Abstract
Most microRNAs (miRNAs) are expressed as a mix of length isoforms (referred to as isomiRs). IsomiR stoichiometry can be differentially impacted upon cell stimulation, as recently evidenced by our group in the context of immune responses induced by type-I interferon (IFN). Here, we revisit published RNA-seq data sets of human and mouse macrophages stimulated with bacterial products at the isomiR level. We demonstrate that for several miRNAs, macrophage stimulation induces changes in isomiR stoichiometry. Critically, we find that changes in miRNA expression can be misinterpreted when miRNAs are quantified by RT-qPCR, as primers directed against canonical miRNA sequences may not equally target the different isomiRs that are regulated endogenously. Beyond the case of phagocyte stimulation, our analyses reinforce the concept that analysis of miRNA expression at the isoform level should become standard practice.
Keywords: microRNA isoform, isomiR, interferon, LPS, RT-qPCR miRNA assays, macrophages, bacterial infection
INTRODUCTION
There is increasing evidence that microRNA (miRNA) length variants are differentially expressed between cell types (Juzenas et al. 2017), cancer types (Telonis et al. 2017), and upon cell stimulation (as seen with stimuli such as PMA [Muiwo et al. 2018] or type-I interferon [IFN] [Nejad et al. 2018b]). Critically, such length variations from what is considered to be the canonical length (as per its definition in miRbase [Kozomara and Griffiths-Jones 2014]) are not restricted to poorly abundant isoforms. Rather, in select cell types, these isomiRs can be the predominant miRNA species such as that seen with the example of the 24–25 nt 3′-end variants of miR-222-3p (miR-222 hereafter) in human monocytes and neutrophils (Juzenas et al. 2017). While little is known of the biological relevance of miRNA length variations, previous reports suggest that 5′-end isomiRs can have different mRNA targets (Tan et al. 2014), while 3′-end isomiRs may also have different cellular functions, although sharing common mRNA targeting (Yu et al. 2017). As such, we have recently demonstrated that long and short miR-222 3′-end variants have a similar function on their targets (with direct miRNA binding sites for miR-222), but may exhibit differential functions through distinct intracellular localization—e.g., nuclear versus cytoplasmic (Yu et al. 2017). In addition, our studies indicated that longer miR-221/222 3′-end variants are preferentially targeted for degradation by enzymes including 3′–5′ exonuclease PNPT1 (Nejad et al. 2018b), suggesting a novel role for 3′-end isoforms in the control of intracellular miRNA concentration. An intriguing recent report also demonstrated a role for 3′-base-pairing with miRNA-response elements located within the protein coding regions of target genes (Zhang et al. 2018). Although canonical (seed-based) targeting is the primary mechanism of target recognition, a subset of genes may therefore be differentially targeted by 3′-end variance.
We have recently reported a concerning bias of commercial RT-qPCR-based quantification toward selected miRNA 3′-end isomiRs, suggesting that assays targeted to amplify the canonical miRBase version of a given miRNA may favor amplification of a minor 3′-isomiR in the cell of interest, depending upon primer design and the isomiR expression profile (Nejad et al. 2018a,b). Using miR-222 as an example, we demonstrated that the stem–loop Taqman assay targeting the 21 nt variant of miR-222 failed to detect a 70% decrease of this miRNA in response to IFN (as determined by isomiR-level RNA-seq), as it was the predominant 24–25 nt isoforms, and not the lowly abundant shorter canonical forms, that were regulated in human fibroblasts (Nejad et al. 2018b). We made a similar observation for miR-221-3p and speculated that unintentional isoform bias may be a common feature of the results obtained with miRNA RT-qPCR approaches (see Fig. 1A for a schematic representation of selective 3′-end amplification by stem–loop and LNA-polyadenylation RT-qPCR approaches). This is of concern given the widespread use of RT-qPCR approaches to detect changes in miRNA expression, which have supplanted older techniques such as northern blot detection (which cannot distinguish 3′ and 5′ isoforms of the same length).
FIGURE 1.
