Abstract
The health impacts of endocrine-disrupting chemicals (EDCs) remain debated, and their tissue and molecular targets are poorly understood. In this study, we leveraged systems biology approaches to assess the target tissues, molecular pathways, and gene regulatory networks associated with prenatal exposure to the model EDC bisphenol A (BPA). Prenatal BPA exposure at 5 mg/kg/d, a dose below most reported no-observed-adverse-effect levels, led to tens to thousands of transcriptomic and methylomic alterations in the adipose, hypothalamus, and liver tissues in male offspring in mice, with cross-tissue perturbations in lipid metabolism as well as tissue-specific alterations in histone subunits, glucose metabolism, and extracellular matrix. Network modeling prioritized main molecular targets of BPA, including Pparg, Hnf4a, Esr1, Srebf1, and Fasn as well as numerous less studied targets such as Cyp51 and long noncoding RNAs across tissues, Fa2h in hypothalamus, and Nfya in adipose tissue. Lastly, integrative analyses identified the association of BPA molecular signatures with cardiometabolic phenotypes in mouse and human. Our multitissue, multiomics investigation provides strong evidence that BPA perturbs diverse molecular networks in central and peripheral tissues and offers insights into the molecular targets that link BPA to human cardiometabolic disorders.
A central concept in the Developmental Origins of Health and Disease states that adverse environmental exposure during early developmental stages is an important determinant for later-onset adverse health outcomes, even in the absence of continuous exposure in adulthood (1–3). Bisphenol A (BPA) is one of the most prevalent environmental metabolic disruptors identified to date, with widespread exposure in human populations, and likely plays a role in Developmental Origins of Health and Disease (3–5). BPA is used in the production of synthetic polymers, including epoxy resins and polycarbonates and, with its advantageous mechanical properties, is ubiquitously found in everyday goods such as plastic bottles and inner coating of canned foods (6, 7). The ability of BPA to leach from these everyday products is a primary route of human exposure (8). BPA has been shown to have the ability to disrupt endocrine signaling important for many biological functions and has been linked to body weight, obesity, insulin resistance, diabetes, metabolic syndrome (MetS), and cardiovascular diseases in both human epidemiologic and animal studies (9–17). Importantly, it has been suggested that the developing fetus is particularly vulnerable to BPA exposure (9, 18). Intrauterine growth retardation has been consistently observed after developmental BPA exposure at intake doses below the suggested human safety level and has been associated with low birth weight, elevated adult fat weight, and altered glucose homeostasis (9, 19–22). As a precaution, BPA has been banned from baby products in Europe, Canada, and the United States. However, BPA is still in use in nonbaby products, renewing concerns about the continuous exposure of populations in addition to the description of its ability to influence health outcomes, including obesity and MetS, over several generations (23–26). Together, these lines of evidence support an intriguing hypothesis that BPA may have been a contributing factor to the rise of MetS and cardiometabolic diseases worldwide in the past decades (27–29).
Despite numerous studies connecting BPA with adverse health outcomes, there remain ample conflicting findings, as summarized by the European Food Safety Agency (30), the BPA Joint Emerging Science Working Group of the US Food and Drug Administration (31), and the recent National Toxicology Program report, CLARITY-BPA, in which functions of multiple organs were examined (32). Although inconsistencies across studies might be attributable to nonmonotonic dose response, exposure window difference, and varying susceptibility among testing models (14, 33), there are also several additional layers of complexity and challenges hindering the full dissection of the biological effects of BPA. First, previous studies examining BPA in various cell types and tissues suggest a broad impact on biological systems (25, 34–36). Second, BPA has been found to modulate multidimensional molecular events, such as gene expression and epigenetic changes, which are functionally important for processes such as metabolism and immune response (37–42). However, due to most studies being designed to focus on one factor at a time as well as noncomparable study designs, it is difficult to directly compare effects across tissues or types of molecular data to derive the molecular rules of sensitivity to BPA exposures. These research gaps in our understanding of the pleiotropy of endocrine-disrupting chemicals and toxicant biological actions necessitated the establishment of the National Institute for Environmental Health Sciences (NIEHS) TaRGET consortium (43) and a more recent call for the research community to systemically interrogate multiple -omics in multiple tissues to accelerate the discovery of key biological fingerprints of environmental exposure (44).
