing the actions of newly appeared C2H2-ZNFs is important to understand human-specific biological activities and is critical to elucidate causes and pathophysiology of human diseases (eg, depression and schizophrenia199,200). Clarifying the regulatory actions of evolutionarily old and new C2H2-type ZNFs on NRs will ultimately improve our understanding on NR actions general to many organisms, but also those relevant and specific to humans.
Another important aspect of C2H2-type ZNFs is their interaction with abundant ncRNAs. Numbers of ncRNAs have been significantly increased in higher organisms following expansion of noncoding-gene area.2 Many of the ncRNAs employ protein molecules as their functional targets and/or mediators of their effects.201 For example, argonaute proteins are necessary for microRNAs (miRNAs) to interact with their target mRNAs, while many long ncRNAs (lncRNAs) interact with various proteins to exert their effects on chromatin/DNA.202,203 C2H2-type ZNFs are among such molecules that mediate the actions of ncRNAs. For example, roX, a drosophila lncRNA essential for dosage compensation of the sex chromosome by stimulating the transcription of single X chromosome in males, communicates with its target DNA sequences through the chromatin-linked adaptor for male-specific lethal (MSL) protein (CLAMP). 204CLAMP is a poly-ZNF with 7 ZFs and is a component of the MSL complex necessary for the dosage compensation by roX. CLAMP binds roX with its ZFs and guides this lncRNA to target DNA sequences.204 CTCF, a poly-ZNFs with 11 ZFs, exerts its insulator function by forming a complex with the DEAD-box RNA helicase p68 and the steroid receptor RNA activator (SRA) lncRNA.205 SRA participates in the transcriptional activity of SRs by interacting with their AF-1 transactivation domain and by forming a complex with p160-type NCoAs205,206; thus, CTCF, SRA, and NRs cooperate with each other to regulate NR-induced transcriptional activity. Similar to SRA, the lncRNAs known to have regulatory actions on NRs, such as the growth arrest-specific 5 (Gas5), prostate cancer–associated noncoding RNA 1 (PRNCR1), and the prostate-specific transcript 1 (PCGEM1)207,208 might exert their effects through the interaction with C2H2-type ZNFs, as some C2H2-ZNFs are known to regulate the transcriptional activity of NRs for which these lncRNAs are also functional. Furthermore, lncRNAs and some C2H2-type ZNFs may play important regulatory roles in tissue-specific gene expression, indicating their functional overlap and potential cooperation in target biological activities. As increasing numbers of the ncRNAs with NR regulatory activities was observed during the last several years, elucidation of their interaction with C2H2-ZNFs may help revealing the regulatory actions of such ncRNAs on NRs.
Footnotes
Author Contributions: RM and TK wrote the article. AK created tables. RM, AKM, AF, and TK edited the article.
Authorship Statement: This article is not under consideration by another journal, and all authors agree to the submission of the article, have approved the final submitted copy, and are aware of and agree to be bound by the editorial policies of Nuclear Receptor Signaling.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Literary work of this article was funded by the Intramural Research Program of the Sidra Medical and Research Center, Doha, Qatar.
