Abstract
PTEN expression is lost in many cancers, and even small changes in PTEN activity affect susceptibility and prognosis in a range of highly aggressive malignancies, such as melanoma and triple-negative breast cancer (TNBC). Loss of PTEN expression occurs via multiple mechanisms, including mutation, transcriptional repression and epigenetic silencing. Transcriptional repression of PTEN contributes to resistance to inhibitors used in the clinic, such as B-Raf inhibitors in BRAF mutant melanoma. We aimed to activate PTEN expression using the CRISPR system, specifically dead (d) Cas9 fused to the transactivator VP64-p65-Rta (VPR). dCas9-VPR was directed to the PTEN proximal promoter by single-guide RNAs (sgRNAs), in cancer cells that exhibited low levels of PTEN expression. The dCas9-VPR system increased PTEN expression in melanoma and TNBC cell lines, without transcriptional regulation at predicted off-target sgRNA binding sites. PTEN activation significantly repressed downstream oncogenic pathways, including AKT, mTOR, and MAPK signaling. BRAF V600E mutant melanoma cells transduced with dCas9-VPR displayed reduced migration, as well as diminished colony formation in the presence of B-Raf inhibitors, PI3K/mTOR inhibitors, and with combined PI3K/mTOR and B-Raf inhibition. CRISPR-mediated targeted activation of PTEN may provide an alternative therapeutic approach for highly aggressive cancers that are refractory to current treatments.
Keywords: human PTEN protein, CRISPR/Cas systems, tumor suppressor genes, melanoma, breast neoplasms
Introduction
Phosphatase and tensin homolog (PTEN) is an important and multifunctional tumor suppressor gene that inhibits a broad range of cellular processes, including survival, cell cycle progression, and migration.1 Loss of PTEN activity contributes to the development and drug resistance of many malignancies associated with poor outcome.1, 2, 3 Canonically, PTEN acts as an antagonist of the phosphoinositide 3-kinase (PI3K)/AKT pathway. By converting phosphatidylinositol-trisphosphate (PIP3) to phosphatidylinositol-disphosphate (PIP2), PTEN prevents the activation of AKT, inhibiting AKT-mediated downstream effects that include cell survival; cell cycle progression; and the regulation of transcription, translation, and metabolism.4 In addition, dephosphorylation of protein substrates by PTEN may be responsible for diverse functions, such as the inhibition of migration, induction of cell-cycle arrest, and inhibition of stem cell self-renewal.4, 5, 6
Loss of PTEN is a very common event in cancer development and progression. Somatic missense mutations, truncating mutations, and deletion of large chromosomal regions in PTEN occur in most cancer types.7 Beyond mutations, PTEN levels are also regulated by an array of transcriptional and post-transcriptional mechanisms. A number of transcription factors are known to repress PTEN expression,8, 9, 10, 11, 12 and epigenetic silencing of PTEN by promoter DNA methylation has been reported in endometrial carcinoma, breast cancer, colorectal cancer, and melanoma, among others.13, 14, 15, 16, 17, 18, 19, 20 Many lines of evidence suggest that PTEN acts as a haploinsufficient tumor suppressor gene, and, consequently, even small changes in PTEN activity can influence cancer development and prognosis,6, 21, 22 which has the potential to be exploited therapeutically.
PTEN expression is reduced or lost in approximately 40% of primary melanomas and 60% of metastases.17, 23, 24 However, genetic PTEN alterations are only detected in about 10% of melanomas, and PTEN promoter DNA methylation and transcriptional repression may be responsible for PTEN loss in many of the remaining clinical cases.17, 24, 25 In melanomas harboring V600E BRAF mutation, loss of PTEN expression confers intrinsic resistance to B-Raf inhibitors due to the possibility of escape through AKT pathway activation.26, 27, 28, 29, 30, 31 Moreover, melanomas that initially respond to B-Raf inhibitors often acquire resistance over prolonged treatment, which is partly attributable to PTEN transcriptional silencing.32, 33, 34, 35 Combination therapies that target both the PI3K/AKT and Ras/Raf/mitogen-activated protein kinase (MAPK) pathways are being trialed to overcome B-Raf inhibitor resistance in advanced melanoma.33, 36, 37, 38, 39 PTEN also plays an important role in the context of highly aggressive breast cancers, with the loss of PTEN expression via PTEN promoter DNA methylation documented in up to 30% of sporadic breast tumors.19, 40, 41 Diminished PTEN expression in breast cancer is positively correlated with poor prognosis, emergence of metastasis, and stem cell-like maintenance and survival, and it has been described as a signature of the aggressive basal-like breast cancer subtype.3, 42, 43, 44, 45
Since changes in PTEN dose have a significant impact on disease severity6, 21, 22 and PTEN expression can be regulated transcriptionally and epigenetically in the absence of PTEN mutation,8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 we reasoned that transcriptional reactivation of PTEN expression could inhibit progression and increase drug sensitivity in aggressive, PTEN-deficient cancers in which PTEN is not mutated. The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system, adapted from the acquired immune system of Streptococcus pyogenes, has provided a novel, highly efficient method for targeted gene activation.46 CRISPR/Cas9 was initially employed for gene-editing applications, in which the endonuclease Cas9 is directed to a target DNA sequence by a single guide RNA (sgRNA) and subsequently induces double-stranded DNA cleavage.47, 48, 49 The versatility of the CRISPR system was expanded by mutation of the catalytic domains of Cas9 to yield a dead Cas9 (dCas9). dCas9 maintains the ability to be directed to a specific genomic location by altering the sgRNA sequence, but it does not cause DNA double-strand breaks. Instead, dCas9 is fused to a variety of effector domains for transcriptional activation and repression, epigenetic editing, and a range of other functions, many of which have been applied in the field of cancer research.46, 50 One such effector domain consists of three mechanistically distinct transactivators, VP64, p65, and Rta (VPR).51 The dCas9-VPR system has achieved unprecedented functional reactivation of epigenetically silenced tumor suppressor genes.52
Here we establish the efficacy of dCas9-VPR artificial transcription factors (ATFs) to activate PTEN transcription to inhibit the PI3K/AKT/mTOR-signaling axis in melanoma and TNBC cell lines. We demonstrate the utility of this approach as a potential therapy to treat highly aggressive tumors for which no effective treatment is currently available.
Results
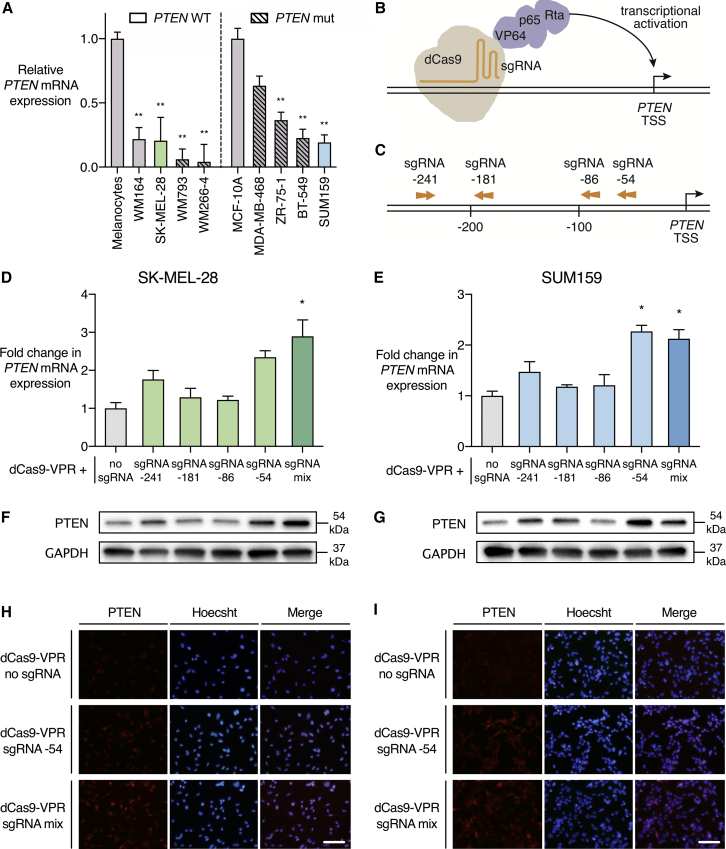
From a panel of melanoma and TNBC human cell lines (Figure 1A), we selected two lines as models for reactivation of PTEN transcription—the melanoma cell line SK-MEL-28, and the TNBC cell line SUM159. These lines carry PTEN wild-type alleles, but they exhibit reduced levels of PTEN expression relative to normal human melanocytes or the normal-like human breast epithelial cell line MCF-10A (Figure 1A; Table S1). SK-MEL-28 possesses a BRAF V600E mutation and shows intrinsic resistance to the B-Raf inhibitor dabrafenib, which is enhanced with prolonged dabrafenib treatment.53 SUM159 is a mesenchymal stem-like TNBC cell line.54 To reactivate PTEN expression in these lines, we exploited the dCas9-VPR transactivation system (Figure 1B). We designed four sgRNAs targeting the PTEN proximal promoter, in a region comprising 300 bp upstream of the PTEN transcription start site (Figure 1C; Table S2). sgRNA target sites were selected for minimal predicted off-target activity and maximal on-target activity, according to established algorithms.55, 56 Previous studies with CRISPR-based ATFs have shown that combined delivery of three to four sgRNAs per target provides increased levels of endogenous gene activation.57, 58, 59, 60
Figure 1.