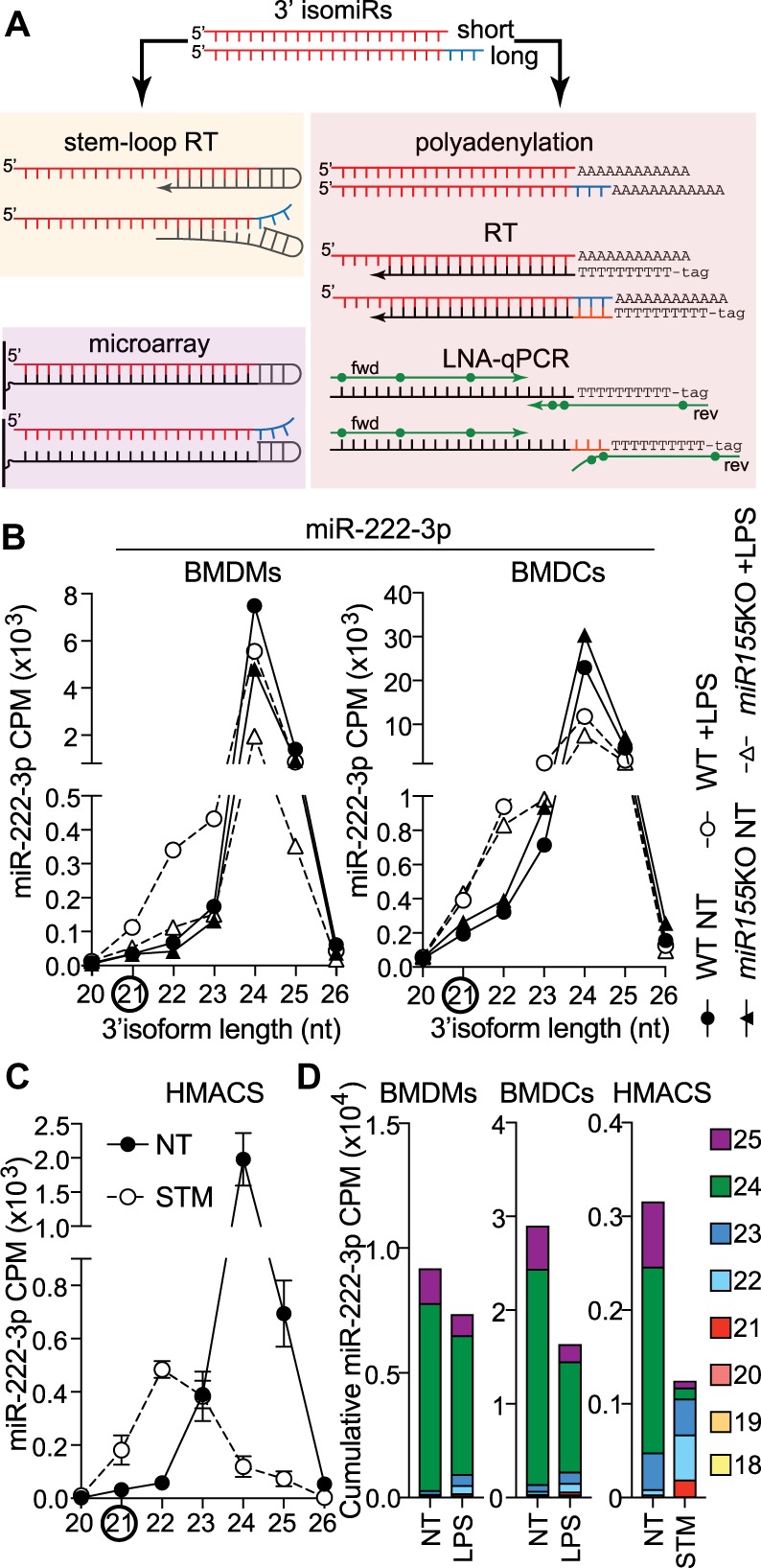
Bacterial-driven modulation of miR-222 isoforms in human and mouse macrophages. (A) Schematic representation of the impact of 3′-end length variation on stem–loop RT-qPCR, LNA-polyadenylation RT-qPCR, and Agilent microarray detection of mature miRNAs. In the case presented here, the assays are targeted to the shorter miRNA isoform, as seen with miR-222-3p. This directly impacts the binding of the reverse transcription stem–loop and efficiency of cDNA synthesis. Similarly, microarray loop-probes will not bind as well to longer isoforms. While it does not affect reverse transcription based on polyadenylation, the isoform selectivity impacts binding of the 3′-end of LNA reverse primer, hampering amplification. (B) Detailed analysis of normalized counts (CPM) for the 3′-end isoforms of miR-222 in primary macrophages (BMDMs) and dendritic cells (BMDCs) from wild-type (WT) and miR-155-deficient mice (Dueck et al. 2014). Cells were treated overnight with 200 ng/mL lipopolysaccharide (LPS) or nontreated (NT). Data shown are from one biological sample for each condition. (C) Detailed analysis of normalized counts (CPM) for the 3′-end isoforms of miR-222 in human monocyte-derived macrophages, infected for 24 h with Salmonella Typhimurium (STM) at an MOI of 5:1, compared to NT cells at 24 h (Pai et al. 2016). Data are averaged from six patients (mean ± standard error of the mean is shown). (D) CPM of the prevalent miR-222 3′-end isoforms from NT and LPS-treated WT BMDMs/BMDCs and STM-infected human macrophages were cumulated to reflect the overall impact of bacterial products on the pool of miR-222 molecules.