References
- 1. Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25-36. [DOI] [PubMed] [Google Scholar]
- 2. Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316-323. [DOI] [PubMed] [Google Scholar]
- 3. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [DOI] [PubMed] [Google Scholar]
- 4. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62. [DOI] [PubMed] [Google Scholar]
- 5. Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592-600. [DOI] [PubMed] [Google Scholar]
- 6. Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chrousos GP, Kino T. Intracellular glucocorticoid signaling: a formerly simple system turns stochastic. Sci STKE. 2005;2005:pe48. [DOI] [PubMed] [Google Scholar]
- 11. Malovannaya A, Lanz RB, Jung SY, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kino T. Glucocorticoid receptor. In: De Groot LJ, Beck-Peccoz P, Chrousos G, et al., eds. Endotext. South Dartmouth MA MDText.com; 2000. [Google Scholar]
- 13. Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685-704. [DOI] [PubMed] [Google Scholar]
- 14. Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid resistance/hypersensitivity syndromes. J Endocrinol. 2001;169:437-445. [DOI] [PubMed] [Google Scholar]
- 15. Simons SS, Jr, Edwards DP, Kumar R. Minireview: dynamic structures of nuclear hormone receptors: new promises and challenges. Mol Endocrinol. 2014;28:173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Renaud JP, Moras D. Structural studies on nuclear receptors. Cell Mol Life Sci. 2000;57:1748-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384-391. [DOI] [PubMed] [Google Scholar]
- 18. Wu DY, Ou CY, Chodankar R, Siegmund KD, Stallcup MR. Distinct, genome-wide, gene-specific selectivity patterns of four glucocorticoid receptor coregulators. Nucl Recept Signal. 2014;12:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giguere V. Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocr Rev. 1994;15:61-79. [DOI] [PubMed] [Google Scholar]
- 21. Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409-435. [DOI] [PubMed] [Google Scholar]
- 22. Knouff C, Auwerx J. Peroxisome proliferator-activated receptor-γ calls for activation in moderation: lessons from genetics and pharmacology. Endocr Rev. 2004;25:899-918. [DOI] [PubMed] [Google Scholar]
- 23. Yin L, Wu N, Lazar MA. Nuclear receptor rev-erbα: a heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal. 2010;8:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Luo XY, Wu DH, Xu Y. ROR nuclear receptors: structures, related diseases, and drug discovery. Acta Pharmacol Sin. 2015;36:71-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jakobsson T, Treuter E, Gustafsson JA, Steffensen KR. Liver X receptor biology and pharmacology: new pathways, challenges and opportunities. Trends Pharmacol Sci. 2012;33:394-404. [DOI] [PubMed] [Google Scholar]
- 26. Chiang JY. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev. 2002;23:443-463. [DOI] [PubMed] [Google Scholar]
- 27. Jurutka PW, Whitfield GK, Hsieh JC, Thompson PD, Haussler CA, Haussler MR. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev Endocr Metab Disord. 2001;2:203-16. [DOI] [PubMed] [Google Scholar]
- 28. Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2002;23:687-702. [DOI] [PubMed] [Google Scholar]
- 29. Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259-266. [DOI] [PubMed] [Google Scholar]
- 30. Sladek R, Giguere V. Orphan nuclear receptors: an emerging family of metabolic regulators. Adv Pharmacol. 2000;47:23-87. [DOI] [PubMed] [Google Scholar]
- 31. Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689-725. [DOI] [PubMed] [Google Scholar]
- 32. Lee YF, Lee HJ, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol. 2002;81:291-308. [DOI] [PubMed] [Google Scholar]
- 33. Benod C, Villagomez R, Webb P. TLX: an elusive receptor. J Steroid Biochem Mol Biol. 2016;157:41-47. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y, Liu HK, Schutz G. Role of the nuclear receptor Tailless in adult neural stem cells. Mech Dev. 2013;130:388-390. [DOI] [PubMed] [Google Scholar]
- 35. Kobayashi M, Takezawa S, Hara K, et al. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci U S A. 1999;96:4814-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsai SY, Tsai MJ. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev. 1997;18:229-240. [DOI] [PubMed] [Google Scholar]
- 37. Zhu XG, Park KS, Kaneshige M, et al. The orphan nuclear receptor Ear-2 is a negative coregulator for thyroid hormone nuclear receptor function. Mol Cell Biol. 2000;20:2604-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358-417. [DOI] [PubMed] [Google Scholar]
