Abstract
Cadmium is a toxic pollutant that in recent decades has become more widespread in the environment due to anthropogenic activities, significantly increasing the risk of exposure. Concurrently, a continually growing body of research has begun to enumerate the harmful effects that this heavy metal has on human health. Consequently, additional research is required to better understand the mechanism and effects of cadmium at the molecular level. The main mechanism of cadmium toxicity is based on the indirect induction of severe oxidative stress, through several processes that unbalance the anti-oxidant cellular defence system, including the displacement of metals such as zinc from its native binding sites. Such mechanism was thought to alter the in vivo enzymatic activity of SOD1, one of the main antioxidant proteins of many tissues, including the central nervous system. SOD1 misfolding and aggregation is correlated with cytotoxicity in neurodegenerative diseases such as amyotrophic lateral sclerosis. We assessed the effect of cadmium on SOD1 folding and maturation pathway directly in human cells through in-cell NMR. Cadmium does not directly bind intracellular SOD1, instead causes the formation of its intramolecular disulfide bond in the zinc-bound form. Metallothionein overexpression is strongly induced by cadmium, reaching NMR-detectable levels. The intracellular availability of zinc modulates both SOD1 oxidation and metallothionein overexpression, strengthening the notion that zinc-loaded metallothioneins help maintaining the redox balance under cadmium-induced acute stress.
Keywords: Cadmium, Superoxide dismutase 1, In-cell NMR, Oxidative stress, Metallothionein
Graphical abstract
Highlights
-
•
Cadmium does not bind to superoxide dismutase 1 (SOD1) in human cells.
-
•
In defect of zinc, cadmium causes the premature oxidation of SOD1.
-
•
Cadmium induces the overexpression of metallothioneins to levels detectable by NMR.
-
•
Zinc modulates metallothionein expression and attenuates SOD1 oxidation.
1. Introduction
Cadmium is a potent environmental and food contaminant that is ranked as the seventh most hazardous substance for human health when considering both toxicity and exposure frequency [1]. Furthermore, it has been classified as a category 1 carcinogen by the International Agency for Research on Cancer [2]. Cadmium can spread in the environment due to natural processes like volcanic eruptions and erosion, but during recent decades anthropogenic activities such as agriculture and industry have significantly boosted its dispersal in soil and water [3]. Cadmium compounds are very soluble in water, facilitating its uptake by plants and consequently human exposure as a food and tobacco contaminant [4], [5]. Cadmium is not a physiological element for any living organism, and furthermore is a cumulative toxin with an extremely long estimated half-life (up to 13.6–23.5 years) due to its very low excretion rate [6]. In mammals, the main accumulation sites of cadmium are kidneys, liver, lungs, bone, testes and brain, where it causes severe oxidative stress and other detrimental effects [7], [8]. Unlike iron and copper, cadmium is not a Fenton-like metal, consequently it is not involved directly in the production of Reactive Oxygen Species (ROS). Rather, it increases their intracellular production and their harmful effects by altering the balance of the cellular anti-oxidative defence system. In fact, cadmium is thought to be responsible for the replacement of metals from their catalytic sites [9], [10], [11], [12]; the depletion of anti-oxidant metabolites, like glutathione, ascorbic acid and vitamin E; the inhibition of the electron transport chain, resulting in mitochondrial damage; and the alteration of the enzymatic activity of anti-oxidant proteins [13], [14], [15]. One of the main cellular defence tools against cadmium are metallothioneins (MTs), a family of highly conserved, cysteine-rich small proteins with several key functions: toxic metal and radical scavenging, maintenance of the intracellular zinc homeostasis, maintenance of the intracellular redox balance and protection against DNA damage [16], [17], [18]. Mammalian MTs have about 60 amino acids, one third of which are cysteines, forming two metal-thiolate clusters. The protein fold is induced by metal ions (such as Zn2+, Cd2+ and Cu+), and cluster formation [19]. Human MTs have 11 functional isoforms divided in 4 subfamilies (MT1-4), which share a high degree of homology and are differentially expressed depending on the tissue [20]. The expression of MTs is known to be induced by Cd2+ exposure through a mechanism mediated by the release of intracellular free Zn2+ [21].
Among the anti-oxidant proteins affected by cadmium intoxication, literature reports the involvement of cadmium in the alteration of the enzymatic activity of superoxide dismutase 1 (SOD1) [13]. SOD1 is a metalloprotein that exerts a fundamental anti-oxidant role catalysing the dismutation of O2•- to H2O2 and O2 with extremely high reaction rates [22], [23], [24]. SOD1 enzymatic activity is essential for the maintenance of the physiological cellular conditions and requires correct protein folding, metal binding and disulfide bond formation. The impairment of SOD1 maturation has been also related to the onset of severe disease states, including amyotrophic lateral sclerosis (ALS) [25], [26], [27], [28], [29].
In vitro, cadmium has been shown to bind to the zinc and copper sites of SOD1 [30], which are necessary for the structural stability and catalytic activity of SOD1, respectively. In vivo, however, opposing effects have been reported on SOD1 activity in response to cadmium exposure, and evidence of alteration at the protein level is lacking [31], [32], [33]. Therefore, it is not clear whether/how cadmium affects SOD1 folding and metallation within the cell, i.e. in the presence of the native SOD1 metal ions and under the control of the cellular metal and redox homeostasis.
Given these contrasting premises, we sought to evaluate the effects of cadmium treatment on the maturation of SOD1 in human cells by in-cell NMR, to determine whether cadmium binds to the zinc and/or copper sites or it affects intracellular SOD1 maturation by other mechanisms. To this aim, in-cell NMR is the ideal technique, as it is able to analyse proteins at atomic resolution directly in living cells. The same technique has been applied previously to observe changes in the intracellular SOD1 folding, metallation and redox state as a consequence of the physiological maturation and/or in response to external stimuli [34], [35], [36], [37].