In the present work, we revisit two published small RNA-seq data sets of human and mouse macrophages infected with bacteria or treated with bacterial products, looking at miRNA variations at the isoform level. We confirm that immune activation of such phagocytes has a broad effect on the isoform stoichiometry of many miRNA families, and illustrate how these effects can lead to mis-interpretation of results that are reliant upon RT-qPCR approaches.
RESULTS AND DISCUSSION
We recently reported that the stoichiometry of select miRNA isoforms was significantly altered by type-I IFN stimulation of human fibroblasts, with a specific focus on miR-222 (Nejad et al. 2018b). Since type-I IFN can be produced by macrophages upon Toll-like receptor 4 (TLR4) activation by the bacterial product lipopolysaccharide (LPS), we re-analyzed a small RNA-seq data set of wild-type (WT) and miR-155-deficient mouse bone marrow-derived dendritic cells (BMDCs) and macrophages (BMDMs) stimulated overnight by LPS (Dueck et al. 2014). We were prompted to carry out these analyses following the recent report by Seeley et al. that miR-222 could be induced by LPS in mouse macrophages, to result in transcriptional silencing of a number of inflammatory genes in the context of bacterial infections (Seeley et al. 2018). We speculated that the observation of miR-222 induction made by Seeley et al. was biased by the isoform selective RT-qPCR approach they used based on our recent observations for this miRNA (Nejad et al. 2018b; Seeley et al. 2018).
Accordingly, in LPS-stimulated BMDMs and BMDCs, the bulk of miR-222 isoform populations was represented by its longer 3′-end variants (>23 nt), and these longer isoforms were not increased but rather decreased following LPS treatment—independent of miR-155 expression (Fig. 1B; Supplemental Tables S1, S2). Importantly, and aligning with the RT-qPCR assay, isoforms of miR-222 ranging between 21 and 23 nt were increased by LPS (Fig. 1B). However, since these shorter isoforms only marginally contributed to the pool of miR-222 molecules, they did not compensate for the decrease of the more abundant longer isoforms (Fig. 1B).
Since Seeley et al. also reported an induction of miR-222 levels in human monocytes following bacterial infection (Seeley et al. 2018), we next revisited an independent small RNA-seq data set of human monocyte-derived macrophages infected by Salmonella enterica serovar Typhimurium for 24 h (which also activates TLR4) (Pai et al. 2016). Analyses of these samples at the isoform level revealed a shift of prevalent miR-222 isoforms from 24 to 22 nt, with an overall decrease of miR-222 molecules (Fig. 1C,D; Supplemental Table S3). These results are similar to our previous analyses of Salmonella Typhimurium-infected human monocyte-derived dendritic cells (Nejad et al. 2018b), and confirm that changes in miR-222 isoform stoichiometry are conserved between human and mouse phagocytes exposed to bacterial products. We have recently demonstrated that long and short miR-222 3′-end variants have a similar function on their targets (with direct miRNA binding sites for miR-222) (Yu et al. 2017). We would therefore predict that regulation of mRNA targets by miR-222 would be independent of the length of the isoform considered, and that in the context of macrophage bacterial stimulation, miR-222 function is rather modestly decreased.
Thus, while the results from Seeley et al. clearly substantiated an interplay between miR-222 and transcriptional silencing of pro-inflammatory responses (Seeley et al. 2018), we believe that their interpretation of induced miR-222 levels as a negative feedback to LPS treatment and bacterial infection in macrophages, is muddied by the technique used to measure the miRNAs and does not account for variations of miR-222 at the isoform level induced by LPS, which rather decreases global miR-222 levels (Fig. 1D). We note that their study relies on the use of predesigned LNA-based RT-qPCRs, targeted to the miRBase canonical miR-222 (21 nt) (Seeley et al. 2018). We have shown that this predesigned LNA miR-222 assay does not amplify the 24–25 nt isoform of miR-222 as well as its 21–23 nt isoforms, based on amplification of synthetic miR-222 isoforms (Nejad et al. 2018a).