Here, we address some of the aforementioned limitations of past studies by using a highly integrative approach. We conducted a multitissue, multiomics systems biology study to examine the systems-level influence of prenatal BPA exposure using modern integrative genomics and network modeling approaches in a mouse model (Fig. 1A). We first used next-generation sequencing technologies to characterize perturbations in both the transcriptome and the epigenome across three tissues (white adipose tissue, hypothalamus, and liver) in mouse offspring that had experienced in utero exposure to BPA. These tissues were chosen due to their important role in energy and metabolic homeostasis. The hypothalamus is the central regulator of endocrine and metabolic systems and plays a critical role in the regulation of nutrient and energy sensing, feeding, and energy expenditure (45); liver is critical for glucose and cholesterol homeostasis (46); and white adipose tissue is essential for energy and lipid storage, serves as a endocrine organ secreting numerous hormones related to metabolic regulation, and contributes to inflammatory processes by releasing various cytokines (47, 48). These tissues interact with one another to coordinately regulate metabolism and energy balance. Based on mounting evidence that genes operate in highly complex tissue-specific regulatory networks, we hypothesized that prenatal BPA exposure induces genomic and epigenomic reprogramming in the offspring by affecting the organization and functions of tissue-specific gene networks (49–52). Using both transcription factor (TF) and Bayesian networks (BNs), we modeled the dynamics of transcriptomic and epigenomic signatures and predicted potential regulators that govern the actions of BPA. Furthermore, the transcriptome, epigenome, and network information were layered upon metabolic phenotypes such as body weight, adiposity, circulating lipids, and glucose levels in the mouse offspring to evaluate disease association. Lastly, to assess the relevance of the BPA molecular targets identified in our mouse model for human diseases, we applied integrative genomics to bridge the mouse molecular signatures and genetic disease association data from human studies. Our study represents a comprehensive data-driven, systems-level investigation of the molecular and health impact of BPA.
Figure 1.
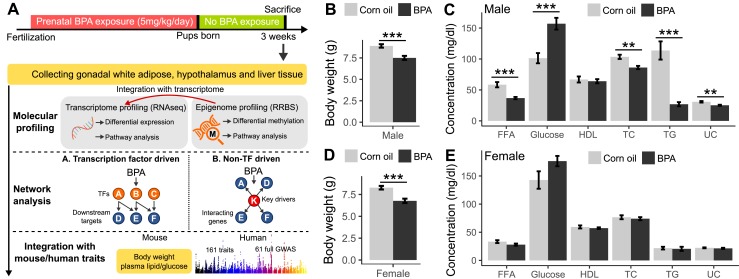
Overall study design and the measurements of metabolic traits in male and female offspring. (A) Framework of multiomics approaches to investigate the impact of prenatal BPA exposure. (B and C) Comparison of body weight, serum lipids, and glucose level in male mice between BPA and control groups at weaning age. (D and E) Comparison of body weight, serum lipids, and glucose level in female mice between BPA and control groups at weaning age. n = 9 to 13 mice (3 to 4 litters from different dams) per group. **P < 0.01; ***P < 0.001 by two-sided Student t test. FFA, free fatty acid; HDL, high-density lipoprotein; TC, total cholesterol; TG, triglyceride; UC, unesterified cholesterol.
Materials and Methods
Ethics statement
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines. Animal studies and procedures were approved by the Chancellor’s Animal Research Committee of the University of California, Los Angeles.
Mouse model of prenatal BPA exposure
Inbred C57BL/6J mice were maintained on a special diet (5V01; LabDiet, St. Louis, MO), certified to contain <150 ppm estrogenic isoflavones, and housed under standard housing conditions (room temperature 22°C to 24°C) with a 12:12-hour light/dark cycle before mating at 8 to 10 weeks of age. Upon mating, female mice were randomly assigned to either the BPA treatment group or the control group. From 1-day postconception to 20 days postconception, BPA (Sigma-Aldrich, St. Louis, MO) dissolved in corn oil was administered to pregnant female mice via oral gavage (mimicking the common exposure route in humans) at 5 mg/kg/d on a daily basis. The dosage is situated below most reported no-observed-adverse-effect levels according to toxicity testing (https://comptox.epa.gov/dashboard/dsstoxdb/results?search=Bisphenol+A) and was typically used in previous studies (25, 53–55). We chose this dosage as a proof-of-concept for our systems biology study design and to facilitate comparison with previous studies. Control mice were fed the same amount of corn oil as vehicle. We chose corn oil over other solvents for BPA because BPA is water insoluble, and corn oil was found to be the least toxic compared with other commonly used solvents (56), but cannot exclude potential confounding from the combined effects of corn oil and BPA. Polycarbonate-free water bottles and cages were used to minimize any unintended exposure to BPA. Both parents and offspring from each treatment were maintained on a low-phytoestrogen special diet (5V01; LabDiet). Offspring in the vehicle- and BPA-treated groups were derived from three and four litters by different dams, respectively, to help assess and adjust for litter effects (57).
