Summary
It is proposed that conformational changes induced in proteins by oxidation can lead to loss of activity or protein aggregation through exposure of hydrophobic residues and alteration in surface hydrophobicity. Because increased oxidative stress and protein aggregation are consistently observed in amyotrophic lateral sclerosis (ALS), we used a BisANS photo-labeling approach to monitor changes in protein unfolding in vivo in skeletal muscle proteins in ALS mice. We find two major proteins, creatine kinase (CK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), conformationally affected in the ALS G93A mouse model concordant with a 43 and 41% reduction in enzyme activity, respectively. This correlated with changes in conformation and activity that were detected in CK and GAPDH with in vitro oxidation. Interestingly, we found that GAPDH, but not CK, is conformationally and functionally affected in a longer-lived ALS model (H46R/H48Q) exhibiting a 22% reduction in enzyme activity. We proposed a reaction mechanism for BisANS with nucleophilic amino acids such as lysine, serine, threonine, and tyrosine; and BisANS was found to be primarily incorporated to lysine residues in GAPDH. We identified the specific BisANS incorporation sites on GAPDH in non-transgenic (NTg), G93A- and H46R/H48Q- mice using LC-MS/MS analysis. Four BisANS containing sites (K52, K104, K212, K248) were found in NTg GAPDH, while three out of four of these sites were lost in either G93A or H46R/H48Q GAPDH. Conversely, eight new sites (K2, K63, K69, K114, K183, K251, S330, and K331) were found on GAPDH for G93A, including one common site (K114) for H46R/H48Q, was detected that is not found in GAPDH from NTg mice. These data show that GAPDH is differentially affected structurally and functionally in vivo in accordance with the degree of oxidative stress associated with these two models of ALS.
Keywords: Protein Oxidation, Amyotrophic lateral sclerosis, BisANS, protein unfolding, GAPDH, Creatine Kinase
Introduction
Oxidative modification of proteins has been observed to increase in a large number of patho-physiological conditions, including neurodegenerative diseases1 and aging2. All amino acid side chains are susceptible to oxidation3, and to date, more than 274 types of verified protein modifications have been submitted to online databases such as unimod (www.unimod.org)4. Conservative estimates suggest that on average, an amino acid modification exists in every tenth amino acid in a particular protein in vivo5.
Because proteins play an essential role in a variety of cellular functions (e.g., signal transduction, mitosis, chaperone activity, protein degradation), oxidative damage to critical amino acid residue(s) in the active sites of proteins and in residues important for maintaining structure could lead to reduced function of the protein. In vitro studies show that the oxidative modification of a single histidine in glutamate synthetase leads to loss of enzyme activity6. Alteration of function can also occur allosterically via induction of conformational changes to the protein by the oxidation of amino acid residues outside of the active site7; 8. Mild protein oxidation also can result in protein misfolding or unfolding, which can lead to exposure of hydrophobic residues9 and ubiquitin-independent degradation by the 20S proteasome10. In addition, protein aggregates can result from increased exposure of hydrophobic residues resulting from more severe oxidation, which are poor substrates for the proteasome9; 11. For example, Cu/Zn superoxide dismutase 1 (SOD1) mutants are misfolded and are substrates for the proteasome, while aggregates of SOD1 are not12.
Proteins possess ordered hydrophobic domains on their surface, as well as a hydrophobic core that facilitates protein folding13. Surface hydrophobicity has been used to follow changes in protein conformation14 and has been shown to be altered by oxidative stress3. ANS (1,8 anilinoline sulfonate) and various derivatives of ANS (e.g., 4,4’-dianilino-1,1’-binaphthyl-5,5’-disulfonic acid, BisANS) are planar, hydrophobic molecules that bind to surface hydrophobic domains and fluoresce at 490 nm when excited at 385 nm in apolar environments, i.e., when they bind to hydrophobic sites on the protein. Using ANS and BisANS, several investigators have studied the binding and incorporation of BisANS in proteins such as rhodanase15, GROEL16, and tubulin17. Chao et al.18 used ANS binding to rat liver extracts to study the effect of oxidative stress and age on surface hydrophobicity. They observed that in vitro oxidation with iron ascorbate or 2,2’-azobis (2-amidinopropane) dihydrochloride led to a dose-dependent increase in global surface hydrophobicity of proteins in liver extracts as shown by an increase in ANS fluorescence. Consistent with this finding, they also reported that surface hydrophobicity was increased with age in rat liver, i.e., increased fluorescence was observed when ANS was added to liver extracts from old rats compared to young rats. Because protein oxidation had been shown to increase with age in the liver19, Chao et al.18 concluded that the increased surface hydrophobicity with age arose from increased protein oxidation. The major limitation of this study is that the assay only allowed Chao et al.18 to quantify global changes in surface hydrophobicity, i.e., changes in surface hydrophobicity in specific proteins could not be determined. This limitation is important because it has been shown that the sensitivity of proteins to oxidative stress varies considerably from protein to protein, as does a protein’s structural stability20.
Although several studies show that oxidative stress correlates with changes in protein surface hydrophobicity in the test tube, it is not clear if the levels of oxidative stress that occur in vivo can lead to protein unfolding and alterations in protein conformation such as surface hydrophobicity. We recently developed a proteomic approach that allows one to screen complex mixtures of proteins for alterations in protein unfolding in specific proteins by photoincorporation of BisANS14. Using the BisANS assay, we have screened the proteome of skeletal muscle of mice with amyotrophic lateral sclerosis (ALS) for changes in protein unfolding on specific proteins. ALS is a progressive neuromuscular disease that affects both motor neurons and skeletal muscle, eventually leading to paralysis and death. Point mutations in the antioxidant protein Cu/Zn superoxide dismutase (SOD1) have been shown to cause ALS in both humans and transgenic mouse models21; 22. The ALS mouse models show increased oxidative stress in spinal cord and skeletal muscle23; therefore, these mice give us the opportunity to study the effect of oxidative stress in vivo on protein conformation in a neurodegenerative disease. In an ALS mouse overexpressing the G93A SOD1 mutation, which is associated with elevated oxidative stress24, we observed major changes in BisANS binding to two major skeletal muscle proteins: creatine kinase (CK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The decreased BisANS incorporation was associated with decreased activity in these two enzymes. BisANS screening of the proteome of a second ALS mutant model, H46R/H48Q, which shows a more modest progression of ALS and less of an increase in oxidative stress than the G93A mutant, showed changes in GAPDH incorporation of BisANS and reduced GAPDH activity. Using LC-MS/MS, we mapped the surface hydrophobic domain(s) on GAPDH in skeletal muscle of G93A, H46R/H48Q, and non-transgenic (NTg) mice and showed that several domains in the NTg mice are lost in the ALS models and that new domains of surface hydrophobicity are observed in the ALS mutant mice. Our data show that increased oxidative stress in vivo resulted in changes in the conformation of specific proteins.
Results
Changes in protein conformation detected in skeletal muscle of symptomatic G93A ALS mice
Cytosolic proteins were isolated from the skeletal muscle of symptomatic G93A (130 days old) mice and age-matched NTg littermates and photolabeled with BisANS. In addition, cytosolic proteins from the skeletal muscle of transgenic mice overexpressing non-mutant human SOD1 (TgSOD1) were used as a negative control. Figure 1 shows the fluorescence and coomassie staining of the BisANS labeled cytosolic proteins on SDS-gels. A significant decrease (30 +/− 11% p=0.047) in the global BisANS: coomassie ratio was observed in skeletal muscle cytosolic proteins of symptomatic G93A mice compared to age-matched NTg mice. In particular, two major protein bands corresponding to 43 and 35kDa in all G93A mice exhibited a decrease (25 to 30%) in BisANS fluorescence compared to NTg mice; however, multiple proteins could be present within these regions. Overexpression of non-mutant SOD1 (TgSOD1) did not alter BisANS fluorescence, indicating that the decrease in BisANS fluorescence was due to the ALS mutant SOD1, and not simply due to increased SOD1 expression or mass-action effects of increased SOD1 expression, which can be seen migrating at 15kDa.