2. Materials and methods
2.1. In-cell NMR
In-cell NMR experiments have been performed as previously described [38] on living human embryonic kidney cells (HEK293T), under three main experimental conditions: i) exposure to Zn2+ (added in the culture at the time of transfection with SOD1); ii) exposure to Cd2+ (added in the culture 24 h after the transfection with SOD1); iii) exposure to Zn2+and Cd2+ (both added in the culture at the aforementioned times). HEK293T cells were grown on uncoated 75 cm2 plastic flasks at 37 °C in 5% CO2 atmosphere, and were maintained in Dulbecco's Modified Eagle's medium (DMEM; high glucose, D6546, Sigma-Aldrich, St. Louis, MO) supplemented with L-glutamine, antibiotics (penicillin and streptomycin) and 10% foetal bovine serum (FBS) (Gibco-Thermo Fisher Scientific, Waltham, MA). Cells were transiently transfected with the pHLsec plasmid [39] encoding for human SOD1, using polyethylenimine (PEI) in the ratio 1:1 (25 μg each), in 15N labelled media (BioExpress6000, Cambridge Isotope Laboratories, Inc., Tewksbury, MA), supplemented 2% FBS in the presence/absence of Zn2+ as ZnSO4 10 μM. Under these conditions, ~150 μM SOD1 is expressed [38]. To decrease the expression levels of SOD1, the pHLsec encoding SOD1 was mixed 1:1 with empty vector and transfected as above, resulting in the expression of ~65 μM SOD1. 24 h after the transfection, 10 μM of CdCl2 was added to the cell cultures; such concentration was chosen considering previous experiments performed on Hep3B and N2A cells [32], [40]. After 24 h of exposure to cadmium, the cells were washed twice with PBS, trypsinised, spun at 500 g after trypsin inactivation, resuspended once in PBS and spun down again at 500 g. Such procedure allows efficient removal of debris from dead cells and of apoptotic cells, if present. Cell viability was assessed both before and after NMR analysis by counting cells stained with trypan blue using a Burker chamber. Cd2+ treatment caused a reduction of ~40% in the final number of cells analysed by NMR, likely due to cell death/apoptosis. However, the fraction of cells treated with Cd2+ that was recovered and analysed by NMR had the same viability as the Cd2+-untreated cells (>95% trypan blue-negative before the NMR experiments, >90% after the NMR experiments). For NMR analysis, the recovered cells were collected and placed in a 3 mm Shigemi NMR tube. 1H WATERGATE (3-9-19) and 1H–15N SOFAST-HMQC NMR spectra were acquired on living HEK293T cells and on lysates at a 950 MHz Bruker (Billerica, MA) Avance III or at a 900 MHz Bruker Avance HD spectrometer both equipped with a TCI CryoProbe, at 308 K. The cell lysates were obtained by freeze-thaw lysis in phosphate buffered saline (PBS) buffer, pH 7.4, followed by centrifugation at 14,000 rpm. For the [15N]-cysteine selective labelling of MTs, untransfected HEK293T cells were grown in homemade medium containing [15N]-cysteine (Cambridge Isotope Laboratories, Inc.); NMR spectra were acquired on the corresponding cell lysate at 298 K. All NMR spectra were acquired and processed using Bruker Topspin software. The uniformly-15N labelled in-cell NMR spectra were further processed by subtracting a spectrum of cells transfected with empty vector, acquired in the same experimental conditions, to eliminate the signals arising from partial 15N labelling of other cellular components.
The intracellular oxidation state of E,Zn-SOD1 was determined from the intensities of the signals arising from G10 and N53 Nδ1 in each oxidation state in the 1H–15N NMR spectra. The amount of oxidised/reduced E,Zn-SOD1 in each condition are reported as percentages over total E,Zn-SOD1 calculated for two independent cell samples.
2.2. In vitro NMR
Recombinant SOD1 was demetallated as previously described [41] by repeated dialyses against 10 mM EDTA in 50 mM acetic acid at pH 3.5. The buffer was then replaced with PBS, pH 7.4. Reduction of SOD1 disulfide bond was performed by incubating apo-SOD1SS with 50 mM of DTT for 40 min at 37 °C. DTT was then removed in oxygen-free PBS buffer.
In vitro NMR titration experiments were performed at 950 MHz in anaerobic conditions. E,Zn-SOD1SH was obtained by titrating 150 μM apo-SOD1SH in PBS buffer, pH 7.4, with 1 equivalent of ZnCl2. Titration with Cd2+ was performed by addition of increasing equivalents of CdCl2 to a sample of 150 μM apo-SOD1SH. 1H WATERGATE (3-9-19) and 1H–15N SOFAST-HMQC NMR spectra were acquired at 298 K after each addition. Disulfide-oxidised E,Zn-SOD1 was prepared by titration of apo-SOD1SS with one equivalent of Zn2+ monitored via NMR.
2.3. Real-time PCR
Gene expression analysis by Real-Time PCR has been performed using the 2^(-ΔΔCT) comparative method of quantification [42]. Briefly, total RNA (1 µg), extracted with TriReagent™ (Sigma-Aldrich) was reverse transcribed using iScript™ cDNA Synthesis Kit for RT-qPCR (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. Human specific TaqMan Gene Expression Assays employed for gene expression studies were purchased from Thermo Fisher Scientific and listed in Table S1. The quantification of target gene mRNA levels was performed employing TaqMan Universal Master Mix (Thermo Fisher Scientific) with the automated ABI Prism 7500 Sequence Detector System (Thermo Fisher Scientific) as described previously [43]. Each measurement was carried out in triplicate, by analysing three independent cell samples. The quantification was performed by simultaneous amplification of the target gene together with the housekeeping (GAPDH) in order to normalise expression data. Results were analysed by ABI Prism Sequence Detection Systems software, version 1.7 (Applied Biosystems, Foster City, CA) and graphical representations of data as mean ± SD of three independent experiment were obtained by GraphPad Prism 5.0 (GraphPad Software, San Diego, CA).
