Abstract
Purpose
To explore top-ranked plasma proteins related to neovascular age-related macular degeneration (AMD) and geographic atrophy (GA), and explore pathways related to neovascular AMD and GA.
Methods
We conducted a pilot study of patients with neovascular AMD (n = 10), GA (n = 10), and age-matched cataract controls (n = 10) who were recruited into an AMD registry. We measured 4001 proteins in ethylenediaminetetraacetic acid plasma samples using an aptamer-based proteomic technology. Relative concentrations of each of 4001 proteins were log (base 2) transformed and compared between cases of neovascular AMD and GA versus controls using linear regression. Pathway analysis was conducted using pathways downloaded from Reactome.
Results
In this pilot study, higher levels of vinculin and lower levels of CD177 were found in patients with neovascular AMD compared with controls. Neuregulin-4 was higher and soluble intercellular adhesion molecule-1 was lower in patients with GA compared with controls. For neovascular AMD, cargo trafficking to the periciliary membrane, fibroblast growth factor receptor 3b ligand binding and activation, and vascular endothelial growth factor–related pathways were in the top ranked pathways. The top-ranked pathways for GA included several related to ErbB4 signaling.
Conclusions
We found different proteins and different pathways associated with neovascular AMD and GA. Vinculin and some of the top-ranked pathways have been previously associated with AMD, whereas others have not been described.
Translational Relevance
Biomarkers identified in plasma likely reflect systemic alterations in protein expression and may improve our understanding of the mechanisms leading to AMD.
Keywords: proteomics, geographic atrophy, neovascular AMD, aptamer-based technologies
Introduction
Age-related macular degeneration (AMD) is a progressive degenerative disease of the macula found in patients who are generally over 60 years of age. AMD accounts for 8.7% of blindness worldwide.1 This devastating, potentially blinding disease is characterized by the deposition of extracellular drusen,2,3 which are found between the basal surface of the retinal pigment epithelium (RPE) and Bruch's membrane. AMD is distinguished by two advanced forms of the disease, neovascular AMD and geographic atrophy (GA).2 Neovascular AMD and GA are characterized by choroidal neovascularization and atrophy of the RPE with overlying photoreceptor cell atrophy, respectively.4 Targeted treatment with intravitreal injections of drugs that suppress vascular endothelial growth factor (VEGF) are used in the treatment of neovascular AMD. However, the response to treatment is unpredictable. There is currently no effective treatment for GA.5
The public health impact of AMD in the aging population is, therefore, important. There is currently a great need for the discovery of biomarkers related to the prediction, cause, diagnosis, progression, regression, and/or outcome of treatment of AMD.6,7 However, apart from the finding of AMD-related complement polymorphisms and several nongenetic-related biomarkers,7 discovering new biomarkers that will lead to a personalized form of care for patients with AMD has not been achieved.
We have used a novel aptamer-based proteomic technology to investigate the role of circulating plasma proteins in select eye8 and noneye-related diseases.9 In the pilot study described herein, we expand our research to investigate the role of circulating proteins in patients with neovascular AMD, GA, and age-matched cataract controls. The purpose of our research was to use this technology to (1) explore top-ranked proteins related to neovascular AMD and GA, and (2) explore pathways related to neovascular AMD and GA. To address these objectives, we conducted a pilot study of patients with the advanced forms of AMD and cataract controls who were recruited into an AMD registry at an academic center in Aurora, Colorado.
Methods
This pilot study was approved by the Colorado Multiple institutional review board. The research followed the tenets of the Declaration of Helsinki. We conducted the study using plasma samples from patients with AMD and cataract controls with no AMD recruited into an AMD registry developed by the Department of Ophthalmology at the University of Colorado School of Medicine. The purpose of the registry was to develop a state-of-the-art clinical database linked with image data and biological samples of patients with AMD and control patients with age-related cataract without AMD. Enrollment into the registry started in July 2014 and is ongoing. At the time of conducting this pilot study (July 2017), 456 AMD patients and 180 cataract controls with no AMD were enrolled in the registry. The methods for recruiting into this registry are explained in detail elsewhere.10 We excluded patients who were terminally ill. Ocular exclusion criteria for cases and controls were as follows: panretinal photocoagulation or anti-VEGF injections for diabetic retinopathy, branch and central retinal vein occlusion, any active ocular inflammatory disease, or a severe decrease in visual acuity secondary to a preexisting severe retinal disease other than AMD. We also excluded controls with dense cataract that precluded optical imaging of the retina.
