Abstract
The prevalence of comorbid diseases poses a major health issue for millions of people worldwide and an enormous socio-economic burden for society. The molecular mechanisms for the development of comorbidities need to be investigated. For this purpose, a workflow system was developed to aggregate data on biomedical entities from heterogeneous data sources. The process of integrating and merging all data sources of the workflow system was implemented as a semi-automatic pipeline that provides the import, fusion, and analysis of the highly connected biomedical data in a Neo4j database GenCoNet. As a starting point, data on the common comorbid diseases essential hypertension and bronchial asthma was integrated. GenCoNet (https://genconet.kalis-amts.de) is a curated database that provides a better understanding of hereditary bases of comorbidities.
Keywords: Database, Workflow, Pipeline, Asthma, Hypertension, Comorbidity
1. Introduction
One important challenge of molecular medicine is the analysis and treatment of comorbid diseases in humans. Comorbid diseases are one or more additional diseases co-occuring in the presence of a primary disease. The urgency of this issue is determined by the need to establish the molecular mechanisms for the development of complex multifactorial diseases (MDs) and the growing attention to the issue of comorbidity. Most common MDs in human populations are complex and their development is based on the interaction between genetic and environmental factors [1]. Taking this into account, the transition from the genetic analysis of specific diseases to the study of their systems, developing on the basis of common metabolic and physiological pathways, will provide a better understanding of hereditary bases of MDs. Furthermore, this will allow the identification of genetic factors that determine the characteristics of the joint manifestations of the disease and will form the basis for the discovery of new drug targets for combined pathological phenotypes. It is shown that the diseases tend to exhibit comorbidity if they share susceptibility genes [2] and a significant comorbidity is observed between diseases with similar evolutionary influences [3].
In this context asthma is a chronic respiratory disease, the prevalence of which has increased significantly in recent years [4]. One of the features of the clinical course of asthma is its comorbidity with cardiovascular disease. According to epidemiological studies, asthma has a decisive influence on the subsequent risk of cardiovascular disease. Essential hypertension and bronchial asthma are considered as an example of common comorbid diseases [5].
Previous research suggests that drug interactions and drug contraindications represent a very important aspect of the issue of comorbid diseases, especially in polypharmacy [6]. Furthermore, the existence of common molecular mechanisms underlying the pathology of comorbid diseases may have a significant impact on drug interactions [2]. For the detection of drug interactions, different information systems are available and in use [7]. For this project, we integrate the relevant data of asthma and hypertension to support the analysis of the comorbidity of asthma and hypertension.
2. Related Works
Integration of independent data sources of the same type helps to improve data completeness and quality. In addition, the integration of different, but complementary data sources allows achieving a more complete view of biological processes and is necessary for hypothesis generation.
The rapid increase in the volume, variety, heterogeneity, and complexity of biological data made storage, search, and access to these data a challenging task. Several large data integration resources with flexible and user-friendly graphical interfaces were created in order to facilitate access, manipulation and integration to these data for biologists without programming skills. Here, we focus on data warehouses and web-portals developed to study human genetics and genomics in the context of diseases [8], [9], [10], [11].
InterMine [8] (http://intermine.org) is a software system facilitating the creation of biological data warehouses with a flexible API and web interface. Many species-specific data integration resources are built on the basis of InterMine. HumanMine [8] (http://www.humanmine.org) aggregates various information about human genes, diseases, variants and other biological entities. It recognizes many identifiers and allows constructing and manipulating the list of entities and widgets calculates enrichments, e.g. pathway or tissue enrichments for lists of genes. Also, InterMine provides pre-defined queries facilitating mappings, e.g. genes to orthologues, proteins to pathways, expressions to tissues. Additionally, InterMine enables automatic generation of script templates in different programming languages. TargetMine [9] (http://targetmine.mizuguchilab.org) is a similar resource based on InterMine but focused on the discovery of potential drugs and drug targets. It integrates specific evidence sources, providing associations between drugs and genes, e.g. DrugBank [12] and ChEMBL [13].
Heterogeneous biological data can be represented either as table data or knowledge network. Some data integration tools, such as KnetMiner and Hetionet, utilize network structure instead of relational, which is especially convenient for storage of sparse data and for applying algorithms determining node relevance in networks.
