Abstract
Precise knowledge of Mg2+ inner-sphere binding site properties is vital for understanding the structure and function of nucleic acid systems. Unfortunately, the PDB, which represents the main source of Mg2+ binding sites, contains a substantial number of assignment issues that blur our understanding of the functions of these ions. Here, following a previous study devoted to Mg2+ binding to nucleobase nitrogens, we surveyed nucleic acid X-ray structures from the PDB with resolutions ≤2.9 Å to classify the Mg2+ inner-sphere binding patterns to nucleotide carbonyl, ribose hydroxyl, cyclic ether, and phosphodiester oxygen atoms. From this classification, we derived a set of “prior-knowledge” nucleobase Mg2+ binding sites. We report that crystallographic examples of trustworthy nucleobase Mg2+ binding sites are fewer than expected since many of those are associated with misidentified Na+ or K+. We also emphasize that binding of Na+ and K+ to nucleic acids is much more frequent than anticipated. Overall, we provide evidence derived from X-ray structures that nucleobases are poor inner-sphere binders for Mg2+ but good binders for monovalent ions. Based on strict stereochemical criteria, we propose an extended set of guidelines designed to help in the assignment and validation of ions directly contacting nucleobase and ribose atoms. These guidelines should help in the interpretation of X-ray and cryo-EM solvent density maps. When borderline Mg2+ stereochemistry is observed, alternative placement of Na+, K+, or Ca2+ must be considered. We also critically examine the use of lanthanides (Yb3+, Tb3+) as Mg2+ substitutes in crystallography experiments.
Keywords: magnesium, monovalent ions, ribozyme, ribosome, lanthanides
INTRODUCTION
As established during recent decades, Mg2+ is of high relevance to the molecular ecosystem regulating nucleic acids folding, architecture, and function (Cate et al. 1997; Draper 2004, 2013; Klein et al. 2004; Woodson 2005; Freisinger and Sigel 2007; Auffinger et al. 2011; Bowman et al. 2012; Erat et al. 2012; Sigel and Sigel 2013; Marcia and Pyle 2014; Nierhaus 2014; Zhou et al. 2017). Still, for an exact understanding of the roles played by Mg2+, a precise structural knowledge of its binding modes is required. This knowledge is typically derived from solution and crystallographic experiments. However, solution studies are unable to locate Mg2+ with atomic precision and poorly distinguish between Mg2+ inner- and outer-sphere binding that are both important to nucleic acid structure and function (Draper 2004). Thus, crystallographic structures deposited to the PDB (Berman et al. 2016) remain the main source of information regarding Mg2+ binding modes. Nonetheless, correctly assigning an ion to an experimental electron density pattern is notoriously difficult and, unfortunately, the PDB embeds a significant number of well- and not so well-documented assignment errors (Williams 2005; Wlodawer et al. 2008, 2013, 2018; Kleywegt 2009; Cooper et al. 2011; Joosten et al. 2012; Pozharski et al. 2013; Dauter et al. 2014; Echols et al. 2014; Jain et al. 2015; Weichenberger et al. 2015; Minor et al. 2016; Raczynska et al. 2016; Rupp 2016; van Beusekom et al. 2016; Richardson et al. 2018). Incorrectly identified ions considerably bias database analysis results. As such, some efforts to correct these issues have been previously described (Zheng et al. 2015). The authors of this PDB survey (September 2014) established that only ≈15% of the ≈100,000 Mg2+ binding sites identified in RNA crystallographic structures should be considered as trustworthy. Later, it was recognized that a significant portion of the remaining Mg2+ binding sites does not satisfy the strict stereochemical criteria associated with Mg2+ (see below). Hence, it has been suggested that Mg2+ binding sites should be reexamined in the light of revised validation checklists (Leonarski et al. 2017).
Here, we pursue efforts to assess the reliability of Mg2+ assignments in crystal structures of RNA, DNA, and nucleobase-containing metabolites with resolutions ≤2.9 Å by examining the binding of Mg2+ to nucleobase and ribose oxygen atoms (Leonarski et al. 2017). This study aims to establish a “prior knowledge” data set of Mg2+ binding modes to be used to validate existing ion attributions in crystallographic but also NMR and cryo-EM structures, and to “limit” future solvent density pattern misinterpretations. This has been done by enforcing stereochemical criteria derived from the “almost” invariable Mg2+ octahedral coordination geometry (Chen et al. 2015). Indeed, even if some binding sites could at first glance seem well suited for Mg2+, the absence of trustworthy structural references in PDB structures of appropriate resolution should arouse reasonable doubts regarding their legitimacy and make one wonder if these binding sites would not better accommodate monovalent ions (Na+, K+) or transition metals (Leonarski et al. 2017) as emphasized by some nonambiguous examples presented in this study.
Throughout, we use the Mg2+ binding site nomenclature described in Zheng et al. (2015). O2/O4/O6, N1/N3/N7, O2′/O3′/O4′/O5′, OP1/2 atoms are respectively labeled Ob, Nb, Or, and Oph atoms; their combination (Ob.Nb, 2Ob or cis-2Oph.Ob, …) leads to the naming of binding sites. The direct binding of Mg2+ to oxygen atoms of phosphate groups that represent the primary nucleic acid binding locations (Klein et al. 2004; Sigel and Sigel 2010; Zheng et al. 2015) will be covered elsewhere.
RESULTS AND DISCUSSION
PDB overview of direct Mg2+ to carbonyl oxygen atom (Ob) contacts
Here, we investigate the potential of Mg2+ to establish direct (inner-sphere) contacts to nucleobase O2/O4/O6 (Ob) oxygen atoms of carbonyl groups. In a set of ≈5250 nucleic acid structures with resolution ≤2.9 Å, we identified ≈64,500 Mg2+ ions with a 1.0 occupancy and B-factors in the 1–79 Å2 range. Out of those, 9325 (≈14.5%) and 664 (≈1%) establish at least one contact with a d(Mg2+…Ob) ≤ 3.5 Å and a d(Mg2+…Ob) ≤ 2.3 Å coordination distance, respectively. The largest part of these Mg2+ is found in RNA and only a few were assigned to DNA (Table 1).
TABLE 1.
Number of nonredundant Mg2+/Na+…Ob contacts in PDB structures (resolution ≤2.9 Å)
In RNA, most of the 658 Mg2+ with d(Mg2+…Ob) ≤ 2.3 Å bind to (G)O6 and (U)O4 atoms and only a small proportion to (C)O2 and (U/T)O2 atoms (70%, 25%, 4%, and 1% contacts, respectively). With this distance criterion, only ≈2% of these Mg2+ establish more than one contact to Ob atoms. Note that we categorized ≈40% of them as redundant (see Materials and Methods) leaving a relatively small sample of nonredundant Mg2+ binding sites involving Ob atoms to be analyzed (≈400 in total). In the following, we focus on nonredundant binding sites.
Although less frequently assigned than Mg2+, monovalent ions such as Na+ and K+ were described to be part of the RNA and DNA solvation shell (Table 1). Among others, these ions recurrently contact (G)O6 atoms within quadruplex structures (Largy et al. 2016) and, as inferred from molecular dynamics simulations, they also frequently contact (G)O6 atoms belonging to the major groove of CpG steps within helical motifs (Auffinger and Westhof 2000; Pan et al. 2014; Auffinger et al. 2016; Šponer et al. 2018).
d(Mg2+…Ob) distance histograms highlight recurrent Mg2+ misidentifications
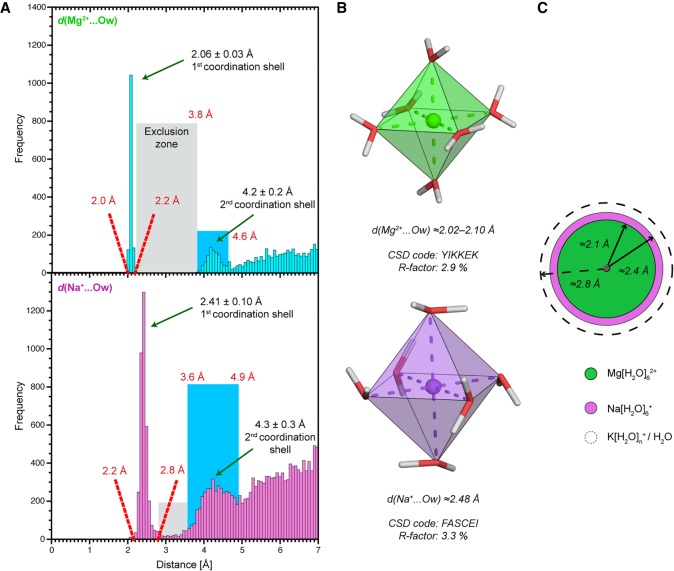
From a stereochemical point of view, if Mg2+ were strongly interacting with C=Ob groups, the sharp peak around 2.1 Å seen in the d(Mg2+…Ow) histogram derived from the CSD (Fig. 1A; CSD or Cambridge Structural Database: a repository for crystallographic structures of small molecules [Groom and Allen 2014; Groom et al. 2016]) would also appear in the d(Mg2+…Ob) PDB-derived histogram (Fig. 2). Instead, the latter displays a broad peak centered around 2.9 Å that overlaps with the 2.3–3.8 Å oxygen atom exclusion zone, a zone where, in principle, the second coordination shell oxygen atoms should not penetrate (see Materials and Methods and Fig. 1A). Therefore, this histogram exposes important ion misidentification issues (Leonarski et al. 2017).
FIGURE 1.
Mg2+/Na+ first hydration shells obey strict stereochemical rules. (A) Distance histograms for d(Mg2+…Ow) (top) and d(Na+…Ow) (bottom) derived from the Cambridge Structural Database (CSD; version 5.38; R-factors ≤5%) (Groom and Allen 2014). No disordered, error containing, polymeric or powder structures were considered. The water exclusion zones and the second coordination shells are marked by gray and blue rectangles, respectively. (B) Ultra-high-accuracy X-ray structures of Mg(H2O)62+ (top) and Na(H2O)6+ (bottom) illustrating similarities between Mg2+ and Na+ octahedral first hydration shells (Gerasimchuk and Dalley 2004; Hennings et al. 2013). (C) In scale schematic representation of the radius of the Mg(H2O)62+ (green) and Na(H2O)6+ (magenta) first cordination shells. The dashed circle marks the ≈2.8 Å d(H2O…Ow) average distance and the radius of the less well-defined K(H2O)n+ first hydration shell.
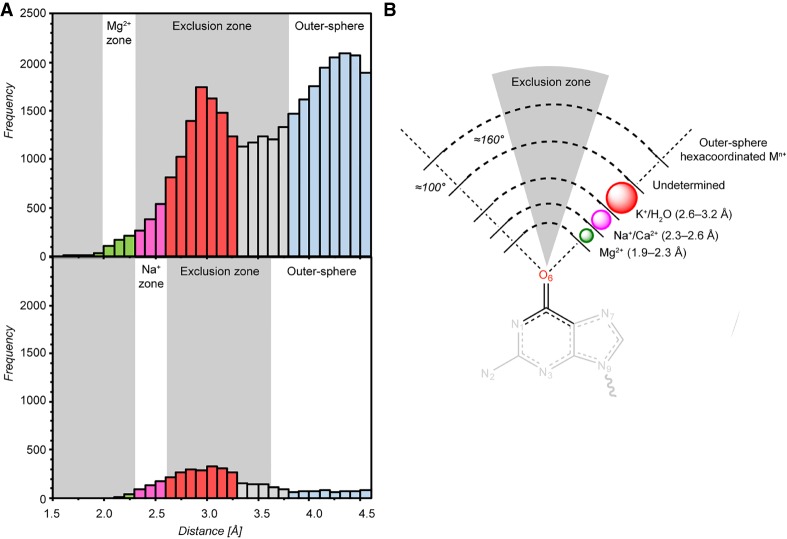
FIGURE 2.
Mg2+/Na+ distance histograms to nucleobase O2/O4/O6 (Ob) carbonyl oxygens. (A) Top and bottom: d(Mg2+…Ob) and d(Na+…Ob) histograms derived from PDB structures with resolution ≤2.9 Å. Only ions with 1.0 occupancies and B-factors in the 1–79 Å2 range were considered. The different ion binding and oxygen atom exclusion zones are colored in accordance to Figures 1A, 2B. Note that the provided boundaries are indicative. (B) Scheme showing ion binding and exclusion zones to nucleobase Ob atoms. The (G)O6 atom is taken as an example. The exclusion zone (gray) illustrates the fact that ions are rarely observed close to the C = Ob axis (see C = Ob…Mg2+ angle values in Figs. 3, 4; Supplemental Fig. S1). Thus, ions placed in this conical exclusion zone should be considered with caution.
To analyze more precisely these histograms (Fig. 2), we note that Mg2+ with coordination distances in the 2.3–2.6 Å range may correspond to misidentified Na+ ions that are characterized by ≈2.4 Å coordination distances and a well-defined octahedral coordination shell (Fig. 1). In the same distance range, Mg2+ could also correspond to misidentified Ca2+ that are sometimes part of crystallization buffers. The latter ion usually adopts an irregular coordination shell comprising 7/8 atoms (Marcus 1988) or, more rarely, an octahedral coordination shell similar to that of Na+/Mg2+ (Kennedy et al. 2004; Kolev et al. 2018).