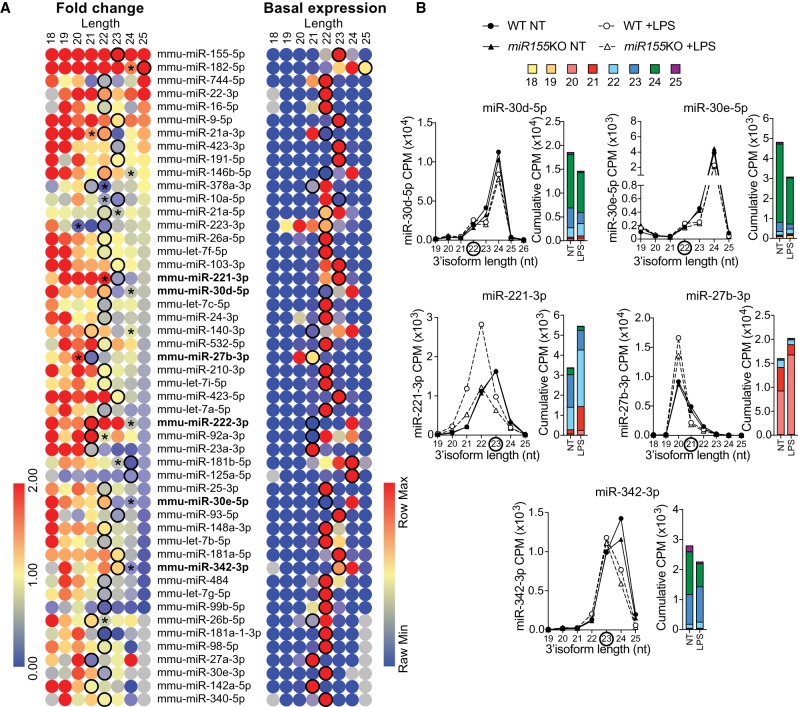
Given the prevalent use of RT-qPCR to measure miRNA levels and that many researchers do not have access to isoform-level RNA-seq, we anticipated that the situation presented here for miR-222 and the work of Seeley et al. (2018) was far from unique, and likely represented a common problem for researchers in the field. Since many miRNAs have been reported to be differentially impacted by LPS in macrophages (Nejad et al. 2018c), we next performed a detailed isoform analysis of the 50 major miRNA families expressed in WT BMDMs, relying on the data set used in Figure 1 (Dueck et al. 2014). As shown in Figure 2A (left panel), isoforms of most miRNAs were differently impacted by LPS simulation. With the exception of a few miRNAs such as miR-155-5p and miR-182-5p for which all the isoforms where robustly induced by LPS, and miR-99b-5p and miR-125a-5p which were broadly suppressed by LPS, longer isoforms were generally decreased and shorter forms increased—with variations between miRNA families. This was reminiscent of what we had previously observed in STM-infected human monocyte-derived dendritic cells (Nejad et al. 2018b), and supports that a complex regulation of isomiR processing/degradation operates upon macrophage activation.
FIGURE 2.
Broad effect of LPS on miRNA isoforms in mouse BMDMs. (A) Heat maps representing the LPS-induced fold change expression of each isoform for the top 50 most abundant miRNA families (right panel), in wild-type (WT) BMDMs, and the basal expression of each isoform within a family in untreated cells (left panel). Fold changes greater than twofold are shown in red (right panel) (values are based on Supplemental Table S4). Black circles highlight miRBase canonical isoforms, while stars refer to abundant isoforms when different from miRBase definition (seen in the right panel). Absent values are shown in gray. miRNA families analyzed further in Figure 1 (i.e., miR-222-3p) or Figure 2B are shown in bold. (B) Detailed analysis of CPM and cumulative CPM for the 3′-end isoforms of selected miRNA families in BMDMs from WT and miR-155-deficient mice (Dueck et al. 2014). For cumulative CPM, the data shown are from WT BMDMs. Data shown are from one biological sample for each condition.