KnetMiner [10] (http://knetminer.rothamsted.ac.uk) is developed for investigating relationships between genes and phenotypes and for eQTL search. KnetMiner is based on Ondex [14], now integrates over 25 data sources and covers many organisms including human. Also, KnetMiner can be used for gene prioritization: takes phenotype and optionally a list of candidates and scores them on the basis of previous knowledge.
Hetionet [11] (http://neo4j.het.io/browser) is a graph database implemented in Neo4j. It represents a heterogeneous network including 11 types of nodes representing biological entities and 24 types of edges interactions. The first version of Hetionet was used for prioritizing of genes from loci associated with multiple sclerosis in GWAS [11]. Later, the updated version of Hetionet was successfully applied to in-silico drug re-purposing [15]. The same approach can be used for the discovery of novel associations between biological entities of any type.
While HumanMine, TargetMine, and KnetMiner all integrate several entities of interest like drugs, diseases and genes and their relationships, none of them integrates a combined dataset on all entities relevant to the detection of comorbidities. Hetionet, on the other hand, integrates many relevant entities and relationships from very different sources and varying quality including predictions. In contrast, the goal of GenCoNet is to only integrate data sources of high quality, specifically for the analysis of comorbidities and the reduction of the database to relevant connected entities. This allows for a more precise analysis of drug-disease relationships in comorbid diseases.
3. Workflow
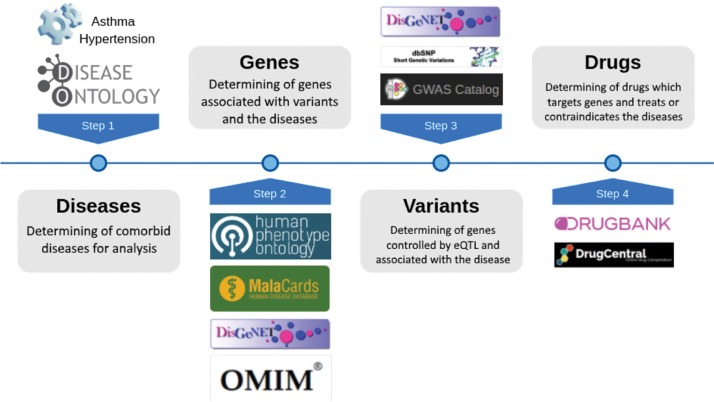
The investigation of multifactorial diseases requires the consideration of the most important biomedical entities: diseases, genes, variants, and drugs. The data on these entities is spread across several data sources. These data sources need to be researched, identified, and integrated into a suitable database format. For this purpose, a workflow system was developed to extract, transform, and load reliable data on these entities into a uniform graph database GenCoNet for network analysis of gene-disease associations. The workflow system is divided into four steps as illustrated in Figure 1.
Figure 1:
Four steps of the workflow system for generating GenCoNet semi-automatically.
First, the comorbidities of high interest in molecular medicine were determined. Essential hypertension and bronchial asthma are considered as an example of common comorbid diseases. Disease Ontology terms associated with genes were obtained from Osborne et al. [16]. Second, human genes that are associated with diseases and variants were obtained from Human Phenotype Ontology (HPO) [17], MalaCards [18], DisGeNet [19], and OMIM [20]. In particular, human genes were extracted from these databases that may cause familial syndromes (Mendelian forms). HPO and DisGeNet provide several database subsets from which the “FREQUENT_FEATURES” marked data for HPO and “curated” marked data for DisGeNet were used. MalaCards does not provide a download of the database, so information was manually extracted and integrated from the website. From the OMIM gene map, information was extracted with the phenotype “Asthma, susceptibility to”, “Hypertension, essential, susceptibility to” and adjacent annotation. In addition, altered expression data of genes associated with high blood pressure or severe asthma were manually curated and integrated [21], [22]. Third, genes that are controlled by eQTL in blood [23], codes variants, and gene associations supported by at least two studies were obtained from GWAS Catalog [24], dbSNP [25], and DisGeNet. These genetic variants were in turn associated with the comorbid diseases. Fourth, drugs and their target genes in humans were extracted from DrugBank [12] using the full database XML version 5.1.0 including meta information like known actions. Additional drugs, that are indicated, contraindicated or induced in asthma and hypertension were extracted from the DrugCentral 2017-08-29 database dump [7].