- 39. Koehler KF, Helguero LA, Haldosen LA, Warner M, Gustafsson JA. Reflections on the discovery and significance of estrogen receptor β. Endocr Rev. 2005;26:465-478. [DOI] [PubMed] [Google Scholar]
- 40. Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J Mol Endocrinol. 2003;31:349-357. [DOI] [PubMed] [Google Scholar]
- 41. Funder JW. Minireview: aldosterone and mineralocorticoid receptors: past, present, and future. Endocrinology. 2010;151:5098-5102. [DOI] [PubMed] [Google Scholar]
- 42. Graham JD, Clarke CL. Physiological action of progesterone in target tissues. Endocr Rev. 1997;18:502-519. [DOI] [PubMed] [Google Scholar]
- 43. Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276-308. [DOI] [PubMed] [Google Scholar]
- 44. Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271-321. [DOI] [PubMed] [Google Scholar]
- 45. Bassett MH, White PC, Rainey WE. A role for the NGFI-B family in adrenal zonation and adrenocortical disease. Endocr Res. 2004;30:567-574. [DOI] [PubMed] [Google Scholar]
- 46. Hsu HC, Zhou T, Mountz JD. Nur77 family of nuclear hormone receptors. Curr Drug Targets Inflamm Allergy. 2004;3:413-423. [DOI] [PubMed] [Google Scholar]
- 47. Ranhotra HS. The NR4A orphan nuclear receptors: mediators in metabolism and diseases. J Recept Signal Transduct Res. 2015;35:184-188. [DOI] [PubMed] [Google Scholar]
- 48. McMorrow JP, Murphy EP. Inflammation: a role for NR4A orphan nuclear receptors? Biochem Soc Trans. 2011;39:688-693. [DOI] [PubMed] [Google Scholar]
- 49. Perlmann T, Wallen-Mackenzie A. Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 2004;318:45-52. [DOI] [PubMed] [Google Scholar]
- 50. Mullican SE, Zhang S, Konopleva M, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730-735. [DOI] [PubMed] [Google Scholar]
- 51. Ozisik G, Achermann JC, Jameson JL. The role of SF1 in adrenal and reproductive function: insight from naturally occurring mutations in humans. Mol Genet Metab. 2002;76:85-91. [DOI] [PubMed] [Google Scholar]
- 52. Stein S, Schoonjans K. Molecular basis for the regulation of the nuclear receptor LRH-1. Curr Opin Cell Biol. 2015;33:26-34. [DOI] [PubMed] [Google Scholar]
- 53. Zechel C. The germ cell nuclear factor (GCNF). Mol Reprod Dev. 2005;72:550-556. [DOI] [PubMed] [Google Scholar]
- 54. El-Khairi R, Martinez-Aguayo A, Ferraz-de-Souza B, Lin L, Achermann JC. Role of DAX-1 (NR0B1) and steroidogenic factor-1 (NR5A1) in human adrenal function. Endocr Dev. 2011;20:38-46. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812:893-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11-26. [DOI] [PubMed] [Google Scholar]
- 57. Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol Cell Biol. 2003;23:238-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen SJ, Zelent A, Tong JH, et al. Rearrangements of the retinoic acid receptor α and promyelocytic leukemia zinc finger genes resulting from t(11;17)(q23;q21) in a patient with acute promyelocytic leukemia. J Clin Invest. 1993;91:2260-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14:369-376. [DOI] [PubMed] [Google Scholar]
- 60. Krylova IN, Sablin EP, Moore J, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343-355. [DOI] [PubMed] [Google Scholar]
- 61. Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol. 2007;14:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eick GN, Thornton JW. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol Cell Endocrinol. 2011;334:31-38. [DOI] [PubMed] [Google Scholar]
- 63. Gardiner JR, Shima Y, Morohashi K, Swain A. SF-1 expression during adrenal development and tumourigenesis. Mol Cell Endocrinol. 2012;351:12-18. [DOI] [PubMed] [Google Scholar]
- 64. Pascual A, Aranda A. Thyroid hormone receptors, cell growth and differentiation. Biochim Biophys Acta. 2013;1830:3908-3916. [DOI] [PubMed] [Google Scholar]
- 65. Samarut E, Rochette-Egly C. Nuclear retinoic acid receptors: conductors of the retinoic acid symphony during development. Mol Cell Endocrinol. 2012;348:348-360. [DOI] [PubMed] [Google Scholar]
- 66. Wagner RT, Cooney AJ. Minireview: the diverse roles of nuclear receptors in the regulation of embryonic stem cell pluripotency. Mol Endocrinol. 2013;27:864-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xie X, Tang K, Yu CT, Tsai SY, Tsai MJ. Regulatory potential of COUP-TFs in development: stem/progenitor cells. Semin Cell Dev Biol. 2013;24:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866-1870. [DOI] [PubMed] [Google Scholar]