In brief, AMD patients attending the retina clinics at the Sue Anschutz-Rodgers University of Colorado Health Eye Center were referred by a health care provider to our research team for explanation of the purpose of the registry and to obtain consent. Informed consent was obtained from the subjects after explanation of the nature and possible consequences of the study. Each patient was consented for (1) review of the medical history, (2) collection of an ethylenediaminetetraacetic acid (EDTA) plasma sample, and (3) review of image data to include color fundus photography, fundus autofluorescence (FAF), optical coherence tomography (OCT), near infrared reflectance (NIR), and fluorescein angiography, if clinically indicated. The images were reviewed by two authors (MTM and FSS) with discrepancies resolved by a third (NM).
Cases and Controls
For this pilot study, we interrogated the plasma proteome of samples collected from the first 10 patients with bilateral neovascular AMD and bilateral GA recruited into the registry. We age-matched the cases of advanced AMD to 10 cataract controls without AMD.2 Neovascular AMD was defined as the presence of choroidal neovascularization based on OCT. GA was defined as circumscribed areas of atrophy, which reflect cell death in the RPE, outer retina, and choriocapillaris in patients with AMD, but no other retinal disease. The diagnosis of neovascular AMD and GA was confirmed using multimodal imaging (FAF, NIR, and OCT).
Collection and Processing of the Plasma Sample
Following phlebotomy, the EDTA tube was spun at 3000 revolutions per minute in a cooled centrifuge (4°C) for 10 minutes to isolate plasma. The plasma was transferred into another tube for a similar second spin. This second supernatant was aliquoted into cryovials of 250 to 500 μL each depending on the sample volume available. All samples were stored at −80°C. Proteomic analysis was conducted on an aliquot using the SOMAscan assay at the laboratories of SomaLogic, Inc. (Boulder, CO).
SomaLogic Proteomic Technology and Analysis
The SOMAscan proteomic assay is described in detail elsewhere.9,11,12 In brief, a plasma sample in each well of a 96-well plate was incubated with a mixture of the 4001 SOMAmer reagents. Two sequential bead-based immobilization and washing steps, coupled with kinetic challenge with polyanionic competitors, eliminated unbound or nonspecifically bound proteins and the unbound SOMAmer reagents, leaving only protein target-bound SOMAmer reagents. These remaining SOMAmer reagents were isolated, and each reagent was quantified simultaneously on a custom Agilent (Santa Clara, CA) hybridization array. The amount of each SOMAmer measured was quantitatively proportional to the protein concentration in the original sample as described elsewhere.9,11,12
For the current version of the SOMAscan assay, the coefficients of variation (CV) for all analytes, including intra- and interassay variation, were measured in plasma using triplicate technical replicates for three different clinical samples across 15 independent runs. The median CV for all analyte measurements in plasma is 5.0% and 90% of analytes have CVs less than 12.0%, obtained by averaging CV results over the three clinical samples. In an independent study, the SOMAscan assay with plasma samples has recently demonstrated a high degree of stability and reproducibility.13
Statistical Analysis
Relative concentrations for each of 4001 proteins were log (base 2) transformed and compared between cases of neovascular AMD versus controls and between cases of GA versus controls using linear regression. We focused on the top-ranked proteins defined as those with a P value less than 0.001, corresponding to a conservative Bonferroni adjustment for multiple comparisons,14 yet still providing some consideration of type II errors given this is a small pilot study (see online Supplementary File S1).
Pathway analysis was conducted using pathways downloaded from Reactome15,16 and a functional class scoring approach, appropriate for platforms where proteins are selected a priori,17 using the P values as the protein-level statistics for the 4001 proteins that were measured.18 This functional class scoring approach differs from an enrichment analysis in that it does not specifically test whether the pathways are enriched with a larger than expected number of significant proteins and therefore also does not require a cut-off to be applied to each protein. The underlying inference from the functional class scoring approach is testing whether the pathway contains at least one measured protein that significantly differed between groups or whether a subset of proteins in the pathway have coordinated differences. Pathways were ranked based on the unadjusted P values calculated using a permutation approach that appropriately accounts for the correlation between proteins, permuting group labels using 1000 permutations.19
Results
We show in Table 1 the characteristics of the patients with neovascular AMD, GA, and cataract controls. There was no difference in any of the variables across the three groups. None of the patients had a history of ocular trauma or stroke. All 10 neovascular AMD patients had treatment in both eyes, one ranibizumab, six bevacizumab, and three aflibercept. All patients in the neovascular AMD group received their research blood draw before their anti-VEGF injection.
Table 1.