Characterization of cardiometabolic phenotypes and tissue collection
Male and female offspring were examined for a spectrum of metabolic phenotypes. In the male set, the control group had n = 9, and the BPA group had n = 11. For females, the control group had n = 9, and the BPA group had n = 13. There were two to three mice from each of the three to four different litters for each treatment group (57). We chose the weaning age to investigate early molecular and phenotypic changes in the offspring, which may predispose the offspring to late-onset diseases. Body weight of offspring was measured daily from postnatal day 5 up to the weaning age of 3 weeks. Mice were fasted overnight before being euthanized, and plasma samples were collected through retro-orbital bleeding. Serum lipid and glucose traits, including total cholesterol, high-density lipoprotein (HDL) cholesterol, unesterified cholesterol, triglycerides (TGs), free fatty acids (FFAs), and glucose, were measured by enzymatic colorimetric assays at the University of California, Los Angeles GTM Mouse Transfer Core as previously described (50). The liver, hypothalamus, and gonadal white adipose tissues were collected from each animal. The whole hypothalamus was collected by first carefully dissecting out the brain and placing it onto an ice-cold dissection board with the ventral side up. Using curved forceps, the tissue was held down, and the whole hypothalamus was gently pinched and spooned out. For the liver dissection, all liver lobes were first dissected out, then a portion of the distal part of the right lobe was collected for molecular profiling, and the rest of the liver was stored separately. For white adipose tissue, we chose the gonadal depot mainly due to its similarity to abdominal fat, established relevance to cardiometabolic risks, tissue abundance, and the fact that it is the most well-studied adipose tissue in mouse models. The gonadal fat depot was carefully dissected around the gonads, avoiding contamination of the gonads. After each tissue collection, the samples were flash frozen in liquid nitrogen and stored at −80°C. All mouse experiments were conducted in accordance with and approved by the Institutional Animal Care and Use Committee at University of California, Los Angeles.
Paired-end RNA-sequencing and data analysis
A total of 18 RNA samples were isolated from gonadal adipose, hypothalamus, and liver tissues (n = 3 per group per tissue; for each group, mice were randomly selected from litters of different dams in independent cages) from male offspring using the AllPrep DNA/RNA Mini Kit (Qiagen GmbH, Hilden, Germany). The sample size was chosen based on previous RNA-sequencing (RNA-seq) studies that demonstrate sufficient reproducibility (50, 58–61). We focused on profiling male tissues because of stronger phenotypes observed in males (Fig. 1). Samples were processed for library preparation using the TruSeq RNA Library Preparation Kit (Illumina, San Diego, CA) for poly-A selection, fragmentation, and reverse transcription using random hexamer primers to generate first-strand cDNA. Second-strand cDNA was generated using RNase H and DNA polymerases, and sequencing adapters were ligated using the Illumina Paired-End sample prep kit. Library products of 250-bp to 400-bp fragments were isolated, amplified, and sequenced. Paired-end read sequencing was performed on an Illumina Hiseq2500 System. After quality control using FastQC (62), the HISAT-StringTie pipeline (63) was used for sequence alignment and transcript assembly. Identification of differentially expressed genes (DEGs) was conducted using DEseq2 (64). Sequenced reads were trimmed for adaptor sequence, masked for low-complexity or low-quality sequence, and then mapped to GRCm38/mm10 whole genome using HISAT v0.1.6. Default paired-end reads alignment parameters for HISAT were used with option -p 8. To account for multiple testing, we used the q-value method (65). After excluding genes with extremely low expression levels (fragments per kilobase of transcript per million mapped reads <1), only DEGs demonstrating differential expression comparing the BPA and control groups per tissue at a false discovery rate (FDR) <5% were used for biological pathway analysis, network analysis, and phenotypic data integration, as described later. The RNAseq quality matrix showing the number of sequencing reads and mapping rate for each sample is provided in an online repository (57). The number of reads aligned with genome for each sample varies between 30 million and 65 million, which satisfies the recommended number of reads needed for differential expression profiling (61).