In accordance with the concept that isoform processing is impacted by the cell type (Juzenas et al. 2017), the dominant isoform was not concordant with its canonical miRBase annotation for 15 out of the 50 miRNA families (Fig. 2A, right panel, compare expression with the defined canonical isomiR, denoted by a black circle). This raised specific concerns for miRNAs where the miRBase annotated canonical isoform responded differently to the prevalent isomiRs. Response to LPS in WT and miR-155-deficient BMDMs was analyzed further for five miRNA families (miR-221-3p, miR-30d-5p, miR-30e-5p, miR-27b-3p and miR-342-3p—Fig. 2B). The responses of miR-30d-5p, miR-30e-5p and miR-342-3p were similar to that of miR-222-3p, since their canonical annotation targets a shorter isoform that was modestly induced by LPS, when the major isoform of these families was rather decreased by LPS stimulation (Fig. 2B). Conversely, the response of miR-27b-3p was similar to that of miR-221-3p with its major isoform being induced or unchanged (Fig. 2B).
Given that commercial RT-qPCR assays to quantify miRNAs are designed to amplify the canonical variants defined in miRBase, these observations suggested that for the five miRNAs selected here, RT-qPCR approaches could lead to the misinterpretation of results. In line with this, we previously reported that RT-qPCR amplification of miR-221-3p with the Taqman stem–loop assay directed against its 23 nt isoform led to the detection of a strong decrease of miR-221-3p in LPS-treated BMDMs (Nejad et al. 2018b), when it is clear at the isoform level that the overall population of miR-221-3p molecules is rather increased in BMDMs due to a shift of the predominant isoform from 23 to 22 nt (Fig. 2B). Similarly, we found that miR-27b-3p was decreased in LPS-treated cells by low-density Taqman RT-qPCR array ([Nejad et al. 2018b] and not shown), when this miRNA is in fact rather increased by stimulation (Fig. 2B). These results are not surprising since miR-221-3p and miR-27b-3p Taqman stem–loop assays are one base longer than their predominant isoform in BMDMs, and we have shown (using synthetic miR-222-3p variants) that stem–loop and polyadenylation RT-qPCR assays designed to target longer miRNAs failed to detect shorter isoforms (Nejad et al. 2018a,b).
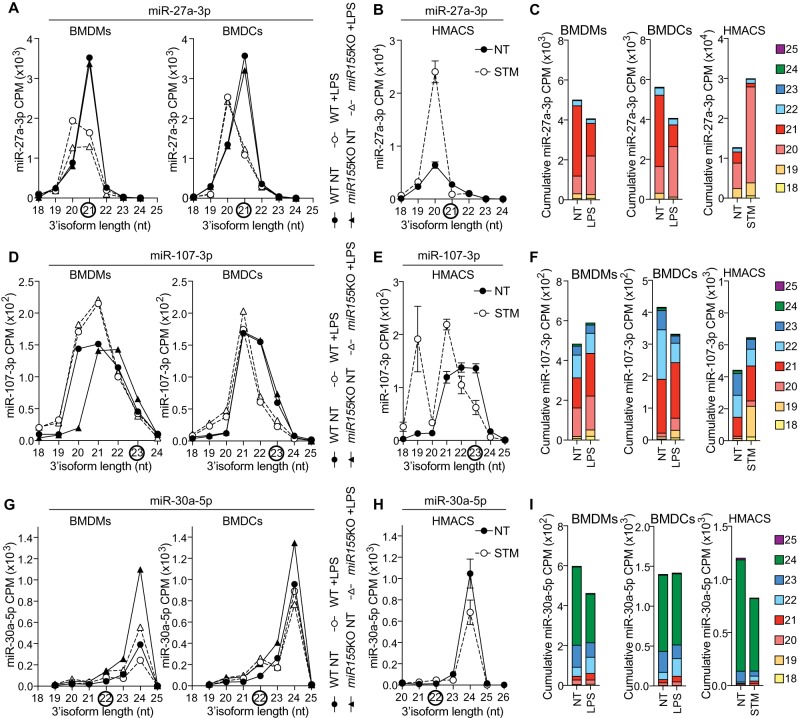
We next searched the literature for other miRNAs reported to be decreased in mouse macrophages upon LPS stimulation, which might have been attributed to an amplification bias of the RT-qPCR assays of a longer minor isoform. Our analyses of miR-27a-3p in the human macrophages and mouse BMDM/BMDC data sets (Dueck et al. 2014; Pai et al. 2016), demonstrated that the stoichiometry of its isoforms was impacted by bacterial products, but that this did not substantially decrease the overall intracellular concentration of this miRNA family, as the 20 nt isoform was induced in all three cell models (Fig. 3A–C, which shows the cumulative effect of LPS on this miRNA family). This contrasts with a study relying on the Taqman stem–loop miRNA assay targeted to the 21 nt isoform of miR-27a-3p, which reported that miR-27a-3p was decreased by more than 50% in mouse and human macrophages stimulated with LPS at 24 h (Xie et al. 2014)—nonetheless aligning with the effect of LPS seen on this specific 21 nt isoform (Fig. 3A–C). We note that an independent study of Mycobacterium tuberculosis infected human dendritic cells based on Agilent Human miRNA microarrays also reported miR-27a-3p to be down-regulated (Siddle et al. 2014). This illustrates that the miRBase isoform bias is not restricted to RT-qPCR approaches, and can also been seen with certain microarray probes with 3′-end selectivity (see Fig. 1A).