Characteristics of Patients With Neovascular AMD, GA, and Cataract Controls
| Status |
Neovascular AMD |
GA |
Cataract Controls |
P Value |
| Total | 10 | 10 | 10 | |
| Age, yr, mean (SD) | 80.5 (6.4) | 82.3 (5.4) | 78.8 (6.0) | 0.43 |
| Caucasian race | 10 (100%) | 10 (100%) | 9 (90%) | 0.99 |
| Female Sex | 6 (60%) | 3 (30%) | 5 (50%) | 0.53 |
| Former smoker | 5 (63%) | 8 (80%) | 4 (40%) | 0.39 |
| Body mass index mean (SD) | 26.2 (5.1) N = 9 | 26.6 (2.5) N = 6 | 26.8 (5.9) N = 9 | 0.97 |
| Duration of AMD, yr | ||||
| 2–5 | 0 | 2 (20%) | 0.55 | |
| 5–10 | 5 (56%) | 4 (40%) | ||
| 10+ | 4 (44%) N = 9 | 4 (40%) | ||
| History of treated hypertension | 8 (80%) | 6 (60%) | 7 (70%) | 0.88 |
| History of kidney disease | 2 (20%) | 1 (10%) | 1 (10%) | 0.99 |
| History of cardiac disease | 4 (40%) | 8 (80%) | 7 (70%) | 0.25 |
| Treatment for ocular hypertension | 2 (20%) | 2 (20%) | 0 | 0.51 |
| Status of the lens | ||||
| Cataract | 4 (40%) | 2 (20%) | 0 | 0.14 |
| Pseudophakic | 5 (50%) | 8 (80%) | 8 (80%) | |
| Pseudophakic/cataract | 1 (10%) | 0 | 2 (20%) | |
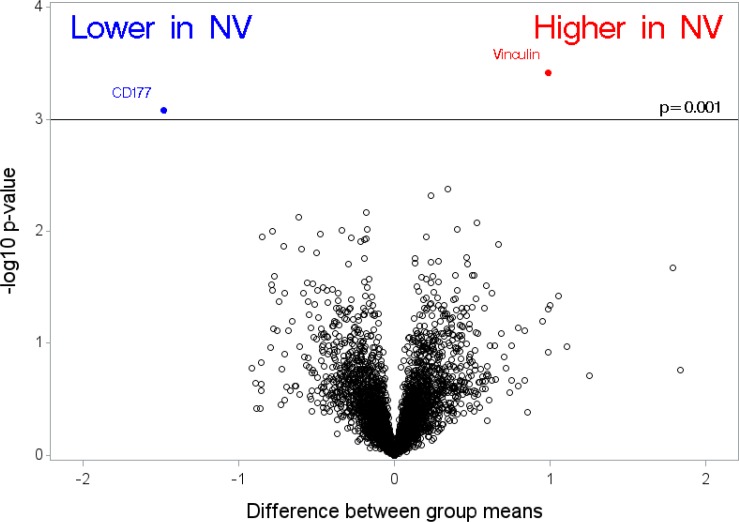
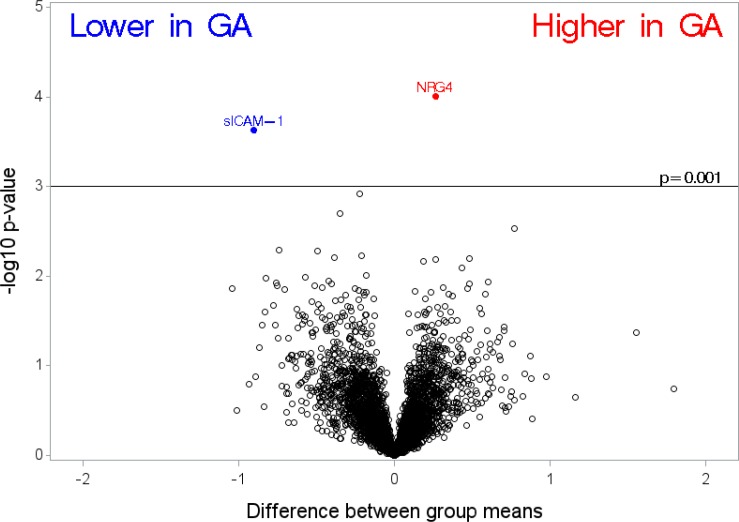
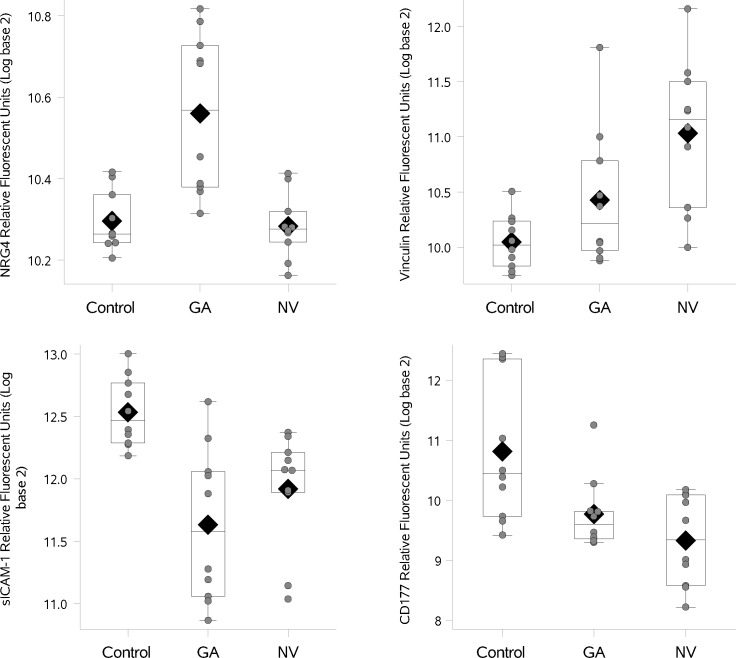
In Figure 1, we compare differences in group means between cases of neovascular AMD compared with cataract controls (where a difference of 1 on log [base 2] scale corresponds to a fold change of 2). Using the unadjusted P value less than 0.001 as a cut-off for identifying proteins in this pilot study, vinculin was higher in patients with neovascular AMD compared with controls. Another protein, CD177 (also known as CD177 antigen), was lower in cases of neovascular AMD compared with controls based on our cutoff criteria. In Figure 2, we show differences in the mean levels of top-ranked proteins in cases of GA compared with cataract controls. We found only one protein, neuroregulin 4 (NRG4), higher in cases of GA compared with controls. We found one protein, soluble intercellular adhesion molecule-1 (sICAM-1), to be lower in patients with GA compared with controls. We also examined the relationship between cases with neovascular AMD compared with cases with GA. Only one protein NRG4 (higher in GA compared with neovascular AMD) was identified based on our cutoff (estimate −0.28, standard error = 0.06, P < 0.001). Two supplemental tables of all proteins examined are appended (Supplementary File S2 and Supplementary File S3). In Figure 3 (box plots) we show the relative levels of the four top ranked proteins in patients with GA, neovascular AMD, and controls. We display (1) a difference in relative levels of NRG4 and sICAM-1 between GA cases and the control and neovascular groups, and (2) a gradient across the groups for vinculin and CD177. For the four analyte measurements reported here, the CVs for technical replicates are 6.2% for CD177, 3.8% for NRG4, 5.2% for sICAM-1, and 3.0% for vinculin.
Figure 1.
Differences between group means in cases of neovascular (NV) AMD compared with cataract controls, where a difference of 1 on log2 scale corresponds to a fold change of two.
Figure 2.
Differences between group means in cases of geographic atrophy (GA) compared with cataract controls, where a difference of 1 on log2 scale corresponds to a fold change of two.
Figure 3.
Distribution of select proteins in patients with GA, NV AMD, and cataract controls. The box indicates the interquartile range (IQR; 25th–75th percentile) and the median and mean are indicated by the lines and diamonds, respectively. Whiskers indicate data within 1.5 times the IQR, and points indicate individual data values.
The pathway analyses detect coordinated changes in levels of proteins in the same pathway and are useful to provide mechanistic insights by evaluating the combined association of groups of proteins that function in the same pathway. The top 15 pathways from the pathway analyses for neovascular AMD and GA are shown in Tables 2 and 3, respectively. For neovascular AMD, (Table 2) cargo trafficking to the periciliary membrane, fibroblast growth factor receptor (FGFR) 3b ligand binding and activation, and VEGF-related pathways were in the top-ranked pathways. As demonstrated, in Table 3, the top-ranked pathways for GA included several related to ErbB4 signaling. More details on the pathways are found in Supplementary File S4 and Supplementary File S5.
Table 2.