Figure 1. Detection of Changes in Protein Folding in Skeletal Muscle Cytosolic Proteins from G93A Mice.
The cytosolic proteins isolated from skeletal muscle of 130-day-old G93A mice were photolabeled with BisANS as described in the Methods section. Equal amounts of proteins (20 μg) of BisANS-labeled proteins were subject to SDS-PAGE and visualized the labeled proteins under UV light followed by staining with coomassie blue. Fluorescent (upper panels) and coomassie (lower panels) images are shown for G93A (left), WT SOD1 (right) and their respective NTg littermates. Regions sensitive to conformational alteration in G93A are indicated by arrows.
In order to identify the two major structurally altered cytosolic proteins from muscle extracts of symptomatic G93A that exhibited a significant loss of fluorescence, two-dimensional gel electrophoresis of BisANS labeled skeletal muscle cytosolic protein was performed, and these data are shown in Figure 2. Two regions on the 2D gels at 43 and 35kDa containing multiple pI variants showed a dramatic decrease in BisANS fluorescence from the muscle of G93A mice compared to NTg mice. Using densitometric ratio analysis, the two regions containing the pI variants showed a decrease of approximately 70% in the ratio of BisANS:Sypro Ruby fluorescence in G93A mice compared to NTg controls. Several spots in these regions were taken from the gels and identified as creatine kinase (CK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by MALDI-TOF/MS (Table 1). The BisANS fluorescence on CK and GAPDH in TgSOD1 mice (130 days old) were identical to NTg littermates, ruling out the possibility that the loss of BisANS fluorescence in CK and GAPDH resulted from the overexpression of WT SOD1. Changes in BisANS incorporation in CK and GAPDH could be detected as early as 90 days (data not shown), which is consistent with the point in time when increases in mitochondrial ROS can be observed in the G93A mice25.
Figure 2. 2D Gel Separation of BisANS Labeled Skeletal Muscle Cytosolic Proteins from G93A Mice.
The cytosolic proteins isolated from skeletal muscle of 130-day-old G93A mice were separated by 2D gel electrophoresis (150 μg), and visualized under UV light (upper panels) followed by overnight staining of the proteins with Sypro Ruby (lower panels). Spots (circled) from two regions of interest were obtained and identified by mass spectrometry.
Table 1:
MALDI-TOF/MS Identification Summary
| Spot # | ID | Mrcalc | Peptides Searched | Peptides Matched | Percent Coverage | Expect | Mowse Score |
|---|---|---|---|---|---|---|---|
| 1 | Creatine Kinase, Muscle | 43018 | 47 | 19 | 48 | 2.3e–13 | 177 |
| 2 | Creatine Kinase, Muscle | 43018 | 50 | 24 | 64 | 1.2e–16 | 210 |
| 3 | Creatine Kinase, Muscle | 43018 | 33 | 17 | 43 | 7.3e–14 | 182 |
| 4 | Creatine Kinase, Muscle | 43018 | 43 | 18 | 42 | 1.8e–12 | 168 |
| 5 | GAPDH | 35787 | 57 | 24 | 63 | 4.3e–09 | 133 |
| 6 | GAPDH | 35787 | 90 | 28 | 59 | 2.9e–11 | 156 |
| 7 | GAPDH | 35787 | 101 | 21 | 58 | 3.4e–06 | 104 |
| 8 | GAPDH | 35787 | 90 | 23 | 57 | 2.7e–07 | 115 |
Spot numbers correspond to spots indicated in Figure 2. Mrcalc: calculated molecular weight.
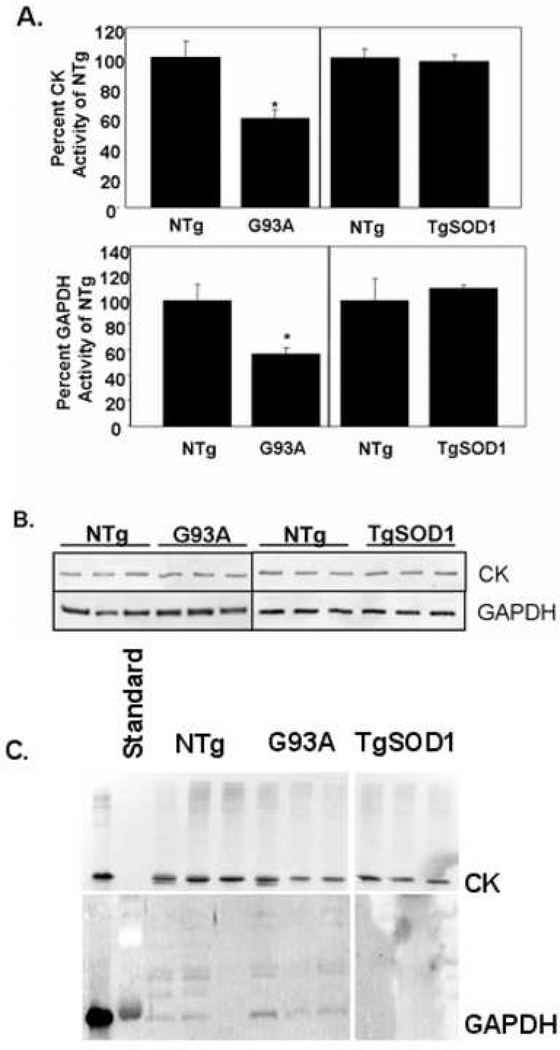
To determine if the changes in BisANS incorporation to CK and GAPDH in the G93A mice were accompanied with a loss of function, the enzymatic activities of CK and GAPDH were measured. The data in Figure 3A show that CK and GAPDH activities were significantly reduced by 40.5 +/− 5.5 and 43 +/− 4.7%, respectively, in G93A skeletal muscle compared to NTg littermates. Overexpression of WT human SOD1 had no effect on the activity of either enzyme in TgSOD1 mice. In addition, the observed decrease in activity was not due to a decrease in protein levels, as determined by Western blot analysis shown in Figure 3B. Because the changes in BisANS binding suggest protein folding is altered in the G93A mice, we were concerned that changes in the aggregation of CK and GAPDH could occur. Therefore, we determined whether CK and GAPDH were enriched in detergent-insoluble fractions of skeletal muscle from G93A mice by sequentially extracting the insoluble pellets from the 100,000 g centrifugation with detergents of increasing ionic strength. However, neither CK nor GAPDH were differentially represented in tissue fractions containing detergent insoluble proteins from G93A mice, as shown by immunoblotting the P3 fraction of skeletal muscle for CK and GAPDH following differential detergent extraction (Figure 3C).
Figure 3. Creatine Kinase and GAPDH Activity in Skeletal Muscle from G93A Mice.
Graph A) the activities of CK (upper panel) and GAPDH (lower panel) were measured in skeletal muscle cytosol from G93A, TgSOD1, and their respective NTg littermates as described in Methods. Values are expressed as the average percent activity of NTg and represent the mean of 3 animals per group ± standard deviation *= p<0.05. Graph B) Western blots for CK and GAPDH are shown. Graph C) Western blot for CK (upper panel) and GAPDH (lower panel) in the detergent-insoluble P3 fraction of skeletal muscle lysates.