PTEN Is Activated by the dCas9-VPR System in SK-MEL-28 Melanoma and SUM159 TNBC Cell Lines
(A) PTEN mRNA expression in a panel of BRAF mutant melanoma and TNBC cell lines. PTEN wild-type (WT) or mutant (mut) status is indicated with clear and hatched bars, respectively. Data are presented as PTEN mRNA expression level relative to normal human melanocytes or normal-like immortalized breast epithelial cells (MCF-10A). SK-MEL-28 and SUM159 were selected as cell line models with wild-type but transcriptionally downregulated PTEN. *p < 0.05, **p < 0.01; n = 3; error bars show SEM. (B) Schematic representation of the sgRNA-dCas9-VPR complex. dCas9 is in direct C-terminal fusion with transactivators VP64, p65, and Rta. sgRNAs bind upstream of the PTEN transcription start site (TSS) to activate gene expression. (C) sgRNA target sites in the PTEN proximal promoter region. sgRNA numbering refers to the distance in base pairs from the TSS of PTEN mRNA transcript variant 1. Arrows indicate whether the sgRNA targets the forward or reverse DNA strand. (D–I) dCas9-VPR was stably expressed in SK-MEL-28 and SUM159 cell lines with no sgRNA, individual sgRNAs targeting the PTEN proximal promoter, or a mix of all four PTEN-targeting sgRNAs. (D and E) Fold change in PTEN mRNA expression in qRT-PCR relative to dCas9-VPR with no sgRNA in SK-MEL-28 (D) and SUM159 (E). *p < 0.05, **p < 0.01; n = 3; error bars show SEM. (F and G) Immunoblot of PTEN and GAPDH in SK-MEL-28 (F) and SUM159 (G) cell lines. Conditions correspond to qRT-PCR data labeled above. (H and I) Immunofluorescence of PTEN and Hoechst-stained cell nuclei in SK-MEL-28 (H) and SUM159 (I) cell lines. Scale bars represent 100 μm.
The dCas9-VPR System Specifically Activates PTEN mRNA and Protein Expression
The dCas9-VPR activation system (Figure S1) was stably expressed in SK-MEL-28 and SUM159 cell lines by lentiviral transduction followed by antibiotic selection. dCas9-VPR was delivered with no sgRNA, with individual PTEN-targeting sgRNAs, or with a mix of all four sgRNAs targeting the PTEN proximal promoter. qRT-PCR and Western blot were performed to assess mRNA and protein expression (Figures 1D and 1E). In SK-MEL-28, PTEN was significantly activated by the mix of sgRNAs (2.89-fold, p < 0.05) relative to no sgRNA. In SUM159, PTEN was significantly activated by sgRNA −54 (2.27-fold, p < 0.05) and the mix of sgRNAs (2.13-fold, p < 0.05). Western blot results corresponded to the observed changes in mRNA expression. Immunofluorescence confirmed the activation of PTEN in both cell lines with the delivery of either sgRNA −54 or a mix of sgRNAs (Figures 1H and 1I). Delivery of dCas9 with no effector domain along with PTEN sgRNAs or dCas9-VPR with sgRNAs targeting the unrelated gene MASPIN resulted in no significant alteration of PTEN mRNA expression (Figure S2).
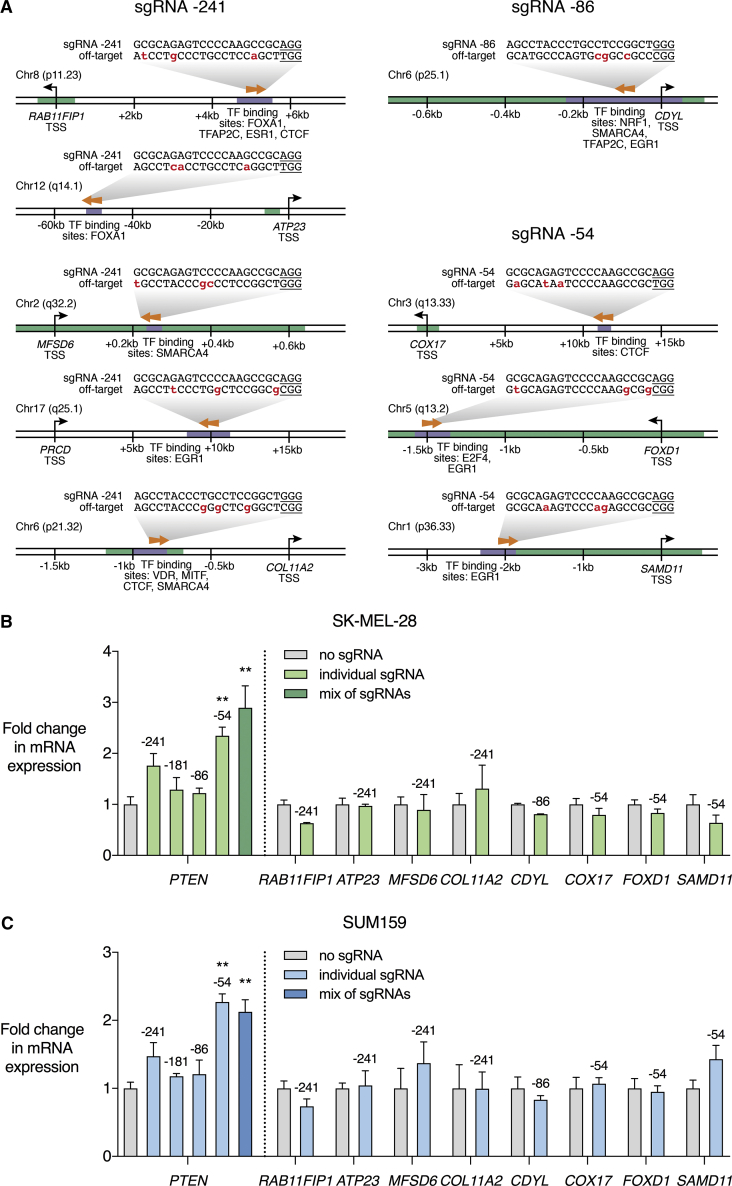
To investigate the possibility of off-target gene regulation, we analyzed PTEN sgRNA sequences to compile potential genome-wide off-target sgRNA binding sites.61 We then identified those off-target sequences located in regulatory regions with the potential to modulate gene expression. Nine potential off-target binding sites with proximity to regulatory elements were identified (Figure 2A; Table S3). qRT-PCR was conducted to assess regulation of these targets by dCas9-VPR, comparing the relevant PTEN-targeting sgRNA to dCas9-VPR with no sgRNA (Figure 2). There was no significant effect on the expression of any of the potential off-target genes in either SK-MEL-28 or SUM159 cell lines (Figures 2B and 2C). Previous studies have also demonstrated negligible impact of CRISPR ATFs and epigenetic editors on gene expression or epigenetic modifications at off-target sites.59, 60, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71
Figure 2.
PTEN Activation by dCas9-VPR Does Not Affect the Expression of Predicted Off-Target Genes
(A) The program Cas-OFFinder was used to identify genomic sequences with 3 mismatches or less to the cognate sequence of any of the 4 PTEN-targeting sgRNAs and upstream of the S. pyogenes NGG protospacer-adjacent motif (PAM). Of the identified off-target sites, nine sites were found in proximity to regulatory elements and were associated with greater potential to modulate gene expression. The potential off-target genes were RAB11FIP1, ATP23, MFSD6, PRCD, COL11A2, CDYL, COX17, FOXD1, and SAMD11. Mismatches between cognate and off-target sequences are highlighted in red and PAM sequences are underlined. Arrows indicate whether the sgRNA targets the forward or reverse DNA strand at the potential off-target binding site. Numbering on the DNA indicates the distance from the transcription start site (TSS) of the indicated gene. Green-shaded regions of DNA represent CpG islands annotated in the UCSC Genome Browser. Purple-shaded regions of DNA represent the location of transcription factor (TF)-binding sites according to literature-curated PAZAR and JASPAR databases. (B and C) dCas9-VPR was stably expressed in SK-MEL-28 (B) and SUM159 (C) cell lines with no sgRNA, individual PTEN sgRNAs, or a mix of all four sgRNAs targeting the PTEN proximal promoter. Data are shown as fold change in mRNA expression in qRT-PCR relative to dCas9-VPR with no sgRNA, for PTEN and each of the nine potential off-target genes. Numbers above the bars indicate the relevant PTEN-targeting sgRNA. There was no significant effect on mRNA expression of any of the predicted off-target genes. Data for PRCD are not shown, as PRCD expression was not reliably detected in either control or sgRNA-treated samples. *p < 0.05; n = 3; error bars show SEM.