- 69. Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid receptors and associated diseases. Essays Biochem. 2004;40:137-155. [DOI] [PubMed] [Google Scholar]
- 70. Kumar R, Tebben PJ, Thompson JR. Vitamin D and the kidney. Arch Biochem Biophys. 2012;523:77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201-220. [DOI] [PubMed] [Google Scholar]
- 72. Chaudhuri G. Nuclear receptors and female reproduction: a tale of 3 scientists, Jensen, Gustafsson, and O’Malley. Reprod Sci. 2008;15:110-120. [DOI] [PubMed] [Google Scholar]
- 73. Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365-376. [DOI] [PubMed] [Google Scholar]
- 74. Huang W, Glass CK. Nuclear receptors and inflammation control: molecular mechanisms and pathophysiological relevance. Arterioscler Thromb Vasc Biol. 2010;30:1542-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem. 2011;3:623-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63-70. [DOI] [PubMed] [Google Scholar]
- 77. Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Berg JM. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988;85:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989;245:635-637. [DOI] [PubMed] [Google Scholar]
- 80. Neuhaus D, Nakaseko Y, Schwabe JW, Klug A. Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of β-sheet. J Mol Biol. 1992;228:637-651. [DOI] [PubMed] [Google Scholar]
- 81. Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809-817. [DOI] [PubMed] [Google Scholar]
- 83. Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1-21. [DOI] [PubMed] [Google Scholar]
- 84. Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94-98. [DOI] [PubMed] [Google Scholar]
- 85. Frietze S, O’Geen H, Littlepage LE, et al. Global analysis of ZNF217 chromatin occupancy in the breast cancer cell genome reveals an association with ERα. BMC Genomics. 2014;15:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liew CK, Simpson RJ, Kwan AH, et al. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci U S A. 2005;102:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat Struct Biol. 1997;4:122-132. [DOI] [PubMed] [Google Scholar]
- 90. Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213-231. [DOI] [PubMed] [Google Scholar]
- 91. Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A. 2009;106:22358-22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Freilich S, Massingham T, Bhattacharyya S, Ponsting H, Lyons PA, Freeman TC, Thornton JM. Relationship between the tissue-specificity of mouse gene expression and the evolutionary origin and function of the proteins. Genome Biol. 2005;6:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang W, Landback P, Gschwend AR, Shen B, Long M. New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. 2015;16:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kolla V, Litwack G. Upregulation of mineralocorticoid- and glucocorticoid-receptor gene expression by Sp-I. Mol Cell Biol Res Commun. 1999;1:44-47. [DOI] [PubMed] [Google Scholar]
- 96. Kolla V, Robertson NM, Litwack G. Identification of a mineralocorticoid/glucocorticoid response element in the human Na/K ATPase α1 gene promoter. Biochem Biophys Res Commun. 1999;266:5-14. [DOI] [PubMed] [Google Scholar]
- 97. Suehiro T, Kaneda T, Ikeda Y, Arii K, Kumon Y, Hashimoto K. Regulation of human glucocorticoid receptor gene transcription by Sp1 and p53. Mol Cell Endocrinol. 2004;222:33-40. [DOI] [PubMed] [Google Scholar]
- 98. McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90:1337-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Oishi Y, Manabe I, Tobe K, et al. SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ. Nat Med. 2008;14:656-666. [DOI] [PubMed] [Google Scholar]
- 100. Simmen RC, Pabona JM, Velarde MC, Simmons C, Rahal O, Simmen FA. The emerging role of Krüppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol. 2010;204:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Deng Z, Wan M, Cao P, Rao A, Cramer SD, Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene. 2009;28:3746-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nem D, Baranyai D, Qiu H, Godtel-Armbrust U, Nestler S, Wojnowski L. Pregnane X receptor and yin yang 1 contribute to the differential tissue expression and induction of CYP3A5 and CYP3A4. PLoS One. 2012;7:e30895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Raval-Pandya M, Dhawan P, Barletta F, Christakos S. YY1 represses vitamin D receptor-mediated 25-hydroxyvitamin D(3)24-hydroxylase transcription: relief of repression by CREB-binding protein. Mol Endocrinol. 2001;15:1035-1046. [DOI] [PubMed] [Google Scholar]