To verify that oxidative stress could induce changes in BisANS incorporation to CK and GAPDH and reduce the activity of these two enzymes, purified CK and GAPDH were oxidized in vitro with iron/ascorbate and probed for changes in BisANS incorporation and enzyme activity. Oxidative stress induced a decrease in BisANS incorporation in both enzymes in vitro; however, BisANS incorporation to GAPDH was more sensitive to oxidative stress (Figure 4A and B). We also observed a decrease in activity of both enzymes in response to iron/ascorbate induced oxidative stress (Figure 4C). GAPDH activity was decreased by 90% in response to 40 μM iron sulfate and 10 mM ascorbate, while CK activity was decreased less than 50% (Figure 4C). The difference between GAPDH and CK sensitivity to oxidative stress can be observed at concentrations as low as 10 μM iron sulfate, 2.5 mM ascorbate where GAPDH activity is reduced 40%, while CK is reduced less than 20%. Interestingly, the loss of activity and change in BisANS incorporation observed at this level of in vitro stress correlates with the loss of activity and BisANS incorporation that we observed in vivo for G93A mice. These data support the possibility that increased oxidative stress associated with the G93A mutation is responsible for the changes in BisANS incorporation and enzymatic activity in CK and GAPDH.
Figure 4. Effect of Oxidative Stress In Vitro on the Activity and Conformation of GAPDH and CK.
Graph A) purified rabbit muscle CK and GAPDH (1 mg/ml) were oxidized separately with various concentrations of iron (0, 0.2, 0.4, 1.0, 2.0, 3.0, 4.0, 6.0, 8.0, 10, 20, and 40 μM) and ascorbate (0, 0.05, 0.1, 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 5 and 10 mM) for 1 hr at 37°C in the dark followed by labeling with BisANS (100 μM). CK and GAPDH (5 μg, each) were mixed and separated by SDS-PAGE and the fluorescence of BisANS was visualized as described in the Methods. Shown are the CK and GAPDH BisANS fluorescence (upper panel) and Coomassie images from the gel (lower panel). Graph B) the ratio of BisANS intensity: Coomassie intensity for CK (closed circles) and GAPDH (open circles) was quantitated by densitometry and expressed as percent intensity of control (no oxidation). Graph C) enzyme activity of iron and ascorbate oxidized CK (closed circles) and GAPDH (open circles) was measured and expressed as percent activity of control (no stress). The data represent the mean of 3 experiments, +/− standard deviation.
To determine if CK and GAPDH were oxidized in the skeletal muscle of G93A mice, we measured the presence of two types of oxidative damage in CK and GAPDH as we have described previously 20; 26: carbonyl groups and the irreversible oxidation of cysteine. We found no difference in the carbonyl content of either CK or GAPDH in muscle from G93A and WT mice (data not shown); however, an increase in irreversible cysteine oxidation was observed. As shown in Figure 5, we found a decrease in the level of dithiothreitol-reducible cysteines in protein spots on 2-D gels corresponding to CK and GAPDH. In this assay, unfolded, reduced proteins react with 6-iodoacetamido fluorescein, and a decrease in fluorescein incorporation is indicative of irreversible cysteine oxidation of these proteins as shown in Figure 5.
Figure 5. 2D Gel Separation of 6-IAF Labeled Skeletal Muscle Cytosolic Proteins from G93A Mice.
The cytosolic proteins isolated from skeletal muscle of H46R/H48Q were labeled with 6IAF as described in Methods. 6-IAF labeled skeletal muscle cytosolic proteins (150 μg) were separated by 2D gel electrophoresis, and the region containing CK and GAPDH is shown. Three separate animals were analyzed per group, and shown are representative gels from each group.
GAPDH is commonly affected in G93A and H46R/H48Q models of ALS
We studied the effect of ALS on BisANS incorporation to CK and GAPDH in a second mouse model of ALS; the H46R/H48Q mouse overexpresses a mutant SOD1 that does not bind copper in the active site and lacks SOD activity. The H46R/H48Q mice develop symptoms around 180 days, and they do not exhibit muscle atrophy until near the end of their lifespan, which is around 250 days compared to G93A mice which live until 150 days. Thus, the H46R/H48Q model exhibits a milder disease course and is also associated with a lesser degree of oxidative stress25. Figures 6A and B show that BisANS incorporation to CK changed very little (less than 8%) in the skeletal muscle of late symptomatic (230 days) H46R/H48Q mice; however, BisANS incorporation to GAPDH decreased almost 25%. The data in Figure 6C show that the CK activity in skeletal muscle extracts from H46R/H48Q mice was not significantly altered compared to NTg mice. However, the activity of GAPDH was significantly decreased (22%) in the H46R/H48Q mice compared to NTg mice. The protein levels of CK and GAPDH were similar for NTg and H46R/H48Q mice (Figure 6D). Thus, the H46R/H48Q mice, which have less oxidative stress than G93A mice, do not show alterations in either BisANS incorporation or enzymatic activity of CK, but do show reduced incorporation of BisANS and enzymatic activity for GAPDH. These data are consistent with our in vitro stress data (Figure 4), showing that GAPDH was more sensitive to oxidative stress compared to CK.
Figure 6. Separation of BisANS Labeled Skeletal Muscle Cytosolic Proteins from H46R/H48Q Mice.
Graph A) The cytosolic proteins isolated from skeletal muscle of H46R/H48Q were photolabeled with BisANS as described in Methods. Equal amounts of BisANS-labeled proteins (20 μg) were subject to SDS-PAGE, visualized under UV and stained with coomassie blue. Graph B) BisANS labeled skeletal muscle cytosolic proteins (150 μg) were separated by 2D gel electrophoresis, and the region containing CK and GAPDH is shown. Graph C) CK (upper panel) and GAPDH (lower panel) activity are shown, and the values are expressed as the average percent activity of WT. The data represent the mean of 3 animals per group +/− standard deviation, and the values that are significantly different from WT are shown (*= p<0.05). D) Western blots for CK and GAPDH are shown.
Identification of BisANS incorporation sites in GAPDH peptides from skeletal muscle of NTg, G93A, and H46R/H48Q mice.
Because the BisANS incorporation to GAPDH is markedly altered in both G93A and H46R/H48Q, we were interested in mapping the BisANS-labeled peptides in GAPDH from skeletal muscle cytosol of NTg, G93A, and H46R/H48Q mice to identify potential hydrophobic domains on GAPDH that are altered in both ALS mouse models. BisANS-labeled cytosolic proteins from NTg, G93A, and H46R/H48Q mice were separated by 2D gel electrophoresis to resolve GAPDH, followed by digestion with trypsin and analysis by LC-MS/MS. Although some reports have limited the amino acid incorporation site of BisANS binding in various proteins to a 45 amino acid long peptide16, there is currently no information on the photoreaction mechanism responsible for the cross-linking of BisANS to a protein, i.e., no information on either the specific site(s) of BisANS cross-link or the chemical structure of the final peptide product. Photo-incorporation of BisANS most likely involves formation of a radical cation that is highly reactive and is targeted by nucleophilic amino acids such as lysine, serine, threonine and tyrosine. Studies conducted on compounds similar to BisANS, such as diphenylamine, have shown that the primary product of flash photolysis is a radical cation.27 Based on these data we proposed a reaction mechanism of BisANS photoincorporation to lysine (shown in Figure 7A), which involves reaction of the nucleophilic side chain of lysine to the radical cation formed on the meta carbon of the BisANS aniline ring. A representative spectrum of peptide AENGKLVINGKPITIFQER from GAPDH cross-linked with BisANS at K63 is shown (Figure 7B). The mass of BisANS can be calculated by subtracting the mass of an unlabeled peptide ion from the mass of the singly charged b (5) fragment ion according to the proposed reaction mechanism (Figure 7A). LC MS/MS spectra from tryptic digests of BisANS labeled GAPDH proteins from NTg, G93A, and H46R/H48Q mice were screened using a variable modification for BisANS to lysine, serine, threonine, and tyrosine. The peptides that incorporated BisANS are given in Table 2 for GAPDH isolated from skeletal muscle of NTg, G93A, and H46R/H48Q mice. We first identified four BisANS incorporation sites in GAPDH from NTg mice; their sequences are indicated in Table 2 and their location in GAPDH is modeled on the X-Ray crystal structure of human GAPDH shown in Figure 8A and B. These four incorporation sites have BisANS bound through lysine residues. In GAPDH from H46R/H48Q mice, only one of these four sites was found. In addition, a new BisANS incorporation site on K114 was observed that is not present in GAPDH from NTg mice. GAPDH isolated from G93A mice showed an absence of BisANS labeling to K52, K104, and K212 found in NTg mice; however, a large number of new sites labeled with BisANS, which were not found in the NTg mice, were found in GAPDH isolated from G93A mice. BisANS was found to bind K114, which also was found in H46R/H48Q mice. In addition, seven new sites were found to be labeled with BisANS in G93A mice. Interestingly, two peptides were found that contained two BisANS molecules bound to the peptides (K248 & K251, S330 & K331), suggesting a high-affinity BisANS incorporation site. All of the new BisANS incorporation sites in the G93A mice had BisANS incorporated to a lysine residue, except for the S330 site, which had BisANS linked through a serine residue. The location of the new BisANS incorporation sites in the GAPDH molecule and their correlation to levels of oxidative stress and GAPDH activity are shown in Figure 9.