Mg2+ modeled with coordination distances in the 2.6–3.2 Å range may correspond to misidentified K+/NH4+/H2O since all have coordination distances around 2.8 Å (Auffinger et al. 2016). In order to differentiate them, it is important to note that water and NH4+ have a coordination number of four while that of K+ ranges from six to eight (Page and Di Cera 2006; Harding et al. 2010). Mg2+ in the 3.2–3.8 Å distance range could correspond to misassigned anions or to crystallographic artifacts (Auffinger et al. 2004a; D'Ascenzo and Auffinger 2016). Finally, the broad peak around 4.2 Å (Figs. 1A, 2A) is attributable to ions—not necessarily Mg2+—establishing water-mediated contacts named outer-sphere or second shell contacts. The peaks in the 3.2–4.2 Å may also be attributable to other solvent molecules present in crystallization buffers (Weichenberger et al. 2015).
To further refine our stereochemical criteria, we identified an angular exclusion “cone” with C = Ob…Mn+ angles in the 160–180° range (Fig. 2), suggesting that binding of Mg2+/Na+ with C = Ob…Mn+ angles >160° should be interpreted with caution. Indeed, in the CSD (Supplemental Fig. S1) and PDB examples described below, the C = Ob…Mg2+/Na+ angle values in the 100°–160° range support the validity of this coordination criterion.
Incidentally, we note that identification issues are not restricted to ions in direct contact with nucleic acids but were also observed for hexahydrated ions that bind through water-mediated contacts, even in structures with resolutions ≤2.0 Å (Supplemental Fig. S2). Supplemental Figure S2C shows Na(H2O)6+, with d(Na+…Ow) in the 2.08–2.15 Å range, probably assigned in place of Mg(H2O)62+ (Wang et al. 2016). On the opposite, Supplemental Figure S2D shows Mg(H2O)62+ with d(Mg2+…Ow) ≈ 2.4 Å that is more likely to be Na(H2O)6+. These data underscore that assignment errors are not limited to divalent ions but can affect all ionic species (Supplemental Fig. S2E,F) and highlight two assignment errors related to high-resolution CSD structures pointing to the unfortunate fact that no database is error free and, thus, completely trustworthy (Spek 2009; Minor et al. 2016).
Direct binding of Mg(H2O)52+ to carbonyl oxygen atoms (Ob) is rare
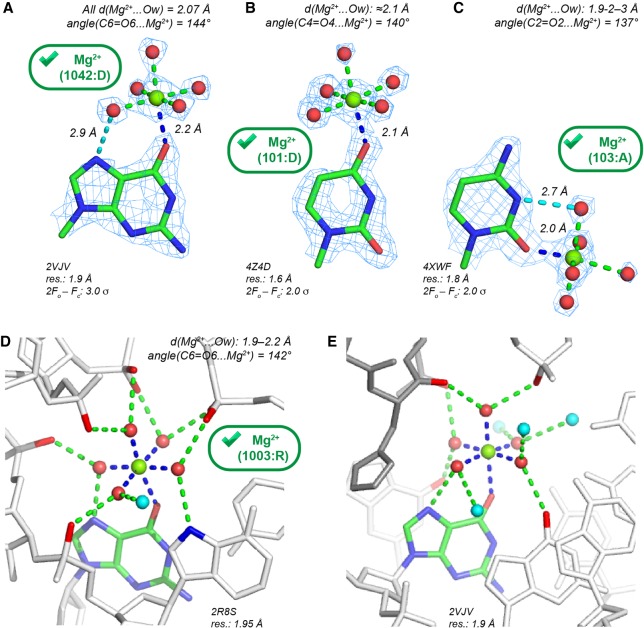
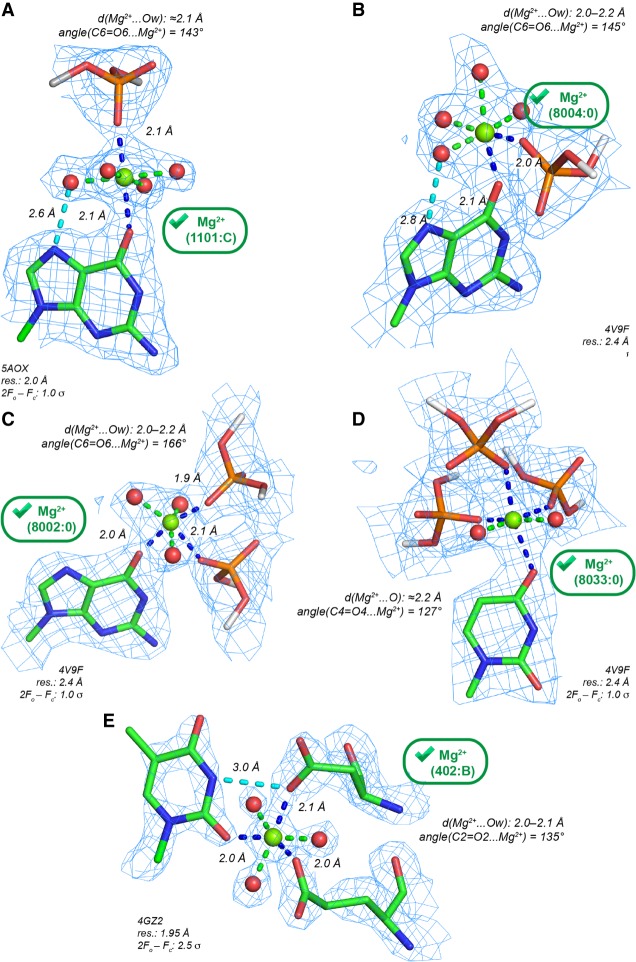
With d(Mg2+…Ob) ≤ 2.3 Å, we identified a limited number of nonredundant binding sites (96 occurrences) where Mg2+ binds to an Ob atom. Among those, 72, 13, 9, and 2 Mg2+ are at direct contact distance with (G)O6, (U)O4, (C)O2, and (U)O2 atoms, respectively. Only 19 of them are pentahydrated with d(Mg2+…Ow) ≤ 2.3 Å. This number does not change when a 2.4 Å distance criterion is applied. Eight of these Mg(H2O)52+ appear in structures with resolution ≤2.0 Å (Fig. 3) and with Mg2+ close to the nucleobase plane. These Mg(H2O)52+ present well-defined solvent densities forming a complete octahedral coordination shell and are, therefore, candidates for “prior-knowledge” Mg2+ binding sites.
FIGURE 3.
Pentahydrated Mg2+ contacting O2/O4/O6 (Ob) carbonyl oxygen atoms with d(Mg2+…O) ≤ 2.3 Å showing extensive H-bond networks. The green marks indicate Mg2+ with appropriate density patterns and stereochemistry. Only 11 Mg(H2O)52+ with d(Mg2+…O) ≤ 2.3 Å were characterized in structures with resolution ≤2.0 Å—see PDBid: 4XWF, 4Z4D, 4Z4E, 4W5O, 1DFU, 2VJV, 2R8S. (A) Mg2+ binding to a Hoogsteen edge (G)O6 atom. A water molecule establishes an H-bond (dashed cyan line) with the (G)N7 atom. (B) Mg2+ binding to a Hoogsteen edge (U)O4 atom. (C) Mg2+ binding to a Watson–Crick edge (C)O2 atom. A symmetry-related first shell water molecule establishes an H-bond (dashed cyan line) with the (C)N3 atom. (D) View of a dense H-bond network stabilizing a group I intron core motif (Ye et al. 2008); see also Figure 5B in Juneau et al. (2001). (E) Extended view of the H-bond network at a DNA/protein interface around the Mg2+ shown in A (see also Supplemental Fig. S3). The protein residues are gray. In D and E, the first and second shell H-bonds are, respectively, blue and green, and the second shell waters are cyan.
In the 2.0–2.9 Å resolution range, 11 Mg(H2O)52+ nonredundant binding sites were identified. All are in ribosomes (Supplemental Table S1) with Mg2+ displaying a distorted coordination shell. Four Mg(H2O)52+ (E. coli; 2.8–2.9 Å resolution) present proper coordination distances to Ob atoms. However, at the same location in a 2.1 Å resolution E. coli structure (Noeske et al. 2015), a d(Mg2+…Ob) ≈ 2.5 Å distance underlines the difficulties of assigning ions with confidence in ribosomes; see Figure 5 in Leonarski et al. (2017).
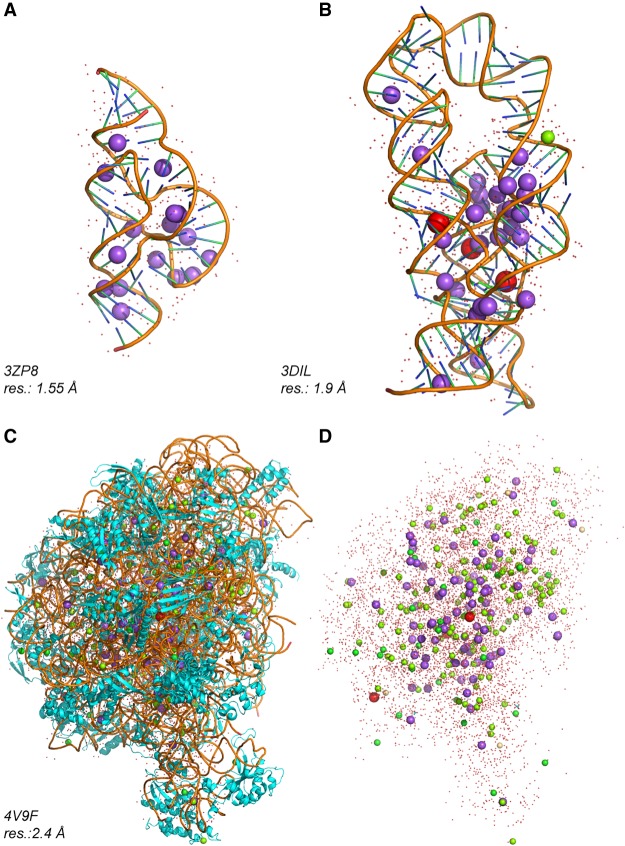
FIGURE 5.
RNA structures with a high Na+ content. Na+, K+, and Mg2+ are shown in magenta, red, and green, respectively. Ob bound Na+ can be found in the following structures with resolution ≤2.0 Å: PDBid: 1J8G, 1J6S, 3ZP8, 2R21, 5B2O/P, 3DIL, 4RKV, 4RNE, 4RJ1. (A) A 1.55 Å resolution hammerhead structure with 16 Na+ (Anderson et al. 2013). (B) A 1.9 Å resolution lysine riboswitch structure with 29 Na+, 3 K+, and 1 Mg2+ (Serganov et al. 2008). (C) A 2.4 Å resolution structure of the Haloarcula marismortui 50S with 85 Na+, 3 K+, and 138 Mg2+ (Klein et al. 2004; Gabdulkhakov et al. 2013). (D) The rRNA and protein chains shown in C were hidden to highlight the ionic content of this structure.
A few pentahydrated Mg2+ ions are involved in large H-bond networks
The few contacts described above in structures with resolution ≤2.0 Å (Fig. 3A–C) are part of large H-bond networks involving ion-coordinated water molecules. In a group I intron P4-P6 domain (Ye et al. 2008), a Mg(H2O)52+ is almost completely isolated from the surrounding solvent (Fig. 3D). The five first shell waters form 10 H-bonds (seven to phosphate groups, two to N7 atoms, and one to water). Intriguingly, this Mg2+ contributes to the stabilization of a local fold by holding five phosphate groups through second shell contacts. This particular configuration suggests that other energetic factors overrule local charge neutralization effects, as also inferred for sulfate-binding proteins where the anion is recognized through the formation of regular H-bonds to neutral amino acid groups (Pflugrath and Quiocho 1988; Hirsch et al. 2007; D'Ascenzo and Auffinger 2016). Thus, it appears that the ability of an ion to establish water-mediated second shell contacts can overrule electrostatic considerations (Auffinger et al. 2003, 2004b; Bowman et al. 2012).
A second Mg2+ binding pattern involving a single direct (G)O6 contact appears at a DNA protein interface (Fig. 3E). The H-bond network involves only one water-mediated contact to a phosphate group. In other structures, comparable Mg(H2O)52+ binding patterns to ApUpG steps were noted (Supplemental Fig. S3). Hence, Mg(H2O)52+ to Ob binding, although rare, is possible when the solvated ion occupies a tight binding pocket formed by RNA and/or protein residues establishing multiple H-bonds with the Mg2+ hydration shell.
Is tetrahydrated Mg2+ coordination to two carbonyl oxygens (2Ob) relevant?
2Ob binding sites are not frequent in PDB structures. We identified only eight nonredundant Mg2+ with two or more Ob contacts and d(Mg2+…Ob) ≤ 2.3 Å. The only 2Ob site observed in a nonribosomal structure, i.e., a group I intron (Ye et al. 2008), displays both d(Mg2+…Ob) below 2.3 Å while all distances to waters are in the 2.3–2.5 Å range, suggesting the presence of Na+ (Supplemental Fig. S4). No other Mg2+ binding to dinucleotide steps with appropriate coordination distances was observed. Thus, if existing, this binding mode is certainly highly uncommon. Elsewhere, we reached similar conclusions regarding the Ob.Nb binding mode that implies monovalent cations rather than Mg2+; see Figure 6B in Leonarski et al. (2017). A rare trans-2Ob binding motif (Supplemental Fig. S5) was identified in the CSD where the crystallization conditions often drastically differ from those used for biomolecular systems (Marino et al. 2016).