3. Results
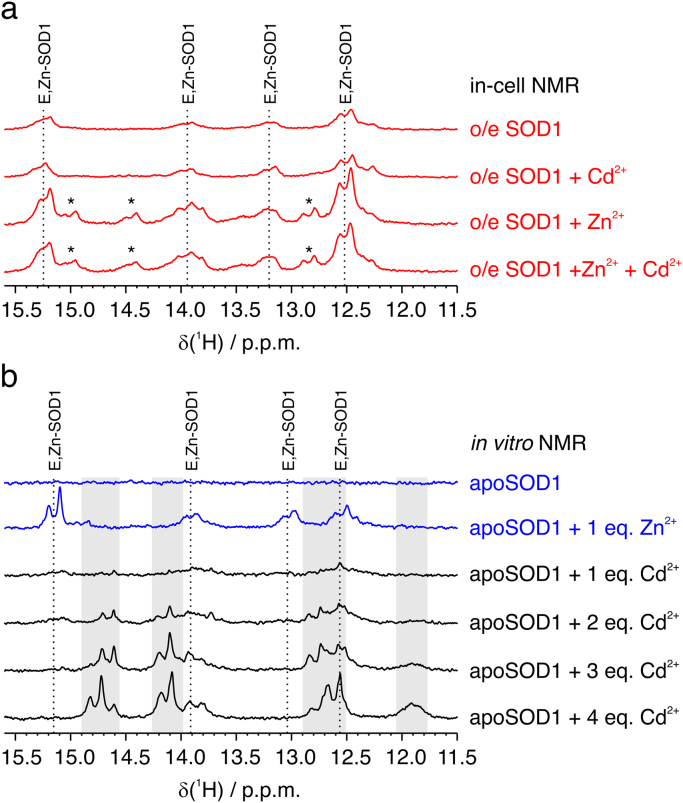
SOD1 overexpressed in human cells was analysed by in-cell NMR to determine its metallation and redox state under different conditions, by comparing the in-cell spectra with in vitro spectra of the protein in known states. The metallation state was determined from the chemical shift of the metal binding histidines in imino region of the 1H NMR spectra. Under basal conditions (i.e. without addition of metals), a fraction of overexpressed SOD1 binds the Zn2+ available in the growth medium while the remaining fraction is in the apo state (apo-SOD1, which gives no signals in the imino region), whereas addition of Zn2+ allows the complete Zn2+ binding to SOD1 with one Zn2+ per monomer (E,Zn-SOD1), as previously reported [34] (Fig. 1a). Exposure to Cd2+ for 24 h did not cause any Cd2+ binding to SOD1, as determined by comparison with the in vitro NMR spectra. Indeed, the histidine signals of intracellular SOD1 in the Cd2+-treated cells did not change with respect to the untreated cells, matched those of apo-SOD1 metallated with Zn2+ in vitro and differed from those of apo-SOD1 metallated in vitro with increasing equivalents of Cd2+ per SOD1 monomer (Fig. 1b). Moreover, the titration performed in vitro denotes that Cd2+ binds to SOD1 with lower affinity than Zn2+, as two equivalents or more of Cd2+ are necessary to observe spectral changes, suggesting that Cd2+ binding to apo-SOD1 or replacement of Zn2+ with Cd2+ is unlikely to occur in the cells.
Fig. 1.
Intracellular SOD1 binds Zn2+, not Cd2+. Imino region of (a) in-cell 1H NMR spectra of cells expressing [U-15N] labelled SOD1 and treated with different metals (red); (b) in vitro1H NMR spectra of disulfide-reduced apo-SOD1 (120 μM) alone (blue), titrated with 1 equivalent per monomer of Zn2+ (blue) and titrated with 2, 3 and 4 equivalents per monomer of Cd2+ (black). The histidine signals corresponding to the E,Zn-SOD1 form are shown with dashed lines and labelled. Signals likely arising from the non-native Zn,Zn-SOD1 form are labelled with asterisks. Spectral regions containing signals arising from Cd2+ binding to SOD1 are shown in grey.
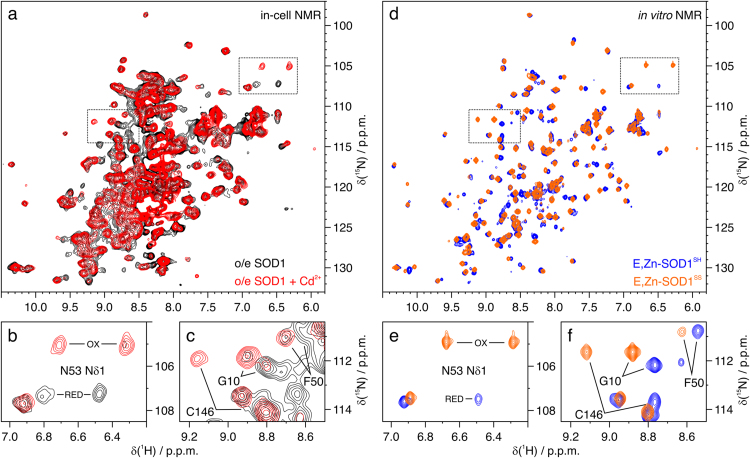
Instead, Cd2+ treatment caused the oxidation of a sizable fraction of intracellular E-Zn,SOD1, i.e. the formation of an intramolecular disulfide bond between C57 and C146. In defect of Zn2+, the metal-free protein is fully disulfide-reduced (apo-SOD1SH), while the form with one Zn2+ bound per monomer is mostly in the reduced state (60–70% E,Zn-SOD1SH over total E,Zn-SOD1, Fig. 2a). When Cd2+ was added to the cells in defect of Zn2+, a sizable fraction (65–75%) of the zinc-bound form was oxidised (E,Zn-SOD1SS), as revealed by the changes in relative signal intensity in the 2D NMR spectra, while the apo form was unaffected, i.e. it showed a NMR spectrum superimposable to that of the untreated sample, indicating that it is in the disulfide-reduced state (apo-SOD1SH) (Fig. 2a). The in-cell spectral changes induced by Cd2+ matched those observed in vitro between the reduced and oxidised forms, i.e. E,Zn-SOD1SH and E,Zn-SOD1SS (Fig. 2b). Notably, the same effect was observed in cells expressing SOD1 at lower levels, where Cd2+ caused the oxidation of ~63% of E,Zn-SOD1 (Fig. S1).
Fig. 2.
Cd2+induces intracellular SOD1 oxidation. (a) Overlay of in-cell 1H–15N NMR spectra acquired on cells expressing [U-15N] labelled SOD1 in defect of Zn2+ either untreated (black) or treated (red) with Cd2+; (d) overlay of in vitro1H–15N NMR spectra acquired on disulfide-reduced E,Zn-SOD1 (blue) and disulfide—oxidised E,Zn-SOD1 (orange); (b, c, e, f) enlarged areas of the same spectra (dotted rectangles in a, c). Representative signals affected by the formation of the C57-C146 disulfide bond are labelled.