Figure 7. Hypothetical Reaction Scheme of BisANS Reaction with Nucleophilic Amino Acids.
A) Nucleophilic amino acids such as lysine, serine, threonine, and tyrosine undergo a nucleophilic attack on the resonance structure of the aniline ring of BisANS, forming a BisANS cross-linked amino acid. B) MS/MS analysis of peptide AENGKLVINGKPIT-IFQER from GAPDH. All of the “y” and “b” ions are assigned in the spectrum. C) Diagram showing the “y” and “b” ion map superimposed on the peptide sequence. The BisANS lysine is labeled in the sequence.
Table 2:
BisANS Incorporation Sites in NTG, G93A, and H46R/H48Q GAPDH
| # | NTG GAPDH BisANS Incorporation | Span | BisANS Site | m/z |
|---|---|---|---|---|
| 1a | K.VE*IVAIND*PFID*LNYM•VYM•FQYD*STHGK°FNGTVK*.A | 24 – 59 | K(52) | (1177.57)4 |
| 1b | K.VE*IVAIND*PFID*LNYM•VYMFQYD*STHGK°FNGTVK*.A | 24 – 59 | K(52) | (1173.32)4 |
| 2 | K.WGEAGAE*YVVE*STGVFTTM•EK°AGAHLK.G | 83 – 111 | K(104) | (881.78)4 |
| 3 | R.D*GRGAAQNIIPASTGAAK°AVGK*.V | 195 – 216 | K(212) | (898.43)3 |
| 4a | K.LTGM•A FRVPTPNVSVVD*LTCRLE*K°PAK*.Y | 225 – 251 | K(248) | (920.22)4 |
| # | G93A GAPDH BisANS Incorporation Sites | Span | BisANS Site | m/z |
| 5a | VK°VGVNGFGR*.I | 1 – 11 | K(2) | (450.27)3 |
| 5b | VK°VGVNGFGR*.I | 1 – 11 | K(2) | (413.24)4 |
| 5c | VK°VGVNGFGR.I | 1 – 11 | K(2) | (543.65)3 |
| 6 | K.AENGK LVINGK°PITIFQE.R | 59 – 77 | K(63) | (908.97)3 |
| 7a | K.LVINGK°PITIFQERD*PTNIK.W | 63 – 84 | K(69) | (972.17)3 |
| 7b | K.LVINGK°PITIFQE*R*.D | 63 – 78 | K(69) | (1134.40)2 |
| 7c | K.LVINGK°PITIFQERD*PTNIK.W | 63 – 84 | K(69) | (972.17)3 |
| 8a | K.AGAHLKGGAK°R.V | 104 – 116 | K(114) | (831.50)2 |
| 8b | K.GGAK°R.V | 110 – 116 | K(114) | (1106.04)1 |
| 8c | K.GGAK°RVIISAPSADAPMFVMGVNHEK.Y | 110 – 137 | K(114) | (1093.56)3 |
| 8d | K.GGAK°RVIISAPSADAPMFVMGVNHEK.Y | 110 – 137 | K(114) | (820.43)4 |
| 9a | K.VIHD*NFGIVE*GLMTTVHAITATQK°TVD*GPSGK.L | 159 – 192 | K(183) | (1333.80)3 |
| 9b | K.VIHD*NFGIVE*GLM•TTVHAITATQK°TVDGPSGKLWR.D | 159 – 195 | K(183) | (1112.60)4 |
| 4b | R.LEK°PAK°YDDIK.K | 245 – 257 | K(248), K(251) | (837.90)3 |
| 4c | R.LE*K°PAK°YD*DIKK.V | 245 – 258 | K(248), K(251) | (888.13)3 |
| 4d | R.LE*K°PAK.Y | 245 – 252 | K(248) | (435.10)3 |
| 4e | R.LE*K°PAK°YD*DIKK.V | 245– 258 | K(248), K(251) | (888.13)3 |
| 10a | R.VVDLM•AYMAS°K°E.- | 320 – 332 | S(330) ,K(331) | (855.77)3 |
| 10b | R.VVD*LM•AYM•AS°K°E*.- | 320 – 332 | S(330) ,K(331) | (875.79)3 |
| 10c | K.LISWYD*NE*YGYSNRVVD*LMAYM•ASK°E.- | 306 – 332 | K(331) | (949.50)4 |
| 10d | K.LISWYD*NE*YGYSNRVVD*LMAYM•ASK°E.- | 306 – 332 | K(331) | (949.48)4 |
| # | H46R/H48Q GAPDH BisANS Incorporation Sites | Span | BisANS Site | m/z |
| 2 | K.WGEAGAE*YVVE*STGVFTTM•EK°AGAHLK.G | 83 – 111 | K(104) | (881.78)4 |
| 8b | K.GGAK°R*.V | 110 – 116 | K(114) | (1106.04)1 |
| 8e | K.GGAK°RVIISAPSADAPMFVMGVNHEK.Y | 110 – 137 | K(114) | (1093.55)3 |
m/z = mass to charge ratio, tR = retention time in minutes
AA* = sodiated amino acid, K° = lysine cross-linked to a BisANS, S° = Serine cross-linked to a BisANS, T° = Threonine cross-linked to BisANS, Y° = Tyrosine cross-linked to BisANS, M • = methionine oxidized to methionine sulphoxide
Figure 8. X-Ray Crystal Structure of Human GAPDH with BisANS Incorporation Sites Modeled in NTg, G93A, and H46R/H48Q Mice.
BisANS cross-linked sites found by LC-MS/MS in mouse GAPDH are indicated in the stereoview of the crystal structure of tetrameric human GAPDH (pdb code 1u8f) described by Jenkins and Tanner51 followed by the corresponding human sites in parentheses. BisANS sites incorporated to GAPDH are shown only on one of the four subunits corresponding to the mouse amino acid number A) BisANS sites found in the NTg mouse GAPDH by LC/MS/MS are indicated by their mouse amino acid number. Amino acids involved in enzyme activity are indicated by black dots, and sites lost by both mutants are indicated by green, lost only in G93A by red, and only in H46R/H48Q in orange. B) Sites gained in G93A are indicated by red spheres, and those gained by both G93A and H46R/H48Q are indicated in cyan.
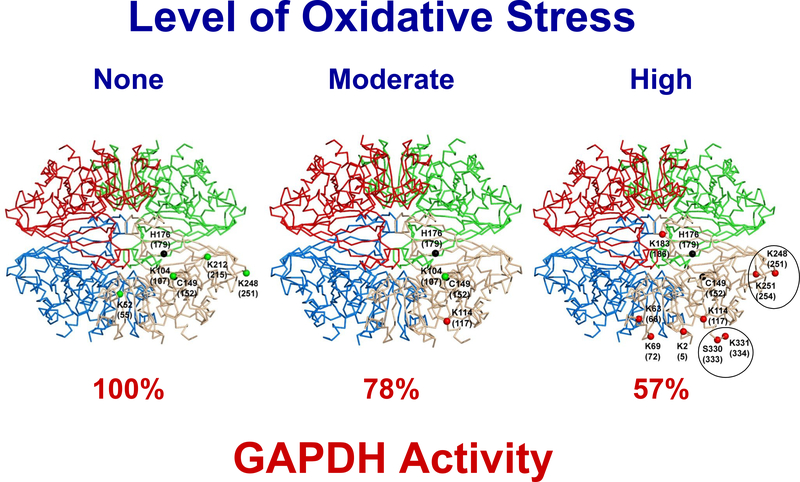
Figure 9. Summary of Functional and Conformational Changes Observed in GAPDH in Response to Oxidative Stress.