FIGURE 6.
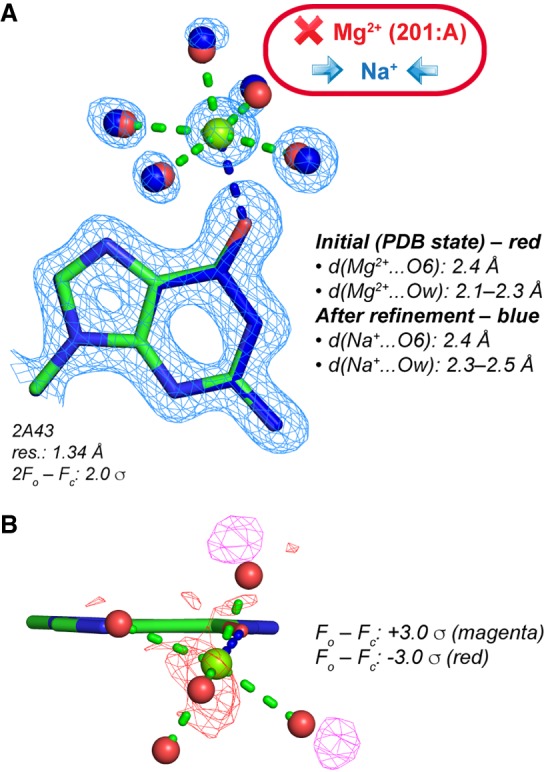
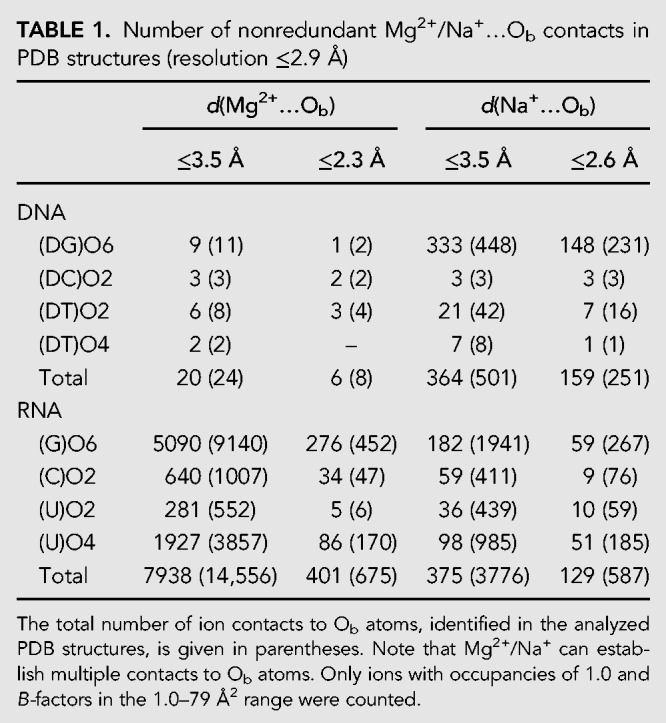
Re-refinement of a Mg2+ binding site in a high-resolution luteoviral pseudoknot fragment (Pallan et al. 2005). (A) Note that the original distances are not in agreement with the presence of a Mg2+, and the coordinated water molecule positions do not coincide with electron density peaks. A subsequent unrestrained refinement with Na+ led to more appropriate distances for this ion. (B) Positive and negative peaks in the original (PDB deposited) Fo – Fc maps around the pentahydrated ion hint to refinement issues. Such peaks should not appear in high-resolution structure Fo – Fc maps.
This finding looks surprising since it is well appreciated that Mg2+ ions are often bridging oxygen atoms (Oph) of adjacent phosphate groups and are known to form bidendate RNA-Mg2+ clamps (Petrov et al. 2011). This discrepancy can be rationalized if we consider that anionic groups (Oph) are better Mg2+ binders than carbonyl groups (Ob atoms; see below) and that the binding of Mg(H2O)42+ at a 2Ob site formed by consecutive nucleotides would prevent, in most instances, the formation of optimal ion-coordinated water-mediated contacts such as those shown in Figure 3D,E.
Mg2+ binding to Ob and phosphate/carbonyl anionic oxygens
In proteins and nucleic acids, anionic oxygen atoms are commonly considered as primary Mg2+ binders (Zheng et al. 2008, 2015, 2017; Bowman et al. 2012). Patterns involving Ob and Oph atoms with d(Mg2+…O) ≤ 2.3 Å are slightly more frequent than those involving Ob/Nb atoms since we identified 48 (Oph.Ob), 36 (2Oph.Ob), and 4 (3Oph.Ob) nonredundant binding sites. For Oph.Ob, a simple pattern is observed when Mg2+ coordinates in trans (trans-Oph.Ob) to an OP and a (G)O6 atom (Fig. 4A). In ribosomes, few examples of cis-Oph.Ob and cis-2Oph.Ob patterns were noted (Fig. 4B,C). Potential fac-3Oph.Ob binding sites are infrequent (Fig. 4D). Fac- refers to the isoform where the three Oph atoms are on the same side of the coordination octahedron (Zheng et al. 2015).
FIGURE 4.
Mg2+ contacting both carbonyl (Ob) and anionic oxygen atoms (Oph, Ocoo) with d(Mg2+…O) ≤ 2.3 Å. The green marks indicate Mg2+ with appropriate densities and stereochemistry. (A) Trans-Oph.Ob: Mg2+ binding to a guanine Hoogsteen edge. A water molecule establishes an H-bond (dashed cyan line) with the (G)N7 atom. (B) Cis-Oph.Ob: Mg2+ binding to a guanine Hoogsteen edge. A water molecule establishes an H-bond (dashed cyan line) with the (G)N7 atom. (C) A ribosomal cis-2Oph.Ob binding site. (D) A ribosomal fac-3Oph.Ob binding site. Note the coordination distances ≈2.2 Å that suggest the use of restraints (Supplemental Table S2). (E) A cis-2Ocoo.Ob binding site at a protein–RNA interface. The contact with a first shell water molecule and a (U)N3-H group, that was considered as difficult to establish (see Fig. 3C), is here replaced by a (U)N3-H…O=C(Asp) H-bond. (C–E) For nomenclature, see Zheng et al. (2015); Ocoo corresponds to oxygen atoms of carboxylate groups.
In nucleic acid/protein complexes, carbonyl groups belonging to carboxyl(ate) Asp, Glu (Ocoo), or carboxamide Asn and Gln side chains (Ocno) as well as to the peptidyl backbone C=O group (Obb) may also contact Mg2+. In a DNA/protein complex, Mg2+ establishes three contacts to water molecules, two to Ocoo atoms, and one to an (U)O2 atom (Fig. 4E). At present, an exhaustive classification of binding sites involving Ob and anionic oxygen atoms is out of reach given the high level of uncertainty associated with these binding modes, the ion identity and the rarity of reliable Mg2+ binding site in structures with appropriate resolution.
No evidence for Mg2+ binding to ribose and backbone oxygen (Or) atoms
Besides the binding of Mg2+ to Ob atoms, we also explored Mg2+ binding to Or atoms defined as oxygen atoms belonging to the nucleic acid ribose group (O2′, O4′) and backbone (O3′, O5′). In the ≤2.9 Å resolution range, we found no Mg2+-to-Or binding site with appropriate coordination distances. In support of this observation, it has been reported that direct interactions between divalent metals and hydroxyl groups are very weak and that the strength of the hydroxyl–metal interaction increases with the decreasing charge of the coordinating atom, suggesting that monovalent ions would better interact with hydroxyl groups than Mg2+ (Al-Sogair et al. 2011). Many studies targeted at assessing the involvement of metals in catalytic mechanisms have been conducted. For instance, the participation of a metal ion in the ribosomal catalytic mechanism that would facilitate nucleophilic attack by binding to a 2′-OH group has been investigated through ion substitution experiments (Schmeing et al. 2005). The authors concluded that their crystallographic data are most consistent with a model where a water molecule and not a mono or divalent ion interacts with an active site ribose O2′ atom. These data question the involvement of divalent metal ions interacting with hydroxyl groups in at least some ribozymes (Lilley 2011; Ward et al. 2014).
Critical evaluation of the use of Yb3+ and other lanthanide ions as Mg2+ substitutes
In contrast to what is reported above, a peculiar and rare example of Mg2+ binding to the O3′ group of a terminal guanine is seen in a 1.7 Å resolution protein/RNA complex; see Supplemental Figure S6A (Gan et al. 2008). In this specific context, the O3′ hydroxyl group may take an anionic form and turn into a more appropriate ligand for Mg2+. Similar examples are exceptional in the CSD/PDB. However, in a recent group II intron lariat structure (PDBid: 5J01, resolution: 3.4 Å); based on Yb3+ anomalous signals, a Mg2+ was modeled at a bonding distance of O2′/O3′ ribose atoms of a terminal uridine; see Supplemental Figure S6B (Costa et al. 2016). Similar Mn2+ and lanthanide-based strategies were used to explore the RNA ionic landscape (Adams et al. 2004; Stahley and Strobel 2005; Toor et al. 2008; Kazantsev et al. 2009; Wang 2010; Marcia and Pyle 2012, 2014; Bénas et al. 2014; Robart et al. 2014). In these crystallographic studies, it was assumed that Yb3+ is a good Mg2+ mimic. However, the coordination distance of Yb3+ to water (≈2.3 Å) is closer to that of Na+ (≈2.41 Å) than to that of Mg2+ (2.06 Å). Moreover, the Yb3+ coordination number derived from high-resolution CSD structures is dominantly eight, sometimes seven or nine (Cossy et al. 1989; Thuéry 2009) and exceptionally six as observed in a handful of specific chemical contexts; see Supplemental Figure S6C,D (Lundberg et al. 2010). Thus, the observation that Yb3+ may bind to group I/II introns and other ribozyme hydroxyl groups does not warrant that Mg2+ is present at these sites.
However, it can also be hypothesized that this binding mode occurs only in structural contexts unique to ribozymes. In the group II intron mentioned above (Supplemental Fig. S6B), Mg2+ is bound to three phosphates and its charge may be sufficiently delocalized to allow binding of two (see Supplemental Fig. S6B), eventually deprotonated, 2′-OH groups belonging to a terminal ribose. Yet, we suggest that, in the absence of high-resolution data, caution should be exerted in interpreting crystallographic ion binding motifs such as those encountered at ribozyme catalytic sites since binding principles of Mg2+, lanthanides, and other ions are still incompletely understood.
Mg2+ does not bind to carbonyl oxygen (Ob) atoms of metabolites containing nucleobases
By using the Relibase+ program to search the PDB (Hendlich et al. 2003), we checked if Mg2+ to Ob binding is associated with nucleobase-containing metabolites. In the ≤2.9 Å resolution range, ≈11,000 metabolites (with G/C/U/T nucleobases) were identified with only one binding site with d(Mg2+…Ob) ≤ 2.3 Å. This unique site occurs in a structure of a human signaling protein involving GDP (guanine-diphosphate; Supplemental Fig. S7). Unfortunately, this structure and the four related PDB releases (2005/6) from the same group are not currently associated with a publication record, an issue addressed by Wlodawer et al. (2018). Moreover, this structure contains 12 Ca2+ and five Mg2+. Most of the latter display a high electron density peak and a tetrahedral coordination with ≈2.1 Å coordination distances that better match a transition metal such as Zn2+. Therefore, this Mg2+ binding site has to be interpreted with caution given the serious solvent attribution issues reported for this structure. The fact that no obvious Mg2+ binding to nucleobase metabolite Ob or Nb atoms, as shown previously (Leonarski et al. 2017), could be characterized, supports the claim that the nucleobase Mg2+ binding potential is poor, as also inferred from solution studies on nucleoside and nucleotides (Sigel and Kapinos 2000; Sigel and Sigel 2010).
Direct Na+ binding to carbonyl oxygens (Ob) is possible
Since we established that reliable instances of Mg2+ binding to Ob atoms are few, we propose potential hexacoordinated substitutes for these ions. As already mentioned, the d(Mg2+…Ob) histograms (Fig. 2) suggest that Na+ or K+ binding to Ob atoms is more likely than Mg2+ binding. First, we provide a few examples of Na+ to Ob binding. Yet, since misattributions are also an issue for monovalent ions (Supplemental Fig. S2), we focus on structures with resolution ≤2.0 Å.