4. Implementation
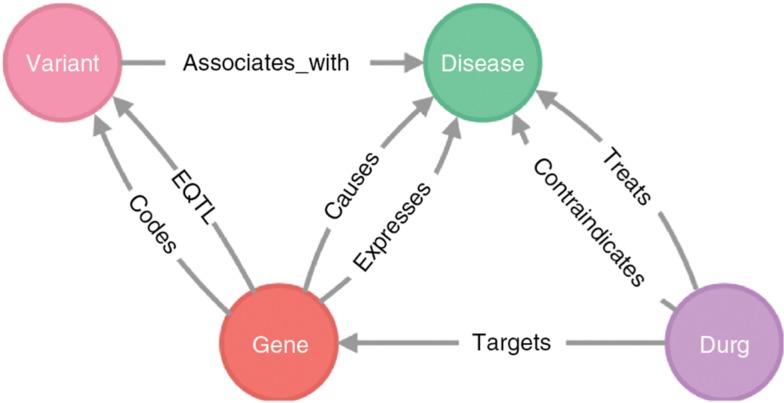
The process of integrating all data sources and merging of the entities of the aforementioned workflow system is an iterative process. Therefore, a semi-automatic pipeline was implemented for the import, fusion, and analysis of data in a highly connected database structure, in this case being Neo4j. This pipeline provides custom and very fast import Python scripts and Cypher queries for generating a new Neo4j database from large data. The Neo4j instance is running in a Docker container to simplify the setup process. Each step of the workflow system is executed in a separate Python script in order to have a better overview and to be able to execute specific steps on demand. Some of the data sources need to be preprocessed due to the higher information density or more complicated file formats than others. Afterwards, the data sources are processed and the basic connections are formed. Following nodes representing the same kind of entities are merged into fusion nodes. Finally, the results were imported into the new GenCoNet database with the respective entities and relationships generated as illustrated in Figure 2.
Figure 2:
Data model of the GenCoNet database.
As summarized in Table 1, GenCoNet is a curated graph database which contains reliable data for identifying novel candidate genes that can support in elucidating the molecular mechanisms underlying the comorbid conditions of asthma and hypertension. Furthermore, the database can be used for genotyping and determining new drug targets without potential comorbidities. This allows completing paths from drugs over genes to diseases to be formed by indirectly related information, just by connecting different entities and identifiers. Aside from a browser interface, GenCoNet can be accessed and manipulated via programming language libraries using the Cypher query language for Neo4j. GenCoNet is fully accessible and available at https://genconet.kalis-amts.de.
Table 1:
Total number of nodes and relationships.
| Node labels | Total |
|---|---|
| Disease | 2 |
| Drug | 235 |
| Gene | 1774 |
| Variant | 5192 |
| Relationship types | Total |
| associates_with | 520 |
| eQTL | 1163 |
| codes | 39 |
| causes | 31 |
| expresses | 1041 |
| targets | 1003 |
| contraindicates | 95 |
| Treats | 151 |
5. Application
The prevalence of comorbidity is increasing and leads to a corresponding polypharmacy, which in turn is the prime risk factor for drug-related problems. In particular, drug contraindications with any disease and drug-induced diseases have to be considered in the context of treatment by healthcare professionals. Using GenCoNet, these risks may be detected as shown in the following use cases.