- 104. Goodyer P, Dehbi M, Torban E, Bruening W, Pelletier J. Repression of the retinoic acid receptor-α gene by the Wilms’ tumor suppressor gene product, wt1. Oncogene. 1995;10:1125-1129. [PubMed] [Google Scholar]
- 105. Kim J, Prawitt D, Bardeesy N, Torban E, Vicaner C, Goodyer P, Zabel B, Pelletier J. The Wilms’ tumor suppressor gene (wt1) product regulates Dax-1 gene expression during gonadal differentiation. Mol Cell Biol. 1999;19:2289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA. Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445-454. [DOI] [PubMed] [Google Scholar]
- 107. Reizner N, Maor S, Sarfstein R, et al. The WT1 Wilms’ tumor suppressor gene product interacts with estrogen receptor-α and regulates IGF-I receptor gene transcription in breast cancer cells. J Mol Endocrinol. 2005;35:135-144. [DOI] [PubMed] [Google Scholar]
- 108. Zaia A, Fraizer GC, Piantanelli L, Saunders GF. Transcriptional regulation of the androgen signaling pathway by the Wilms’ tumor suppressor gene WT1. Anticancer Res. 2001;21:1-10. [PubMed] [Google Scholar]
- 109. Borud B, Mellgren G, Lund J, Bakke M. Cloning and characterization of a novel zinc finger protein that modulates the transcriptional activity of nuclear receptors. Mol Endocrinol. 2003;17:2303-2319. [DOI] [PubMed] [Google Scholar]
- 110. Barz T, Hoffmann A, Panhuysen M, Spengler D. Peroxisome proliferator-activated receptor γ is a Zac target gene mediating Zac antiproliferation. Cancer Res. 2006;66:11975-11982. [DOI] [PubMed] [Google Scholar]
- 111. Huang SM, Stallcup MR. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol Cell Biol. 2000;20:1855-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Qin Z, Ren F, Xu X, et al. ZNF536, a novel zinc finger protein specifically expressed in the brain, negatively regulates neuron differentiation by repressing retinoic acid-induced gene transcription. Mol Cell Biol. 2009;29:3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Carroll JS, Meyer CA, Song J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 114. Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ross-Innes CS, Brown GD, Carroll JS. A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics. 2011;12:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Awad TA, Bigler J, Ulmer JE, et al. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092-27098. [DOI] [PubMed] [Google Scholar]
- 117. Lutz M, Baniahmad A, Renkawitz R. Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor. Biochem Soc Trans. 2000;28:386-389. [PubMed] [Google Scholar]
- 118. Lopez-Garcia J, Periyasamy M, Thomas RS, et al. ZNF366 is an estrogen receptor corepressor that acts through CtBP and histone deacetylases. Nucleic Acids Res. 2006;34:6126-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gupta RK, Arany Z, Seale P, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Huang S, Laoukili J, Epping MT, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ingle JN, Liu M, Wickerham DL, et al. Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov. 2013;3:812-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89-97. [DOI] [PubMed] [Google Scholar]
- 123. Martin PJ, Delmotte MH, Formstecher P, Lefebvre P. PLZF is a negative regulator of retinoic acid receptor transcriptional activity. Nucl Recept. 2003;1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ward JO, McConnell MJ, Carlile GW, Pandolfi PP, Licht JD, Freedman LP. The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D(3)-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D(3) receptor. Blood. 2001;98:3290-3300. [DOI] [PubMed] [Google Scholar]
- 125. Wasim M, Mansha M, Kofler A, Awan AR, Babar ME, Kofler R. Promyelocytic leukemia zinc finger protein (PLZF) enhances glucocorticoid-induced apoptosis in leukemic cell line NALM6. Pak J Pharm Sci. 2012;25:617-621. [PubMed] [Google Scholar]
- 126. Shin JH, Ko HS, Kang H, et al. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yu EJ, Kim SH, Kim MJ, et al. SUMOylation of ZFP282 potentiates its positive effect on estrogen signaling in breast tumorigenesis. Oncogene. 2013;32:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kino T, Pavlatou MG, Moraitis AG, Nemery RL, Raygada M, Stratakis CA. ZNF764 haploinsufficiency may explain partial glucocorticoid, androgen, and thyroid hormone resistance associated with 16p11.2 microdeletion. J Clin Endocrinol Metab. 2012;97:E1557-E1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Fadda A, et al. Genome-wide Regulatory Roles of the C2H2-type Zinc Finger Protein ZNF764 on the Glucocorticoid Receptor. Sci Rep. 2017;7:41598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Conroy AT, Sharma M, Holtz AE, Wu C, Sun Z, Weigel RJ. A novel zinc finger transcription factor with two isoforms that are differentially repressed by estrogen receptor-α. J Biol Chem. 2002;277:9326-9334. [DOI] [PubMed] [Google Scholar]