In this resolution range, 25 RNA and 105 RNA/protein structures containing Na+ were identified. Among those, 13 contain Na+ with d(Na+…Ob) in the 2.3–2.6 Å range (Fig. 1). One hexacoordinated Na(H2O)4+ at a 2Ob site (Supplemental Fig. S8A) was identified in a 1.55 Å resolution hammerhead ribozyme structure (Anderson et al. 2013) that contains a total of 16 Na+ and no divalent ion (Fig. 5A). This best resolution PDB hammerhead structure is at odds with the remaining 22 hammerhead structures that contain no or only divalent/trivalent ions. It could be argued that the crystallization buffer (1.7 M Na+ malonate and 10 mM MgCl2) plays a significant part in displacing Mg2+ in favor of Na+. However, an RNA tridecamer (resolution: 1.3 Å; Supplemental Fig. S8B) and a lysine riboswitch (resolution: 1.9 Å; Fig. 5B) were crystallized in buffers containing Mn2+ and 100 mM NaCl or Mg2+, K+ and 100 mM Na+ citrate, respectively (Timsit and Bombard 2007; Serganov et al. 2008). Out of a total of 29 Na+, the latter structure contains 17 hexacoordinated Na+ with 10 displaying d(Na+…O) in the 2.3–2.6 Å range. Hence, these structures and that of the hammerhead ribozyme demonstrate that Na+ can bind to nucleic acid systems with a well-defined octahedral coordination. Though, the reasons as to why these structures that were crystallized in the presence of Mg2+ display so many Na+ are not understood. The usual explanation stating that Mg2+ easily displace monovalent cations does not hold here. Interestingly, the fact that Na+ can take over the role of Mg2+, even at strong binding sites, was recognized very early (Jack et al. 1977; Quigley et al. 1978). In contrast, Z-DNA structures with resolutions <1.3 Å derived from crystals with a 200 mM MgCl2 or CaCl2 content do not show evidence of bound mono or divalent ions (Luo et al. 2017; Harp et al. 2018). Supplemental Figure S8C,D displays a further misattribution case where a Mg2+ bridges two Ob atoms belonging to stacked nucleobases with 2.6 Å coordination distances that better characterize Na+ but may also correspond to K+ given a high σ peak value at the binding location.
A particularity of a few Na+-containing structures relates to the occurrence of metallic clusters recruiting two or more Na+ with d(Na+…Na+) in the 3.1–3.7 Å range (Supplemental Figs. S8E, S9). These Na+ clusters (Timsit and Bombard 2007; Serganov et al. 2008) represent a neglected category of two-metal binding motifs (Glusker et al. 2001). An example of a Na+ cluster mistaken for a Mg2+ cluster in a protein is given in Figure S6D of Wlodawer et al. (2018), stressing the probable widespread occurrence of dimetallic Na+ clusters in biomolecular systems, a fact to remember during the electron density interpretation process.
The monovalent ion count in ribosomes is underestimated
Overall, the examples we reported show that monovalent ions such as Na+ are identifiable in nucleic acid systems even in presence of Mg2+. They back up the results of a seminal study by Klein and Steitz regarding the ionic distribution of a 2.4 Å resolution H. marismortui 50S ribosome structure (Klein et al. 2004). The authors stated that, besides Mg2+, ribosomes are surrounded by monovalent ions that can be clearly distinguished from divalent ions, based on their coordination patterns and the anomalous signals of Rb+/Cs+ derivatives (Fig. 5C,D). To establish a primary ion-binding classification, these authors claimed that both Na+ and K+ lack preferred coordination geometries. If this is reasonable for K+, which shows irregular binding patterns with coordination numbers ranging from six to eight, it is less reasonable for Na+. Hence, Na+ can easily be mistaken for Mg2+.
A quick survey of the data presented by Klein and Steitz shows that several Mg2+ were assigned to octahedral coordination patterns with d(Mg2+…O) in the 2.3–2.8 Å range (see Supplemental Fig. S10A,B), while Na+ were assigned to irregular coordination patterns better matching K+ (see Supplemental Fig. S10C,D). In a subsequent refinement (PDBid: 4V9F; resolution: 2.4 Å; Gabdulkhakov et al. 2013) of the original H. marismortui 50S structure, the ion coordination patterns and distances remained unchanged.
Mg2+ assignments can obliterate monovalent ions—a case for re-refining ion binding sites
As inferred from above, Na+ is a better match than Mg2+ to hexacoordinated solvent electron densities when d(Mg2+…O) is in the 2.3–2.6 Å range. However, no clear-cut ion identification rule can be provided even with d(Mn+…O) ≤ 2.3 Å. This is illustrated by the 1.34 Å resolution luteoviral pseudoknot structure that contains two modeled Mg2+, one coordinating to a G(O6) atom (Fig. 6) and the other being hexahydrated (Pallan et al. 2005). While for the first ion, water coordination distances are in the 2.1–2.3 Å range, d(Mg2+…O6) is close to 2.4 Å. Based on deposited electron density maps, we observed that the positions of ion coordinated waters do not overlap with electron density peaks resulting in positive and negative blobs in the Fo – Fc maps. Therefore, we suspected that the d(Mg2+…Ow) in the 2.1–2.3 Å range were inappropriate and performed a basic “unrestrained” refinement of the structure with phenix.refine using default settings (Afonine et al. 2012). While d(Mg2+…O6) remained unchanged at 2.4 Å, d(Mg2+…Ow) drifted toward 2.36–2.54 Å suggesting to swap the originally assigned Mg2+ for Na+. For the hexahydrated ion, d(Mg2+…Ow) also drifted toward the 2.39–2.52 Å range, a distance consistent with the presence of Na+, this ion being present in the crystallization buffers (50 mM Na+ cacodylate). Interestingly, the latest version of PDB_REDO, a web service devoted to improving the fit of old and new models to crystallographic data, confirms the coordination distances we obtained but does not yet propose alternative ions that would better fit the data issued from subsequent refinements (Joosten et al. 2009, 2012, 2014; van Beusekom et al. 2018).
K+, a ubiquitous but difficult to assign ion that binds to Ob atoms
K+ is unfrequently assigned in crystallographic structures, although it is generally considered as the dominant intracellular monovalent cation (Nierhaus 2014; Auffinger et al. 2016). K+ is known to bind to specific RNA pockets, as inferred from several nucleic acid X-ray structures (Basu et al. 1998; Batey and Doudna 2002; Conn et al. 2002; Klein et al. 2004; Auffinger et al. 2016). However, its detection remains difficult since K+ lacks a well-defined and regular coordination pattern and because its ≈2.8 Å coordination distance overlaps with those of water and NH4+ molecules (Zheng et al. 2017). Therefore, besides direct observation of K+ anomalous signals (Tereshko et al. 2001; Egli et al. 2002; Ennifar et al. 2003; Stahley et al. 2007), substitution strategies involving Tl+, Rb+, and Cs+ have sometimes been used (Klein et al. 2004; Marcia and Pyle 2014). Yet, these strategies did not prevent ion misidentifications such as those shown in Supplemental Figure S10C,D.
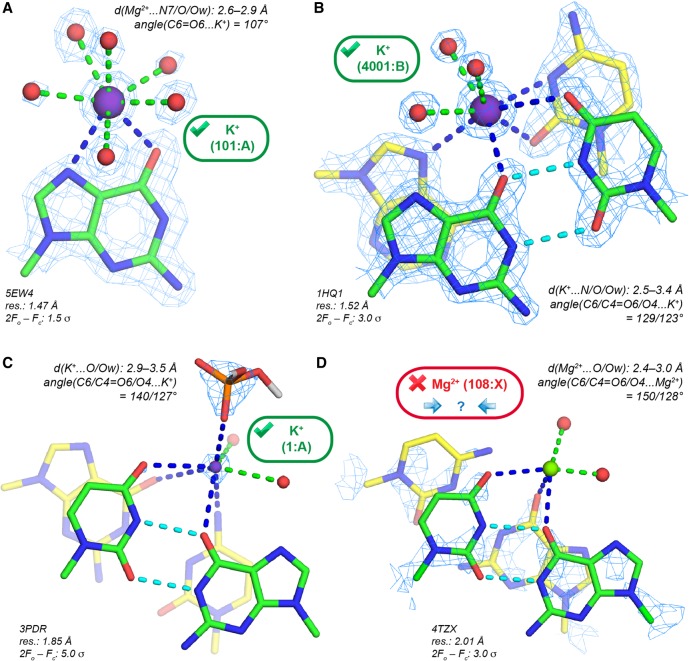
For instance, it has been found that the major groove cleft of the cis-WC G•U pair (Fig. 7C) is often occupied by water (Auffinger and Westhof 1998; Mueller et al. 1999), but also by K+ (Klein et al. 2004; Fan et al. 2005). However, Mg2+ establishing direct contacts with G(O6) and U(O4) atoms, was sometimes unduly modeled at this location. Figure 7D shows d(Mg2+…O) in the 2.6–3.3 Å range that point to the presence of K+. In a re-refinement of Cas9–RNA/DNA structures (Anders et al. 2014; Olieric et al. 2016), it was observed that a fraction of the eight ions originally modeled as Mg2+ display anomalous peaks above the 10 σ level (Supplemental Fig. S11). It was not possible to unambiguously identify the corresponding ion from the anomalous peak height or the local stereochemistry. However, X-ray fluorescence (XRF) analysis confirmed the presence of K+ in crystals that were obtained by using a buffer containing 250 mM KCl, 300 mM KSCN, and 5 mM Mg2+. Based on this evidence, six Mg2+ were reassigned as K+. However, three uncorrected structures with Mg2+ assignments (PDBid: 4UN3/4/5) remain in the PDB, jeopardizing subsequent reinterpretations of the ionic environment of this system (Minor et al. 2016; Rupp et al. 2016; van Beusekom et al. 2016).
FIGURE 7.
K+ binding to Ob atoms in the major groove of cis-WC G•U pairs. (A) K+ binding to a guanine Hoogsteen edge; coordination number: 8. (B) K+ binding to the major groove of a G•U pair; coordination number: 8. (C) K+ binding to the major groove of a G•U pair. To highlight the 5.0 σ level K+ density, the radius of the K+ sphere (magenta) has been reduced. (D) Mg2+ modeled in the major groove of a G•U pair that could correspond to a monovalent cation. The electron density pattern is too imprecise for an unambiguous assignment. (B,C,D) (Green) G•U carbons; (cyan) G•U H-bonds; (yellow) other carbon atoms.
Ions in the ribosomal peptidyl and decoding sites: Mg2+ or K+?
In ribosomes, Mg2+ located close to important functional elements (Hsiao and Williams 2009; Bowman et al. 2012; Petrov et al. 2012) are of particular interest. In the first H. marismortui 50S X-ray structures, a K+ bridging two guanine Hoogsteen edges (Fig. 8A) was modeled close to the peptidyl transferase center or PTC (Nissen et al. 2000; Klein et al. 2004). It was proposed but not confirmed that this ion plays a role in the catalytic mechanism by stabilizing tautomeric nucleotide forms. The coordination distances for this ion with (G)O6 and (G)N7 are in the 2.8–3.3 Å range and unambiguously point to the presence of K+. This K+ has been consistently assigned to the same location in 44 H. marismortui PDB structures (Supplemental Table S1). Overall, this unique K+ binding site demonstrates the monovalent ion ability to fit within well-defined structural notches (Auffinger et al. 2016).
FIGURE 8.
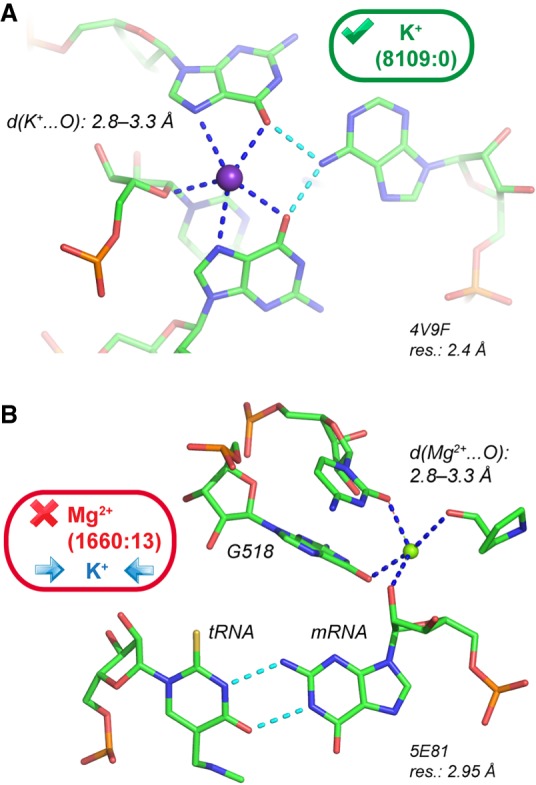
rRNA K+ binding sites. (A) K+ close to the peptidyl transferase center has been recurrently assigned in H. marismortui 50S structures (Supplemental Table S1). The guanine K+ coordination is comparable to that shown in Fig. 7A. (B) A recurrently assigned Mg2+ with K+ characteristics in the ribosomal decoding center of several Thermus thermophilus structures.
At odds, an ion close to the decoding center with similar coordination distances to oxygens (Fig. 8B) has been systematically assigned to Mg2+ (Murphy and Ramakrishnan 2004; Weixlbaumer et al. 2007; Rozov et al. 2015, 2016a). This ion assignment is made in structures with resolutions in the 2.95–3.30 Å range from which it is really problematic to infer light ion binding. However, its coordination distance to the (C518)O2 and (G530)O6 atoms in the 2.7–3.3 Å range point to the presence of K+. In a recent review, it was stated that these ions were modeled as Mg2+ but that their precise identity could not be established (Rozov et al. 2016b). To us, the likelihood for this ion to be K+ is high, although it has been labeled Mg2+ in crystal structures or “M” for metal in related publications. This binding site is representative of ion misattributions in ribosomes (Klein et al. 2004; Noeske et al. 2015).