Tetrameric human GAPDH (pdb code 1u8f) is shown with BisANS sites incorporated to GAPDH shown only on one of the four subunits. The changes in BisANS incorporation sites that correlate with the level of oxidative stress and GAPDH activity in the NTg (left), H46R/H48Q (middle), G93A (right) models are shown. The amino acids involved in enzyme activity are indicated in black. All NTg sites are indicated by green dots, and sites gained with increased oxidative stress are indicated by red dots. The sites corresponding to doubly labeled peptides are circled.
Discussion
The functional state of a protein is important in maintaining normal homeostasis in cells and tissues. Oxidation of key amino acids in proteins in vitro can lead to alterations in protein structure and function; however, there is essentially no information on the effect of oxidative stress in vivo on protein structure. Using an unbiased approach to identify specific proteins showing altered protein folding, we observed a major loss of BisANS incorporation in two proteins, CK and GAPDH, in skeletal muscle of the G93A mice, an ALS mouse model that expresses a “wild-type like” functional SOD1 enzyme that acts as a pro-oxidant28; 29; 30. Although there is a possibility that the changes in BisANS incorporation could arise from increased oxidation of these proteins during isolation, all samples were treated identically and measures were taken to minimize reactions with ROS during isolation. Therefore, we believe the changes in BisANS incorporation are indicative of changes that occur in protein folding in vivo. Although there is still the possibility that factors other than oxidation could occur during ALS that would alter protein folding (e.g., denervation), several investigators have shown increases in protein oxidation and oxidative stress in the spinal cord and skeletal muscle of G93A mice using “redox proteomic” approaches 25; 31; 32. We showed that CK and GAPDH from the muscle of G93A mice have increased oxidative damage and that oxidation of GAPDH or CK in vitro resulted in reduced BisANS binding. Thus, our data are consistent with the hypothesis that protein unfolding is induced by oxidative stress in vivo.
We found that the alterations in protein folding were paralleled by a decrease in the activities of CK and GAPDH in the skeletal muscle of G93A mice but not protein aggregation, suggesting that the changes in protein folding we observed were functionally important. Wendt et al33 previously reported a decrease in CK activity in spinal cord tissues of G93A mice. Our cell-free experiments confirmed that oxidative stress in vitro concomitantly alters protein folding and enzyme activity of purified CK and GAPDH, which confirms previous reports showing that these enzymes are sensitive to oxidative stress in vitro34; 35. Our cell-free experiments also suggest that GAPDH was more sensitive to oxidative stress than CK, which was confirmed in our studies with the H46R/H48Q SOD1 mutant ALS mouse model, which lacks a functional copper binding site and dismutase activity and is associated with less oxidative stress than observed in the G93A mouse model25. No change in either protein folding or activity was observed in CK in the skeletal muscle of the H46R/H48Q mutant mouse; however, a modest, but significant decrease in BisANS incorporation and enzyme activity was observed for GAPDH.
The different levels of oxidative stress associated with the G93A and H46R/H48Q ALS models allow us to examine changes in GAPDH conformation under different levels of oxidative stress in vivo. Using LC/MS/MS, we compared BisANS incorporation sites in GAPDH from the skeletal muscle of G93A and H46R/H48Q ALS mice to normal, NTg mice. This technology allows us to detect changes in the presence or absence of BisANS incorporation sites in GAPDH, but does not allow us to assess the levels of these BisANS sites in GAPDH. In addition, changes in BisANS incorporation could also occur in a protein when conformational changes bring reactive amino acids closer to hydrophobic domains where BisANS is bound. Figure 9 summarizes the data we have obtained with the G93A and H46R/H48Q mouse models. In skeletal muscle from NTg mice, we observed four BisANS incorporation sites in GAPDH: K52, K104, K212, and K248. In the H46R/H48Q mice, which show a modest increase in oxidative stress, three of the four BisANS sites (K52, K212, K248) in the GAPDH of NTg mice were lost, and a new BisANS site (K114) was observed. The loss of these three sites and the gain of K114 were correlated with a 22% decrease in GAPDH activity. However, in the G93A mice, which show a substantial increase in oxidative stress 23; 24, the fourth NTg BisANS site was lost (K104), while the K248 site, which was lost in the H46R/H48Q mice, was present. Seven new BisANS incorporation sites also appeared in the GAPDH from G93A mice (K2, K63, K69, K183, K251, S330, and K331). The changes in BisANS incorporation were correlated with a 43% decrease in GAPDH activity, supporting the observation that GAPDH from G93A experience more substantial changes in conformation (protein folding) due to increased oxidative stress. We propose that the K248 BisANS incorporation site is lost and not available on the surface of the GAPDH under low levels of oxidative stress (H46R/H48Q mice); under higher levels of oxidative stress, we propose that the conformation of GAPDH is further changed so this site again becomes available for BisANS incorporation. This transient nature of site availability in response to oxidative stress has also been observed in proteins such as α-synuclein36. It should be noted that the K248 site that developed under high oxidative stress appears to have a higher affinity for BisANS, as another site (K251) of BisANS incorporation was observed on the same peptide. One other doubly labeled peptide was found in GAPDH from G93A mice (S330, K331), and Figure 9 shows the sites of these two peptides in the GAPDH. The incorporation of two BisANS molecules to a single peptide suggests that these regions have high-affinity for BisANS, i.e., perhaps due to a higher degree of protein unfolding.
As shown in Figure 9, the new BisANS sites found in G93A are clustered primarily on the periphery of GAPDH (K2, K63, K69, S330, and K331). This indicates that the conformation of GAPDH is altered in such a way with oxidative stress to increase protein unfolding. This is supported by the observation that two, doubly labeled peptide species were found at the surface of GAPDH. In contrast, the K183 BisANS incorporation site found in the GAPDH of the G93A mice is located at the junction of the four subunits in GAPDH. Thus, the conformation of GAPDH is altered in such a way under high oxidative stress that this site becomes available for BisANS incorporation, e.g., the GAPDH tetramer could dissociate in vivo under oxidative stress. Schmalhausen et al35 have shown that the oxidation of the cysteine 149 in the active site of GAPDH to sulfenic acid occurs readily in vitro, and this oxidation reduces its dehydrogenase activity and converts GAPDH tetramers to a dimeric or monomeric form37. Consistent with this, we have observed that GAPDH in the skeletal muscle of G93A mice exhibits a reduction in DTT-reducible cysteines.
In summary, our study shows for the first time that oxidative stress in vivo is associated with a change in the conformation of specific proteins in skeletal muscle in ALS. GAPDH was one of the two proteins identified, a protein that plays an important role in glycolysis. Recently several non-traditional activities for GAPDH have been reported that may relate to neurodegenerative diseases38; 39. For example, oxidation-induced conformational changes occur in GAPDH that are associated with RNA and DNA binding activities that may have implications for regulation of mRNA translation and DNA repair, respectively37. Oxidation and conformational alterations of GAPDH also have been shown to promote binding of the ubiquitin ligase Siah1 and facilitate its nuclear translocation and the induction of apoptosis40; 41; 42. The conformational changes observed in this study of GAPDH by measuring changes in BisANS incorporation could lead to these non-traditional activities and cause the functional changes observed under conditions of oxidative stress.