Top-Ranked Pathwaysa in Cases of Neovascular AMD Compared With Cataract Controls
| Rank |
Proteinsb |
Pathway |
P Value |
| 1 | 9 | Cargo trafficking to the periciliary membrane | 0.02 |
| 2 | 10 | FGFR3b ligand binding and activation | 0.03 |
| 3 | 11 | VEGF binds to VEGFR leading to receptor dimerization/VEGF ligand-receptor interactions | 0.04 |
| 4 | 21 | Common pathway of fibrin clot formation | 0.05 |
| 5 | 9 | Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding | 0.08 |
| 6 | 12 | Downregulation of TGF-B receptor signaling | 0.09 |
| 7 | 12 | TFAP2 (AP-2) family regulates transcription of growth factors and their receptors | 0.10 |
| 8 | 14 | Formation of a pool of free 40S subunits | 0.10 |
| 9 | 11 | Transport of bile salts and organic acids, metal ions and amine compounds | 0.10 |
| 10 | 13 | Gamma-carboxylation of protein precursors/transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus | 0.11 |
| 11 | 14 | Dual incision in TC-NER | 0.11 |
| 12 | 14 | Gamma-carboxylation, transport, and amino-terminal cleavage of proteins/Removal of aminoterminal propeptides from gamma-carboxylated proteins | 0.13 |
| 13 | 16 | L13a-mediated translational silencing of Ceruloplasmin expression | 0.13 |
| 14 | 12 | Formation of the ternary complex, and subsequently, the 43S complex | 0.13 |
| 15 | 17 | GTP hydrolysis and joining of the 60S ribosomal subunit | 0.13 |
Limited to the top 15 ranked pathways.
Refers to the number of aptamers included in the pathway that were measured as part of the SOMAscan.
Table 3.
Top Ranked Pathwaysa in Cases of GA Compared With Cataract Controls
| Rank |
Proteinsb |
Pathway |
P Value |
| 1 | 13 | SHC1 events in ERBB4 signaling | <0.01 |
| 2 | 9 | PI3K events in ERBB4 signaling | 0.01 |
| 3 | 20 | SHC1 events in ERBB2 signaling | 0.02 |
| 4 | 16 | GRB2 events in ERBB2 signaling | 0.03 |
| 5 | 13 | Nuclear signaling by ERBB4 | 0.03 |
| 6 | 8 | NADE modulates death signaling | 0.03 |
| 7 | 15 | PI3K events in ERBB2 signaling | 0.03 |
| 8 | 14 | Signaling by BMP | 0.04 |
| 9 | 20 | Interleukin receptor SHC signaling | 0.04 |
| 10 | 12 | Regulation of beta-cell development | 0.04 |
| 11 | 8 | Regulation of commissural axon pathfinding by SLIT and ROBO | 0.04 |
| 12 | 10 | Reversible hydration of carbon dioxide | 0.04 |
| 13 | 15 | A tetrasaccharide linker sequence is required for GAG synthesis | 0.05 |
| 14 | 9 | Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding | 0.05 |
| 15 | 26 | Signaling by ERBB4 | 0.05 |
Limited to the top 15 ranked pathways.
Refers to the number of aptamers included in the pathway that were measured as part of the SOMAscan.
Discussion
We used an aptamer-based proteomic technology to measure 4001 proteins across many protein pathways to determine the top-ranked proteins in cases of neovascular AMD and GA compared with cataract controls. There are two key findings from this exploratory pilot study. First, the top-ranked proteins associated with advanced AMD were vinculin (neovascular AMD), CD177 (neovascular AMD), NRG4 (GA), and sICAM-1 (GA). Second, the top-ranked pathways in neovascular AMD and GA were different. For neovascular AMD we found cargo trafficking to the periciliary membrane, FGFR3b ligand binding and activation, and VEGF related pathways were important. ErbB signaling distinguished pathways related to GA.
Vinculin was elevated in patients with neovascular AMD compared with controls. Vinculin, originally described in 1979, has a key role in cell–matrix and cell–cell adhesion to the actin-based cytoskeleton.20 This adapter protein is also distinguished as a regulator of apoptosis and of other biological functions, such as cell growth, migration, differentiation, and survival. Vinculin also has an important role in embryogenesis and in cardiac function (reviewed in Ref. 21). In agreement with the results of our study, Kim et al.22 also described a role for vinculin as a potential plasma marker for neovascular AMD. Using mass spectrometry, these investigators examined plasma proteomes from 20 neovascular AMD patients and 20 healthy controls. The area under the curve for vinculin was 0.87 for discriminating between AMD cases and controls. They also found that vinculin was elevated in expression in RPE cells of cadaver human cells when these cells were exposed to oxidative stress. The authors suggest that dysfunction in levels of vinculin may reflect RPE damage or degeneration.22
CD177, a neutrophil cell surface receptor was lower in cases of neovascular AMD compared with controls. This protein has been identified as a target antigen in immune-mediated disorders and plays an important role in neutrophil viability.23 CD177 is associated with immune-mediated neutropenia of the newborn,24 Granulomatosis with Polyangiitis,25 (formerly Wegener's) and is upregulated during bacterial infections.26 It has been shown to play a role in neutrophil–endothelial cell interactions, and inflammatory cell recruitment. Through an interaction with PECAM-1 (cell adhesion molecule), it mediates transmigration of a subpopulation of neutrophils into tissue in response to infection.23,25,27 The lower levels of this protein observed in our study may contribute to dysregulation of inflammatory pathways and an impaired inflammatory response in patients with the advanced form of AMD.