FIGURE 3.
Bacterial-driven modulation of miR-27a-3p, miR-107-3p, and miR-30a-5p isoforms in human and mouse macrophages. (A,D,G) Detailed analysis of CPM for the 3′-end isoforms of miR-27a-3p (A), miR-107-3p (D), and miR-30a-5p (G) in BMDMs and BMDCs from wild-type (WT) and miR-155-deficient mice (Dueck et al. 2014). Data shown are from one biological sample for each condition. Black circles highlight miRBase canonical isoforms. (B,E,H) Detailed analysis of CPM for the 3′-end isoforms of miR-27a-3p (B), miR-107-3p (E), and miR-30a-5p (H) in human monocyte-derived macrophages, infected for 24 h with Salmonella Typhimurium (STM) at an MOI of 5:1, compared to NT cells at 24 h (Pai et al. 2016). Data are averaged from six patients (mean ± standard error of the mean is shown). Black circles highlight miRBase canonical isoforms. (C,F,I) CPM of the prevalent miR-27a-3p (C), miR-107-3p (F), and miR-30a-5p (I) 3′-end isoforms from NT and LPS-treated WT BMDMs/BMDCs and STM-infected human macrophages were cumulated to reflect the overall impact of bacterial products on the pool of miRNA molecules.
Similarly, we found that miR-107-3p isoforms displayed differential responses in human macrophages and mouse BMDMs/BMDCs, although variations existed between the cell types (Fig. 3D–F). Overall, miR-107-3p was not decreased in LPS-treated BMDMs and human macrophages (Fig. 3F), and was only marginally decreased in LPS-treated BMDCs (Fig. 3D). Since miR-107-3p assays are designed to target the longer 23 nt isoform, which is decreased in BMDCs and human macrophages but is less abundant that the 21 nt isoform after stimulation, it was not surprising that two independent studies previously identified miR-107-3p as down-regulated by LPS at 24 h relying on the stem–loop Taqman RT-qPCR approach in BMDMs and BMDCs (Hennessy et al. 2011; Xue et al. 2014).
Finally, we searched the literature for miRNAs similar to miR-222, which were reported to be induced due to targeting of a shorter minor isoform induced by LPS. We found that miR-30a-5p which is annotated as a 22 nt isoform in miRBase, was previously shown to be induced in mouse RAW267.4 macrophages stimulated with 24-h LPS treatment, by stem–loop Taqman RT-qPCR (Jiang et al. 2017). Nonetheless, the overall miR-30a-5p levels were not induced by bacterial products in human macrophages and mouse BMDMs/BMDCs in our analyses, since the prevalent isoform of this miRNA, the 24 nt, was not induced (and rather modestly decreased for instance in human and mouse macrophages) (Fig. 3G–I).
Collectively, the analyses presented here underline the need for a better understanding by miRNA researchers of the bias introduced by the use of commercial RT-qPCR assays designed against miRBase canonical isoforms. While such bias may be different according to whether the assay targets a longer or shorter isoform than the prevalent one expressed (Nejad et al. 2018a,b), and the technology used (stem–loop, polyadenylation-based or microarray-based—see Fig. 1A), the few examples discussed in this report call for better scrutiny of technical artifacts introduced by the miRNA detection method used, as they may jeopardize the validity of the findings.