Methods
Animals
Male and female B6SJL Tg (SOD1-G93A)1Gur/J mice43 (G93A) were backcrossed to non-transgenic C57Bl/6J (NTg), C57Bl/6J x C3H Tg (SOD1-H46R/H48Q) mice44(H46R/H48Q), line 139, and C57Bl/6J Tg (NTg SOD1) were used at the ages indicated as the source of whole hindlimb skeletal muscle protein. All procedures for handling animals in this study were reviewed and approved by the IACUC (Institutional Animal Care and Use Committee) of the University of Texas Health Science Center at San Antonio and the IACUC of Audie L. Murphy Memorial Veterans Hospital, San Antonio, Texas.
UV-induced Photoincorporation of BisANS to Proteins
Photoincorporation of BisANS was carried out as described by Pierce et al14. In brief, the tissue was homogenized using a potter elvehjem all-glass tissue grinder in 10-volumes of 50 mM Tris HCl, 10 mM MgSO4, pH 7.4 and protease inhibitor cocktail (500 μM AEBSF, HCl, 150 nM aprotinin, and 1 μM leupeptin hemisulfate). After centrifugation at 100,000 g, protein concentrations were determined by the Bradford method 45, and the clear cytosolic protein extracts were diluted to 1 mg/ml in a labeling buffer containing 50 mM Tris HCl, 10 mM MgSO4, pH 7.4 and protease inhibitors. Additional aliquots were made from the same extracts for enzyme assays and western blots as described below. Then, 100 μM BisANS was added and samples were immediately mixed. Next, 200 μl of the sample was added to a clear 96-well plate and incubated on ice (to minimize local heating) for 1 hr under direct exposure to a 115 V, 0.16 Amp handheld, long-wave UV lamp (365 nm) (Ultra-violet Products, Inc., model UVL-21). Following the photoincorporation of BisANS, proteins were then dissolved in Laemmli buffer46 and subjected to SDS-PAGE. During SDS-PAGE, unbound BisANS in the sample migrates with the dye front and is consequently removed. After electrophoresis, gels were removed and illuminated with 365nm UV light, and BisANS fluorescence was captured with an AlphaImage™ 3400 for quantification. Following capture of BisANS fluorescence, gels were stained in either coomassie blue (SDS-PAGE gels) or Sypro Ruby (2D gels) in order to normalize for protein level.
2D Gel Electrophoresis
For 2D gel electrophoresis, BisANS labeled proteins (control and experimental samples) were treated with an equal volume of 20% TCA for 10 min on ice and centrifuged at 16,000 g for 15 min at 25oC. The pellets were then washed with a mixture of ethanol: ethyl acetate (1:1) (v/v) at least three times to remove the unbound BisANS. Following removal of the BisANS, the pellets were then dissolved in lysis buffer containing 8 M urea, 2% CHAPS, 0.5% IPG buffer, pH 3.0–10. The protein concentration was measured by BCA assay. Approximately 150 μg of protein was loaded in 250 μl of rehydration buffer containing 8 M urea, 2% CHAPS, 0.5% IPG buffer, 1% bromophenol blue, and 2.4 mg/ml DTT. The first dimension of electrophoresis was run on 13 cm strips (pH 3–10) using the IPGphor Isoelectric Focusing System overnight. After the first dimension, strips were washed in equilibration buffer (50 mM Tris buffer pH 8.8, 6 M urea, 30% glycerol, 2% SDS and 0.002% bromophenol blue). Strips were then washed twice for 15 min; first with DTT (100 mg/ml) and second with iodoacetamide (250 mg/ml). The second dimension was run on 12% SDS-PAGE gels. After electrophoresis, gels were removed and illuminated with 365nm UV light, and the image was captured with an AlphaImage™ 3400. Gels were then fixed with 10% methanol and 7% acetic acid for 10 min followed by staining overnight with Sypro Ruby. The stained gels were washed in 10% methanol and 7% acetic acid for 30 min to remove residual dye and then were placed in water. The Typhoon 9400 variable mode imager with excitation at 532 nm and emission filter of 610BP30 was used to scan the gels. Scanned gels for BisANS fluorescence (obtained from AlphaImage™ 3400) and Sypro Ruby fluorescence (obtained from Typhoon 9400 variable mode imager) were quantitated separately from 16-bit grayscale images using Imagequant v5.0 (Molecular Dynamics, Amersham, Piscataway, NJ). The background was corrected for each spot using the object average function. To normalize BisANS fluorescence for protein expressional changes, the pixel intensity for BisANS fluorescence was divided by the pixel intensity of Sypro Ruby fluorescence.
Creatine Kinase and GAPDH activity
The same unlabeled extracts from the BisANS labeling studies were used to measure the enzyme activity of creatine kinase and glyceraldehyde-3-phosphate dehydrogenase. Creatine Kinase (CK) was measured spectrophotometrically in an enzyme coupled system as described by Tanzer and Gilvarg47. Enzyme units were calculated by the extinction coefficient of NADH, where 1 unit is equal to the amount of enzyme required to convert 1μmol creatine to creatine phosphate per minute at 25°C, pH 8.9. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was measured spectrophotometrically as described by Rafter et al.48, and Harting and Velick49. Absorbance was monitored at 340 nm, and enzyme units were calculated by the extinction coefficient of NADH, where 1 unit is equal to the amount of enzyme required to convert 1μmol glyceraldehyde-3-phosphate to 1,3 bisphospho-glycerate per minute at 25°C, pH 8.5. The activity was expressed as percent activity of NTg mice.
Western Blot for Creatine Kinase and GAPDH
Using the same extracts from the BisANS labeling studies, unlabeled skeletal muscle cytosol was boiled in Laemmli buffer46 containing 2% β-mercaptoethanol at 0.1mg/ml. 0.5 and 1μg of protein were loaded per lane for CK and GAPDH western blots, respectively, and separated by SDS-PAGE using 12% gels, followed by electrophoretic transfer to polyvinylidine fluoride membranes (Bio-Rad) in a semi-dry transfer cell at 20 V for 45 min. After blocking for 1 hr at room temperature (RT) with TBS containing Tween-20 (TBST, 20 mmol/L Tris, 137 nmol/L NaCl and 0.4% Tween; pH 7.6) in the presence of 5% nonfat dry milk, the membranes were incubated with polyclonal goat anti-mouse creatine kinase or polyclonal rabbit anti-mouse GAPDH (Alamo Laboratories, San Antonio, TX) in TBST containing 1% nonfat dry milk at 4°C overnight. Secondary antibodies (goat anti-rabbit or rabbit anti-goat IgG) conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA) in TBST containing 1% nonfat dry milk were added for 2 hr at room temperature (1:6000 dilution). After this incubation, the blots were washed five times for 3 min each in TBST containing 0.2% nonfat dry milk. Bound secondary antibodies were detected using an ECL plus detection system (GE Healthcare, Piscataway, NJ) and visualized with the Typhoon variable mode imager (GE Healthcare, Piscataway, NJ) with excitation at 457 nm and the 520bp30 emission filter.
Differential Detergent Extraction
Differential detergent extraction was performed as described by Wang et al.50 by sequentially sonicating tissue pellets with detergents of increasing ionic strength. Following the 100,000 g centrifugation of extracts, the insoluble pellet was sonicated with 50 mM Tris HCl, 10 mM MgSO4, pH 7.4 containing 0.5% Nonidet-P40 (P1 buffer). This sonicate was centrifuged at 100,000 g for 1hr, and the resulting supernatant was termed P1. Subsequent extractions were carried out on the remaining pellet with P1 buffer containing 0.25% SDS, and 0.5% deoxycholate (P2 Buffer), and P1 buffer containing 2% SDS and 0.5% deoxycholate (P3 Buffer). Protein concentration was determined using the BCA Assay (Pierce) and 25μg of protein were boiled in Laemmli buffer and separated by reducing SDS-PAGE for western blot.