We found NRG4, a selective ligand for the ErbB4 receptor tyrosine kinase, to be higher in patients with GA compared with controls. NRG4 is a brown fat-enriched protein28 that has been shown to be downregulated in obese mice and humans. In the liver, NRG4 reduces hepatic lipogenic signaling by activating the ErbB3 and ErbB4 receptors.28 The end result is a decrease in de novo lipogenesis and hepatic steatosis with an increase in insulin sensitivity. Impaired NRG4 signaling may contribute to the development of metabolic disorders, such as type 2 diabetes and nonalcoholic fatty liver disease.28,29 Investigators have also demonstrated that circulating NRG4 levels are inversely associated with subclinical atherosclerosis in obese adults.30 The elevated levels of NRG4 found in this pilot study will need additional study in a larger cohort of patients with GA.
We found lower levels of sICAM-1 in patients with GA compared with controls. sICAM-1 is an intercellular adhesion molecule that facilitates adhesion and transmigration of leucocytes on the vascular endothelial wall to sites of inflammation, an integral part of the inflammatory response.31,32 As discussed by Rothlein and Wegner,31 without these adhesion molecules the inflammatory response would be impaired. Anti–ICAM-1 in various animal models has been shown to attenuate the inflammatory response mediated by granulocytes and lymphocytes.31 The altered levels of this protein found in our small pilot study suggest a compromised response to an inflammatory event in patients with GA.
The top-ranked pathway (the pathway analysis is outlined in detail in the statistics section) in neovascular AMD was cargo trafficking to the periciliary membrane. The primary cilium is a dynamic hair-like membrane projecting from almost every cell type.33 Vesicle trafficking plays an important role in the cilia. This process is important in the retina where trafficking of rhodopsin occurs in the cilia of photoreceptors cells.33,34 Defects in the primary cilium function cause ciliopathies. These disorders are linked with retinal degeneration (reviewed in detail by May-Simera et al.35). Although the pathology of AMD has been isolated to the RPE and choriocapillaris, the role of defective cilia cannot be ruled out as a contributory factor in the pathophysiology of AMD.35 We suggest that this finding should be explored further in a larger AMD cohort. Another top-ranked pathway was FGFR3b ligand binding and activation. This pathway is involved in epithelial cell growth and differentiation.36
Our study is also distinguished by the finding in our pathway analysis of enhanced VEGF signaling in the peripheral blood in patients with neovascular AMD. In the context of this information, it is important to note that all the neovascular AMD patients in this study had their research blood draw conducted before their anti-VEGF injection. To the best of our knowledge links between VEGF signaling pathways in blood and neovascular AMD have not been previously reported. In contrast, it is widely accepted that upregulation of VEGF leads to choroidal neovascularization in the retina in patients with neovascular AMD. Indeed, targeting VEGF signaling with intravitreal injections of anti-VEGF drugs is one of the most effective therapeutic strategies for treating neovascular AMD (reviewed in Refs. 37 and 38).
The ErbB family of transmembrane receptor tyrosine kinases proteins were also important in the pathway analysis in GA. ErbB receptors are expressed in various tissues of epithelial, mesenchymal, and neuronal origin, in which they are involved in the control of diverse biological processes, such as proliferation, differentiation, migration, and apoptosis.39 As discussed above, one of our top signal proteins, NRG4 has links with ErbB3 and ErbB4 receptors.28 ErbB family members have a role in several solid tumors.40 Indeed, targeting ErbB2 (HER2), which is amplified in 20% of breast cancers, results in improved survival for many patients.40 It is also noteworthy that Romano et al.41 recently described a link between select microRNAs with the ErbB signaling pathway in patients with AMD.