Quantitative RT-PCR analysis
RNA from male and female liver, hypothalamus, and adipose tissue samples (n = 3 per tissue per treatment of males and n = 5 per tissue per treatment of females) were extracted using the MiRNeasy Mini Kit purchased from Qiagen following the manufacturer’s instruction. Concentration and quality of the RNA were measured using the Thermo Fisher Scientific NanoDrop instrument (Thermo Fisher Scientific, Waltham, MA). cDNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Waltham, MA) following the manufacturer’s protocol with minor modification by adding RNaseOUT Recombinant Ribonuclease Inhibitor (20 U/μL) from Invitrogen (Carlsbad, CA). Following the addition of reverse-transcription components, samples were incubated at the following thermocycler conditions: 10 minutes at 25°C, 120 minutes at 37°C, and then 5 minutes at 85°C. Upon completion, the cDNA was stored at −20°C until quantitative RT-PCR (qPCR) was performed. qPCR was done using the PowerUP SYBR Green Master Mix from Applied Biosystems. For both male and female liver and hypothalamus tissues, 20 ng of cDNA was used for the reaction. For the male and female adipose samples, 10 ng of cDNA was used due to low concentration. For all primers, a final concentration of 0.5 μM was used in the reaction. The qPCR reaction was run using the manufacturer’s instructions followed by a melt-curve analysis. All primer sets displayed a single peak demonstrating specificity. PCR products were also run on a 1.5% agarose gel to validate appropriate amplicon size. Primer sequences are listed in an online repository (57). Gapdh was used as a housekeeping gene to quantify relative expression levels using the ΔΔ threshold cycle analysis. Statistics was performed on the Δ threshold cycle using a t test.
Reduced representation bisulfite sequencing and data analysis
We constructed reduced representation bisulfite sequencing (RRBS) libraries for 18 DNA samples from adipose, hypothalamus, and liver tissues from male offspring (n = 3 per group per tissue from the same set of tissues chosen for transcriptome analysis described previously). The DNA samples were quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific), and 100 ng of DNA was used for library preparation. After digestion of the DNA with the MspI enzyme, samples underwent an end-repair and adenylation process, followed by adapter ligation using the TruSeq barcode adapter (Illumina), size selection using AMPure Beads (Beckman Coulter, Brea, CA), and bisulfite treatment using the Epitect Kit (Qiagen). Bisulfite-treated DNA was then amplified using the TruSeq Library Prep Kit (Illumina) and sequenced with the Illumina Hiseq2500 System. Bisulfite-converted reads were processed and aligned to the reference mouse genome (GRCm38/mm10 build) using the bisulfite aligner BSMAP (66). We then used MOABS (67) for methylation ratio calling and identification of differentially methylated CpGs (DMCs). FDR was estimated using the q-value approach. Loci with methylation level changes of >5% between BPA and control groups and FDR <5% for each tissue were considered statistically significant DMCs. To annotate the locations of the identified DMCs in relation to gene regions and repetitive DNA elements accessed from the UCSC Genome Browser, we used the Bioconductor package “annotatr” (68). Specifically, gene regions were categorized into: (i) 1 to 5 kb upstream of the transcription start site; (ii) promoter (<1 kb upstream of the transcription start site); (iii) 5′ untranslated region; (iv) exons; (v) introns; and (vi) 3′ untranslated region. The “annotatr” package was also used to annotate DMCs for known long noncoding RNAs (lncRNAs) based on GENCODE Release M16. Overrepresentation of DMCs within each category was calculated using a one-sided Fisher exact test. We further evaluated the link between DEGs and their local DMCs (DMCs annotated as any of the six above-mentioned gene regions) by correlating the methylation ratio of DMCs with the expression level of DEGs.
Pathway, network, and disease association analyses of DEGs and DMCs using the Mergeomics R package
To investigate the functional connections among the BPA-associated DEGs or DMCs (collectively referred to as molecular signatures of BPAs) and to assess the potential association of BPA-affected genes with diseases in human populations, we used the Mergeomics package (69), an open-source bioconductor R package (https://bioconductor.org/packages/devel/bioc/html/Mergeomics.html) designed to perform various integrative analyses in multiomics studies. Mergeomics consists of two main libraries, Marker Set Enrichment Analysis (MSEA) and Weighted Key Driver Analysis (wKDA). In the current study, we used MSEA to assess: (i) whether known biological processes, pathways, or TF targets were enriched for BPA molecular signatures as a means to annotate the potential functions or regulators of the molecular signatures; and (ii) whether the BPA signatures demonstrate enrichment for disease associations identified in human genome-wide association studies (GWAS) of various complex diseases (57). wKDA leverages gene network topology (interactions or regulatory relations among genes) and edge weight (strength or reliability of interactions and regulatory relations) information of graphical gene networks to predict potential key regulators of a given group of genes—in this case, the BPA-associated DEGs (57). Both MSEA and wKDA were built around χ2-like statistics (57) that yield robust findings that have been experimentally validated (51, 52, 69). Details of each usage of the Mergeomics package are discussed later.