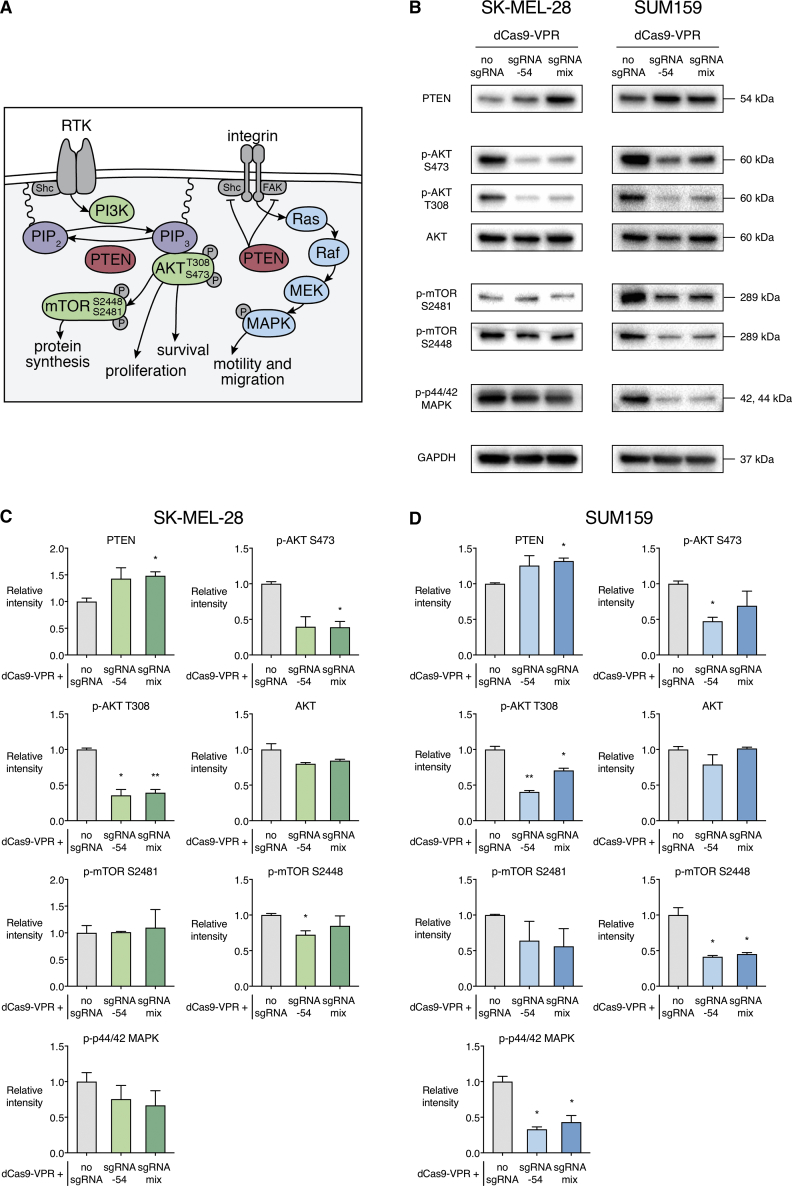
PTEN Activation Inhibits Oncogenic Signaling Pathways
We next assessed the consequence of dCas9-VPR-mediated reactivation of PTEN on downstream oncogenic signaling pathways (Figure 3A). In SK-MEL-28, immunoblotting confirmed a significant reduction in levels of phosphorylated (phospho)-AKT at serine (S)473 when dCas9-VPR was delivered with a mix of four PTEN sgRNAs (0.39-fold, p < 0.05) and a significant reduction in levels of phospho-AKT at threonine (T)308 when dCas9-VPR was delivered with either sgRNA −54 (0.36-fold, p < 0.05) or a mix of sgRNAs (0.39-fold, p < 0.01) (Figures 3B and 3C). There was no significant difference in the levels of total AKT protein between these conditions. SK-MEL-28 also showed a small but significant reduction in levels of phospho-mTOR S2448 when dCas9-VPR was delivered with sgRNA −54 alone (0.72-fold, p < 0.05).
Figure 3.
PTEN Activation Inhibits Downstream AKT/mTOR and MAPK Oncogenic Signaling Pathways
(A) Schematic representation of PTEN-related-signaling pathways. Activation of receptor tyrosine kinases (RTKs) recruits PI3K to convert PIP2 to the second messenger PIP3. PTEN works in direct opposition to PI3K, dephosphorylating PIP3 to PIP2. AKT binds to PIP3 and is activated by phosphorylation on threonine (T)308 by PDK1 and serine (S)473 by mTORC2. AKT phosphorylates and inactivates many downstream targets, resulting in increased cellular survival and proliferation. AKT activity also results in the activation of mTOR at S2448 and S2480, which increases protein synthesis and cell growth. In addition, PTEN inhibits the Ras/Raf/MEK/MAPK pathway through dephosphorylation of FAK and the adaptor protein Shc on integrins and RTKs, leading to reduced cell motility and migration. (B) Immunoblot of proteins in AKT/mTOR and MAPK pathways in SK-MEL-28 and SUM159 cell lines that stably express dCas9-VPR with no sgRNA, PTEN sgRNA −54, or a mix of four sgRNAs targeting the PTEN proximal promoter (each blot is representative of two independent biological replicates). (C and D) Relative quantification of protein abundance normalized to GAPDH, relative to dCas9-VPR with no sgRNA, in SK-MEL-28 (C) and SUM159 (D) cell lines. *p < 0.05, **p < 0.01; n = 2; error bars show SEM.
In SUM159, there was a significant reduction in levels of phospho-AKT S473 when dCas9-VPR was delivered with PTEN sgRNA −54 (0.47-fold, p < 0.05) and a significant reduction in levels of phospho-AKT T308 with both sgRNA −54 (0.40-fold, p < 0.01) and the mix of sgRNAs (0.70-fold, p < 0.05) (Figures 3B and 3D). Again, there was no significant difference in the total AKT protein levels between conditions. In addition, SUM159 showed a significant reduction in levels of phospho-mTOR S2448 when dCas9-VPR was delivered with either sgRNA −54 (0.41-fold, p < 0.05) or a mix of sgRNAs (0.45-fold, p < 0.05). As well as inhibiting the AKT/mTOR pathway, PTEN has been shown to reduce MAPK activation via dephosphorylation of the adaptor protein Shc on growth factor receptor tyrosine kinases and integrins.72 In SUM159, there was a significant reduction in phospho-p44/42 MAPK with both dCas9-VPR with sgRNA −54 alone (0.33-fold, p < 0.05) and with a mix of sgRNAs (0.43-fold, p < 0.05).