The small sample size was a limitation of our study. The SOMAscan we used measures 4001 proteins of the roughly 25,000 known human proteins. Because the focus was on the measured proteins in the pathway (see Supplementary File S1), rather than the pathways themselves containing an overrepresentation of significant proteins, the fact that we are not evaluating the entire proteome was lessened. A strength of the technology is the inclusion of low abundance proteins that are difficult to detect in other high-dimensional proteomic platforms. We feel we accomplished the objective of this small pilot study, which was to primarily determine the feasibility and potential usefulness of large-scale proteomics in AMD. Moreover, as a result of conducting this project we found some interesting associations of several proteins for neovascular AMD and GA. We chose to further illustrate the association with the top four proteins graphically but suggest that there may be other proteins worth pursuing in follow-up studies. Furthermore, establishing whether the observed changes are biologically relevant will require additional experiments. We feel the next step of our research is to conduct a larger proteomic study with validation of all high signal proteins using another method of measuring proteins, such as an enzyme-linked immunosorbent assay (ELISA). It will also be important moving forward with this research to correlate findings of altered protein levels in the blood with evaluation of protein levels and profiles in the vitreous, aqueous,42 and tears.43 We found no overlap in the proteins described in a recent review describing AMD biomarkers42 and the proteins described in our small pilot study. However, the study groups were different with regard to sample size, inclusion/exclusion criteria, proteins examined, and the technology used compared with our study group. Although AMD manifests as a local disease, useful biomarkers in the plasma may be found and reflect altered pathways based on the individual's genetics that predispose to the degenerative processes within the eye.
Pathway analysis suggests that the signaling pathways related to neovascular AMD and GA are different. The top-ranked proteins identified in this pilot study were informative and merit further research. It is of interest that we found one protein, vinculin, and several pathways that have previously been described by other investigators as having a role in AMD. Based on our pilot data, analysis of larger numbers of patients may provide further detail into the differences between patients who develop vision-threatening AMD and those who do not. When coupled with analysis of genetic polymorphisms, a more complete picture of the risks and pathogenesis of AMD may become clear.
Supplementary Material
Acknowledgments
Supported by a Challenge Grant to the Department of Ophthalmology from Research to Prevent Blindness, Inc. and the Frederic C. Hamilton Macular Degeneration Center.
Paper presented at the Association for Research in Vision and Ophthalmology (ARVO), Annual meeting, April 29–May 3, Honolulu, Hawaii, USA.
SOMAmer reagent and SOMAscan assay are registered trademarks of SomaLogic, Inc.
Disclosure: A.M. Lynch, None; B.D. Wagner, None; S.J. Weiss, SomaLogic, Inc. (F,E); K.M. Wall, SomaLogic, Inc. (F,E); A.G. Palestine, None; M.T. Mathias, None; F.S. Siringo, None; J.N. Cathcart, None; J.L. Patnaik, None; D.W. Drolet, SomaLogic, Inc. (F,E); N. Janjic, SomaLogic, Inc. (F,E); N. Mandava, SomaLogic, Inc. (S)
References
- 1.Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106–e16. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Ferris FL, III, Wilkinson CP, Bird A, et al. Clinical classification of age-related macular degeneration. Ophthalmology. 2013;120:844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crabb JW. The proteomics of drusen. Cold Spring Harb Perspect Med. 2014;4:a017194. doi: 10.1101/cshperspect.a017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293. doi: 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 5.Advancing Therapeutic Development for Dry Age-Related Macular Degeneration (AMD): Workshop in Brief. Institute of Medicine. 2015. [PubMed]
- 6.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert NG, El Shelmani H, Singh MK, et al. Risk factors and biomarkers of age-related macular degeneration. Prog Retin Eye Res. 2016;54:64–102. doi: 10.1016/j.preteyeres.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch AM, Wagner BD, Mandava N, et al. the relationship of novel plasma proteins in the early neonatal period with retinopathy of prematurity. Invest Ophthalmol Vis Sci. 2016;57:5076–5082. doi: 10.1167/iovs.16-19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch AM, Wagner BD, Deterding RR, et al. The relationship of circulating proteins in early pregnancy with preterm birth. Am J Obstet Gynecol. 2016;214:517.e1–517.e8. doi: 10.1016/j.ajog.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch AM, Patnaik JL, Cathcart JN, et al. Colorado Age-Related Macular Degeneration Registry: design and clinical risk factors of the cohort. Retina. 2018 doi: 10.1097/IAE.0000000000002023. published online ahead of print January 31. [DOI] [PubMed]
- 11.Rohloff JC, Gelinas AD, Jarvis TC, et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim CH, Tworoger SS, Stampfer MJ, et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep. 2018;8:8382. doi: 10.1038/s41598-018-26640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efron B. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. J Am Stat Assoc. 2004;99:96–104. [Google Scholar]