Functional annotation of DEGs and DMCs
To infer the functions of the DEGs and DMCs affected by BPA, we used MSEA to annotate the DEGs or local genes adjacent to the DMCs with known biological pathways curated from the Kyoto Encyclopedia of Genes and Genomes (70) and Reactome (71). In brief, we extracted the differential expression P values of genes in each pathway from the differential expression or methylation analyses and compared these P values against the null distribution of P values from random gene sets with matching gene numbers. If genes in a given pathway collectively show more significant differential expression or differential methylation P values compared with random genes based on a χ2-like statistic, we annotate the DEGs or DMCs using that pathway (57). DEGs and DMCs can have multiple overrepresented pathways.
Identification of TF hot spots perturbed by BPA
To dissect the regulatory cascades of BPA, we first assessed whether BPA-associated DEGs were downstream targets of specific TFs. The hypothesis behind this analysis is that BPA first affects TFs, which in turn regulate the expression of downstream genes. We used TF regulatory networks for adipose, brain, and liver tissue retrieved from the FANTOM5 database (72). Note that only a whole brain (instead of hypothalamus) TF network was available, which may only partially represent hypothalamic gene regulation. Each TF network was processed to keep the edges with high confidence (57). To identify TFs for which targets were perturbed by BPA, the downstream nodes of each TF in the network were pooled as the target genes for that TF. We then assessed the enrichment for BPA exposure–related DEGs among the target genes of each TF using MSEA. TFs with FDR <5% were considered statistically significant. Cytoscape software was used for TF network visualization (73).
BN and wKDA to identify potential non-TF regulators
To further identify non-TF regulators that sense BPA and then perturb downstream genes, we used BNs of adipose, hypothalamus, and liver tissues constructed from genetic and transcriptomic data from several large-scale mouse and human studies (57). wKDA was used to identify network key drivers (KDs), which are defined as network nodes for which neighboring subnetworks are significantly enriched for BPA-associated DEGs. Briefly, wKDA takes gene set G (i.e., BPA DEGs) and directional gene network N (i.e., BNs) as inputs. For every gene K in network N, neighboring genes within 1-edge distance were tested for enrichment of genes in G using χ2-like statistics followed by FDR assessment by permutation (57). Network genes that reached FDR <5% were reported as potential KDs.
Association of BPA DEGs and DMCs with mouse phenotypes and human diseases/traits
To assess whether the BPA molecular signatures were related to phenotypes examined in the mouse offspring, we calculated the Pearson correlation coefficient among expression level of DEGs, methylation ratio of DMCs, and the measurement of metabolic traits. For human diseases or traits, we accessed the GWAS catalog database (74) and collected the lists of candidate genes reported to be associated with 161 human traits/diseases (P < 1e-5). These genes were tested for enrichment of the BPA DEGs and DMCs in our mouse study using MSEA. We further curated all publicly available full summary statistics for 61 human traits/diseases from various public repositories (57). This allowed us to apply MSEA to comprehensively assess the enrichment for human disease association among BPA transcriptomic signatures using the full spectrum of large-scale human GWAS. For each tissue-specific gene signature, we used the single nucleotide polymorphisms (SNPs) within a 50-kb chromosomal distance as the representing SNPs for that gene. The trait/disease association P values of the SNPs were then extracted from each GWAS and compared with the P values of SNPs of random sets of genes to assess whether the BPA signatures were more likely to show stronger disease association in human GWAS (57). This strategy has been successfully used in our previous animal model studies to assess the connection of genes affected by environmental perturbations such as diets and trauma to various human diseases (50, 59).
Data availability
Supplemental methods, figures, and tables are available at Figshare (doi.org/10.6084/m9.figshare.7451069.v2) (57). RNA-seq and RRBS data have been submitted to the Gene Expression Omnibus (GEO) under accession numbers GSE121603 (for RNA-seq) and GSE121604 (for RRBS).
Results
Prenatal BPA exposure induces lower body weight and alterations in cardiometabolic phenotypes
We exposed pregnant C57BL/6J mice to BPA during gestation via oral gavage at the dosage of 5 mg/kg/d and examined the male and female offspring for a spectrum of metabolic phenotypes at weaning age. Compared with the control group, both male and female offspring from the BPA group showed significantly lower body weight (Fig. 1B and 1D). There were also considerable decreases in serum lipid parameters and an increase in serum glucose level in males (Fig. 1C), but not in females (Fig. 1E). The phenotypic differences between BPA and control groups are not the results of litter effect, as offspring from different dams in each group showed similar patterns (57).