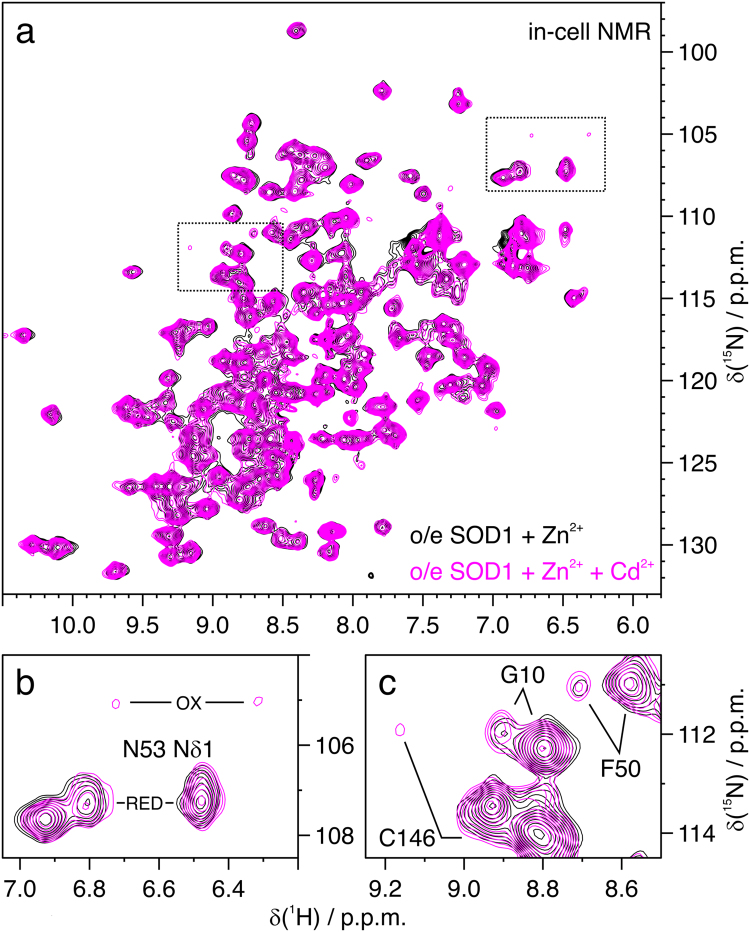
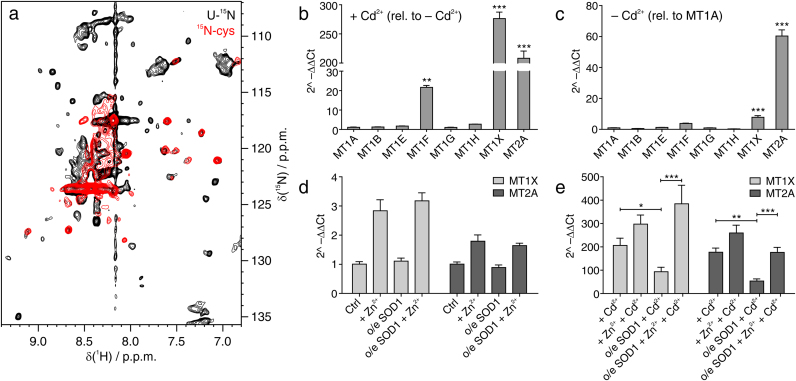
The oxidation of SOD1 induced by Cd2+ was largely attenuated by the addition of Zn2+. In the presence of excess Zn2+, intracellular E,Zn-SOD1 was mostly found in the reduced state (80–90% E,Zn-SOD1SH), as previously reported [34], [36]. When both Zn2+and Cd2+ were supplemented, a smaller fraction (25–45%) of SOD1 was oxidised (E,Zn-SODSS), while the rest of the protein remained reduced, as that observed in the Zn2+-treated cell sample (Fig. 3). The considerably smaller fraction of oxidised SOD1, compared to the sample supplemented only with Cd2+, suggested that a protective mechanism against oxidative stress had been activated by supplementing Zn2+. We hypothesised that such effect could have been exerted by MTs, considering their protective role against cadmium, their involvement in zinc homeostasis and their intracellular anti-oxidant action. Indeed, MTs are well known to be overexpressed in cell lines and tissues as a consequence of cadmium exposure [44], [45]. Interestingly, analysing the NMR data, several additional amide crosspeaks not originating from any known state of SOD1 were observed in the 2D spectra of cells treated with both Cd2+ and Zn2+ (Fig. S2). To further investigate the origin of these signals, we performed additional in-cell NMR experiments on cells not transfected with the plasmid encoding for SOD1, that were grown in either uniform-15N or [15N]-cysteine labelled medium and subsequently exposed to Cd2+. The unknown crosspeaks were also present in the resulting NMR spectra (Fig. 4a), indicating that they did not arise from SOD1 but from some other protein(s) upregulated upon Cd2+ treatment. Strikingly, many signals (~20) were also present in the [15N]-cysteine labelled sample (Fig. 4a and Fig. S3). Such a high ratio of cysteine residues is typical of MTs. Therefore, MT overexpression could explain the additional crosspeaks in Cd2+-treated cells, and the lower amount of oxidised SOD1.
Fig. 3.
Excess of Zn2+prevents Cd2+-induced SOD1 oxidation. (a) Overlay of in-cell 1H–15N NMR spectra acquired on cells expressing [U-15N] labelled SOD1 in excess of Zn2+ either untreated (black) or treated (magenta) with Cd2+; (b, c) enlarged areas of the same spectra (dotted rectangles in a). Representative signals affected by the formation of the C57-C146 disulfide bond are labelled.
Fig. 4.
Cd2+treatment induces MT overexpression to NMR-detectable levels. (a) Overlay of 1H–15N NMR spectra acquired on untransfected cells treated with Cd2+ either [U-15N] labelled (black) or [15N-cysteine] labelled (red); (b–e) Real-Time PCR analysis: (b) expression of different MT isoforms in Cd2+-treated untransfected cells, relative to basal (i.e. untransfected cells not treated with Cd2+). Significant values are marked with asterisks (Student's t-test, ** = P < 0.01; *** = P < 0.001). (c) Basal expression of different MT isoforms relative to that of MT1A. Significant values are marked with asterisks (1-way ANOVA followed by Bonferroni post-hoc test, *** = P < 0.001). (d) Expression of MT1X and MT2A in cells not treated with Cd2+, overexpressing SOD1 in defect/excess of Zn2+, relative to basal. (e) Expression of MT1X and MT2A in cells treated with Cd2+, overexpressing SOD1 in defect/excess of Zn2+, relative to basal. Significant differences are marked with asterisks (2-way ANOVA followed by Bonferroni post-hoc test, * = P < 0.05, ** = P < 0.01, *** = P < 0.001). Ctrl: untransfected, untreated cells; +Zn2+: cells treated with excess Zn2+; +Cd2+: cells treated with Cd2+; o/e SOD1: cells overexpressing SOD1.