To avoid these issues, miRNA researchers should consider the sequences of the miRNAs that are actually being expressed or, if RNA-seq is not available, at least consider if the miRNA of interest has been reported to exhibit significant 3′-end variance between cells or tissues. Rapidly expanding isomiR data sets are available to assist with this (Juzenas et al. 2017; Telonis et al. 2017). If resorting to predesigned commercial RT-qPCR assays, researchers need to know what isoforms they are targeted to, and to determine whether the amplification will be impacted by such 3′-end variations. In our hands, assays targeted to longer isoforms (i.e., >22 nt), generally failed to amplify isoforms shorter by two or more bases, for both stem–loop and polyadenylation strategies (Nejad et al. 2018a,b). Equally, some probes targeted to shorter isoforms (i.e., 20–21 nt) did not amplify longer isoform as efficiently (as depicted in Fig. 1A), as seen with LNA assay targeting miR-222 (Nejad et al. 2018a). In the instance where shifts of predominant 3′-end isoforms have previously been evidenced, the researchers should therefore decide whether they are interested in quantifying the overall concentration of the miRNA, independent of the isoform length, or whether they are specifically interested in one of the 3′-end isoforms. Our comparison of stem–loop Taqman, polyadenylation, and LNA-RT-qPCR approaches showed that modified polyadenylation RT-qPCR relying on simple/inexpensive DNA primers was the most flexible approach, offering the capacity to amplify overall miRNA levels, and more specific 3′-end isoforms (Nejad et al. 2018a). Nonetheless, polyadenylation RT-qPCR can come at the cost of lower specificity toward select miRNA from closely related sequences, which should also be taken into account. Beyond the concern of results misinterpretation, changes of isomiR stoichiometry can present novel opportunities for miRNA biomarkers analyzed by RT-qPCR-based approaches, as illustrated here for miR-221-3p. We propose that the use of strategies relying on more selective isoform amplification such as the modified polyadenylation strategy we recently reported (Nejad et al. 2018a), may indeed facilitate the identification of disease specific isoforms, and their use in clinical context, as for instance suggested for the use of miR-222 in septic patients (Seeley et al. 2018).
Whether changes of isomiR stoichiometry shown here are restricted to phagocytes is unlikely since we saw similar responses in human fibroblasts (Nejad et al. 2018a,b), which are functionally very different from phagocytes. In addition, a recent study reported that PMA treatment of the chronic myelocytic leukemia cell line K562 promoted changes of isomiR stoichiometry in 7% of the 133 miRNA families analyzed (Muiwo et al. 2018). Similar to what we describe here with LPS, the isoforms of miR-27b-3p were differently impacted by PMA in this study, and the prevalent isoform shifted from the 21 to 20 nt long 3′-isomiR (Muiwo et al. 2018). However, further studies are needed to define whether the broad changes of isomiRs seen in the context of pathogen infection are also seen with other stimuli, and to define how such changes impact miRNA function. As such, the possibility that variation of miRNA 3′-end helps modulate intracellular localization and turnover is probably only scratching the surface of a more complex modulation of miRNA activity (Yu et al. 2017; Nejad et al. 2018b).
MATERIALS AND METHODS
Bioinformatics analyses of miRNA isoforms
Raw sequencing reads were downloaded from NCBI Sequence Read Archive for Studies SRP026382 (Dueck et al. 2014) and SRP064235 (Pai et al. 2016). Data were processed essentially as described for analysis of miRNA isoforms in Nejad et al. (2018b) except using cutadapt v1.14 (Martin 2011) and BWA v0.7.12 (Li and Durbin 2010) against the human (build hg19) or mouse (build mm10) reference genomes. Prior to normalization, raw microRNA isoform counts were filtered to retain only isoforms with at least three reads in at least one sample and counts were normalized to counts per million (CPM) using EdgeR (CalcNormFactors parameter “method”: TMM [Robinson et al. 2010]). Only isoforms with canonical 5′-end positions were used to generate the figures, which comprised all of the most abundant isoforms (see Supplemental Tables S1–S3 for details of all isoforms identified). The heatmaps shown in Figure 2A were generated using the Morpheus tool (https://software.broadinstitute.org/morpheus), and values normalized to NT are shown in Supplemental Table S4.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the Australian National Health and Medical Research Council (1062683 to M.P.G., 1069128 to C.P.B. and G.J.G., and fellowship 1026191 to G.J.G.), the Australian Research Council (140100594 Future Fellowship to M.P.G.), and the Victorian Government's Operational Infrastructure Support Program.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.069047.118.