Prenatal BPA exposure induces tissue-specific transcriptomic alterations in male weaning offspring
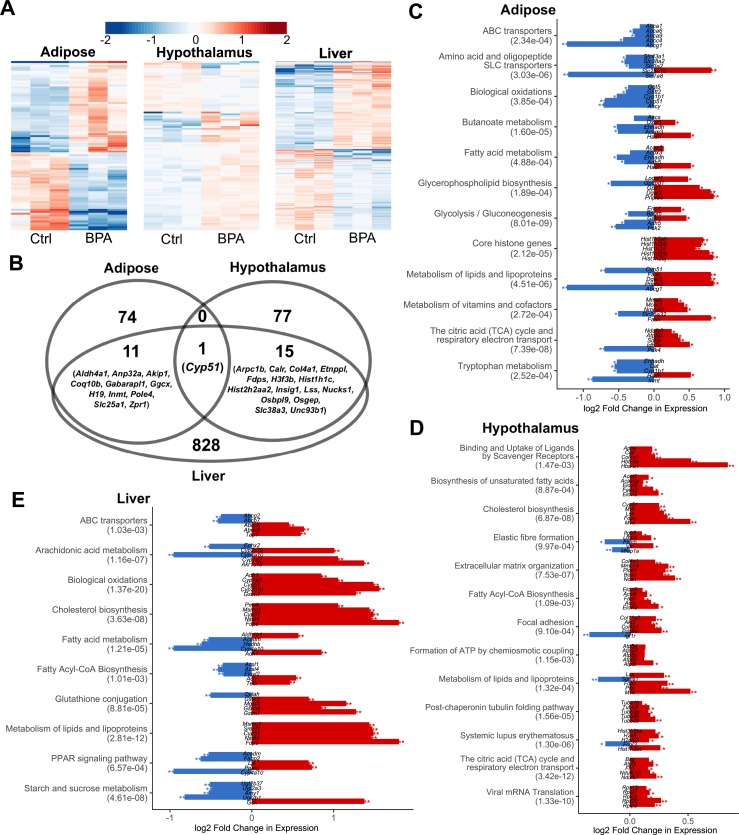
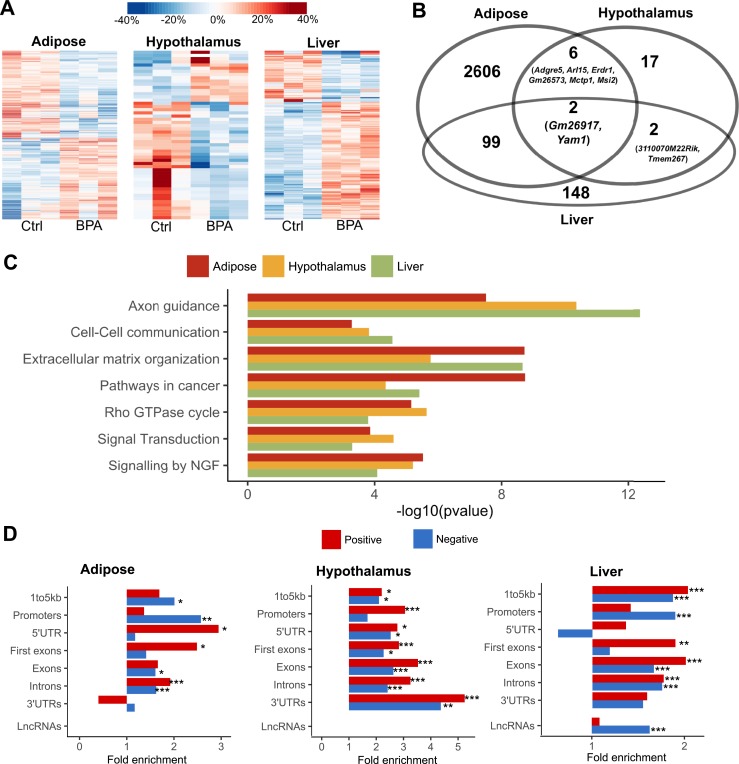
To explore the molecular basis underlying the potential health impact of prenatal BPA exposure, we collected three key metabolic tissues including white adipose tissue, hypothalamus, and liver from male offspring (due to the stronger observed phenotypes) at 3 weeks. We used RNA-seq to profile the transcriptome and identified 86, 93, and 855 DEGs in the adipose tissue, hypothalamus, and liver tissue, respectively, at FDR <5% (57). This supports the ability of prenatal BPA exposure to induce large-scale transcriptomic disruptions in offspring, with the impact appearing to be more prominent in liver. The DEGs show distinct expression patterns between the control and BPA groups, and samples within each group generally agree with one another on the upregulation or downregulation (Fig. 2A). The DEGs were highly tissue specific, with only 12 out of the 86 adipose DEGs and 16 out of the 93 hypothalamus DEGs being found in liver. Interestingly, the hypothalamic DEGs are predominantly upregulated in the BPA group, whereas the other two tissues did not show such direction bias (57). Only one gene, Cyp51 (sterol 14-α demethylase), was shared across all three tissues but with different directional changes (upregulated in hypothalamus and liver and downregulated in adipose) (Fig. 2B).