Measurement of Irreversible Oxidation of Cysteine Residues in Proteins
The level of irreversibly oxidized cysteines in proteins was measured according to the method described by Pierce et al.26. In brief, whole hindlimb skeletal muscle was homogenized in 20 mM potassium phosphate buffer containing 1mM EDTA, 0.5 mM MgCl2 pH 7.9 with a cocktail of protease inhibitors (EMD Chemicals,NJ). After homogenization (Tissumizer, Tekmar) for 30 sec, a cytosolic extract was obtained by centrifugation at 100,000 g for 1 hr at 4°C; the protein concentration was measured by the Bradford assay (Bio-Rad, Hercules, CA). Proteins were diluted to 1 mg/ml in homogenization buffer containing 1 mM dithiothreitol (DTT) and 6 M urea, pH 7.9 for 1 hr at 37°C to reduce mixed disulfides (but not sulfenic, sulfinic, and sulfonic acids) to their free thiol form. The free thiols in cysteines were then labeled by incubation with a thiol-specific fluorescent probe, 1 mM 6-iodoacetamidofluorescein (6IAF) (Invitrogen, Carlsbad, CA), for 1 hr at 37°C. Labeled proteins were precipitated and cleaned using the 2D cleanup kit (Amersham Biosciences, Piscataway,NJ). Final pellets were solubilized in 8 M urea after brief sonication and allowed to dissolve in the dark for 1 hr at room temperature for 2D gel electrophoresis. Protein concentration was determined by the Bradford method.
LC-MS/MS analysis
Protein spots of GAPDH from NTg, G93A, and H46R/H48Q were excised from the 2D gel, dehydrated with acetonitrile, and digested with 0.825 μg of modified trypsin (Promega, Madison, WI) in 40 mM NH4HCO3 and analyzed using an Agilent 1100 series HPLC-PE Sciex QSTAR™ hybrid LC-MS/MS Quadrupole TOF mass spectrometer. Peptide mixtures were separated on an Agilent zorbax C18 column (75μm × 150 mm) using an Agilent 1100 series instrument (Agilent Technologies, Inc. Palo Alto, CA) at 0.2 μL/min. Solvent A was 0.01% TFA in deionized H2O (dI H2O) and solvent B was 95% CH3CN/0.01% TFA in dI H2O. The flow from the column was directed to the QSTAR workstation (Applied Biosystems, Framingham, MA) equipped with an ESI source. Peptides were separated in a 60 min linear gradient (from 0% B to 60% B). MS/MS spectra were obtained in the positive ion mode, 4300 volt of ionization voltage, 5 units of curtain gas at a sampling rate of one spectrum per second.
Spectral analysis.
MS/MS data were analyzed using the MASCOT database search engine. The following filters were applied to address potential false positives: 1) ion mass must fit a tryptic peptide from GAPDH; 2) if the cross-linking occurred via a lysine, the peptide had to be miss-cleaved at that site, because trypsin will not recognize crosslinked lysine; 3) the same site was found cross-linked in several different peptides; and 4) because sodiation is an ESI artifact, sodiated and non-sodiated forms of the same peptide must co-elute. All searches were conducted against a mouse proteome database. The new mass for BisANS cross-linked lysine, serine, threonine, and tyrosine were used as variable modifications. Other variable modifications searched were sodiation of acidic groups and methionine oxidation. Because pure spots were cut from gel, the MS/MS search was conducted using a combination of peptide finger-printing and MS/MS analysis.
Oxidative Modification of Proteins
Oxidation of CK and GAPDH was carried out essentially as described by Chao et al.18. Proteins were diluted to 1 mg/ml in various concentrations of oxidizing solution ranging from (0–40 μM) FeSO4 and (0–10 mM) ascorbate, maintaining a constant 1:250 ratio of FeSO4 to ascorbate in 100 mM KCl, 100 mM MgSO4, and 50 mM Hepes pH 7.2. Oxidation was carried out for 1 hr at 37°C in the dark. Solutions containing oxidized or unoxidized proteins were directly applied to a Centricon 10 microconcentrator (Amicon, Inc., MA) to remove the excess FeSO4 and ascorbate from oxidized samples and diluted in equi-volumes of 50 mM Tris HCl and 10 mM MgSO4, pH 7.4 before proceeding to measure the enzyme activity as described above. Protein concentrations were determined by the Bradford method 45.
Statistics
The data were analyzed using the student’s t test with the Winstat add-in (Microsoft Excel) or using one-way ANOVA with Dunett’s post hoc test for multiple comparisons. Statistical significance was set at the p≤0.05 level.
Acknowledgements
We would like to thank Dr. David Borchelt for kindly providing us with the H46R/H48Q mice for use in this study. We would also like to thank Dr. Susan Weintraub and the Mass Spectrometry Core for access to their facility and their assistance in the identification of proteins by MALDI-TOF MS.
This work was supported by VA-VISN, ALSA, and VA Merit grants (A.C.); VA-merit and MDA 3879 (H.V.R); NIH grant AG025362 (F.R., H.V.R.); NIH grants AG23843 and R37 AG26557 (AR); a REAP grant from the Department of Veteran Affairs (to A.R., H.V.R., and A.C.), and a VA CDA-2 grant (to AP).
Footnotes
Competing Financial Interests Statement: N/A
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Widmer R, Ziaja I & Grune T (2006). Protein oxidation and degradation during aging: role in skin aging and neurodegeneration. Free Radic Res 40, 1259–68. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman ER (2006). Protein oxidation and aging. Free Radic Res 40, 1250–8. [DOI] [PubMed] [Google Scholar]
- 3.Stadtman ER (2001). Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci 928, 22–38. [DOI] [PubMed] [Google Scholar]
- 4.Khidekel N & Hsieh-Wilson LC (2004). A ‘molecular switchboard’--covalent modifications to proteins and their impact on transcription. Org Biomol Chem 2, 1–7. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen ML, Savitski MM & Zubarev RA (2006). Extent of modifications in human proteome samples and their effect on dynamic range of analysis in shotgun proteomics. Mol Cell Proteomics 5, 2384–91. [DOI] [PubMed] [Google Scholar]
- 6.Levine RL (1983). Oxidative modification of glutamine synthetase. I. Inactivation is due to loss of one histidine residue. J Biol Chem 258, 11823–7. [PubMed] [Google Scholar]
- 7.Sharp JS, Sullivan DM, Cavanagh J & Tomer KB (2006). Measurement of multisite oxidation kinetics reveals an active site conformational change in Spo0F as a result of protein oxidation. Biochemistry 45, 6260–6. [DOI] [PubMed] [Google Scholar]
- 8.Meucci E, Mordente A & Martorana GE (1991). Metal-catalyzed oxidation of human serum albumin: conformational and functional changes. Implications in protein aging. J Biol Chem 266, 4692–9. [PubMed] [Google Scholar]
- 9.Grune T, Merker K, Sandig G & Davies KJ (2003). Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun 305, 709–18. [DOI] [PubMed] [Google Scholar]
- 10.Giulivi C, Pacifici RE & Davies KJ (1994). Exposure of hydrophobic moieties promotes the selective degradation of hydrogen peroxide-modified hemoglobin by the multicatalytic proteinase complex, proteasome. Arch Biochem Biophys 311, 329–41. [DOI] [PubMed] [Google Scholar]
- 11.Shringarpure R & Davies KJ (2002). Protein turnover by the proteasome in aging and disease. Free Radic Biol Med 32, 1084–9. [DOI] [PubMed] [Google Scholar]
- 12.Kabashi E & Durham HD (2006). Failure of protein quality control in amyotrophic lateral sclerosis. Biochim Biophys Acta 1762, 1038–50. [DOI] [PubMed] [Google Scholar]
- 13.Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT & Hunt JF (2004). The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J Mol Biol 344, 549–65. [DOI] [PubMed] [Google Scholar]
- 14.Pierce A, Dewaal E, Vanremmen H, Richardson A & Chaudhuri A (2006). A Novel Approach for Screening the Proteome for Changes in Protein Conformation. Biochemistry 45, 3077–3085. [DOI] [PubMed] [Google Scholar]