To confirm that MTs are overexpressed as a consequence of cadmium exposure and to identify which MT isoforms are upregulated the most, we performed Real-Time PCR experiments on mRNA extracts from cells either untreated or treated with Cd2+ and/or Zn2+. Real-Time PCR analysis revealed that, indeed, expression of MTs was greatly increased upon Cd2+ treatment in our experimental conditions (Fig. 4b and Table S2). Specifically, in cells treated with Cd2+ MT-1X expression was increased ~280-fold compared to the untreated cell sample, while MT-2A expression was increased ~210-fold; MT-1F was also overexpressed to a lesser extent (~20-fold). Considering that in the control sample MT-1X and MT-2A are expressed respectively ~8 and ~60 times more than MT-1A (taken as reference, Fig. 4c and Table S2), the levels of MT-1X and MT-2A in Cd2+-treated cells reached up to ~2200 and ~12500 times the basal MT-1A levels, respectively, by far the highest isoforms, thus suggesting that the observed NMR signals arise from these isoforms. In cells treated with both Cd2+ and Zn2+, an increase in the levels of MT-1X and MT-2A was observed (respectively, ~300- and ~260-fold), while in cells treated only with Zn2+ a much lower MT overexpression was observed (Fig. 4d,e and Table S3).
Real-Time PCR MTs expression analysis was also performed on cells overexpressing SOD1 as it occurs in the in-cell NMR samples upon supplementation of Cd2+ and/or Zn2+. Interestingly, in cells expressing SOD1 a different behaviour was observed in defect of Zn2+ versus excess of Zn2+, upon Cd2+ treatment. In cells expressing SOD1 in defect of Zn2+, Cd2+ treatment still induced the expression of MT-1X and MT-2A, but at significantly lower levels (~90- and ~50-fold, respectively, compared to basal levels) than in untransfected cells, while excess Zn2+ in SOD1-expressing cells significantly restored the Cd2+-induced expression of MT-1X and MT-2A to levels (~380- and ~180-fold, respectively) similar to those reached in untransfected cells (Fig. 4e and Table S3). These data are consistent with the fact that MTs were not detected by NMR in cells expressing SOD1 in defect of Zn2+ (Fig. S4).
Upregulation of MTs was much lower in cells (both untransfected and transfected with SOD1) treated with Zn2+ in excess but not exposed to Cd2+ (Fig. 4d). Taken together, these data indicate that cadmium is the main trigger of MT overexpression and that SOD1 and zinc supplementation can modulate Cd2+-induced MT overexpression in opposite ways.
4. Discussion
In-cell NMR allows a direct assessment of the intracellular state of SOD1 upon Cd2+ treatment, and therefore can provide a close-to-physiological picture of the effect of cadmium exposure on SOD1 in human cells. Previous in vitro NMR studies demonstrated that Cd2+ is able to bind to the zinc and copper sites of SOD1 [30], while studies on animal models showed that Cd2+ affects the superoxide dismutase activity in several tissues [31], [32], [33]. We therefore sought to determine whether direct binding of Cd2+ to SOD1 can occur in living cells. By in-cell NMR, we did not observe any direct Cd2+ binding to SOD1, as SOD1 was present only with one Zn2+ bound per monomer or in the apo state. Indeed, the chemical shifts of the signals arising from the metal-coordinating histidines observed in the Cd2+-treated cells were identical to those of E,Zn-SOD1 in-cell and in vitro, and differed from those observed when apo-SOD1 was titrated in vitro with increasing equivalents of Cd2+ (Fig. 1). Moreover, different species were simultaneously present during the titration and residual apo-SOD1 was still present after up to two equivalents of Cd2+. This behaviour is consistent with Cd2+ binding with low selectivity to either the zinc or the copper site of SOD1. At a protein concentration of 120 µM, the Kd can be estimated in the order of ~10-4 M, strikingly higher than that of Zn2+ binding to its native site (Kd = 4.2 ×10-14 M [46]), indicating that Cd2+ binding to intracellular SOD1 in place of Zn2+ would not be thermodynamically favoured.
Despite the lack of direct binding to intracellular SOD1, a clear effect of Cd2+ treatment was the oxidation of SOD1 cysteines, i.e. the formation of the intrasubunit disulfide bond between C57 and C146. This outcome is likely a consequence of an intracellular redox unbalance caused by Cd2+. The disulfide bond formation is required for the correct maturation of SOD1, as it stabilises the protein structure and is essential for its activity [47], [48]. Moreover, the ability of an exogenous redox-reactive compound to stabilise SOD1 by promoting disulfide oxidation has also been reported [37]. While cysteine oxidation contributes to stabilise the protein, it could interfere with the native maturation pathway, as SOD1 needs to interact with its partner copper chaperone for SOD1 (CCS) in order to bind copper [49], [50]. SOD1 interacts with CCS more efficiently in the reduced state [51], [52]. Upon interaction, CCS delivers copper to SOD1 and catalyses its oxidation, thus producing mature SOD1 [53], [54]. The premature oxidation of SOD1 could therefore prevent the interaction with CCS and impair copper delivery, as shown previously in yeast [55] and with human SOD1 in vitro [54]. Such effect would be consistent with the decrease of superoxide dismutase activity reported in vivo [13], [31]. Cd2+-mediated SOD1 oxidation did not occur in the presence of excess Zn2+, thus suggesting that Zn2+ could modulate the extent of the effect of Cd2+. Notably, this mechanism cannot involve mature Cu,Zn-SOD1, as the disulfide bond is already formed and Cd2+ would not be able to replace either zinc or copper, which is bound to SOD1 with even higher affinity.
Furthermore, we observed that Cd2+ treatment strongly induced the overexpression of the metallothionein isoforms MT-1X and MT-2A. Interestingly, while MT overexpression is a well known consequence of Cd2+ exposure [44], [45], MT-1X and MT-2A reached such high levels in our experimental conditions that they could be observed by in-cell NMR. Based on the estimated detection limit of the technique of ~5 µM for small folded proteins and on previous in-cell NMR analyses of other proteins [36], the concentration of MTs in the NMR tube was estimated around 5–10 µM. While in principle the high spectral resolution should allow separating the signals of MT-1X and MT-2A, only one set of resonances could be identified due to the low signal-to-noise ratio and likely because most signals have the same chemical shift due to the high structural similarity of the two isoforms, which have an overall sequence identity > 90% and share a central stretch of 38 identical residues. Strong MT induction required the presence of Cd2+, and was also observed in HEK293T not transfected with the SOD1 gene, where Zn2+ supplementation further potentiated the MT induction. Interestingly, SOD1 overexpression negatively modulated the level of Cd2+-mediated MT induction, and indeed in such experimental conditions, where SOD1 was partially oxidised upon Cd2+ treatment, MT-1X/MT-2A did not reach sufficiently high levels to be detected in the NMR spectrum. Conversely, supplementation of both Cd2+ and Zn2+ to cells overexpressing SOD1, i.e. in the experimental conditions where SOD1 was mostly reduced, Cd2+-mediated MT induction was restored, and high MT-1X/MT-2A levels were observed in the NMR spectrum.