Although at first glance, such issues may not seem important for the interpretation of crystal structures, one could envisage that they may change the outcome of molecular dynamics simulations since the Mg2+ stabilization effect is much greater than that of K+ (Hayatshahi et al. 2017). Henceforth, it is highly probable that a decoding site modeled with one or the other ion would behave in significantly different ways and change our perception of the energetics and dynamics of the ribosomal decoding center (Lind et al. 2017). Thus, it is recommended to avoid using structures that contain ions with poor stereochemistry for initiating modeling studies (Hashem and Auffinger 2009).
Remarks regarding the MgRNA database and proposals for a revised set of “prior-knowledge” Mg2+ binding sites
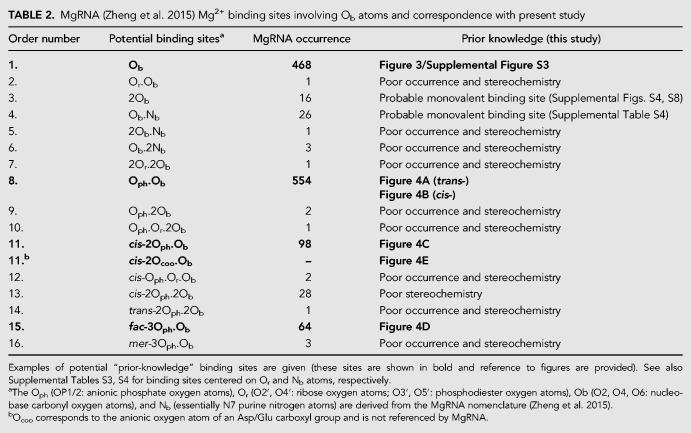
The MgRNA database was designed to build an exhaustive and comprehensive classification of Mg2+ binding sites (Zheng et al. 2015). In its present state, MgRNA lists 41 inner-sphere coordination patterns among which 16 are associated with Ob atoms. It has already been documented that MgRNA significantly overestimates the binding of Mg2+ to nucleobase Nb atoms, a category that regroups the N1/N3/N7 atoms (Leonarski et al. 2017). The same issues originating from too lenient selection criteria are observed for Mg2+ to Ob binding. These issues are related to (i) the inclusion in the analyzed sample of structures with resolution >3.0 Å and sometimes >4.5 Å, from which it is impossible to infer the position of light ions or water molecules; (ii) the fact that Mg2+-to-Ob coordination distances significantly exceeding 2.3 Å were not discarded; (iii) that artificially restrained ions with d(Mg2+…Ow) = 2.18 Å (see Materials and Methods section and Supplemental Table S2) were not excluded; (iv) that several binding modes were described based on only a handful of ions displaying often inappropriate stereochemistry; (v) that no attempts to consider redundancy were made.
To illustrate the above statements, we note that in the Ob data set (468 occurrences), over 50% of the binding sites are redundant. In the remaining structures with resolution <2.9 Å, 15 nonredundant octahedral coordination sites (≈5%) were identified that display often suboptimal coordination stereochemistry. Besides, MgRNA categorizes some infrequent binding modes and defines nine binding types comprising between one and 10 occurrences. These should not be used to define “prior-knowledge” categories, especially when associated with low resolution and poor stereochemistry. In the same line, the 13 MgRNA binding modes involving the Or atoms (O2′, O3′, O4′, O5′) should not be considered as “prior-knowledge” Mg2+ binding motifs (Rupp 2016). More detailed information on issues related to the current MgRNA version is given in Leonarski et al. (2017). These findings are summarized in Table 2 for the Ob atoms and Supplemental Tables S3, S4 for Or and Nb atoms, respectively.
TABLE 2.
MgRNA (Zheng et al. 2015) Mg2+ binding sites involving Ob atoms and correspondence with present study
About statistics
The few instances of Mg2+ binding sites with appropriate stereochemistry we characterized do not really allow the collection of meaningful statistics. However, we believe that the trends noted by the MgRNA authors are somewhat preserved although the number of binding sites is considerably reduced because many of them, including binding of Mg2+ to hydroxyl groups, need urgently to be discarded. It remains possible that, from forthcoming nucleic acid structures, we may be able to derive slightly different binding principles leading to an extension of those described here.
Mg2+ assignment and validation checklist
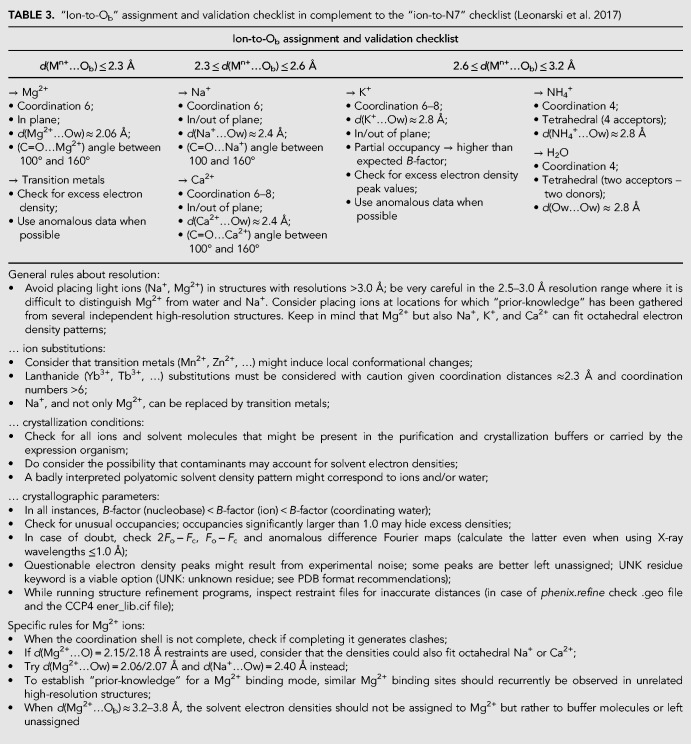
At this point, we hope that it has become clear that the characterization of each Mg2+ to nucleobase binding occurrence needs experimental validation rather than circumstantial evidence derived from low resolution data and limited occurrence. Thus, we feel that it is essential to update existing validation checklists. In Table 3, we adapt our previous Mg2+-to-N7 validation checklist to Ob atoms (Leonarski et al. 2017).
TABLE 3.
“Ion-to-Ob” assignment and validation checklist in complement to the “ion-to-N7” checklist (Leonarski et al. 2017)
As before, we stress that the chosen cutoff distances are merely indicative and may be modulated regarding the specific structural context. Thus, d(Mg2+…O) ≈2.4 Å distances are borderline and must be considered with caution. Marginal ion-to-oxygen distances may also find their origin in hidden crystallographic disorder (multiple conformations, partial/mixed occupancies…), the presence of unexpected solvent molecules contaminating purification or crystallization buffers (Borek et al. 2003; Giegé 2013; Dauter et al. 2014; Weichenberger et al. 2015; Niedzialkowska et al. 2016; Moon et al. 2017), or even inappropriate refinement protocols that remain to be documented.
The use of restraints during crystallographic refinement often introduces major interpretational biases (Leonarski et al. 2017). In particular, we strongly discourage the use of the widespread 2.15/2.18 Å d(Mg2+…O) default distance restraint values in the CCP4 ener_lib.cif library (Supplemental Table S2). When distance restraints are used during refinement, it becomes problematic to assign with confidence Mg2+ or Na+ to a given electron density pattern (Zheng et al. 2014, 2017; Leonarski et al. 2017). When needed, one should use CSD derived distance restraints (Zheng et al. 2014, 2017; Leonarski et al. 2017) since distances from the PDB were shown not to be reliable (Fig. 2). Unfortunately, structures refined by using inappropriate restraints remain in the PDB and represent a serious hazard for the less experienced users (Cooper et al. 2011; Dauter et al. 2014; Minor et al. 2016; Leonarski et al. 2017).
It is also important to mention that the use of restraints for structures in the >2.5 Å resolution range is often justified by the fact that diffraction data are unable to determine metal-to-N/O coordination distances with the required precision (Harding 2001). However, this should be avoided, especially when the risk of Na+/Mg2+ misidentification is high. In such instances, crystallographers should consider that Na+/K+/Ca2+ are rightful alternatives to the placement of Mg2+ and refrain from drawing firm conclusions regarding ion identity. When ion identity remains ambiguous and when, nevertheless, the electron density pattern points to the presence of a metal, the “M” marker should be used (Rozov et al. 2016b).
In the future, significant help in assigning solvent molecules should stem from better use of anomalous signals (Leonarski et al. 2017). Thanks to continual improvements in anomalous difference measurements through specialized beamlines, constant accuracy improvement of X-ray detectors, and more efficient software (Storoni et al. 2004; Thorn and Sheldrick 2011; Weinert et al. 2015; Olieric et al. 2016; Wagner et al. 2016; Leonarski et al. 2018), it might become possible to make use of weak Na+/Mg2+ signals when high resolution is available. With greater likelihood, the detection of the anomalous signals of heavier ions such as K+, Ca2+, Cl−, and SO42− will be facilitated (Ennifar et al. 2003; Auffinger et al. 2004a; Mueller-Dieckmann et al. 2007; Thorn and Sheldrick 2011; Echols et al. 2014; D'Ascenzo and Auffinger 2016). In the meantime, we advocate for the deposition of diffraction images for all relevant X-ray measurements, including heavy atom soaks, to allow reprocessing of the data and to check for weak anomalous signals. We suggest also to define a marker that would help to differentiate ions that were placed based on native data from those that were modeled by using anomalous signals from different ions (Grabowski et al. 2016). Indeed, as discussed earlier (Leonarski et al. 2017), ion substitution experiments are not always sufficiently reliable to confirm Mg2+ binding sites since the binding preference of lanthanides or soft ions like Mn2+ does not systematically match those of the harder Mg2+. This has already been suggested in an early study of tRNA ion binding where the authors noted in a sobering manner: “A comparison of the magnesium, cobalt and manganese binding sites gives reason to doubt the idea that these last two mimic magnesium in their binding properties” (Jack et al. 1977).
In order to get clues about the ionic composition of the crystals, the systematic use of X–ray fluorescence (XRF) analysis should be encouraged (Olieric et al. 2016) as well as the exploration of the local environment of a metal by extended X-ray fine absorption structure (EXAFS) techniques (Hensley et al. 2011; Hummer and Rompel 2013). Interestingly, inductively coupled plasma emission spectroscopy (ICP-ES) has been used to eliminate the presence of possible divalent metal contaminants (Mn, Ni, Zn, Pb, and Cd) in tRNA crystals (Jovine et al. 2000).
SUMMARY AND CONCLUDING REMARKS
The combined data presented in this and an earlier investigation (Leonarski et al. 2017) interrogate recurrent assumptions made in interpreting solvent electron density maps within nucleic acid structures. These assumptions or “disruptive nudges”—to divert a concept made popular by Richard Thaler (Thaler 2000; De Bondt et al. 2018)—have led in our opinion to the deposition in the PDB of a significant number of nucleic structures with exaggerated Mg2+ contents at the expense of the assignment of monovalent cations (Na+, K+) and other small solvent molecules. Here, we stress that the possibility that other ions (Na+, K+, Ca2+, …) could fit solvent electron density patterns should be systematically envisaged, especially for sites displaying borderline stereochemistry and that ion substitution experiments should be interpreted with caution given the rising number of documented instances emphasizing deceiving effects associated with ion replacement strategies (Jack et al. 1977; Leonarski et al. 2017).
We suggest that, before inferring ion binding from low-resolution crystallographic structures, a set of trustworthy binding sites derived from high-resolution structures or, in other words, a set of “prior-knowledge” binding sites must be defined. This has been tentatively proposed in Table 2 for Ob atoms and Supplemental Tables S3, S4 for Or and Nb atoms. In the current state of the art, “prior-knowledge” binding sites are difficult to collect since Mg2+/Na+/K+ misattributions are observed even in structures with resolutions <2.0 Å. In the current PDB data set, through replication of errors, the recurrence of a given binding site may unfortunately not warrant its reliability and, therefore, it is important to keep redundancy issues in mind. More specifically, present data imply that the assertion that “the coordination of Mg2+ by nucleobases should be considered as a significant factor in the stabilization of RNA structure” (Zheng et al. 2015) must be approached with caution and should not be used to support claims regarding the implication of nucleobase carbonyl groups in catalytic mechanisms (Liu et al. 2017).
This study raises the following interrogation: Why, given the high number of accessible nucleic acid carbonyl groups, do we observe such a small number of Mg2+ binding sites involving nucleobases? We propose that carbonyl groups (as well as hydroxyl groups) are poor Mg2+ binders but excellent monovalent binders. For instance, both quadruplexes and K+ ion channels use the same principles based on monovalent binding to carbonyl groups to fulfill their function (Auffinger et al. 2016).