- 131. Song KH, Park YY, Kee HJ, et al. Orphan nuclear receptor Nur77 induces zinc finger protein GIOT-1 gene expression, and GIOT-1 acts as a novel corepressor of orphan nuclear receptor SF-1 via recruitment of HDAC2. J Biol Chem. 2006;281:15605-15614. [DOI] [PubMed] [Google Scholar]
- 132. Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci U S A. 2012;109:17507-17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551-556. [DOI] [PubMed] [Google Scholar]
- 134. Harrison SM, Houzelstein D, Dunwoodie SL, Beddington RS. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev Biol. 2000;227:358-372. [DOI] [PubMed] [Google Scholar]
- 135. Azizkhan JC, Jensen DE, Pierce AJ, Wade M. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit Rev Eukaryot Gene Expr. 1993;3:229-254. [PubMed] [Google Scholar]
- 136. Knoedler JR, Denver RJ. Krüppel-like factors are effectors of nuclear receptor signaling. Gen Comp Endocrinol. 2014;203:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog Nucleic Acid Res Mol Biol. 2004;77:1-36. [DOI] [PubMed] [Google Scholar]
- 138. Curtin D, Jenkins S, Farmer N, et al. Androgen suppression of GnRH-stimulated rat LHβ gene transcription occurs through Sp1 sites in the distal GnRH-responsive promoter region. Mol Endocrinol. 2001;15:1906-1917. [DOI] [PubMed] [Google Scholar]
- 139. Lu S, Jenster G, Epner DE. Androgen induction of cyclin-dependent kinase inhibitor p21 gene: role of androgen receptor and transcription factor Sp1 complex. Mol Endocrinol. 2000;14:753-760. [DOI] [PubMed] [Google Scholar]
- 140. Sugawara A, Uruno A, Kudo M, et al. Transcription suppression of thromboxane receptor gene by peroxisome proliferator-activated receptor-γ via an interaction with Sp1 in vascular smooth muscle cells. J Biol Chem. 2002;277:9676-9683. [DOI] [PubMed] [Google Scholar]
- 141. Sugawara T, Saito M, Fujimoto S. Sp1 and SF-1 interact and cooperate in the regulation of human steroidogenic acute regulatory protein gene expression. Endocrinology. 2000;141:2895-2903. [DOI] [PubMed] [Google Scholar]
- 142. Grunewald M, Johnson S, Lu D, et al. Mechanistic role for a novel glucocorticoid-KLF11 (TIEG2) protein pathway in stress-induced monoamine oxidase A expression. J Biol Chem. 2012;287:24195-24206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sevilla LM, Latorre V, Carceller E, et al. Glucocorticoid receptor and Klf4 co-regulate anti-inflammatory genes in keratinocytes. Mol Cell Endocrinol. 2015;412:281-289. [DOI] [PubMed] [Google Scholar]
- 144. Das A, Fernandez-Zapico ME, Cao S, et al. Disruption of an SP2/KLF6 repression complex by SHP is required for farnesoid X receptor-induced endothelial cell migration. J Biol Chem. 2006;281:39105-39113. [DOI] [PubMed] [Google Scholar]
- 145. Bagamasbad P, Ziera T, Borden SA, et al. Molecular basis for glucocorticoid induction of the Krüppel-like factor 9 gene in hippocampal neurons. Endocrinology. 2012;153:5334-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Chinenov Y, Coppo M, Gupte R, Sacta MA, Rogatsky I. Glucocorticoid receptor coordinates transcription factor-dominated regulatory network in macrophages. BMC Genomics. 2014;15:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kino T. Stress, glucocorticoid hormones, and hippocampal neural progenitor cells: implications to mood disorders. Front Physiol. 2015;6:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kino T, Chrousos GP. Glucocorticoid effect on gene expression. In: Steckler T, Kalin NH, Reul JMHM, eds. Handbook on Stress and the Brain Part 1. Amsterdam, The Netherlands: Elsevier BV; 2004;295-312. [Google Scholar]
- 149. Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1:81-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Guo B, Aslam F, van Wijnen AJ, et al. YY1 regulates vitamin D receptor/retinoid X receptor mediated transactivation of the vitamin D responsive osteocalcin gene. Proc Natl Acad Sci U S A. 1997;94:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bergad PL, Towle HC, Berry SA. Yin-yang 1 and glucocorticoid receptor participate in the Stat5-mediated growth hormone response of the serine protease inhibitor 2.1 gene. J Biol Chem. 2000;275:8114-8120. [DOI] [PubMed] [Google Scholar]