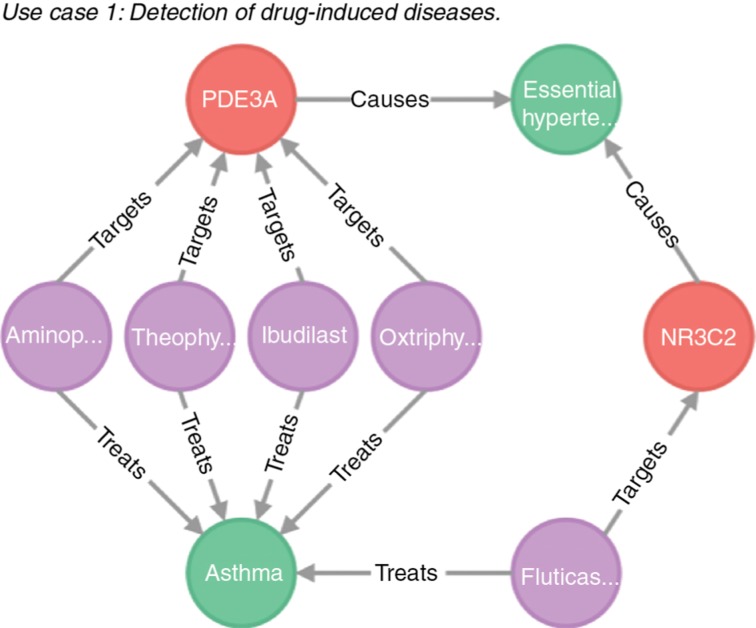
The web interface represents the database entities (diseases, drugs, and proteins) as nodes in a network with edges representing the relationships between them. As illustrated in Figure 3, the network shows drugs (violet) which are prescribed for the treatment of the comorbid diseases (green) but also target genes (red) which may cause the diseases. For instance, the anti-asthmatic drug “Ibudilast” may induce hypertension by targeting the gene PDE3A. As a consequence, these drugs have to be avoided in order to reduce the risk of induced diseases.
Figure 3:
Network analysis of drug-induced diseases.
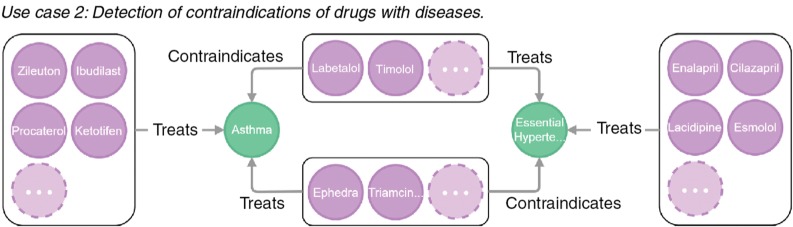
Figure 4 shows a network of diseases (green) encircled by drugs (violet) that can be prescribed for their treatment and a vertical central line of drugs that are contraindicated. For instance, triamcinolone can be used to treat asthma but worsening hypertension.
Figure 4:
Network analysis for contraindications of drugs.
6. Result
The diversity of information is overwhelming even for healthcare professionals due to the excessive time pressure in the daily medical practice. Based on GenCoNet, we offer a positive and negative list of drugs with respect to comorbidity that can be considered in different decision-making situations for appropriate treatment. The positive list (Supplementary Table 1) includes 95 drugs that are presumed to treat the indicated diseases and do not have any impact on the comorbidity. In contrast, the negative list (Supplementary Table 2) includes 51 drugs that are presumed to treat the indicated diseases and do have a negative impact on the comorbidity, e.g. contraindication or induction. The assignment of drugs with diseases in the resulting tables was reviewed by healthcare professionals among the authors.
7. Discussion
The discovery of shared molecular players and mechanisms in the pathogenesis of comorbid diseases is still complicated and nevertheless, necessary for decision-making of the most appropriate treatment strategy. To address this ongoing need, the Neo4j database GenCoNet was developed which integrates various associations between diseases, genes, variants and drugs for the diseases bronchial asthma and essential hypertension that have a strong molecular-genetic component and demonstrate the comorbidity. While information of highest quality is preferred for integration in GenCoNet, false positives, although rare, cannot be ruled out. However, the use cases and lists already emphasized the potential of applicability in daily medical practice. GenCoNet is meant to be a qualitative resource that facilitates researchers access to the relevant information for the network analysis of comorbidities. Therefore, GenCoNet is planned to be extended by data on further diseases as diabetes, Alzheimer’s disease, and preeclampsia using the implemented pipeline.
Availability and Requirements
GenCoNet is available at https://genconet.kalis-amts.de. To fully access all features of the database, an up-to-date browser version should first be installed on your PC or mobile device.
Supporting Information
Supplementary Material
The online version of this article offers supplementary material (DOI: jib-2018-0049).
Funding Statement
This work was supported by the International DFG Research Training Group GRK Funder Id: 10.13039/501100001659, 1906/1 and the Volkswagen Foundation Trilateral Partnerships Funder Id: 10.13039/501100001663, 90335 – Cooperation Projects between Scholars and Scientists from Ukraine, Russia, and Germany “In silico screening and experimental validation of new drug targets for the treatment of co-morbid multifactorial diseases”.