- 15.Fabregat A, Sidiropoulos K, Garapati P, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milacic M, Haw R, Rothfels K, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers. 2012;4:1180–1211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole W, Gibbs DL, Shmulevich I, Bernard B, Knijnenburg TA. Combining dependent P-values with an empirical adaptation of Brown's method. Bioinformatics. 2016;32:i430–i436. doi: 10.1093/bioinformatics/btw438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman D, Lane D. A nonstochastic interpretation of reported significance levels. J Bus Econom Statst. 1983;1:292–298. [Google Scholar]
- 20.Geiger B. A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18:193–205. doi: 10.1016/0092-8674(79)90368-4. [DOI] [PubMed] [Google Scholar]
- 21.Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, Woo SJ, Suh EJ, et al. Identification of vinculin as a potential plasma marker for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2014;55:7166–7176. doi: 10.1167/iovs.14-15168. [DOI] [PubMed] [Google Scholar]
- 23.Xie Q, Klesney-Tait J, Keck K, et al. Characterization of a novel mouse model with genetic deletion of CD177. Protein Cell. 2015;6(2):117–126. doi: 10.1007/s13238-014-0109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalezari P, Murphy GB, Allen FH., Jr NB1, a new neutrophil-specific antigen involved in the pathogenesis of neonatal neutropenia. J Clin Invest. 1971;50:1108–1115. doi: 10.1172/JCI106582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu N, Westra J, Huitema MG, et al. Coexpression of CD177 and membrane proteinase 3 on neutrophils in antineutrophil cytoplasmic autoantibody-associated systemic vasculitis: anti-proteinase 3-mediated neutrophil activation is independent of the role of CD177-expressing neutrophils. Arthritis Rheum. 2009;60:1548–1557. doi: 10.1002/art.24442. [DOI] [PubMed] [Google Scholar]
- 26.Gohring K, Wolff J, Doppl W, et al. Neutrophil CD177 (NB1 gp, HNA-2a) expression is increased in severe bacterial infections and polycythaemia vera. Br J Haematol. 2004;126:252–254. doi: 10.1111/j.1365-2141.2004.05027.x. [DOI] [PubMed] [Google Scholar]
- 27.Sachs UJ, Andrei-Selmer CL, Maniar A, et al. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31) J Biol Chem. 2007;282:23603–23612. doi: 10.1074/jbc.M701120200. [DOI] [PubMed] [Google Scholar]
- 28.Wang GX, Zhao XY, Meng ZX, et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med. 2014;20:1436–1443. doi: 10.1038/nm.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Wang GX, Ma SL, et al. Nrg4 promotes fuel oxidation and a healthy adipokine profile to ameliorate diet-induced metabolic disorders. Mol Metab. 2017;6:863–872. doi: 10.1016/j.molmet.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J, Lin M, Xu Y, et al. Circulating neuregulin 4 levels are inversely associated with subclinical cardiovascular disease in obese adults. Sci Rep. 2016;6:36710. doi: 10.1038/srep36710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothlein R, Wegner C. Role of intercellular adhesion molecule-1 in the inflammatory response. Kidney Int. 1992;41:617–619. doi: 10.1038/ki.1992.94. [DOI] [PubMed] [Google Scholar]
- 32.Mackay CR, Imhof BA. Cell adhesion in the immune system. Immunol Today. 1993;14:99–102. doi: 10.1016/0167-5699(93)90205-Y. [DOI] [PubMed] [Google Scholar]
- 33.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchison HM, Valente EM. Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 35.May-Simera HL, Wan Q, Jha BS, et al. Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep. 2018;22:189–205. doi: 10.1016/j.celrep.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanai M, Goke M, Tsunekawa S, Podolsky DK. Signal transduction pathway of human fibroblast growth factor receptor 3. Identification of a novel 66-kDa phosphoprotein. J Biol Chem. 1997;272:6621–6628. doi: 10.1074/jbc.272.10.6621. [DOI] [PubMed] [Google Scholar]
- 37.Amadio M, Govoni S, Pascale A. Targeting VEGF in eye neovascularization: what's new?: a comprehensive review on current therapies and oligonucleotide-based interventions under development. Pharmacol Res. 2016;103:253–269. doi: 10.1016/j.phrs.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 38.Kim LA, D'Amore PA. A brief history of anti-VEGF for the treatment of ocular angiogenesis. Am J Pathol. 2012;181:376–379. doi: 10.1016/j.ajpath.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olayioye MA. Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Res. 2001;3:385–389. doi: 10.1186/bcr327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romano GL, Platania CBM, Drago F, et al. Retinal and circulating miRNAs in age-related macular degeneration: an in vivo animal and human study. Front Pharmacol. 2017;8:168. doi: 10.3389/fphar.2017.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kersten E, Paun CC, Schellevis RL, et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv Ophthalmol. 2018;63:9–39. doi: 10.1016/j.survophthal.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Winiarczyk M, Kaarniranta K, Winiarczyk S, Adaszek L, Winiarczyk D, Mackiewicz J. Tear film proteome in age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2018;256:1127–1139. doi: 10.1007/s00417-018-3984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.