- 152. Miller-Hodges E, Hohenstein P. WT1 in disease: shifting the epithelial-mesenchymal balance. J Pathol. 2012;226:229-240. [DOI] [PubMed] [Google Scholar]
- 153. Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868-876. [DOI] [PubMed] [Google Scholar]
- 154. Rampal R, Figueroa ME. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica. 2016;101:672-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Maurer U, Jehan F, Englert C, et al. The Wilms’ tumor gene product (WT1) modulates the response to 1,25-dihydroxyvitamin D3 by induction of the vitamin D receptor. J Biol Chem. 2001;276:3727-3732. [DOI] [PubMed] [Google Scholar]
- 156. Wagner KD, Wagner N, Sukhatme VP, Scholz H. Activation of vitamin D receptor by the Wilms’ tumor gene product mediates apoptosis of renal cells. J Am Soc Nephrol. 2001;12:1188-1196. [DOI] [PubMed] [Google Scholar]
- 157. Michiels JF, Perrin C, Leccia N, Massi D, Grimaldi P, Wagner N. PPARβ activation inhibits melanoma cell proliferation involving repression of the Wilms’ tumour suppressor WT1. Pflugers Arch. 2010;459:689-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Proudhon C, Hao B, Raviram R, Chaumeil J, Skok JA. Long-range regulation of V(D)J recombination. Adv Immunol. 2015;128:123-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lin CY, Vega VB, Thomsen JS, et al. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 2007;3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Weth O, Weth C, Bartkuhn M, Leers J, Uhle F, Renkawitz R. Modular insulators: genome wide search for composite CTCF/thyroid hormone receptor binding sites. PLoS One. 2010;5:e10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Spengler D, Villalba M, Hoffmann A, et al. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Varrault A, Ciani E, Apiou F, et al. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci U S A. 1998;95:8835-8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Hoffmann A, Barz T, Spengler D. Multitasking C2H2 zinc fingers link Zac DNA binding to coordinated regulation of p300-histone acetyltransferase activity. Mol Cell Biol. 2006;26:5544-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Hoffmann A, Spengler D. A new coactivator function for Zac1’s C2H2 zinc finger DNA-binding domain in selectively controlling PCAF activity. Mol Cell Biol. 2008;28:6078-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Tanner MM, Tirkkonen M, Kallioniemi A, et al. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res. 1995;1:1455-1461. [PubMed] [Google Scholar]
- 168. Turner J, Crossley M. Basic Krüppel-like factor functions within a network of interacting haematopoietic transcription factors. Int J Biochem Cell Biol. 1999;31:1169-1174. [DOI] [PubMed] [Google Scholar]
- 169. Turner J, Crossley M. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236-240. [DOI] [PubMed] [Google Scholar]
- 170. Harder L, Puller AC, Horstmann MA. ZNF423: transcriptional modulation in development and cancer. Mol Cell Oncol. 2014;1:e969655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Holzel M, Huang S, Koster J, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zhou L, Zhong Y, Yang FH, et al. Kaiso represses the expression of glucocorticoid receptor via a methylation-dependent mechanism and attenuates the anti-apoptotic activity of glucocorticoids in breast cancer cells. BMB Rep. 2016;49:167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Chen Z, Brand NJ, Chen A, et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-α locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166-172. [DOI] [PubMed] [Google Scholar]
- 175. Fahnenstich J, Nandy A, Milde-Langosch K, Schneider-Merck T, Walther N, Gellersen B. Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Mol Hum Reprod. 2003;9:611-623. [DOI] [PubMed] [Google Scholar]
- 176. Melnick A, Licht JD. Deconstructing a disease: rARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167-3215. [PubMed] [Google Scholar]
- 177. Zelent A, Guidez F, Melnick A, Waxman S, Licht JD. Translocations of the RARα gene in acute promyelocytic leukemia. Oncogene. 2001;20:7186-7203. [DOI] [PubMed] [Google Scholar]