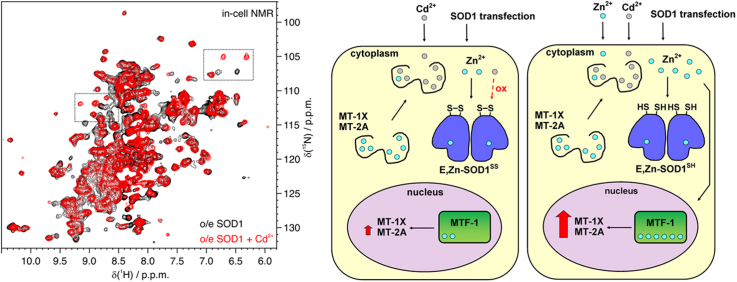
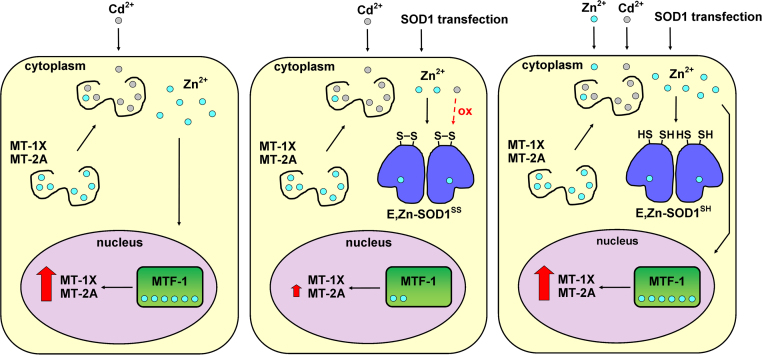
Such interplay between Zn2+, MT expression and SOD1 oxidation can be rationalised by considering the role of MTs in the cellular homeostasis of zinc. Zinc is an essential metal for the correct functionality and health of the cell. However, due to its high affinity for biological molecules, its intracellular availability has to be regulated to avoid uncontrolled binding to non-native acceptors. MTs, along with the other high affinity zinc-binding proteins, contribute to buffering the intracellular Zn2+ concentration [18]. Furthermore, both MTs and Zn2+ are involved in the cellular redox homeostasis [18], [56]. When intracellular Zn2+ increases, it binds to the metal regulatory transcription factor 1 (MTF-1), activating it and triggering the transcription of more MTs [21]. Consequently, when Cd2+ is added to the cells it displaces Zn2+ from MTs and other proteins, releasing it in the cytoplasm and causing it to activate MTF-1, inducing MTs overexpression (Fig. 5, left). In turn, overexpressed MTs protect the cells by directly binding Cd2+ and possibly by exerting other antioxidant functions [18]. When Cd2+ is added to cells overexpressing SOD1, the released Zn2+ is immediately sequestered by SOD1, which has a much higher affinity than MTF-1 and MTs, preventing the activation of MTF-1 and decreasing MTs overexpression as a consequence (Fig. 5, centre). Instead, when both Zn2+ and Cd2+ are supplemented, overexpressed SOD1 is fully metallated by the excess of Zn2+, so that the amount released by the MTs can still bind to MTF-1, restoring the high induction of MTs to NMR-detectable levels (Fig. 5, right). Finally, the different oxidation state of SOD1 observed when supplementing both Zn2+ and Cd2+ can be explained with the fact that in cells with higher levels of MTs the cellular redox balance is partially restored, preventing the premature oxidation of SOD1.
Fig. 5.
Cd2+-induced SOD1 oxidation is modulated by Zn2+and MT levels. Schematic drawing of the proposed mechanism of SOD1 oxidation as a function of Cd2+, Zn2+ and MTs. Left panel: when SOD1 is not overexpressed, Zn2+ (cyan) is displaced by Cd2+ (grey) and activates MTF-1 (green), which strongly induces MTs expression; the high levels of MTs contribute to restore the cellular redox balance by sequestering Cd2+. Centre panel: when SOD1 is overexpressed, it sequesters the Zn2+ displaced by Cd2+ causing a decreased MTs induction; Cd2+ is not efficiently sequestered by MTs and causes redox imbalance, leading to SOD1 oxidation. Right panel: when excess Zn2+ is supplemented, overexpressed SOD1 is metallated and the strong induction of MTs is restored, thus preventing SOD1 oxidation.
5. Conclusions
In this work, the effect of cadmium treatment on the metallation and redox state of SOD1 was investigated directly in human cells by in-cell NMR. Our study revealed an interesting relationship between the SOD1 redox state, the availability of Zn2+ and the Cd2+-induced overexpression of MTs. While Cd2+ itself does not bind SOD1 in place of Zn2+, it is able to induce the premature formation of SOD1 intramolecular disulfide bond, interfering with its correct maturation pathway. Furthermore, Cd2+ alters the cellular zinc homeostasis causing a strong induction of MTs. Such induction is negatively modulated by the overexpression of SOD1 itself, likely because the increased SOD1 levels interfere with the activation of the MT transcription factor MTF-1. While in our experiments the levels of SOD1 are artificially increased, our findings suggest that in cells with endogenously high levels of SOD1 (~40–100 µM), such as motor neurons and erythrocytes [57], [58], under certain conditions, SOD1 itself could sensitise the cells to Cd2+ toxicity, by sequestering the Zn2+ displaced by Cd2+, inhibiting the cellular response against Cd2+ and further altering the cellular redox balance.
Finally, as an interesting remark from the methodological point of view, we report that the strong induction of MTs resulted in the observation of NMR signals from MT-1X/MT-2A. To the best of our knowledge, this is the first time that signals from a protein synthesised by genomic DNA, i.e. not delivered into the cells nor ectopically expressed, could be detected by solution NMR in human cells.
Acknowledgements
Recombinant SOD1 was kindly provided by Dr. L. Barbieri (CERM/CIRMMP, Italy). This work has been supported by iNEXT, grant number 653706, funded by the Horizon 2020 programme of the European Commission. The authors acknowledge the support and the use of resources of Instruct-ERIC, a Landmark ESFRI project, and specifically the CERM/CIRMMP Italy Centre.
Acknowledgments
Author contributions
P.P., E.L. and L.B. conceived the work; P.P. and E.L. designed the experiments; P.P. maintained the cells and produced the samples for NMR and RT-PCR; E.L. performed the NMR experiments and analysed the data; F.C. performed the RT-PCR experiment and analysed the data; C.D. and L.B. supervised the study; P.P., F.C., C.D., E.L. and L.B. wrote the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101102.
Contributor Information
Enrico Luchinat, Email: eluchinat@cerm.unifi.it.