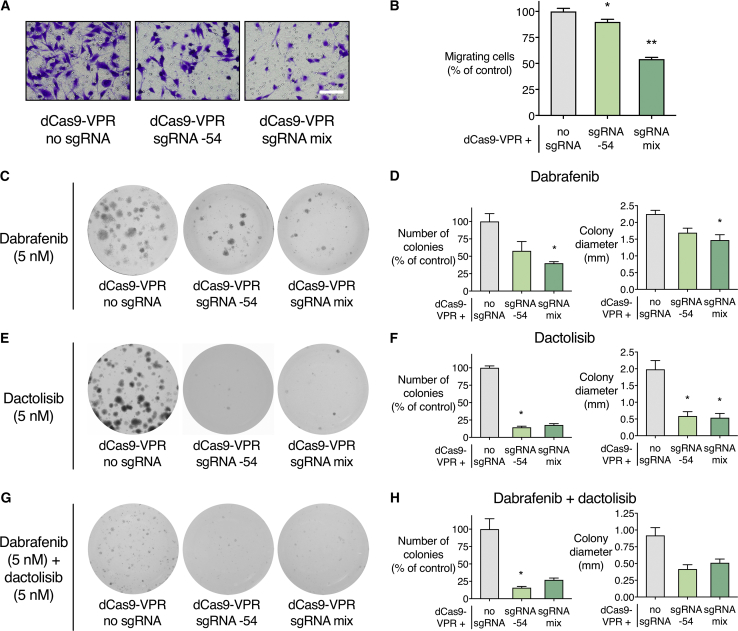
PTEN Activation Reduces the Migration of SK-MEL-28 Melanoma Cells
After demonstrating significant inhibition of the AKT/mTOR pathway by forced PTEN reactivation, we investigated phenotypic changes in PTEN-activated SK-MEL-28 cells. PTEN inhibits cell migration via PI3K/AKT pathway antagonism and its protein phosphatase activity.73 We used a transwell migration assay to assess the potential of SK-MEL-28 cells to migrate across a membrane toward a serum chemoattractant. There were significantly fewer migrating cells when dCas9-VPR was delivered with a mix of four PTEN-targeting sgRNAs (54.1% migrating cells relative to no sgRNA, p < 0.05) (Figures 4A and 4B). We also conducted immunofluorescence staining for Ki67 to identify actively proliferating cells. We observed a small but significant reduction in the percentage of Ki67+ SK-MEL-28 cells when dCas9-VPR was delivered with a mix of sgRNAs (85.8% Ki67+ cells compared to 95.2% Ki67+ cells with no sgRNA, p < 0.05) (Figure S3).
Figure 4.
PTEN Activation Inhibits Migration and Confers Increased Sensitivity to Combined B-Raf and PI3K/mTOR Inhibition in the SK-MEL-28 Melanoma Cell Line
dCas9-VPR was stably expressed in SK-MEL-28 with no sgRNA, PTEN sgRNA −54, or a mix of four sgRNAs targeting the PTEN proximal promoter. (A and B) Representative images of migrating SK-MEL-28 cells are displayed (A), along with quantification of the number of migrating cells relative to dCas9-VPR with no sgRNA (B). *p < 0.05, **p < 0.01; n = 3; error bars show SEM. (C–H) Long-term colony formation assay with SK-MEL-28 cells grown in dabrafenib (C and D), dactolisib (E and F), or combined dabrafenib and dactolisib (G and H) for a period of 3 weeks. Representative images of wells are displayed (C, E, and G), along with the average number of colonies per well (relative to dCas9-VPR with no sgRNA) and average colony diameter (D, F, and H). *p < 0.05, **p < 0.01; n = 3; error bars show SEM.
PTEN Activation Reduces the Resistance of SK-MEL-28 Melanoma Cells to B-Raf and PI3K/mTOR Inhibitors
Loss of PTEN expression has been shown to confer innate and acquired resistance to B-Raf inhibitors in BRAF mutant melanoma through PI3K/AKT activation.26, 27, 28, 29, 30, 31, 32, 33, 34, 35 SK-MEL-28 exhibits both innate and acquired resistance to B-Raf inhibitors.53 We treated PTEN-activated SK-MEL-28 cells with dabrafenib, an FDA-approved B-Raf inhibitor for the treatment of patients with BRAF V600E mutation-positive advanced melanoma.74 With 72 h dabrafenib treatment, we observed a small but significant increase in the maximum inhibition of cell viability in PTEN-activated SK-MEL-28 cells (51.5% viability for no sgRNA compared to 42.3% viability for sgRNA −54 and 43.0% viability for the mix of sgRNAs, p < 0.01) (Figure S4).
We subsequently determined the capacity of SK-MEL-28 to form colonies in the presence of dabrafenib. Cells were seeded at low density and grown in 5 nM dabrafenib over a period of 3 weeks. As expected, there were significantly fewer colonies per well on average when dCas9-VPR was delivered with a mix of sgRNAs (40.0% of the number of colonies with no sgRNA, p < 0.05) (Figures 4C and 4D). Average colony size was also significantly reduced when dCas9-VPR was expressed with a mix of sgRNAs (1.47-mm diameter compared to 2.25-mm diameter with no sgRNA, p < 0.05). These results indicate the possibility of using PTEN activation to reduce the propensity for B-Raf inhibitor resistance that has been observed in many melanomas in the clinical setting.
We next investigated whether PTEN activation could enhance sensitivity to PI3K/mTOR inhibition. Dactolisib is a dual pan-PI3K/mTOR inhibitor that has been administered in combination with B-Raf inhibitors for the treatment of BRAF mutant melanoma in preclinical studies.75 We again seeded PTEN-activated SK-MEL-28 cells at low density and grew them in 5 nM dactolisib for 3 weeks. There were significantly fewer colonies per well when dCas9-VPR was stably expressed with sgRNA −54 (14.5% of the number of colonies with no sgRNA, p < 0.05) (Figures 4E and 4F). In addition, colony diameter was significantly reduced with sgRNA −54 or a mix of sgRNAs (0.59- and 0.53-mm diameter, respectively, compared to 2.0-mm diameter with no sgRNA; p < 0.05).
Colony formation was also significantly inhibited in PTEN-activated SK-MEL-28 cells subjected to combined B-Raf and PI3K/mTOR inhibition. When cells were grown in 5 nM dabrafenib and 5 nM dactolisib for 3 weeks, there were fewer colonies per well when dCas9-VPR was delivered with sgRNA −54 (15.8% of the number of colonies with no sgRNA, p < 0.05) (Figures 4G and 4H). While SK-MEL-28 cells treated with the mix of sgRNAs also produced fewer colonies than no sgRNA control when subjected to combination dabrafenib and dactolisib treatment, this effect was not statistically significant. Overall, these results suggest that PTEN activation could potentially sensitize BRAF mutant melanoma to B-Raf and PI3K/mTOR inhibitors.
Discussion
PTEN is a key tumor suppressor protein whose silencing promotes metastatic behavior and therapy resistance in highly aggressive cancers, including BRAF mutant metastatic melanoma and basal-like TNBCs.3, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 42, 43, 44, 45 PTEN is lost in these cancers not only by mutation but also through transcriptional and epigenetic silencing in up to 60% of melanomas and 30% of breast tumors.17, 19, 23, 24, 25, 40, 41 Here we demonstrated the efficacy of the CRISPR/dCas9 system to specifically activate PTEN expression in BRAF mutant melanoma and TNBC cell lines, resulting in significant inhibition of downstream oncogenic signaling pathways involved in cell proliferation and migration. Activation of PTEN was associated with a significant reduction in the levels of phosphorylated AKT (S473 and T308) in both cell lines tested. A greater reduction of phosphorylated mTOR (S2448) was observed in the SUM159 cell line than in SK-MEL-28. SUM159 also showed a significant inhibition of phosphorylated p44/42 MAPK, whereas SK-MEL-28 did not. However, PTEN activation significantly reduced the migratory potential of SK-MEL-28 melanoma cells and increased sensitivity to B-Raf and PI3K/mTOR inhibitors. CRISPR-treated SK-MEL-28 cells exhibited fewer and smaller colonies than control when grown in the presence of the B-Raf inhibitor dabrafenib, and this effect was even more pronounced for both the PI3K/mTOR inhibitor dactolisib and for the combination dabrafenib and dactolisib treatment.
By dephosphorylating phosphoinositides and protein substrates on receptor tyrosine kinases and integrins, PTEN is a master regulator of downstream PI3K/AKT/mTOR- and Ras/Raf/MAPK-signaling pathways, controlling survival, motility, and drug resistance mechanisms.4, 5, 6, 7, 76 As PTEN possesses intrinsic enzymatic activity, even small changes in functional PTEN levels in cancer cells can elicit profound alterations in cancer progression.6, 21, 22 Although previous CRISPR activation studies have demonstrated strong activation of multiple loci,52 the critical importance of PTEN in orchestrating multiple oncogenic signaling pathways may enable significant reprogramming of the cancer kinome and overall cancer phenotype, even with relatively modest levels of PTEN transcriptional activation.
Genome-based CRISPR activation offers several advantages that could be harnessed for precision medicine and for the future treatment of PTEN-deficient cancers. While the frequency of CRISPR-mediated genome editing by homology-directed repair can often be lower than 20%,49, 77, 78, 79 the efficiency of epigenetic reactivation with dCas9-VPR is highly efficient, as shown by our immunofluorescence analyses of PTEN activation in the transduced cell population. It is also important to note that reactivation of PTEN could benefit patients who have lost one PTEN allele through mutation or deletion, by increasing transcription of the intact allele. Loss of a single PTEN allele is frequently observed during early stages of tumorigenesis.80 Combining CRISPR activation of PTEN with conventional small molecule inhibitors could provide even more effective tumor inhibition and limit drug resistance, for example, to PI3K/mTOR or B-Raf inhibitors. In addition, CRISPR activation has demonstrated negligible off-target effects,59, 60, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71 and combining CRISPR with chemotherapy could potentially allow for a reduction in the drug dose and associated toxicity in patients.