Figure 2.
Prenatal BPA exposure induced transcriptomic alterations in adipose, hypothalamus, and liver. (A) Heat map of expression changes in adipose, hypothalamus, and liver for the DEGs affected by BPA. Color indicates fold change of expression, with red and blue indicating upregulation and downregulation by BPA, respectively. (B) Venn diagram demonstrating tissue-specific and shared DEGs between tissues. (C–E) Significantly enriched pathways (FDR <5%) among DEGs from each tissue. Enrichment P value (shown in parentheses following the name of functional annotation) is determined by MSEA. The fold change and statistical significance for the top five DEGs in each pathway are shown. *P < 0.05; **FDR <5% in differential expression analysis using DEseq2.
Replication of the DEG signatures using both qPCR and independent studies
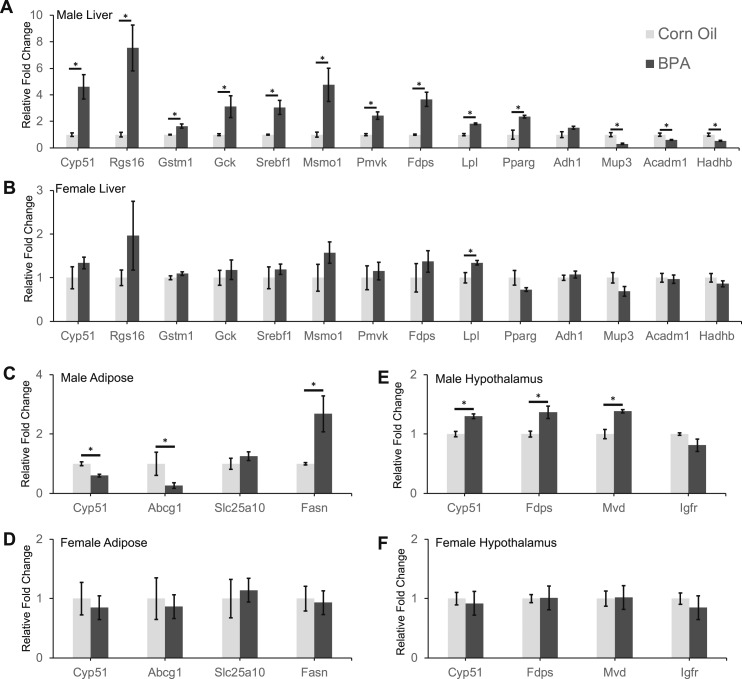
To validate the identified DEGs in the RNA-seq analysis of the male samples, we selected 22 genes (14 from the liver, 4 from the hypothalamus, and 4 from the adipose tissue) for qPCR analysis. We found that the majority (19 out of 22; 86%) of the genes tested in the male samples were significantly altered in the BPA samples in our qPCR data (Fig. 3A, 3C, and 3E). All 22 genes tested via qPCR showed consistent directions in expression changes as observed in our RNA-seq analysis (Fig. 3), supporting the accuracy of our RNA-seq data.
Figure 3.
qPCR analysis on identified DEGs in male and female offspring. Relative fold change in expression levels of 14 genes in liver samples of (A) male and (B) female offspring. Relative fold change in expression of four genes in gonadal adipose samples of (C) male and (D) female offspring. Relative fold change in expression of four genes in hypothalamus samples of (E) male and (F) female offspring. Data presented as mean ± SE of independent replicates. n = 3 per tissue per group for males and n = 5 per tissue per group for females. *P < 0.05 by two-sided Student t test.
Next, to evaluate the effect of BPA in different sexes, we also analyzed the expression of the same 22 select genes in the female cohort. We found that only one gene, Lpl, in the liver was significantly affected in the female offspring exposed to BPA (Fig. 3B). The liver expression of Rgs16, Msm01, Pparg, and Mup3 in the exposed females also showed very similar trends to the BPA male group (Fig. 3B). These subtle changes in expression levels are consistent with the weaker phenotypic data observed in females (Fig. 1D and 1E).