Lucia Banci, Email: banci@cerm.unifi.it.
Appendix A. Supplementary material
Supplementary material
References
- 1.Substance Priority List | ATSDR, 2018. 〈https://www.atsdr.cdc.gov/spl/index.html〉. (Accessed 3 August 2018), 2018.
- 2.List of classifications, Volumes 1–122 – IARC, n.d. 〈https://monographs.iarc.fr/list-of-classifications-volumes/〉. (Accessed 3 August 2018).
- 3.Järup L., Akesson A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 2009;238:201–208. doi: 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Satarug S., Moore M.R. Adverse health effects of chronic exposure to low-level cadmium in foodstuffs and cigarette smoke. Environ. Health Perspect. 2004;112:1099–1103. doi: 10.1289/ehp.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarwar N., Saifullah, Malhi S.S., Zia M.H., Naeem A., Bibi S., Farid G. Role of mineral nutrition in minimizing cadmium accumulation by plants. J. Sci. Food Agric. 2010;90:925–937. doi: 10.1002/jsfa.3916. [DOI] [PubMed] [Google Scholar]
- 6.Suwazono Y., Kido T., Nakagawa H., Nishijo M., Honda R., Kobayashi E., Dochi M., Nogawa K. Biological half-life of cadmium in the urine of inhabitants after cessation of cadmium exposure. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2009;14:77–81. doi: 10.1080/13547500902730698. [DOI] [PubMed] [Google Scholar]
- 7.Bertin G., Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88:1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Nair A.R., DeGheselle O., Smeets K., Van Kerkhove E., Cuypers A. Cadmium-induced pathologies: where is the oxidative balance lost (or not)? Int. J. Mol. Sci. 2013;14:6116–6143. doi: 10.3390/ijms14036116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonomi F., Ganadu M.L., Lubinu G., Pagani S. Reversible and non-denaturing replacement of iron by cadmium in Clostridium pasteurianum ferredoxin. Eur. J. Biochem. 1994;222:639–644. doi: 10.1111/j.1432-1033.1994.tb18907.x. [DOI] [PubMed] [Google Scholar]
- 10.Moulis J.-M. Cellular mechanisms of cadmium toxicity related to the homeostasis of essential metals. BioMetals. 2010;23:877–896. doi: 10.1007/s10534-010-9336-y. [DOI] [PubMed] [Google Scholar]
- 11.Malgieri G., Zaccaro L., Leone M., Bucci E., Esposito S., Baglivo I., Del Gatto A., Russo L., Scandurra R., Pedone P.V., Fattorusso R., Isernia C. Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2 His2 zinc finger induces structural rearrangements of typical DNA base determinant positions. Biopolymers. 2011;95:801–810. doi: 10.1002/bip.21680. [DOI] [PubMed] [Google Scholar]
- 12.Malgieri G., Palmieri M., Esposito S., Maione V., Russo L., Baglivo I., de Paola I., Milardi D., Diana D., Zaccaro L., Pedone P.V., Fattorusso R., Isernia C. Zinc to cadmium replacement in the prokaryotic zinc-finger domain. Metallomics. 2014;6:96–104. doi: 10.1039/c3mt00208j. [DOI] [PubMed] [Google Scholar]
- 13.Thévenod F. Cadmium and cellular signaling cascades: to be or not to be? Toxicol. Appl. Pharmacol. 2009;238:221–239. doi: 10.1016/j.taap.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Gobe G., Crane D. Mitochondria, reactive oxygen species and cadmium toxicity in the kidney. Toxicol. Lett. 2010;198:49–55. doi: 10.1016/j.toxlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Cuypers A., Plusquin M., Remans T., Jozefczak M., Keunen E., Gielen H., Opdenakker K., Nair A.R., Munters E., Artois T.J., Nawrot T., Vangronsveld J., Smeets K. Cadmium stress: an oxidative challenge. Biomet. Int. J. Role Met. Ions Biol. Biochem. Med. 2010;23:927–940. doi: 10.1007/s10534-010-9329-x. [DOI] [PubMed] [Google Scholar]
- 16.Cai L., Klein J.B., Kang Y.J. Metallothionein inhibits peroxynitrite-induced DNA and lipoprotein damage. J. Biol. Chem. 2000;275:38957–38960. doi: 10.1074/jbc.C000593200. [DOI] [PubMed] [Google Scholar]
- 17.Chiaverini N., De Ley M. Protective effect of metallothionein on oxidative stress-induced DNA damage. Free Radic. Res. 2010;44:605–613. doi: 10.3109/10715761003692511. [DOI] [PubMed] [Google Scholar]
- 18.Kimura T., Kambe T. The functions of metallothionein and ZIP and ZnT transporters: an overview and perspective. Int. J. Mol. Sci. 2016;17:336. doi: 10.3390/ijms17030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutherland D.E.K., Stillman M.J. The “magic numbers” of metallothionein. Met. Integr. Biomet. Sci. 2011;3:444–463. doi: 10.1039/c0mt00102c. [DOI] [PubMed] [Google Scholar]
- 20.Coyle P., Philcox J.C., Carey L.C., Rofe A.M. Metallothionein: the multipurpose protein. Cell. Mol. Life Sci. CMLS. 2002;59:627–647. doi: 10.1007/s00018-002-8454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Günther V., Lindert U., Schaffner W. The taste of heavy metals: gene regulation by MTF-1. Biochim. Biophys. Acta. 2012;1823:1416–1425. doi: 10.1016/j.bbamcr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 22.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 23.Tainer J.A., Getzoff E.D., Beem K.M., Richardson J.S., Richardson D.C. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- 24.Getzoff E.D., Cabelli D.E., Fisher C.L., Parge H.E., Viezzoli M.S., Banci L., Hallewell R.A. Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature. 1992;358:347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- 25.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J.P., Deng H.-X., Rahmani Z., Krizus A., McKenna-Yasek D., Cayabyab A., Gaston S.M., Berger R., Tanzi R.E., Halperin J.J., Herzfeldt B., den Bergh R.V., Hung W.-Y., Bird T., Deng G., Mulder D.W., Smyth C., Laing N.G., Soriano E., Pericak–Vance M.A., Haines J., Rouleau G.A., Gusella J.S., Horvitz H.R., R.H.B Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 26.Lindberg M.J., Tibell L., Oliveberg M. Common denominator of Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state. Proc. Natl. Acad. Sci. USA. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasinelli P., Belford M.E., Lennon N., Bacskai B.J., Hyman B.T., Trotti D., Brown R.H. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 28.Valentine J.S., Doucette P.A., Potter S. Zittin. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 29.Banci L., Bertini I., Durazo A., Girotto S., Gralla E.B., Martinelli M., Valentine J.S., Vieru M., Whitelegge J.P. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc. Natl. Acad. Sci. USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kofod P., Bauer R., Danielsen E., Larsen E., Bjerrum M.J. 113Cd-NMR investigation of a cadmium-substituted copper, zinc-containing superoxide dismutase from yeast. Eur. J. Biochem. 1991;198:607–611. doi: 10.1111/j.1432-1033.1991.tb16057.x. [DOI] [PubMed] [Google Scholar]