In both cell lines tested, the sgRNA designed against a binding site closest to the PTEN transcription start site was most effective. A mix of four sgRNAs activated PTEN expression slightly more than the individual most potent sgRNA in the SK-MEL-28 cell line; however, the combination of sgRNAs did not produce more potent activation than this individual sgRNA in SUM159. The results described here only pertain to a single gene, so they cannot be relied upon to predict how CRISPR activation will function for other target genes. However, our results demonstrate that a single sgRNA can perform equally as well as a combination of sgRNAs, at least for PTEN activation. CRISPR activation systems present the future possibility of multiplex gene activation, through the simultaneous delivery of several sgRNAs with each targeting a different cancer-related gene. Such an approach could be used to activate PTEN along with other inhibitors of oncogenic pathways, offering an advantage over traditional gene overexpression from exogenous cDNA. Combining multiple sgRNAs targeting a single gene could remain a worthwhile strategy, as a significant level of gene activation could be achieved while delivering less of each individual sgRNA.81 This could feasibly reduce the likelihood of transcriptional regulation at off-target binding sites of individual sgRNAs, although such an effect was not experimentally verified here.
Lastly, there is potential for this system to be applied in vivo. Zinc fingers and transcription activator-like effectors (TALEs) have been extensively developed for the activation of tumor suppressor genes and silencing of oncogenes for cancer therapy, and these ATFs have achieved tumor suppression in mouse xenograft tumor models.82, 83, 84, 85, 86 A hit-and-run approach using transient delivery systems rather than viruses for in vivo applications could lessen safety concerns and reduce off-target effects from the sustained expression of CRISPR editing components. Nanoparticles have been developed to deliver CRISPR components as plasmid DNA, mRNA, and ribonucleoproteins,78, 87, 88, 89, 90, 91, 92, 93 and polymeric formulations encapsulating chemotherapeutic drugs could be adapted to also carry the CRISPR molecular constituents for combination therapy.94 Whether the efficiency and longevity of PTEN transcriptional upregulation by dCas9-VPR is sufficient to maintain the effects on the downstream phenotype in vivo remains to be established. It is unclear how long the transcriptional upregulation achieved by dCas9-VPR would be maintained in vivo in the absence of continued effector expression, and this issue cannot be addressed by the in vitro lentiviral delivery model described here. Alternative effector domains that have demonstrated stable epigenetic editing and transcriptional upregulation could be explored to increase the longevity of PTEN activation.70, 95 Additionally, limitations of transient in vivo delivery methods mean that activation constructs are unlikely to reach all, or perhaps even a majority, of cells in the tumor. However, PTEN has an alternative long isoform that is preferentially secreted, and ubiquitinated PTEN can be exported in exosomes, both of which result in tumor-suppressive effects in the surrounding cells.96, 97 This suggests the possibility of a paracrine effect of PTEN activation systems on neighboring tumor cells that have not themselves received CRISPR activation components. PTEN transcriptional activation using the CRISPR/dCas9 system has the potential to reduce invasiveness and improve the response to existing chemotherapeutics in aggressive and currently incurable drug-resistant tumors.
Materials and Methods
Cell Culture
SK-MEL-28, WM164, WM793, and WM266-4 cell lines were a gift from Peter Leedman, and they were cultured in RPMI-1640 medium without phenol red (produced by the Harry Perkins Institute of Medical Research, Perth, Australia; formulated to ATCC specifications), supplemented with 10% heat-inactivated HyClone fetal bovine serum (FBS; GE Healthcare Life Sciences, Chicago, IL) and 1% antibiotic-antimycotic (Anti-Anti; Gibco, Fisher Scientific, Hampton, NH). Normal human melanocytes were a gift from Mel Ziman, and they were cultured in melanocyte growth medium M2 (PromoCell, Heidelberg, Germany). SUM159 cells were cultured in F-12 Nutrient Mixture (Gibco) supplemented with 5% FBS, 0.6% 1 M HEPES (Gibco), 5 μg/mL insulin, human recombinant, zinc solution (Gibco), 1 μg/mL hydrocortisone (Merck, Kenilworth, NJ), and 1% Anti-Anti. MCF-10A cells were cultured in DMEM/F-12 medium (Gibco), supplemented with 5% Normal Horse Serum (Life Technologies, Carlsbad, CA), 20 ng/mL EGF Recombinant Human Protein (Gibco), 0.01 mg/mL insulin, human recombinant, zinc solution, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin (Sigma-Aldrich, St. Louis, MO), and 1% Anti-Anti. BT-549, ZR-75-1, and MDA-MB-468 cell lines were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% Anti-Anti.
sgRNA Target Design and Off-Target Identification
Candidate sgRNA sequences for PTEN activation were identified using the Benchling CRISPR design tool (https://benchling.com/), which provides a score indicating the predicted targeting specificity and off-target binding sites of each sgRNA according to established algorithms.55, 56 sgRNAs were only considered if they had a specificity score greater than 60 and an efficiency score greater than 30. 47 putative sgRNAs were available in the 250 bp immediately upstream of the TSS of PTEN mRNA transcript variant 1. From these, 4 sgRNAs were selected that had specificity and efficiency scores above the designated thresholds and that were relatively evenly spaced across the target region. Previous studies with CRISPR-based ATFs have shown the combined delivery of three to four sgRNAs per target provides increased levels of endogenous gene activation.57, 58, 59, 60 The four sgRNA target sequences chosen for PTEN activation, along with their specificity and efficiency scores, are listed in Table S2.
To identify potential off-target sgRNA binding sites with the potential to modulate gene expression, the software program Cas-OFFinder61 was used to search for genomic sequences that were highly similar to any of the 4 PTEN sgRNAs and upstream of the S. pyogenes NGG protospacer-adjacent motif (PAM). The search was restricted to off-target sites with 3 mismatches or less to the corresponding cognate sgRNA sequence. The location of each potential off-target site was compared to UCSC Genome Browser and Encyclopedia of DNA Elements (ENCODE) data to identify proximity to annotated NCBI RefSeq genes, promoters, enhancers, CpG islands, DNase I hypersensitive regions, and transcription factor-binding sites, which would indicate greater potential for the dCas9-VPR complex to modulate gene expression. Nine off-target sequences were found to fall in close proximity to potential regulatory elements (Table S3), and they were assessed by qRT-PCR as described below.
Plasmids
The third-generation lentiviral transfer plasmid pLV_dCas9-VPR_sgRNA was cloned by replacing the KRAB domain from pLV_hU6-sgRNA_hUbC-dCas9-KRAB-T2A-Puro (Addgene plasmid 71236, a gift from Charles Gersbach)98 with the VPR domain from SP-dCas9-VPR (Addgene plasmid 63798, a gift from George Church).51 For each sgRNA, sense and anti-sense custom DNA oligonucleotides (Integrated DNA Technologies) containing the recognition sequence were annealed and ligated into pLV_dCas9-VPR_sgRNA using T4 DNA ligase (Promega, Madison, WI), as described previously.99 pLV_dCas9-VPR_sgRNA with no inserted sgRNA recognition sequence was used as a control. The third-generation lentiviral packaging plasmids pMDLg/pRRE (Addgene plasmid 12251) and pRSV-Rev (Addgene plasmid 12253) and envelope plasmid pMD2.G (Addgene plasmid 12259; all gifts from Didier Trono) were used for lentiviral production.
Transient Transfection
4 × 105 SUM159 cells were seeded in each well of a 6-well plate 24 h prior to transfection, in culture medium without Anti-Anti. The next day, each well was transfected using 4.8 μL Lipofectamine 2000 (Thermo Fisher Scientific) in 250 μL Opti-MEM (Gibco) with 2.4 μg total plasmid DNA, according to the manufacturer’s protocol. Transfection mix was removed after 4 h and replaced with complete culture medium. RNA was collected from cells 48 h post-transfection.
Lentiviral Production and Transduction
All experiments were approved by the Australian Office of the Gene Technology Regulator (OGTR) under Notifiable Low Risk Dealing (NLRD) approval number 004/2017, and they were conducted in the PC2 laboratory at the Harry Perkins Institute of Medical Research. Lentivirus was produced by transfection of HEK293T cells with third-generation lentiviral transfer, packaging, and envelope plasmids, as described previously.100 To select for transduced cells, puromycin dihydrochloride (Gibco) was added to the culture medium at a concentration of 1.25 μg/mL for SUM159 and 0.625 μg/mL for SK-MEL-28 over a period of 6 days.