Footnotes
Expression Quantitative Trait Locus.
Neo4j is a Database Management System implementing the graph database model.
Cypher is Neo4j’s graph query language.
Docker is a computer program to run software packages called containers.
Conflict of interest statement
Authors state no conflict of interest. All authors have read the journal’s Publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Ober C. Asthma genetics in the Post-GWAS Era. Ann Am Thorac Soc. 2016;13(1):85–90. doi: 10.1513/AnnalsATS.201507-459MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA. 2007;104(28):11694–9. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Park S, Yang JS, Kim J, Shin YE, Hwang J, Park J. et al. Evolutionary history of human disease genes reveals phenotypic connections and comorbidity among genetic diseases. Sci Rep. 2012;2:757. doi: 10.1038/srep00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pelkonen MK, Notkola IK, Laatikainen TK, Jousilahti P. 30-year trends in asthma and the trends in relation to hospitalization and mortality. Respir Med. 2018;142:29–35. doi: 10.1016/j.rmed.2018.07.012. [DOI] [PubMed] [Google Scholar]
- [5].Aung T, Bisognano JD, Morgan MA. Allergic respiratory disease as a potential co-morbidity for hypertension. Cardiol J. 2010;17(5):443–7. [PubMed] [Google Scholar]
- [6].Dumbreck S, Flynn A, Nairn M, Wilson M, Treweek S, Mercer SW. et al. Drug-disease and drug-drug interactions: systematic examination of recommendations in 12 UK national clinical guidelines. Br Med J. 2015;350:h949. doi: 10.1136/bmj.h949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ursu O, Holmes J, Knockel J, Bologa CG, Yang JJ, Mathias SL. et al. DrugCentral: online drug compendium. Nucleic Acids Res. 2017;45(Database issue):D932–9. doi: 10.1093/nar/gkw993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J. et al. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42(Web Server issue):W468–72. doi: 10.1093/nar/gku301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen YA, Tripathi LP, Mizuguchi K. An integrative data analysis platform for gene set analysis and knowledge discovery in a data warehouse framework. Database (Oxford); 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hassani-Pak K, Castellote M, Esch M, Hindle M, Lysenko A, Taubert J. et al. Developing integrated crop knowledge networks to advance candidate gene discovery. Appl Transl Genom. 2016;11:18–26. doi: 10.1016/j.atg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Himmelstein DS, Baranzini SE. Heterogeneous network edge prediction: a data integration approach to prioritize disease-associated genes. PloS Comput Biol. 2015;11(7):e1004259. doi: 10.1371/journal.pcbi.1004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D. et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(Database issue):D945–54. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Taubert J, Hassani-Pak K, Castells-Brooke N, Rawlings CJ. Ondex Web: web-based visualization and exploration of heterogeneous biological networks. Bioinformatics. 2014;30(7):1034–5. doi: 10.1093/bioinformatics/btt740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Himmelstein DS, Lizee A, Hessler C, Brueggeman L, Chen SL, Hadley D. et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017;6:e26726. doi: 10.7554/eLife.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Osborne JD, Flatow J, Holko M, Lin SM, Kibbe WA, Zhu L. et al. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10(Suppl 1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S. et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017;45(D1):D865–76. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J. et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45(D1):D877–87. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–9. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD), 28.04.2018. World Wide Web URL: https://omim.org.
- [21].Huan T, Esko T, Peters MJ, Pilling LC, Schramm K, Schurmann C. et al. A meta-analysis of gene expression signatures of blood pressures and hypertension. PloS Genet. 2015;11(3):e1005035. doi: 10.1371/journal.pgen.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bigler J, Boedigheimer M, Schofield JPR, Skipp PJ, Corfield J, Rowe A. et al. A severe asthma signature from gene expression profiling of peripheral blood from BIOPRED cohorts. Am J Respir Crit Care Med. 2017;195(10):1311–20. doi: 10.1164/rccm.201604-0866OC. [DOI] [PubMed] [Google Scholar]
- [23].Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(Database issue):D896–901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sherry ST, Ward MH, Kholodov M. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.