A more speculative rationale for the poor binding occurrence of Mg2+ to Ob atoms can be proposed. If these quite frequent oxygen atoms would be linked to efficient Mg2+ binding sites, such binding occurrences would be particularly abubdant imposing a high Mg2+ consumption by ribosomes in the cell. Thus, a limited occurrence of Mg2+ binding to nucleobase atoms may be required in order not to impede crucial folding and assembly steps and allowing structural fluidity at critical regions of these molecular machines.
As such, we advocate for a greater awareness of the fact that monovalent ions can easily be mistaken for Mg2+. We strongly believe that strict compliance to well-established stereochemical rules (Fig. 1) may lead to less misidentifications. Indeed, such issues were shown to considerably blur our understanding of nucleic acid ion binding principles Therefore, we must correct our perception of the existing ionic equilibrium around nucleic acids.
Artificial intelligence or machine learning technologies could certainly help to disentangle these difficult ion assignment issues provided that their algorithms are nurtured by sound data (Kowiel et al. 2018). At least, such techniques may help to recognize that current structural databases are far from an error free state and suggest reprocessing some of the underlying experimental data. With current statistics, we might reach counterproductive conclusions regarding the roles of ions in nucleic acids (Zheng et al. 2015). This might significantly impact domains related to the development of molecular dynamics force fields that have to rely on a rigorous interpretation of experimental data for calibration purposes (Panteva et al. 2015; Lemkul and MacKerell 2016; Casalino et al. 2017; Li and Merz 2017) and domains related to the automatic detection and classification of ion binding sites (Brylinski and Skolnick 2011; Lemkul et al. 2016; Casalino et al. 2017; Cunha and Bussi 2017; Sun et al. 2017). For these strategies to be successful, a “prior-knowledge” database of validated Mg2+ to nucleic acid binding modes derived from high-resolution structures is urgently needed.
MATERIALS AND METHODS
All ≈5250 nucleic acid crystal structures deposited to the Protein Data Bank (PDB; February 2017) with resolution ≤2.9 Å were searched for Mg2+ binding to purine and pyrimidine O2/O4/O6 carbonyl oxygen atoms, hereafter named Ob atoms (Zheng et al. 2015). It is well established that Mg2+ has an octahedral coordination sphere with a stringent d(Mg2+…Ow) ≈ 2.06 ± 0.03 Å coordination distance and a second hydration shell around 4.2 Å that is marked by a shallow peak in the CSD distance histogram (see Fig. 1; Markham et al. 2002; Harding et al. 2010). The clearly identifiable gap in the 2.3–3.8 Å range, between the first and second coordination shell peaks, defines an oxygen atom “exclusion zone.”
To account for crystallographic inaccuracies, we used a rather tolerant d(Mg2+…Ob/Ow) ≤ 2.3 Å criterion for our PDB searches. This distance criterion is more stringent than the d(Mg2+…Nb) ≤ 2.4 Å criterion used in an earlier study (Leonarski et al. 2017). The latter cutoff choice was based on the fact that d(Mg2+…Nb) is often assumed to be ≈0.1 Å longer than d(Mg2+…O) (Harding et al. 2010; Leonarski et al. 2016). The present d(Mg2+…Ob/Ow) ≤ 2.3 Å cutoff has the added benefit to allow for a better differentiation of Mg2+ versus Na+ oxygen binding given that d(Mg2+/Na+…O) ≈2.06/2.41 Å (Fig. 1A). For Na+, as for Mg2+, a shallow second coordination peak ≈4.3 Å is observed that is associated with a less marked oxygen “exclusion zone.” Remarkably, Na+ displays in numerous instances a well-defined octahedral coordination shell (Fig. 1B), a fact that is not always fully appreciated (Klein et al. 2004). A pentahydrated coordination for Mg2+ is excessively rare and may not be observed in biological contexts (Chattopadhyay et al. 2009) with the exception of chlorophyll, where Mg2+ coordination requires the assistance of chelatase enzymes (Chen et al. 2015).
To increase the reliability of our structural sample, we downsized our resolution cutoff from 3.0 to 2.9 Å. This 0.1 Å shift resulted in the exclusion of ≈400 (roughly 10% of the total) structures including ≈40 redundant ribosomes (Supplemental Table S1). This is advisable since resolutions ≥3.0 Å are by far not ideal for accurate light ion placement (Z ≤ 12; i.e., Na+ or Mg2+). Other authors selected even more cautious cutoff criteria by stating that, at resolutions >2.5 Å, unambiguous placement of light ions is not reasonable (Harding et al. 2010; Harding and Hsin 2014).
Ions with B-factors ≥79 Å2 were excluded from the statistics since such high B-factors do not warrant unambiguous ion characterization (see Supplemental Material). We further excluded ions with B-factors ≤1.0 Å2 and occupancies ≠ 1.0 unless otherwise specified since the assignment of such ions is not reliable—see a 3.0 Å resolution rRNA structure (PDBid: 1FJG) that displays Mg2+ occupancies in the 0.22–1.47 range (Carter et al. 2000).
As for Mg2+ to N7 binding (Leonarski et al. 2017), we applied a 1.0 Å out-of-nucleobase plane cutoff since Mg2+ tend to bind to the lone pairs of carboxyl oxygen atoms and should therefore lie in the nucleobase plane. On the other hand, monovalent cation binding is not restricted to the nucleobase plane as exemplified by K+ binding to quadruplex structures (Largy et al. 2016). Finally, for all ions for which crystallographic assignment is unclear, 2Fo − Fc and Fo − Fc electron density maps were inspected (Gutmanas et al. 2014; Velankar et al. 2016). For a quick assessment of ion binding stereochemistry, we also used the CheckMyMetal website that permits to rapidly visualize the coordination of all ions present in a PDB structure and suggests meaningful ion replacements but does not allow to observe electron densities and does not read files in the mmCIF format that are associated with the large ribosomal structures (Zheng et al. 2014, 2017).
Nonredundant Mg2+ binding sites were tagged as follows. Two nucleotides from different structures at a comparable Mg2+ binding site and sharing the same residue numbers, chain codes, trinucleotide sequences, ribose puckers, backbone dihedral angle sequences (with the g+, g−, t categorization) and syn/anti conformations, were considered similar and the one with the best resolution was considered nonredundant. In case of matching resolutions, the nucleotide with the lowest B-factor was selected. Similarly, if in a same structure, two nucleotides involved in a comparable Mg2+ binding site and located in different biological units shared the same residue numbers and trinucleotide sequences (with different chain codes) as well as ribose puckers, backbone dihedral angle sequences, and syn/anti conformations, they were considered as similar and the one corresponding to the first biological unit was marked as nonredundant. To further limit redundancy in ribosomal structures, we restricted our analysis to a single biological assembly (see Supplemental Material).
Three nonredundant sets were calculated with 2.3/2.6/3.5 Å cutoffs for d(Mg2+/Na+…Ob) (Table 1). Note that our redundancy criteria were designed for analyzing local structural features and must be distinguished from more global structure-based “nonredundant” criteria (Leontis and Zirbel 2012). Indeed, structures embedding RNA systems with identical sequences are frequent in the PDB. However, these structures often differ by the solvent composition of the buffers in which they were crystallized (Supplemental Table S1). Homemade programs were used to collect data relative to ion binding sites. More precisely, in-house PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC) and Perl scripts were used to download and analyze nucleic acids from the PDB, as well as to extract and categorize information relating to ion binding sites. PyMOL was used to apply symmetry operators and to visualize data. Phenix.refine (Afonine et al. 2012) was used for X-ray refinement, as indicated in the appropriate section.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Professor Eric Westhof for ongoing support and helpful discussions, as well as Dr. Eric Ennifar and Dr. Quentin Vicens for careful reading of the manuscript and for constructive discussions. This work was supported by the French National Research Agency (ANR-15-CE11-0021-01).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.068437.118.
Freely available online through the RNA Open Access option.
REFERENCES
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. 10.1038/nature02642 [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sogair FM, Operschall BP, Sigel A, Sigel H, Schnabl J, Sigel RK. 2011. Probing the metal-ion-binding strength of the hydroxyl group. Chem Rev 111: 4964–5003. 10.1021/cr100415s [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. 10.1038/nature13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M, Schultz EP, Martick M, Scott WG. 2013. Active-site monovalent cations revealed in a 1.55-Å-resolution hammerhead ribozyme structure. J Mol Biol 425: 3790–3798. 10.1016/j.jmb.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 1998. Hydration of RNA base pairs. J Biomol Struct Dyn 16: 693–707. 10.1080/07391102.1998.10508281 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 2000. Water and ion binding around RNA and DNA (C,G) oligomers. J Mol Biol 300: 1113–1131. 10.1006/jmbi.2000.3894 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Bielecki L, Westhof E. 2003. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem Biol 10: 551–561. 10.1016/S1074-5521(03)00121-2 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Bielecki L, Westhof E. 2004a. Anion binding to nucleic acids. Structure 12: 379–388. 10.1016/j.str.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Bielecki L, Westhof E. 2004b. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J Mol Biol 335: 555–571. 10.1016/j.jmb.2003.10.057 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Grover N, Westhof E. 2011. Metal ion binding to RNA. Met Ions Life Sci 9: 1–35. 10.1039/978184973251200001 [DOI] [PubMed] [Google Scholar]
- Auffinger P, D'Ascenzo L, Ennifar E. 2016. Sodium and potassium interactions with nucleic acids. Met Ions Life Sci 16: 167–201. 10.1007/978-3-319-21756-7_6 [DOI] [PubMed] [Google Scholar]
- Basu S, Rambo RP, Strauss-Soukup J, Cate JH, Ferré-D'Amaré AR, Strobel SA, Doudna JA. 1998. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat Struct Biol 5: 986–992. 10.1038/2960 [DOI] [PubMed] [Google Scholar]
- Batey RT, Doudna JA. 2002. Structural and energetics of metal ions essential to SRP signal recognition domain assembly. Biochemistry 41: 11703–11710. 10.1021/bi026163c [DOI] [PubMed] [Google Scholar]
- Bénas P, Auzeil N, Legrand L, Brachet F, Regazzetti A, Riès-Kautt M. 2014. Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry. Acta Crystallogr D Biol Crystallogr 70: 2217–2231. 10.1107/S1399004714011304 [DOI] [PubMed] [Google Scholar]
- Berman HM, Burley SK, Kleywegt GJ, Markley JL, Nakamura H, Velankar S. 2016. The archiving and dissemination of biological structure data. Curr Opin Struct Biol 40: 17–22. 10.1016/j.sbi.2016.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek D, Minor W, Otwinowski Z. 2003. Measurement errors and their consequences in protein crystallography. Acta Crystallogr D Biol Crystallogr 59: 2031–2038. [DOI] [PubMed] [Google Scholar]
- Bowman JC, Lenz TK, Hud NV, Williams LD. 2012. Cations in charge: magnesium ions in RNA folding and catalysis. Curr Opin Struct Biol 22: 262–272. 10.1016/j.sbi.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J. 2011. FINDSITE-metal: integrating evolutionary information and machine learning for structure-based metal-binding site prediction at the proteome level. Proteins 79: 735–751. 10.1002/prot.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. 10.1038/35030019 [DOI] [PubMed] [Google Scholar]
- Casalino L, Palermo G, Abdurakhmonova N, Rothlisberger U, Magistrato A. 2017. Development of site-specific Mg2+-RNA force field parameters: a dream or reality? Guidelines from combined molecular dynamics and quantum mechanics simulations. J Chem Theory Comput 13: 340–352. 10.1021/acs.jctc.6b00905 [DOI] [PubMed] [Google Scholar]
- Cate JH, Hanna RL, Doudna JA. 1997. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol 4: 553–558. 10.1038/nsb0797-553 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay T, Banu KS, Chattopadhyay S, Banerjee A, Mondal S, Suresh E, Das D. 2009. A unique coordination chemistry of sodium. Inorg Chem Commun 12: 26–28. 10.1016/j.inoche.2008.10.015 [DOI] [Google Scholar]
- Chen X, Pu H, Fang Y, Wang X, Zhao S, Lin Y, Zhang M, Dai HE, Gong W, Liu L. 2015. Crystal structure of the catalytic subunit of magnesium chelatase. Nat Plants 1: 15125 10.1038/nplants.2015.125 [DOI] [PubMed] [Google Scholar]
- Conn GL, Gittis AG, Lattman EE, Misra VK, Draper DE. 2002. A compact RNA tertiary structure contains a buried backbone-K+ complex. J Mol Biol 318: 963–973. 10.1016/S0022-2836(02)00147-X [DOI] [PubMed] [Google Scholar]
- Cooper DR, Porebski PJ, Chruszcz M, Minor W. 2011. X-ray crystallography: assessment and validation of protein-small molecule complexes for drug discovery. Expert Opin Drug Discov 6: 771–782. 10.1517/17460441.2011.585154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossy C, Barnes AC, Enderby JE, Merbach AE. 1989. The hydration of Dy3+ and Yb3+ in aqueous solution: a neutron scattering first order difference study. J Chem Phys 90: 3254–3260. 10.1063/1.455878 [DOI] [Google Scholar]