RNA Extraction and Reverse Transcription
RNA was isolated from transfected or transduced cells by phenol-chloroform extraction using QIAzol Lysis Reagent (QIAGEN, Hilden, Germany). 2 μg purified total RNA per sample was used as a template for cDNA synthesis using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA), with the following thermocycling settings: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min.
qRT-PCR
Real-time qRT-PCR for genes PTEN, GAPDH, GUSB, COL11A2, and SAMD11 was performed using TaqMan Gene Expression Assays (Applied Biosystems), listed in Table S4. qRT-PCR for genes RAB11FIP1, ATP23, MFSD6, PRCD, CDYL, COX17, and FOXD1 was performed using QuantiFast SYBR Green PCR Master Mix (QIAGEN) and custom-designed primers (Integrated DNA Technologies), listed in Table S5. qRT-PCR reactions were carried out in the ViiA 7 Real-Time PCR System (Applied Biosystems). TaqMan assays were performed with the following thermocycling settings: 95°C for 20 s, followed by 40 cycles of 95°C for 1 s and 60°C for 20 s. SYBR Green assays were performed with the following thermocycling settings: 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Cycle threshold (Ct) was automatically determined for each well using QuantStudio Real Time PCR Software (version [v.]1.1, Applied Biosystems). Relative quantification of gene expression was carried out according to the comparative (ΔΔ) Ct method,101, 102 with GAPDH and GUSB as housekeeping genes.
Protein Extraction and Quantification
Protein was extracted from transduced cells using Cell Lysis Buffer (Cell Signaling Technology, Danvers, MA). Samples were sonicated for 15 s at 10 mA, centrifuged at 16,000 relative centrifugal force (RCF) for 10 min at 4°C, and the supernatant was transferred to a new tube. Protein lysates were quantified using the DC Protein Assay (Bio-Rad, Hercules, CA). The 750-nm absorbance was detected in the PowerWave XS2 Microplate Spectrophotometer (BioTek, Winooski, VT).
Western Blotting
20 μg protein/well was resolved using Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad) at 100 V for approximately 1.5 h. The proteins were transferred from the gel to a 0.2-μm polyvinylidene fluoride (PVDF) membrane (Trans-Blot Turbo Transfer Pack, Bio-Rad) using the TransBlot Turbo (Bio-Rad). Membranes were blocked with 5% skim milk powder in Tris-buffered saline with Tween-20 (TBS-T, Sigma-Aldrich) for 1 h at room temperature with gentle shaking, then incubated with primary antibody diluted in TBS-T at 4°C overnight (antibodies are listed in Table S6). The next day, membranes were washed and incubated with secondary antibody diluted in TBS-T for 1 h at room temperature, then visualized by enhanced chemiluminescence using Luminata Crescendo Western HRP Substrate (Merck) with the ChemiDoc MP system (Bio-Rad). Membrane pictures were processed and quantified using ImageLab Software (v.5.2, Bio-Rad). Uncropped images of Western blot membranes from Figures 1 and 3 are displayed in Figure S5.
Immunofluorescence
1 × 105 cells were seeded on poly-L-lysine- (Merck) coated coverslips in 24-well plates. 48 h after seeding, cells were fixed with 4% formaldehyde in Dulbecco’s PBS (DPBS) for 20 min at room temperature. Cells were blocked with 5% normal goat serum (Invitrogen, Carlsbad, CA) and 0.3% Triton X-100 (Sigma-Aldrich) in DPBS for 1 h at room temperature, then incubated with primary antibodies in diluent buffer (1% BSA and 0.3% Triton X-100 in DPBS) overnight at 4°C. The next day, cells were washed and incubated with secondary antibody in diluent buffer for 2 h at room temperature, protected from light. Coverslips were mounted on SuperFrost Ultra Plus glass slides (Menzel-Gläser, Thermo Fisher Scientific, Waltham, MA), using SlowFade Diamond Antifade Mountant (Molecular Probes, Eugene, OR). Images were acquired with the Olympus DP71 fluorescent microscope and DP Controller and DP Manager software (Olympus, Shinjuku, Japan). Images were quantified using ImageJ software. Antibodies used for immunofluorescence are listed in Table S7.
Migration Assay
2 × 104 cells were seeded in medium with 0.05% FBS in the inner chamber of Costar Transwell cell culture inserts (6.5-mm diameter, 8-μm pore size; Corning, Corning, NY). Medium with 5% FBS was added to the outer chamber as chemoattractant. Cells were incubated for 22 h, and then chambers were immersed in 0.5% crystal violet and 25% methanol for 10 min. Images of migrating cells were acquired with the Olympus DP71 fluorescent microscope and DP Controller and DP Manager software (Olympus). The number of migrating cells per field of view in each condition was quantified using ImageJ software to analyze 6 separate fields of view each of the 3 replicate wells.
Cell Viability Assay
4 × 103 cells were seeded in each well of 96-well white-bottom plates (Greiner). 24 h later, cells were treated with dabrafenib (Selleck Chemicals, Houston, TX) and/or dactolisib (Selleck Chemicals) at the indicated concentrations. After 72 h, 10 μL CellTiter Glo Luminescent Cell Viability Assay reagent (Promega) was added to each well, and luminescence was measured using the EnVision 2102 Multilabel Reader (PerkinElmer, Waltham, MA) and Wallac Environ software.
Long-Term Colony Formation Assay
1 × 103 cells were seeded in each well of a 6-well plate. 48 h later, medium with dabrafenib and/or dactolisib (each 5 nM) or DMSO vehicle was added. Cells were grown for 3 weeks, replacing the medium containing drugs or DMSO every 3 days. Cells were fixed by immersion in 100% methanol for 10 min, followed by immersion in 0.5% crystal violet and 25% methanol for 10 min. Plates were washed, dried, and photographed, and the average colony diameter (mm) and number of colonies per well were quantified with ImageJ software.
Statistical Analysis
Statistical analyses were performed with Prism 7 (GraphPad, La Jolla, CA). Statistical significance was determined using the non-parametric Kruskal-Wallis test, comparing the mean of each experimental condition to the mean of the control condition with Dunn’s multiple comparisons test. Differences were considered significant at p < 0.05 (*) and p < 0.01 (**). Error bars show SEM.
Author Contributions
Conceptualization, C.M. and P.B.; Methodology, C.M. and P.B.; Formal Analysis, C.M.; Investigation, C.M., F.N., C.B.W., and B.G.-B.; Visualization, C.M.; Writing – Original Draft, C.M.; Writing – Review & Editing, C.M., F.N., A.R.H., and P.B.; Funding Acquisition, P.B.; Resources, P.B.; Supervision, A.R.H. and P.B.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
C.M. was a recipient of the Hackett Postgraduate Research Scholarship from the University of Western Australia. B.G.-B. received financial support from the Comision Nacional de Investigacion Cientifica y Tecnologica (CONICYT) of Chile (CONICYT-PCHA/Doctorado-Nacional/13-21130879 and CONICYT-PAI/7813110022), the School of Medicine and the Vice Rectorate of Investigation at Pontificia Universidad Catolica de Chile, as well as Novartis Chile. P.B. is a recipient of Australian Research Council Future Fellowship FT130101767; the Cancer Council of Western Australia Research Fellowship; National Health and Medical Research Council Project grants APP1165208, APP1069308, APP1147528, and APP1130212; NIH grants R01CA170370 and R01DA036906; and National Breast Cancer Foundation grant NC-14-024.
Footnotes
Supplemental Information includes five figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.12.003.
Supplemental Information
sgRNA numbering refers to start position relative to the transcription start site (TSS) of PTEN mRNA transcript variant 1. The numbering of the start and end of each sgRNA is given according to the Genome Reference Consortium Human Build 38 (GRCh38). Mismatches to the cognate sgRNA sequence are shown in lower case and underlined. Position in relation to TSS refers to annotated RefSeq transcripts. Literature-curated TF binding sites and associated genes are from the PAZAR and JASPAR databases. Chr: chromosome; TF: transcription factor; TSS: transcription start site.