REFERENCES
- Dueck A, Eichner A, Sixt M, Meister G. 2014. A miR-155-dependent microRNA hierarchy in dendritic cell maturation and macrophage activation. FEBS Lett 588: 632–640. 10.1016/j.febslet.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Hennessy EJ, Sheedy FJ, Santamaria D, Barbacid M, O'Neill LA. 2011. Toll-like receptor-4 (TLR4) down-regulates microRNA-107, increasing macrophage adhesion via cyclin-dependent kinase 6. J Biol Chem 286: 25531–25539. 10.1074/jbc.M111.256206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Xu C, Lei F, Liao M, Wang W, Xu N, Zhang Y, Xie W. 2017. MiR-30a targets IL-1α and regulates islet functions as an inflammation buffer and response factor. Sci Rep 7: 5270 10.1038/s41598-017-05560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juzenas S, Venkatesh G, Hübenthal M, Hoeppner MP, Du ZG, Paulsen M, Rosenstiel P, Senger P, Hofmann-Apitius M, Keller A, et al. 2017. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res 45: 9290–9301. 10.1093/nar/gkx706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Muiwo P, Pandey P, Ahmad HM, Ramachandran SS, Bhattacharya A. 2018. IsomiR processing during differentiation of myelogenous leukemic cell line K562 by phorbol ester PMA. Gene 641: 172–179. 10.1016/j.gene.2017.10.025 [DOI] [PubMed] [Google Scholar]
- Nejad C, Pépin G, Behlke MA, Gantier MP. 2018a. Modified polyadenylation-based RT-qPCR increases selectivity of amplification of 3′-microRNA isoforms. Front Genet 9: 11 10.3389/fgene.2018.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejad C, Pillman KA, Siddle KJ, Pépin G, Änkö ML, McCoy CE, Beilharz TH, Quintana-Murci L, Goodall GJ, Bracken CP, et al. 2018b. miR-222 isoforms are differentially regulated by type-I interferon. RNA 24: 332–341. 10.1261/rna.064550.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejad C, Stunden HJ, Gantier MP. 2018c. A guide to miRNAs in inflammation and innate immune responses. FEBS J 285: 3695–3716. 10.1111/febs.14482. [DOI] [PubMed] [Google Scholar]
- Pai AA, Baharian G, Pagé Sabourin A, Brinkworth JF, Nédélec Y, Foley JW, Grenier JC, Siddle KJ, Dumaine A, Yotova V, et al. 2016. Widespread shortening of 3′ untranslated regions and increased exon inclusion are evolutionarily conserved features of innate immune responses to infection. PLoS Genet 12: e1006338 10.1371/journal.pgen.1006338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley JJ, Baker RG, Mohamed G, Bruns T, Hayden MS, Deshmukh SD, Freedberg DE, Ghosh S. 2018. Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature 559: 114–119. 10.1038/s41586-018-0253-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle KJ, Deschamps M, Tailleux L, Nédélec Y, Pothlichet J, Lugo-Villarino G, Libri V, Gicquel B, Neyrolles O, Laval G, et al. 2014. A genomic portrait of the genetic architecture and regulatory impact of microRNA expression in response to infection. Genome Res 24: 850–859. 10.1101/gr.161471.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GC, Chan E, Molnar A, Sarkar R, Alexieva D, Isa IM, Robinson S, Zhang S, Ellis P, Langford CF, et al. 2014. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res 42: 9424–9435. 10.1093/nar/gku656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telonis AG, Magee R, Loher P, Chervoneva I, Londin E, Rigoutsos I. 2017. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res 45: 2973–2985. 10.1093/nar/gkx082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N, Cui H, Banerjee S, Tan Z, Salomao R, Fu M, Abraham E, Thannickal VJ, Liu G. 2014. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol 193: 327–334. 10.4049/jimmunol.1400203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Cao AT, Cao X, Yao S, Carlsen ED, Soong L, Liu CG, Liu X, Liu Z, Duck LW, et al. 2014. Downregulation of microRNA-107 in intestinal CD11c(+) myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. Eur J Immunol 44: 673–682. 10.1002/eji.201343717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Pillman KA, Neilsen CT, Toubia J, Lawrence DM, Tsykin A, Gantier MP, Callen DF, Goodall GJ, Bracken CP. 2017. Naturally existing isoforms of miR-222 have distinct functions. Nucleic Acids Res 45: 11371–11385. 10.1093/nar/gkx788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhang X, Cai Z, Zhou J, Cao R, Zhao Y, Chen Z, Wang D, Ruan W, Zhao Q, et al. 2018. A novel class of microRNA-recognition elements that function only within open reading frames. Nat Struct Mol Biol 25: 1019–1027. 10.1038/s41594-018-0136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.