- Costa M, Walbott H, Monachello D, Westhof E, Michel F. 2016. Crystal structures of a group II intron lariat primed for reverse splicing. Science 354: aaf9258 10.1126/science.aaf9258 [DOI] [PubMed] [Google Scholar]
- Cunha RA, Bussi G. 2017. Unraveling Mg2+-RNA binding with atomistic molecular dynamics. RNA 23: 628–638. 10.1261/rna.060079.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ascenzo L, Auffinger P. 2016. Anions in nucleic acid crystallography. Methods Mol Biol 1320: 337–351. [DOI] [PubMed] [Google Scholar]
- Dauter Z, Wlodawer A, Minor W, Jaskolski M, Rupp B. 2014. Avoidable errors in deposited macromolecular structures: an impediment to efficient data mining. IUCrJ 1: 179–193. 10.1107/S2052252514005442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bondt W, Pfiffelmann M, Roger P. 2018. Richard Thaler: the anomalies of life. Finance 39: 9–34. [Google Scholar]
- Draper DE. 2004. A guide to ions and RNA structure. RNA 10: 335–343. 10.1261/rna.5205404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE. 2013. Folding of RNA tertiary structure: linkages between backbone phosphates, ions, and water. Biopolymers 99: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols N, Morshed N, Afonine PV, McCoy AJ, Miller MD, Read RJ, Richardson JS, Terwilliger TC, Adams PD. 2014. Automated identification of elemental ions in macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr D70: 1104–1114. 10.1107/S1399004714001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M, Minasov G, Su L, Rich A. 2002. Metal ions and flexibility in a viral RNA pseudoknot at atomic resolution. Proc Natl Acad Sci 99: 4302–4307. 10.1073/pnas.062055599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar E, Walter P, Dumas P. 2003. A crystallographic study of the binding of 13 metal ions to two related RNA duplexes. Nucleic Acids Res 31: 2671–2682. 10.1093/nar/gkg350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erat MC, Coles J, Finazzo C, Knobloch B, Sigel RKO. 2012. Accurate analysis of Mg2+ binding to RNA: from classical methods to a novel iterative calculation procedure. Coord Chem Rev 256: 279–288. 10.1016/j.ccr.2011.08.009 [DOI] [Google Scholar]
- Fan Y, Gaffney BL, Jones RA. 2005. RNA GG × UU motif binds K+ but not Mg2+. J Am Chem Soc 127: 17588–17589. 10.1021/ja0555522 [DOI] [PubMed] [Google Scholar]
- Freisinger E, Sigel RKO. 2007. From nucleotides to ribozymes—a comparison of their metal ion binding properties. Coord Chem Rev 251: 1834–1851. 10.1016/j.ccr.2007.03.008 [DOI] [Google Scholar]
- Gabdulkhakov A, Nikonov S, Garber M. 2013. Revisiting the Haloarcula marismortui 50S ribosomal subunit model. Acta Crystallogr D Biol Crystallogr D69: 997–1004. 10.1107/S0907444913004745 [DOI] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X. 2008. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol 67: 143–154. 10.1111/j.1365-2958.2007.06032.x [DOI] [PubMed] [Google Scholar]
- Gerasimchuk NN, Dalley NK. 2004. Demetallation of a Ni(II) tetraazamacrocyclic complex by cyanoxime resulting in the formation of a stereospecific trinuclear compound [Na(H2O)6]+[NaNi2L6]− (L = NC-C(NO)-C(O)NH2−). J Coord Chem 57: 1431–1445. 10.1080/00958970412331312652 [DOI] [Google Scholar]
- Giegé R. 2013. A historical perspective on protein crystallization from 1840 to the present day. FEBS J 280: 6456–6497. 10.1111/febs.12580 [DOI] [PubMed] [Google Scholar]
- Glusker JP, Katz AK, Bock CW. 2001. Two-metal binding motifs in protein crystal structures. Struct Chem 12: 323–341. 10.1023/A:1016636712985 [DOI] [Google Scholar]
- Grabowski M, Langner KM, Cymborowski M, Porebski PJ, Sroka P, Zheng H, Cooper DR, Zimmerman MD, Elsliger MA, Burley SK, et al. 2016. A public database of macromolecular diffraction experiments. Acta Crystallogr D Struct Biol 72: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom CR, Allen FH. 2014. The Cambridge Structural Database in retrospect and prospect. Angew Chem Int Ed Engl 53: 662–671. 10.1002/anie.201306438 [DOI] [PubMed] [Google Scholar]
- Groom CR, Bruno IJ, Lightfoot MP, Ward SC. 2016. The Cambridge Structural Database. Acta Crystallogr B Struct Sci Cryst Eng Mater 72: 171–179. 10.1107/S2052520616003954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmanas A, Alhroub Y, Battle GM, Berrisford JM, Bochet E, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Gore SP, et al. 2014. PDBe: Protein Data Bank in Europe. Nucleic Acids Res 42: D285–D291. 10.1093/nar/gkt1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MH. 2001. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr 57: 401–411. [DOI] [PubMed] [Google Scholar]
- Harding MM, Hsin KY. 2014. Mespeus–a database of metal interactions with proteins. Methods Mol Biol 1091: 333–342. 10.1007/978-1-62703-691-7_23 [DOI] [PubMed] [Google Scholar]
- Harding MJ, Nowicki MW, Walkinshaw MD. 2010. Metals in protein structures: a review of their principal features. Cryst Rev 16: 247–302. 10.1080/0889311X.2010.485616 [DOI] [Google Scholar]
- Harp JM, Coates L, Sullivan B, Egli M. 2018. Cryo-neutron crystallographic data collection and preliminary refinement of left-handed Z-DNA d(CGCGCG). Acta Crystallogr F Struct Biol Commun F74: 603–609. 10.1107/S2053230X1801066X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, Auffinger P. 2009. A short guide to molecular dynamics simulations of RNA systems. Methods 47: 187–197. 10.1016/j.ymeth.2008.09.020 [DOI] [PubMed] [Google Scholar]
- Hayatshahi HS, Roe DR, Galindo-Murillo R, Hall KB, Cheatham TE III. 2017. Computational assessment of potassium and magnesium ion binding to a buried pocket in GTPase-associating center RNA. J Phys Chem B 121: 451–462. 10.1021/acs.jpcb.6b08764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendlich M, Bergner A, Günther J, Klebe G. 2003. Relibase: design and development of a database for comprehensive analysis of protein-ligand interactions. J Mol Biol 326: 607–620. 10.1016/S0022-2836(02)01408-0 [DOI] [PubMed] [Google Scholar]
- Hennings E, Schmidt H, Voigt W. 2013. Crystal structures of hydrates of simple inorganic salts. I. Water-rich magnesium halide hydrates MgCl2 8H2O, MgCl2 12H2O, MgBr2 6H2O, MgBr2 9H2O, MgI2 8H2O and MgI2 9H2O. Acta Crystallogr C 69: 1292–1300. 10.1107/S0108270113028138 [DOI] [PubMed] [Google Scholar]
- Hensley MP, Tierney DL, Crowder MW. 2011. Zn(II) binding to Escherichia coli 70S ribosomes. Biochemistry 50: 9937–9939. 10.1021/bi200619w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AK, Fischer FR, Diederich F. 2007. Phosphate recognition in structural biology. Angew Chem Int Ed Engl 46: 338–352. 10.1002/anie.200603420 [DOI] [PubMed] [Google Scholar]
- Hsiao C, Williams LD. 2009. A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Res 37: 3134–3142. 10.1093/nar/gkp119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer AA, Rompel A. 2013. X-ray absorption spectroscopy: a tool to investigate the local structure of metal-based anticancer compounds in vivo. Adv Protein Chem Struct Biol 93: 257–305. 10.1016/B978-0-12-416596-0.00008-7 [DOI] [PubMed] [Google Scholar]
- Jack A, Ladner JE, Rhodes D, Brown RS, Klug A. 1977. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol 111: 315–328. 10.1016/S0022-2836(77)80054-5 [DOI] [PubMed] [Google Scholar]
- Jain S, Richardson DC, Richardson JS. 2015. Computational methods for RNA structure validation and improvement. Methods Enzymol 558: 181–212. 10.1016/bs.mie.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Joosten RP, Womack T, Vriend G, Bricogne G. 2009. Re-refinement from deposited X-ray data can deliver improved models for most PDB entries. Acta Crystallogr D Biol Crystallogr D65: 176–185. 10.1107/S0907444908037591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten RP, Joosten K, Murshudov GN, Perrakis A. 2012. PDB_REDO: constructive validation, more than just looking for errors. Acta Crystallogr D Biol Crystallogr 68: 484–496. 10.1107/S0907444911054515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten RP, Long F, Murshudov GN, Perrakis A. 2014. The PDB_ REDO server for macromolecular structure model optimization. IUCrJ 1: 213–220. 10.1107/S2052252514009324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine L, Djordjevic S, Rhodes D. 2000. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J Mol Biol 301: 401–414. 10.1006/jmbi.2000.3950 [DOI] [PubMed] [Google Scholar]
- Juneau K, Podell E, Harrington DJ, Cech TR. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA-solvent interactions. Structure 9: 221–231. 10.1016/S0969-2126(01)00579-2 [DOI] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Pace NR. 2009. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 15: 266–276. 10.1261/rna.1331809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AR, Kirkhouse JB, McCarney KM, Puissegur O, Smith WE, Staunton E, Teat SJ, Cherryman JC, James R. 2004. Supramolecular motifs in s-block metal-bound sulfonated monoazo dyes, part 1: structural class controlled by cation type and modulated by sulfonate aryl ring position. Chemistry 10: 4606–4615. 10.1002/chem.200400375 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10: 1366–1379. 10.1261/rna.7390804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ. 2009. On vital aid: the why, what and how of validation. Acta Crystallogr D Biol Crystallogr D65: 134–139. 10.1107/S090744490900081X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev SK, Petkov PS, Rangelov MA, Trifonov DV, Milenov TI, Vayssilov GN. 2018. Interaction of Na+, K+, Mg2+ and Ca2+ counter cations with RNA. Metallomics 10: 659–678. 10.1039/C8MT00043C [DOI] [PubMed] [Google Scholar]
- Kowiel M, Brzezinski D, Porebski PJ, Shabalin IG, Jaskolski M, Minor W. 2018. Automatic recognition of ligands in electron density by machine learning. Bioinformatics 10.1093/bioinformatics/bty626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largy E, Mergny JL, Gabelica V. 2016. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. Met Ions Life Sci 16: 203–258. 10.1007/978-3-319-21756-7_7 [DOI] [PubMed] [Google Scholar]
- Lemkul JA, MacKerell AD Jr. 2016. Balancing the interactions of Mg2+ in aqueous solution and with nucleic acid moieties for a polarizable force field based on the classical Drude oscillator model. J Phys Chem B 120: 11436–11448. 10.1021/acs.jpcb.6b09262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemkul JA, Lakkaraju SK, MacKerell AD Jr. 2016. Characterization of Mg2+ distributions around RNA in solution. ACS Omega 1: 680–688. 10.1021/acsomega.6b00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarski F, D'Ascenzo L, Auffinger P. 2016. Binding of metal ions to purine N7 atoms and implications for nucleic acids: a CSD survey. Inorg Chim Acta 452: 82–89. 10.1016/j.ica.2016.04.005 [DOI] [Google Scholar]
- Leonarski F, D'Ascenzo L, Auffinger P. 2017. Mg2+ ions: do they bind to nucleobase nitrogens? Nucleic Acids Res 45: 987–1004. 10.1093/nar/gkw1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonarski F, Redford S, Mozzanica A, Lopez-Cuenca C, Panepucci E, Nass K, Ozerov D, Vera L, Olieric V, Buntschu D, et al. 2018. JUNGFRAU detector: accurate data for macromolecular crystallography. Nat Methods 15: 799–804. 10.1038/s41592-018-0143-7 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Zirbel CL. 2012. Nonredundant 3D structure datasets for RNA knowledge extraction and benchmarking. In RNA 3D structure analysis and prediction (ed. Leontis NB, Westhof E), pp. 281–298. Springer, Berlin/Heidelberg, Germany. [Google Scholar]
- Li PF, Merz KM Jr. 2017. Metal ion modeling using classical mechanics. Chem Rev 117: 1564–1686. 10.1021/acs.chemrev.6b00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DM. 2011. Mechanisms of RNA catalysis. Philos Trans R Soc Lond B Biol Sci 366: 2910–2917. 10.1098/rstb.2011.0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind C, Esguerra M, Åqvist J. 2017. A close-up view of codon selection in eukaryotic initiation. RNA Biol 14: 815–819. 10.1080/15476286.2017.1308998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen Y, Fierke CA. 2017. Inner-sphere coordination of divalent metal ion with nucleobase in catalytic RNA. J Am Chem Soc 139: 17457–17463. 10.1021/jacs.7b08755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D, Persson I, Eriksson L, D'Angelo P, De Panfilis S. 2010. Structural study of the N,N′-dimethylpropyleneurea solvated lanthanoid(III) ions in solution and solid state with an analysis of the ionic radii of lanthanoid(III) ions. Inorg Chem 49: 4420–4432. 10.1021/ic100034q [DOI] [PubMed] [Google Scholar]
- Luo Z, Dauter Z, Gilski M. 2017. Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA. Acta Crystallogr D Struct Biol D73: 940–951. 10.1107/S2059798317014954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. 10.1016/j.cell.2012.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2014. Principles of ion recognition in RNA: insights from the group II intron structures. RNA 20: 516–527. 10.1261/rna.043414.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Y. 1988. Ionic radius in aqueous solutions. Chem Rev 88: 1475–1498. 10.1021/cr00090a003 [DOI] [Google Scholar]
- Marino N, Armentano D, De Munno G. 2016. Cytosine and 1-methylcytosine Mg(II) complexes: structural insights on the reactivity of magnesium(II) toward nucleic acid constituents. Inorg Chim Acta 452: 229–237. 10.1016/j.ica.2016.02.006 [DOI] [Google Scholar]