References
- 1.Di Cristofano A., Pandolfi P.P. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 2.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Saal L.H., Johansson P., Holm K., Gruvberger-Saal S.K., She Q.B., Maurer M., Koujak S., Ferrando A.A., Malmström P., Memeo L. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc. Natl. Acad. Sci. USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milella M., Falcone I., Conciatori F., Cesta Incani U., Del Curatolo A., Inzerilli N., Nuzzo C.M., Vaccaro V., Vari S., Cognetti F., Ciuffreda L. PTEN: multiple functions in human malignant tumors. Front. Oncol. 2015;5:24. doi: 10.3389/fonc.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hlobilkova A., Guldberg P., Thullberg M., Zeuthen J., Lukas J., Bartek J. Cell cycle arrest by the PTEN tumor suppressor is target cell specific and may require protein phosphatase activity. Exp. Cell Res. 2000;256:571–577. doi: 10.1006/excr.2000.4867. [DOI] [PubMed] [Google Scholar]
- 6.Leslie N.R., Foti M. Non-genomic loss of PTEN function in cancer: not in my genes. Trends Pharmacol. Sci. 2011;32:131–140. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Bonneau D., Longy M. Mutations of the human PTEN gene. Hum. Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Ciuffreda L., Di Sanza C., Cesta Incani U., Eramo A., Desideri M., Biagioni F., Passeri D., Falcone I., Sette G., Bergamo P. The mitogen-activated protein kinase (MAPK) cascade controls phosphatase and tensin homolog (PTEN) expression through multiple mechanisms. J. Mol. Med. (Berl.) 2012;90:667–679. doi: 10.1007/s00109-011-0844-1. [DOI] [PubMed] [Google Scholar]
- 9.Escrivà M., Peiró S., Herranz N., Villagrasa P., Dave N., Montserrat-Sentís B., Murray S.A., Francí C., Gridley T., Virtanen I., García de Herreros A. Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol. Cell. Biol. 2008;28:1528–1540. doi: 10.1128/MCB.02061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Bergami P., Huang C., Goydos J.S., Yip D., Bar-Eli M., Herlyn M., Smalley K.S., Mahale A., Eroshkin A., Aaronson S., Ronai Z. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hettinger K., Vikhanskaya F., Poh M.K., Lee M.K., de Belle I., Zhang J.T., Reddy S.A., Sabapathy K. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007;14:218–229. doi: 10.1038/sj.cdd.4401946. [DOI] [PubMed] [Google Scholar]
- 12.Vasudevan K.M., Burikhanov R., Goswami A., Rangnekar V.M. Suppression of PTEN expression is essential for antiapoptosis and cellular transformation by oncogenic Ras. Cancer Res. 2007;67:10343–10350. doi: 10.1158/0008-5472.CAN-07-1827. [DOI] [PubMed] [Google Scholar]
- 13.Wiencke J.K., Zheng S., Jelluma N., Tihan T., Vandenberg S., Tamgüney T., Baumber R., Parsons R., Lamborn K.R., Berger M.S. Methylation of the PTEN promoter defines low-grade gliomas and secondary glioblastoma. Neuro-oncol. 2007;9:271–279. doi: 10.1215/15228517-2007-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García J.M., Silva J., Peña C., Garcia V., Rodríguez R., Cruz M.A., Cantos B., Provencio M., España P., Bonilla F. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chromosomes Cancer. 2004;41:117–124. doi: 10.1002/gcc.20062. [DOI] [PubMed] [Google Scholar]
- 15.Goel A., Arnold C.N., Niedzwiecki D., Carethers J.M., Dowell J.M., Wasserman L., Compton C., Mayer R.J., Bertagnolli M.M., Boland C.R. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 16.Kang Y.H., Lee H.S., Kim W.H. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab. Invest. 2002;82:285–291. doi: 10.1038/labinvest.3780422. [DOI] [PubMed] [Google Scholar]
- 17.Mirmohammadsadegh A., Marini A., Nambiar S., Hassan M., Tannapfel A., Ruzicka T., Hengge U.R. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 18.Soria J.C., Lee H.Y., Lee J.I., Wang L., Issa J.P., Kemp B.L., Liu D.D., Kurie J.M., Mao L., Khuri F.R. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin. Cancer Res. 2002;8:1178–1184. [PubMed] [Google Scholar]
- 19.Khan S., Kumagai T., Vora J., Bose N., Sehgal I., Koeffler P.H., Bose S. PTEN promoter is methylated in a proportion of invasive breast cancers. Int. J. Cancer. 2004;112:407–410. doi: 10.1002/ijc.20447. [DOI] [PubMed] [Google Scholar]
- 20.Salvesen H.B., MacDonald N., Ryan A., Jacobs I.J., Lynch E.D., Akslen L.A., Das S. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int. J. Cancer. 2001;91:22–26. doi: 10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 21.Carracedo A., Alimonti A., Pandolfi P.P. PTEN level in tumor suppression: how much is too little? Cancer Res. 2011;71:629–633. doi: 10.1158/0008-5472.CAN-10-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikhail M., Velazquez E., Shapiro R., Berman R., Pavlick A., Sorhaindo L., Spira J., Mir C., Panageas K.S., Polsky D., Osman I. PTEN expression in melanoma: relationship with patient survival, Bcl-2 expression, and proliferation. Clin. Cancer Res. 2005;11:5153–5157. doi: 10.1158/1078-0432.CCR-05-0397. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X.P., Gimm O., Hampel H., Niemann T., Walker M.J., Eng C. Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am. J. Pathol. 2000;157:1123–1128. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollander M.C., Blumenthal G.M., Dennis P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nathanson K.L., Martin A.M., Wubbenhorst B., Greshock J., Letrero R., D’Andrea K., O’Day S., Infante J.R., Falchook G.S., Arkenau H.T. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436) Clin. Cancer Res. 2013;19:4868–4878. doi: 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dankort D., Curley D.P., Cartlidge R.A., Nelson B., Karnezis A.N., Damsky W.E., Jr., You M.J., DePinho R.A., McMahon M., Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byron S.A., Loch D.C., Wellens C.L., Wortmann A., Wu J., Wang J., Nomoto K., Pollock P.M. Sensitivity to the MEK inhibitor E6201 in melanoma cells is associated with mutant BRAF and wildtype PTEN status. Mol. Cancer. 2012;11:75. doi: 10.1186/1476-4598-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing F., Persaud Y., Pratilas C.A., Taylor B.S., Janakiraman M., She Q.B., Gallardo H., Liu C., Merghoub T., Hefter B. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31:446–457. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paraiso K.H., Xiang Y., Rebecca V.W., Abel E.V., Chen Y.A., Munko A.C., Wood E., Fedorenko I.V., Sondak V.K., Anderson A.R. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng W., Gopal Y.N., Scott A., Chen G., Woodman S.E., Davies M.A. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment Cell Melanoma Res. 2012;25:248–258. doi: 10.1111/j.1755-148X.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 32.Jang S., Atkins M.B. Which drug, and when, for patients with BRAF-mutant melanoma? Lancet Oncol. 2013;14:e60–e69. doi: 10.1016/S1470-2045(12)70539-9. [DOI] [PubMed] [Google Scholar]
- 33.Atefi M., von Euw E., Attar N., Ng C., Chu C., Guo D., Nazarian R., Chmielowski B., Glaspy J.A., Comin-Anduix B. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS ONE. 2011;6:e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopal Y.N., Deng W., Woodman S.E., Komurov K., Ram P., Smith P.D., Davies M.A. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–8747. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi H., Hugo W., Kong X., Hong A., Koya R.C., Moriceau G., Chodon T., Guo R., Johnson D.B., Dahlman K.B. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villanueva J., Vultur A., Lee J.T., Somasundaram R., Fukunaga-Kalabis M., Cipolla A.K., Wubbenhorst B., Xu X., Gimotty P.A., Kee D. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gopal Y.N., Rizos H., Chen G., Deng W., Frederick D.T., Cooper Z.A., Scolyer R.A., Pupo G., Komurov K., Sehgal V. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma. Cancer Res. 2014;74:7037–7047. doi: 10.1158/0008-5472.CAN-14-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greger J.G., Eastman S.D., Zhang V., Bleam M.R., Hughes A.M., Smitheman K.N., Dickerson S.H., Laquerre S.G., Liu L., Gilmer T.M. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol. Cancer Ther. 2012;11:909–920. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 39.Shi H., Kong X., Ribas A., Lo R.S. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H.Y., Liang F., Jia Z.L., Song S.T., Jiang Z.F. PTEN mutation, methylation and expression in breast cancer patients. Oncol. Lett. 2013;6:161–168. doi: 10.3892/ol.2013.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muggerud A.A., Rønneberg J.A., Wärnberg F., Botling J., Busato F., Jovanovic J., Solvang H., Bukholm I., Børresen-Dale A.L., Kristensen V.N. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12:R3. doi: 10.1186/bcr2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marty B., Maire V., Gravier E., Rigaill G., Vincent-Salomon A., Kappler M., Lebigot I., Djelti F., Tourdès A., Gestraud P. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Knowles E., O’Toole S.A., McNeil C.M., Millar E.K., Qiu M.R., Crea P., Daly R.J., Musgrove E.A., Sutherland R.L. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int. J. Cancer. 2010;126:1121–1131. doi: 10.1002/ijc.24831. [DOI] [PubMed] [Google Scholar]