- 178. Boukarabila H, Saurin AJ, Batsche E, et al. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23:1195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Huntley S, Baggott DM, Hamilton AT, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nowick K, Fields C, Gernat T, Caetano-Anolles D, Kholina N, Stubbs L. Gain, loss and divergence in primate zinc-finger genes: a rich resource for evolution of gene regulatory differences between species. PLoS One. 2011;6:e21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Berg IL, Neumann R, Lam KW, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Bellefroid EJ, Marine JC, Ried T, et al. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 1993;12:1363-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Rosati M, Marino M, Franze A, Tramontano A, Grimaldi G. Members of the zinc finger protein gene family sharing a conserved N-terminal module. Nucleic Acids Res. 1991;19:5661-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Friedman JR, Fredericks WJ, Jensen DE, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067-2078. [DOI] [PubMed] [Google Scholar]
- 185. Vissing H, Meyer WK, Aagaard L, Tommerup N, Thiesen HJ. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153-157. [DOI] [PubMed] [Google Scholar]
- 186. Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci U S A. 1996;93:15299-15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kikuchi M, Okumura F, Tsukiyama T, et al. TRIM24 mediates ligand-dependent activation of androgen receptor and is repressed by a bromodomain-containing protein, BRD7, in prostate cancer cells. Biochim Biophys Acta. 2009;1793:1828-1836. [DOI] [PubMed] [Google Scholar]
- 188. Le Douarin B, Nielsen AL, Garnier JM, et al. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701-6715. [PMC free article] [PubMed] [Google Scholar]
- 189. Le Douarin B, Nielsen AL, You J, Chambon P, Losson R. TIF1α: a chromatin-specific mediator for the ligand-dependent activation function AF-2 of nuclear receptors? Biochem Soc Trans. 1997;25:605-612. [DOI] [PubMed] [Google Scholar]
- 190. Nielsen AL, Ortiz JA, You J, et al. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78-90. [DOI] [PubMed] [Google Scholar]
- 192. Okumura K, Sakaguchi G, Naito K, Tamura T, Igarashi H. HUB1, a novel Krüppel type zinc finger protein, represses the human T cell leukemia virus type I long terminal repeat-mediated expression. Nucleic Acids Res. 1997;25:5025-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537-1549. [DOI] [PubMed] [Google Scholar]
- 194. Kim JH, Yang CK, Stallcup MR. Downstream signaling mechanism of the C-terminal activation domain of transcriptional coactivator CoCoA. Nucleic Acids Res. 2006;34:2736-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Nuttle X, Giannuzzi G, Duyzend MH, et al. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 2016;538:205-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Stabach PR, Thiyagarajan MM, Weigel RJ. Expression of ZER6 in ERα-positive breast cancer. J Surg Res. 2005;126:86-91;discussion 81-82. [DOI] [PubMed] [Google Scholar]
- 197. Mizutani T, Yamada K, Yazawa T, Okada T, Minegishi T, Miyamoto K. cloning and characterization of gonadotropin-inducible ovarian transcription factors (GIOT1 and -2) that are novel members of the (Cys)(2)-(His)(2)-type zinc finger protein family. Mol Endocrinol. 2001;15:1693-1705. [DOI] [PubMed] [Google Scholar]
- 198. Yazawa T, Mizutani T, Yamada K, et al. Involvement of cyclic adenosine 5’-monophosphate response element-binding protein, steroidogenic factor 1, and Dax-1 in the regulation of gonadotropin-inducible ovarian transcription factor 1 gene expression by follicle-stimulating hormone in ovarian granulosa cells. Endocrinology. 2003;144:1920-1930. [DOI] [PubMed] [Google Scholar]
- 199. Subaran RL, Odgerel Z, Swaminathan R, Glatt CE, Weissman MM. Novel variants in ZNF34 and other brain-expressed transcription factors are shared among early-onset MDD relatives. Am J Med Genet B Neuropsychiatr Genet. 2016;171:333-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Takase K, Ohtsuki T, Migita O, et al. Association of ZNF74 gene genotypes with age-at-onset of schizophrenia. Schizophr Res. 2001;52:161-165. [DOI] [PubMed] [Google Scholar]
- 201. Sabin LR, Delas MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell. 2013;49:783-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [DOI] [PubMed] [Google Scholar]
- 203. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Quinn JJ, Ilik IA, Qu K, et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321-344. [DOI] [PubMed] [Google Scholar]
- 207. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]