- 31.Jurczuk M., Brzóska M.M., Moniuszko-Jakoniuk J., Gałażyn-Sidorczuk M., Kulikowska-Karpińska E. Antioxidant enzymes activity and lipid peroxidation in liver and kidney of rats exposed to cadmium and ethanol. Food Chem. Toxicol. 2004;42:429–438. doi: 10.1016/j.fct.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y.-H., Shih C.-M., Huang C.-J., Lin C.-M., Chou C.-M., Tsai M.-L., Liu T.P., Chiu J.-F., Chen C.-T. Effects of cadmium on structure and enzymatic activity of Cu,Zn-SOD and oxidative status in neural cells. J. Cell. Biochem. 2006;98:577–589. doi: 10.1002/jcb.20772. [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Zhang H., Zhang T., Zhang R., Liu R., Chen Y. Molecular mechanism on cadmium-induced activity changes of catalase and superoxide dismutase. Int. J. Biol. Macromol. 2015;77:59–67. doi: 10.1016/j.ijbiomac.2015.02.037. [DOI] [PubMed] [Google Scholar]
- 34.Banci L., Barbieri L., Bertini I., Luchinat E., Secci E., Zhao Y., Aricescu A.R. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luchinat E., Barbieri L., Rubino J.T., Kozyreva T., Cantini F., Banci L. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 2014;5:5502. doi: 10.1038/ncomms6502. [DOI] [PubMed] [Google Scholar]
- 36.Mercatelli E., Barbieri L., Luchinat E., Banci L. Direct structural evidence of protein redox regulation obtained by in-cell NMR. Biochim. Biophys. Acta. 2016;1863:198–204. doi: 10.1016/j.bbamcr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Capper M.J., Wright G.S.A., Barbieri L., Luchinat E., Mercatelli E., McAlary L., Yerbury J.J., O’Neill P.M., Antonyuk S.V., Banci L., Hasnain S.S. The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation. Nat. Commun. 2018;9:1693. doi: 10.1038/s41467-018-04114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbieri L., Luchinat E., Banci L. Characterization of proteins by in-cell NMR spectroscopy in cultured mammalian cells. Nat. Protoc. 2016;11:1101–1111. doi: 10.1038/nprot.2016.061. [DOI] [PubMed] [Google Scholar]
- 39.Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 40.Lemarié A., Lagadic-Gossmann D., Morzadec C., Allain N., Fardel O., Vernhet L. Cadmium induces caspase-independent apoptosis in liver Hep3B cells: role for calcium in signaling oxidative stress-related impairment of mitochondria and relocation of endonuclease G and apoptosis-inducing factor. Free Radic. Biol. Med. 2004;36:1517–1531. doi: 10.1016/j.freeradbiomed.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 41.Banci L., Barbieri L., Bertini I., Cantini F., Luchinat E. In-cell NMR in E. coli to monitor maturation steps of hSOD1. PloS One. 2011;6:e23561. doi: 10.1371/journal.pone.0023561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 43.Cencetti F., Bernacchioni C., Nincheri P., Donati C., Bruni P. Transforming growth factor-beta1 induces transdifferentiation of myoblasts into myofibroblasts via up-regulation of sphingosine kinase-1/S1P3 axis. Mol. Biol. Cell. 2010;21:1111–1124. doi: 10.1091/mbc.E09-09-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews G.K. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem. Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 45.Klaassen C.D., Liu J., Diwan B.A. Metallothionein protection of cadmium toxicity. Toxicol. Appl. Pharmacol. 2009;238:215–220. doi: 10.1016/j.taap.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crow J.P., Sampson J.B., Zhuang Y., Thompson J.A., Beckman J.S. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J. Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 47.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O’Halloran T.V. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 48.Furukawa Y., O’Halloran T.V. Posttranslational modifications in Cu,Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid. Redox Signal. 2006;8:847–867. doi: 10.1089/ars.2006.8.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casareno R.L., Waggoner D., Gitlin J.D. The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J. Biol. Chem. 1998;273:23625–23628. doi: 10.1074/jbc.273.37.23625. [DOI] [PubMed] [Google Scholar]
- 50.Culotta V.C., Yang M., O’Halloran T.V. Activation of superoxide dismutases: putting the metal to the pedal. Biochim. Biophys. Acta. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright G.S.A., Antonyuk S.V., Hasnain S.S. A faulty interaction between SOD1 and hCCS in neurodegenerative disease. Sci. Rep. 2016;6 doi: 10.1038/srep27691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luchinat E., Barbieri L., Banci L. A molecular chaperone activity of CCS restores the maturation of SOD1 fALS mutants. Sci. Rep. 2017;7:17433. doi: 10.1038/s41598-017-17815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamb A.L., Torres A.S., O’Halloran T.V., Rosenzweig A.C. Heterodimer formation between superoxide dismutase and its copper chaperone. Biochemistry. 2000;39:14720–14727. doi: 10.1021/bi002207a. [DOI] [PubMed] [Google Scholar]
- 54.Banci L., Bertini I., Cantini F., Kozyreva T., Massagni C., Palumaa P., Rubino J.T., Zovo K. Human superoxide dismutase 1 (hSOD1) maturation through interaction with human copper chaperone for SOD1 (hCCS) Proc. Natl. Acad. Sci. USA. 2012;109:13555–13560. doi: 10.1073/pnas.1207493109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furukawa Y., Torres A.S., O’Halloran T.V. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oteiza P.I. Zinc and the modulation of redox homeostasis. Free Radic. Biol. Med. 2012;53:1748–1759. doi: 10.1016/j.freeradbiomed.2012.08.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurobe N., Suzuki F., Okajima K., Kato K. Sensitive enzyme immunoassay for human Cu/Zn superoxide dismutase. Clin. Chim. Acta. 1990;187:11–20. doi: 10.1016/0009-8981(90)90257-s. [DOI] [PubMed] [Google Scholar]
- 58.Pardo C.A., Xu Z., Borchelt D.R., Price D.L., Sisodia S.S., Cleveland D.W. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl. Acad. Sci. USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material