- 44.Zhou J., Wulfkuhle J., Zhang H., Gu P., Yang Y., Deng J., Margolick J.B., Liotta L.A., Petricoin E., 3rd, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daverey A., Drain A.P., Kidambi S. Physical intimacy of breast cancer cells with mesenchymal stem cells elicits trastuzumab resistance through Src activation. Sci. Rep. 2015;5:13744. doi: 10.1038/srep13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waryah C.B., Moses C., Arooj M., Blancafort P. Zinc Fingers, TALEs, and CRISPR Systems: A Comparison of Tools for Epigenome Editing. Methods Mol. Biol. 2018;1767:19–63. doi: 10.1007/978-1-4939-7774-1_2. [DOI] [PubMed] [Google Scholar]
- 47.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moses C., Garcia-Bloj B., Harvey A.R., Blancafort P. Hallmarks of cancer: The CRISPR generation. Eur. J. Cancer. 2018;93:10–18. doi: 10.1016/j.ejca.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., P R Iyer E., Lin S., Kiani S., Guzman C.D., Wiegand D.J. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Bloj B., Moses C., Sgro A., Plani-Lam J., Arooj M., Duffy C., Thiruvengadam S., Sorolla A., Rashwan R., Mancera R.L. Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system. Oncotarget. 2016;7:60535–60554. doi: 10.18632/oncotarget.11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fofaria N.M., Frederick D.T., Sullivan R.J., Flaherty K.T., Srivastava S.K. Overexpression of Mcl-1 confers resistance to BRAFV600E inhibitors alone and in combination with MEK1/2 inhibitors in melanoma. Oncotarget. 2015;6:40535–40556. doi: 10.18632/oncotarget.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jovanović B., Beeler J.S., Pickup M.W., Chytil A., Gorska A.E., Ashby W.J., Lehmann B.D., Zijlstra A., Pietenpol J.A., Moses H.L. Transforming growth factor beta receptor type III is a tumor promoter in mesenchymal-stem like triple negative breast cancer. Breast Cancer Res. 2014;16:R69. doi: 10.1186/bcr3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grimmer M.R., Stolzenburg S., Ford E., Lister R., Blancafort P., Farnham P.J. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 2014;42:10856–10868. doi: 10.1093/nar/gku708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huisman C., van der Wijst M.G., Schokker M., Blancafort P., Terpstra M.M., Kok K., van der Zee A.G., Schuuring E., Wisman G.B., Rots M.G. Re-expression of selected epigenetically silenced candidate tumor suppressor genes in cervical cancer by TET2-directed demethylation. Mol. Ther. 2016;24:536–547. doi: 10.1038/mt.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mendenhall E.M., Williamson K.E., Reyon D., Zou J.Y., Ram O., Joung J.K., Bernstein B.E. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polstein L.R., Perez-Pinera P., Kocak D.D., Vockley C.M., Bledsoe P., Song L., Safi A., Crawford G.E., Reddy T.E., Gersbach C.A. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25:1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morita S., Noguchi H., Horii T., Nakabayashi K., Kimura M., Okamura K., Sakai A., Nakashima H., Hata K., Nakashima K., Hatada I. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 2016;34:1060–1065. doi: 10.1038/nbt.3658. [DOI] [PubMed] [Google Scholar]
- 69.Liu X.S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S., Shu J., Dadon D., Young R.A., Jaenisch R. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247.e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amabile A., Migliara A., Capasso P., Biffi M., Cittaro D., Naldini L., Lombardo A. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell. 2016;167:219–232.e14. doi: 10.1016/j.cell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu J., Tamura M., Pankov R., Danen E.H.J., Takino T., Matsumoto K., Yamada K.M. Shc and FAK differentially regulate cell motility and directionality modulated by PTEN. J. Cell Biol. 1999;146:389–403. doi: 10.1083/jcb.146.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leslie N.R., Yang X., Downes C.P., Weijer C.J. The regulation of cell migration by PTEN. Biochem. Soc. Trans. 2005;33:1507–1508. doi: 10.1042/BST0331507. [DOI] [PubMed] [Google Scholar]
- 74.Hauschild A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M., Rutkowski P., Blank C.U., Miller W.H., Jr., Kaempgen E. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 75.Sweetlove M., Wrightson E., Kolekar S., Rewcastle G.W., Baguley B.C., Shepherd P.R., Jamieson S.M. Inhibitors of pan-PI3K signaling synergize with BRAF or MEK inhibitors to prevent BRAF-mutant melanoma cell growth. Front. Oncol. 2015;5:135. doi: 10.3389/fonc.2015.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamura M., Gu J., Matsumoto K., Aota S., Parsons R., Yamada K.M. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 77.Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Muñoz D.P., Boffelli D. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Govender D., Chetty R. Gene of the month: PTEN. J. Clin. Pathol. 2012;65:601–603. doi: 10.1136/jclinpath-2012-200711. [DOI] [PubMed] [Google Scholar]
- 81.Gregory D.J., Zhang Y., Kobzik L., Fedulov A.V. Specific transcriptional enhancement of inducible nitric oxide synthase by targeted promoter demethylation. Epigenetics. 2013;8:1205–1212. doi: 10.4161/epi.26267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beltran A.S., Russo A., Lara H., Fan C., Lizardi P.M., Blancafort P. Suppression of breast tumor growth and metastasis by an engineered transcription factor. PLoS ONE. 2011;6:e24595. doi: 10.1371/journal.pone.0024595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beltran A.S., Blancafort P. Reactivation of MASPIN in non-small cell lung carcinoma (NSCLC) cells by artificial transcription factors (ATFs) Epigenetics. 2011;6:224–235. doi: 10.4161/epi.6.2.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huisman C., Wisman G.B.A., Kazemier H.G., van Vugt M.A., van der Zee A.G., Schuuring E., Rots M.G. Functional validation of putative tumor suppressor gene C13ORF18 in cervical cancer by Artificial Transcription Factors. Mol. Oncol. 2013;7:669–679. doi: 10.1016/j.molonc.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stolzenburg S., Rots M.G., Beltran A.S., Rivenbark A.G., Yuan X., Qian H., Strahl B.D., Blancafort P. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012;40:6725–6740. doi: 10.1093/nar/gks360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Gun B.T., Huisman C., Stolzenburg S., Kazemier H.G., Ruiters M.H., Blancafort P., Rots M.G. Bidirectional modulation of endogenous EpCAM expression to unravel its function in ovarian cancer. Br. J. Cancer. 2013;108:881–886. doi: 10.1038/bjc.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kretzmann J.A., Ho D., Evans C.W., Plani-Lam J.H.C., Garcia-Bloj B., Mohamed A.E., O’Mara M.L., Ford E., Tan D.E.K., Lister R. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem. Sci. (Camb.) 2017;8:2923–2930. doi: 10.1039/c7sc00097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lara H., Wang Y., Beltran A.S., Juárez-Moreno K., Yuan X., Kato S., Leisewitz A.V., Cuello Fredes M., Licea A.F., Connolly D.C. Targeting serous epithelial ovarian cancer with designer zinc finger transcription factors. J. Biol. Chem. 2012;287:29873–29886. doi: 10.1074/jbc.M112.360768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y., Su H.H., Yang Y., Hu Y., Zhang L., Blancafort P., Huang L. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 92.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller J.B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S.S., Zhu H., Siegwart D.J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang A.Z., Langer R., Farokhzad O.C. Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 95.Cano-Rodriguez D., Gjaltema R.A.F., Jilderda L.J., Jellema P., Dokter-Fokkens J., Ruiters M.H.J., Rots M.G. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun. 2016;7:12284. doi: 10.1038/ncomms12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hopkins B.D., Fine B., Steinbach N., Dendy M., Rapp Z., Shaw J., Pappas K., Yu J.S., Hodakoski C., Mense S. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013;341:399–402. doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Putz U., Howitt J., Doan A., Goh C.P., Low L.H., Silke J., Tan S.S. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci. Signal. 2012;5:ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 98.Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kabadi A.M., Gersbach C.A. Engineering synthetic TALE and CRISPR/Cas9 transcription factors for regulating gene expression. Methods. 2014;69:188–197. doi: 10.1016/j.ymeth.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tiscornia G., Singer O., Verma I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 101.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 102.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sgRNA numbering refers to start position relative to the transcription start site (TSS) of PTEN mRNA transcript variant 1. The numbering of the start and end of each sgRNA is given according to the Genome Reference Consortium Human Build 38 (GRCh38). Mismatches to the cognate sgRNA sequence are shown in lower case and underlined. Position in relation to TSS refers to annotated RefSeq transcripts. Literature-curated TF binding sites and associated genes are from the PAZAR and JASPAR databases. Chr: chromosome; TF: transcription factor; TSS: transcription start site.