- Markham GD, Glusker JP, Bock CW. 2002. The arrangement of first and second-sphere water molecules in divalent magnesium complexes: results from molecular orbital and density functional theory and from structural crystallography. J Phys Chem B 106: 5118–5134. 10.1021/jp020078x [DOI] [Google Scholar]
- Minor W, Dauter Z, Helliwell JR, Jaskolski M, Wlodawer A. 2016. Safeguarding structural data repositories against bad apples. Structure 24: 216–220. 10.1016/j.str.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Pryor JM, Ramsden DA, Kunkel TA, Bebenek K, Pedersen LC. 2017. Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res 45: 9138–9148. 10.1093/nar/gkx527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U, Schübel H, Sprinzl M, Heinemann U. 1999. Crystal structure of acceptor stem of tRNAAla from Escherichia coli shows unique G•U wobble base pair at 1.16 Å resolution. RNA 5: 670–677. 10.1017/S1355838299982304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Dieckmann C, Panjikar S, Schmidt A, Mueller S, Kuper J, Geerlof A, Wilmanns M, Singh RK, Tucker PA, Weiss MS. 2007. On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths. Acta Crystallogr D Biol Crystallogr D63: 366–380. [DOI] [PubMed] [Google Scholar]
- Murphy FV IV, Ramakrishnan V. 2004. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11: 1251–1252. 10.1038/nsmb866 [DOI] [PubMed] [Google Scholar]
- Niedzialkowska E, Gasiorowska O, Handing KB, Majorek KA, Porebski PJ, Shabalin IG, Zasadzinska E, Cymborowski M, Minor W. 2016. Protein purification and crystallization artifacts: the tale usually not told. Protein Sci 25: 720–733. 10.1002/pro.2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH. 2014. Mg2+, K+, and the ribosome. J Bacteriol 196: 3817–3819. 10.1128/JB.02297-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. 10.1126/science.289.5481.920 [DOI] [PubMed] [Google Scholar]
- Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JH. 2015. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol 22: 336–341. 10.1038/nsmb.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olieric V, Weinert T, Finke AD, Anders C, Li D, Olieric N, Borca CN, Steinmetz MO, Caffrey M, Jinek M, et al. 2016. Data-collection strategy for challenging native SAD phasing. Acta Crystallogr D Struct Biol 72: 421–429. 10.1107/S2059798315024110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MJ, Di Cera E. 2006. Role of Na+ and K+ in enzyme function. Physiol Rev 86: 1049–1092. 10.1152/physrev.00008.2006 [DOI] [PubMed] [Google Scholar]
- Pallan PS, Marshall WS, Harp J, Jewett FC, Wawrzak Z, Brown BA II, Rich A, Egli M. 2005. Crystal structure of a luteoviral RNA pseudoknot and model for a minimal ribosomal frameshifting motif. Biochemistry 44: 11315–11322. 10.1021/bi051061i [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Roland C, Sagui C. 2014. Ion distributions around left- and right-handed DNA and RNA duplexes: a comparative study. Nucleic Acids Res 42: 13981–13996. 10.1093/nar/gku1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteva MT, Giambasu GM, York DM. 2015. Force field for Mg2+, Mn2+, Zn2+, and Cd2+ ions that have balanced interactions with nucleic acids. J Phys Chem B 119: 15460–15470. 10.1021/acs.jpcb.5b10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bowman JC, Harvey SC, Williams LD. 2011. Bidentate RNA-magnesium clamps: on the origin of the special role of magnesium in RNA folding. RNA 17: 291–297. 10.1261/rna.2390311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Okafor CD, Tannenbaum E, Stern J, Gaucher E, Schneider D, Hud NV, Harvey SC, et al. 2012. RNA-magnesium-protein interactions in large ribosomal subunit. J Phys Chem B 116: 8113–8120. 10.1021/jp304723w [DOI] [PubMed] [Google Scholar]
- Pflugrath JW, Quiocho FA. 1988. The 2 Å resolution structure of the sulfate-binding protein involved in active transport in Salmonella typhimurium. J Mol Biol 200: 163–180. 10.1016/0022-2836(88)90341-5 [DOI] [PubMed] [Google Scholar]
- Pozharski E, Weichenberger CX, Rupp B. 2013. Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures. Acta Crystallogr D Biol Crystallogr D69: 150–167. 10.1107/S0907444912044423 [DOI] [PubMed] [Google Scholar]
- Quigley GJ, Teeter MM, Rich A. 1978. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci 75: 64–68. 10.1073/pnas.75.1.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska JE, Wlodawer A, Jaskolski M. 2016. Prior knowledge or freedom of interpretation? A critical look at a recently published classification of “novel” Zn binding sites. Proteins 84: 770–776. 10.1002/prot.25024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS, Williams CJ, Hintze BJ, Chen VB, Prisant MG, Videau LL, Richardson DC. 2018. Model validation: local diagnosis, correction and when to quit. Acta Crystallogr D Struct Biol D74: 132–142. 10.1107/S2059798317009834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. 2014. Crystal structure of a eukaryotic group II intron lariat. Nature 514: 193–197. 10.1038/nature13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2015. Structural insights into the translational infidelity mechanism. Nat Commun 6: 7251 10.1038/ncomms8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. 2016a. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun 7: 10457 10.1038/ncomms10457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2016b. New structural insights into translational miscoding. Trends Biochem Sci 41: 798–814. 10.1016/j.tibs.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Rupp B. 2016. Only seeing is believing – the power of evidence and reason. Postepy Biochem 62: 250–256. [PubMed] [Google Scholar]
- Rupp B, Wlodawer A, Minor W, Helliwell JR, Jaskolski M. 2016. Correcting the record of structural publications requires joint effort of the community and journal editors. FEBS J 283: 4452–4457. 10.1111/febs.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. 2005. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell 20: 437–448. 10.1016/j.molcel.2005.09.006 [DOI] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. 2008. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1268. 10.1038/nature07326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel H, Kapinos LE. 2000. Quantification of isomeric equilibria for metal ion complexes formed in solution by phosphate or phosphonate ligands with a weakly coordinating second site. Coord Chem Rev 200: 563–594. [Google Scholar]
- Sigel RK, Sigel H. 2010. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc Chem Res 43: 974–984. 10.1021/ar900197y [DOI] [PubMed] [Google Scholar]
- Sigel RKO, Sigel H. 2013. Metal-ion interactions with nucleic acids and their constituents. In Comprehensive inorganic chemistry II (ed. Reedjik J, Poeppelmeier K), Vol. 3, pp. 623–660. Elsevier, Oxford. [Google Scholar]
- Spek AL. 2009. Structure validation in chemical crystallography. Acta Cryst D65: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šponer J, Bussi G, Krepl M, Banáš P, Bottaro S, Cunha RA, Gil-Ley A, Pinamonti G, Poblete S, Jurečka P, et al. 2018. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem Rev 118: 4177–4338. 10.1021/acs.chemrev.7b00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley MR, Strobel SA. 2005. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science 309: 1587–1590. 10.1126/science.1114994 [DOI] [PubMed] [Google Scholar]
- Stahley MR, Adams PL, Wang J, Strobel SA. 2007. Structural metals in the group I intron: a ribozyme with a multiple metal ion core. J Mol Biol 372: 89–102. 10.1016/j.jmb.2007.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. 2004. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr 60: 432–438. [DOI] [PubMed] [Google Scholar]
- Sun LZ, Zhang D, Chen SJ. 2017. Theory and modeling of RNA structure and interactions with metal ions and small molecules. Annu Rev Biophys 46: 227–246. 10.1146/annurev-biophys-070816-033920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tereshko V, Wilds CJ, Minasov G, Prakash TP, Maier MA, Howard A, Wawrzak Z, Manoharan M, Egli M. 2001. Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments. Nucleic Acids Res 29: 1208–1215. 10.1093/nar/29.5.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler RH. 2000. From Homo economicus to Homo sapiens. JEP 17: 191–202. [Google Scholar]
- Thorn A, Sheldrick GM. 2011. ANODE: anomalous and heavy-atom density calculation. J Appl Crystallogr 44: 1285–1287. 10.1107/S0021889811041768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuéry P. 2009. Lanthanide complexes with cucurbit[n]urils (n=5, 6, 7) and perrhenate ligands: new examples of encapsulation of perrhenate anions. Inorg Chem 48: 4497–4513. 10.1021/ic900328z [DOI] [PubMed] [Google Scholar]
- Timsit Y, Bombard S. 2007. The 1.3 Å resolution structure of the RNA tridecamer r(GCGUUUGAAACGC): metal ion binding correlates with base unstacking and groove contraction. RNA 13: 2098–2107. 10.1261/rna.730207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. 10.1126/science.1153803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beusekom B, Perrakis A, Joosten RP. 2016. Data mining of macromolecular structures. Methods Mol Biol 1415: 107–138. 10.1007/978-1-4939-3572-7_6 [DOI] [PubMed] [Google Scholar]
- van Beusekom B, Touw WG, Tatineni M, Somani S, Rajagopal G, Luo J, Gilliland GL, Perrakis A, Joosten RP. 2018. Homology-based hydrogen bond information improves crystallographic structures in the PDB. Protein Sci 27: 798–808. 10.1002/pro.3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar S, van Ginkel G, Alhroub Y, Battle GM, Berrisford JM, Conroy MJ, Dana JM, Gore SP, Gutmanas A, Haslam P, et al. 2016. PDBe: improved accessibility of macromolecular structure data from PDB and EMDB. Nucleic Acids Res 44: D385–D395. 10.1093/nar/gkv1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Duman R, Henderson K, Mykhaylyk V. 2016. In-vacuum long-wavelength macromolecular crystallography. Acta Crystallogr D Struct Biol D72: 430–439. 10.1107/S2059798316001078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. 2010. Inclusion of weak high-resolution X-ray data for improvement of a group II intron structure. Acta Crystallogr D Biol Crystallogr 66: 988–1000. 10.1107/S0907444910029938 [DOI] [PubMed] [Google Scholar]
- Wang R, Luo Z, He K, Delaney MO, Chen D, Sheng J. 2016. Base pairing and structural insights into the 5-formylcytosine in RNA duplex. Nucleic Acids Res 44: 4968–4977. 10.1093/nar/gkw235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WL, Plakos K, DeRose VJ. 2014. Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem Rev 114: 4318–4342. 10.1021/cr400476k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenberger CX, Afonine PV, Kantardjieff K, Rupp B. 2015. The solvent component of macromolecular crystals. Acta Crystallogr D Biol Crystallogr 71: 1023–1038. 10.1107/S1399004715006045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T, Olieric V, Waltersperger S, Panepucci E, Chen L, Zhang H, Zhou D, Rose J, Ebihara A, Kuramitsu S, et al. 2015. Fast native-SAD phasing for routine macromolecular structure determination. Nat Methods 12: 131–133. 10.1038/nmeth.3211 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Murphy FV IV, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. 2007. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol 14: 498–502. 10.1038/nsmb1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LD. 2005. Between objectivity and whim: nucleic acid structural biology. Top Curr Chem 253: 77–88. 10.1007/b100443 [DOI] [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M. 2008. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J 275: 1–21. 10.1111/j.1742-4658.2007.06178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M. 2013. Protein crystallography for aspiring crystallographers or how to avoid pitfalls and traps in macromolecular structure determination. FEBS J 280: 5705–5736. 10.1111/febs.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Dauter Z, Porebski PJ, Minor W, Stanfield R, Jaskolski M, Pozharski E, Weichenberger CX, Rupp B. 2018. Detect, correct, retract: how to manage incorrect structural models. FEBS J 285: 444–466. 10.1111/febs.14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol 9: 104–109. 10.1016/j.cbpa.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. 2008. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci 105: 82–87. 10.1073/pnas.0709082105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chruszcz M, Lasota P, Lebioda L, Minor W. 2008. Data mining of metal ion environments present in protein structures. J Inorg Biochem 102: 1765–1776. 10.1016/j.jinorgbio.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chordia MD, Cooper DR, Chruszcz M, Müller P, Sheldrick GM, Minor W. 2014. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc 9: 156–170. 10.1038/nprot.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Shabalin IG, Handing KB, Bujnicki JM, Minor W. 2015. Magnesium-binding architectures in RNA crystal structures: validation, binding preferences, classification and motif detection. Nucleic Acids Res 43: 3789–3801. 10.1093/nar/gkv225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Cooper DR, Porebski PJ, Shabalin IG, Handing KB, Minor W. 2017. CheckMyMetal: a macromolecular metal-binding validation tool. Acta Crystallogr D Struct Biol 73: 223–233. 10.1107/S2059798317001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Saran R, Liu J. 2017. Metal sensing by DNA. Chem Rev 117: 8272–8325. 10.1021/acs.chemrev.7b00063 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.