- 15.Horowitz PM & Butler M (1993). Interactive intermediates are formed during the urea unfolding of rhodanese. J Biol Chem 268, 2500–4. [PubMed] [Google Scholar]
- 16.Seale JW, Brazil BT & Horowitz PM (1998). Photoincorporation of fluorescent probe into GroEL: defining site of interaction. Methods Enzymol 290, 318–23. [DOI] [PubMed] [Google Scholar]
- 17.Horowitz P, Prasad V & Luduena RF (1984). Bis(1,8anilinonaphthalenesulfonate). A novel and potent inhibitor of microtubule assembly. J Biol Chem 259, 14647–50. [PubMed] [Google Scholar]
- 18.Chao CC, Ma YS & Stadtman ER (1997). Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc Natl Acad Sci U S A 94, 2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadtman ER (1993). Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu Rev Biochem 62, 797–821. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhuri AR, de Waal EM, Pierce A, Van Remmen H, Ward WF & Richardson A (2006). Detection of protein carbonyls in aging liver tissue: A fluorescence-based proteomic approach. Mech Ageing Dev 127, 849–861. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Caruano-Yzermans A, Rodriguez A, Scheurmann JP, Slunt HH, Cao X, Gitlin J, Hart PJ & Borchelt DR (2007). Disease-associated mutations at copper ligand histidine residues of superoxide dismutase 1 diminish the binding of copper and compromise dimer stability. J Biol Chem 282, 345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX & et al. (1993). Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62. [DOI] [PubMed] [Google Scholar]
- 23.Mahoney DJ, Kaczor JJ, Bourgeois J, Yasuda N & Tarnopolsky MA (2006). Oxidative stress and antioxidant enzyme upregulation in SOD1-G93A mouse skeletal muscle. Muscle Nerve 33, 809–16. [DOI] [PubMed] [Google Scholar]
- 24.Hensley K, Mhatre M, Mou S, Pye QN, Stewart C, West M & Williamson KS (2006). On the relation of oxidative stress to neuroinflammation: lessons learned from the G93A-SOD1 mouse model of amyotrophic lateral sclerosis. Antioxid Redox Signal 8, 2075–87. [DOI] [PubMed] [Google Scholar]
- 25.Muller FL, Song W, Jang YC, Liu Y, Sabia M, Richardson A & Van Remmen H (2007). Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am J Physiol Regul Integr Comp Physiol 293, R1159–68. [DOI] [PubMed] [Google Scholar]
- 26.Pierce AP, de Waal E, McManus LM, Shireman PK & Chaudhuri AR (2007). Oxidation and structural perturbation of redox-sensitive enzymes in injured skeletal muscle. Free Radic Biol Med 43, 1584–93. [DOI] [PubMed] [Google Scholar]
- 27.Johnston LJ & Redmond RW (1997). Triplet state mechanism for diphenylamine photoionization. Journal of Physical Chemistry A 101, 4660–4665. [Google Scholar]
- 28.Wiedau-Pazos M, Goto JJ, Rabizadeh S, Gralla EB, Roe JA, Lee MK, Valentine JS & Bredesen DE (1996). Altered reactivity of superoxide dismutase in familial amyotrophic lateral sclerosis. Science 271, 515–8. [DOI] [PubMed] [Google Scholar]
- 29.Estevez AG, Crow JP, Sampson JB, Reiter C, Zhuang Y, Richardson GJ, Tarpey MM, Barbeito L & Beckman JS (1999). Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science 286, 2498–500. [DOI] [PubMed] [Google Scholar]
- 30.Said Ahmed M, Hung WY, Zu JS, Hockberger P & Siddique T (2000). Increased reactive oxygen species in familial amyotrophic lateral sclerosis with mutations in SOD1. J Neurol Sci 176, 88–94. [DOI] [PubMed] [Google Scholar]
- 31.Poon HF, Hensley K, Thongboonkerd V, Merchant ML, Lynn BC, Pierce WM, Klein JB, Calabrese V & Butterfield DA (2005). Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice--a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med 39, 453–62. [DOI] [PubMed] [Google Scholar]
- 32.Casoni F, Basso M, Massignan T, Gianazza E, Cheroni C, Salmona M, Bendotti C & Bonetto V (2005). Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: possible multifunctional role in the pathogenesis. J Biol Chem 280, 16295–304. [DOI] [PubMed] [Google Scholar]
- 33.Wendt S, Dedeoglu A, Speer O, Wallimann T, Beal MF & Andreassen OA (2002). Reduced creatine kinase activity in transgenic amyotrophic lateral sclerosis mice. Free Radic Biol Med 32, 920–6. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki YJ, Edmondson JD & Ford GD (1992). Inactivation of rabbit muscle creatine kinase by hydrogen peroxide. Free Radic Res Commun 16, 131–6. [DOI] [PubMed] [Google Scholar]
- 35.Schmalhausen EV, Pleten AP & Muronetz VI (2003). Ascorbate-induced oxidation of glyceraldehyde-3-phosphate dehydrogenase. Biochem Biophys Res Commun 308, 492–6. [DOI] [PubMed] [Google Scholar]
- 36.Mirzaei H, Schieler JL, Rochet JC & Regnier F (2006). Identification of rotenone-induced modifications in alpha-synuclein using affinity pull-down and tandem mass spectrometry. Anal Chem 78, 2422–31. [DOI] [PubMed] [Google Scholar]
- 37.Arutyunova EI, Danshina PV, Domnina LV, Pleten AP & Muronetz VI (2003). Oxidation of glyceraldehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem Biophys Res Commun 307, 547–52. [DOI] [PubMed] [Google Scholar]
- 38.Berry MD (2004). Glyceraldehyde-3-phosphate dehydrogenase as a target for small-molecule disease-modifying therapies in human neurodegenerative disorders. J Psychiatry Neurosci 29, 337–45. [PMC free article] [PubMed] [Google Scholar]
- 39.Sirover MA (1999). New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta 1432, 159–84. [DOI] [PubMed] [Google Scholar]
- 40.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH & Sawa A (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7, 665–74. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima H, Amano W, Fujita A, Fukuhara A, Azuma YT, Hata F, Inui T & Takeuchi T (2007). The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J Biol Chem 282, 26562–74. [DOI] [PubMed] [Google Scholar]
- 42.Sunaga K, Takahashi H, Chuang DM & Ishitani R (1995). Glyceraldehyde3-phosphate dehydrogenase is over-expressed during apoptotic death of neuronal cultures and is recognized by a monoclonal antibody against amyloid plaques from Alzheimer’s brain. Neurosci Lett 200, 133–6. [DOI] [PubMed] [Google Scholar]
- 43.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX & et al. (1994). Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–5. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Xu G, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA & Borchelt DR (2002). Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol Dis 10, 128–38. [DOI] [PubMed] [Google Scholar]
- 45.Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248–54. [DOI] [PubMed] [Google Scholar]
- 46.Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–5. [DOI] [PubMed] [Google Scholar]
- 47.Tanzer ML & Gilvarg C (1959). Creatine and creatine kinase measurement. J Biol Chem 234, 3201–4. [PubMed] [Google Scholar]
- 48.Rafter GW, Chaykin S & Krebs EG (1954). The action of glyceraldehyde3-phosphate dehydrogenase on reduced diphosphopyridine nucleotide. J Biol Chem 208, 799–811. [PubMed] [Google Scholar]
- 49.Harting J & Velick SF (1954). Transfer reactions of acetyl phosphate catalyzed by glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem 207, 86778. [PubMed] [Google Scholar]
- 50.Wang J, Slunt H, Gonzales V, Fromholt D, Coonfield M, Copeland NG, Jenkins NA & Borchelt DR (2003). Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum Mol Genet 12, 2753–64. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins JL & Tanner JJ (2006). High-resolution structure of human Dglyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr 62, 290–301. [DOI] [PubMed] [Google Scholar]