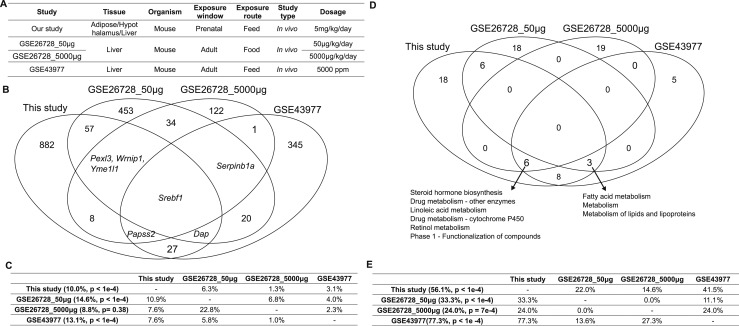
To further assess the reproducibility of the differential expression signatures identified in our study, we examined our DEG signatures using independent expression profiling data deposited on the GEO (57). We identified three GEO datasets related to BPA exposure in mice: two from GSE26728 (75) and one from GSE43977 (76) (Fig. 4A). These publicly available liver transcriptome datasets were derived from studies of BPA exposure during adulthood, as we were not able to identify other publicly available datasets with the same in utero exposure condition tested in our exposure paradigm, making a direct replication difficult. However, we reasoned that if core mechanisms exist for BPA regardless of experimental conditions, consistent signals should be derived. We compared the differential expression signatures from the three existing liver studies against ours and found limited consistency in BPA signatures across datasets, even for the two datasets that were originated from the same study (GSE26728) (Fig. 4C). These results support that BPA has condition-specific activities. Nevertheless, 10% of our DEGs were replicated in the other GEO datasets (P < 1e-4 compared with random expectation via a permutation analysis) (Fig. 4C). Srebf1, encoding a key TF in lipid metabolism, was consistent across all four datasets, along with numerous additional genes consistent in two or more studies (Fig. 4B).
Figure 4.
Comparison of the liver DEGs and their functional annotations against publicly available datasets relevant to BPA exposure in GEO. (A) Descriptions of the study design of different datasets. (B) Venn diagram of the DEGs identified in different datasets. DEGs were determined by Limma at P < 0.01. (C) The percentage of DEGs from the datasets in each row header that are replicated by the datasets in each column header. Numbers in parentheses indicate the percentage of DEGs that are replicated by at least one independent study and the significance of the replication percentage determined by permutation test. (D) Venn diagram of the functional annotations for the DEGs identified in different datasets. Functional annotations were determined by MSEA at FDR <5%. (E) The percentage of functional annotations from the datasets in each row header that are replicated by the datasets in each column header. Numbers in parentheses indicate the percentage of annotations that are replicated by at least one independent study and the significance of the replication percentage determined by permutation test.
Due to the major difference in the exposure window between our study and the previously noted publicly available GEO datasets, we expanded our search by collecting the BPA signatures that were reported in published mouse studies on developmental BPA exposure. We collected 24 unique DEGs from 7 mouse studies (22, 77–82), which evaluated BPA effects on liver during development (57) and found that 6 (Cyp17a1, Fasn, FdpsGstt3, Pparg, and Scd1) of the 24 DEGs were also significantly affected in our study. Similarly, we compared our hypothalamic DEGs with those derived from three mouse and rat studies with similar BPA exposure window and found three overlapping genes with our study (Akt2, Hip1r, and Ndufb7) (83–85). Recent adipose tissue gene expression studies of developmental BPA exposure revealed very few DEGs, and none overlapped with those in our study (22, 37, 40, 86). These comparisons support certain consistencies in liver and hypothalamus DEGs between studies but limited overlaps for adipose DEGs.
Functional annotation of DEGs in adipose, hypothalamus, and liver tissues
To better understand the biological implications of the BPA exposure–related DEGs in individual tissues, we evaluated the enrichment of DEGs for known biological pathways and functional categories using the Mergeomics package (69) (Fig. 2C–2E; full results in an online repository) (57). We observed strong enrichment for pathways related to lipid metabolism (lipid transport, fatty acid metabolism, and cholesterol biosynthesis) and energy metabolism (biological oxidation and tricarboxylic acid cycle) across all three tissues. Most of these pathways appeared to be upregulated in all three tissues, except that genes involved in biological oxidation in adipose tissue were downregulated (Fig. 2C–2E). Individual tissues also showed perturbations of unique pathways: peroxisome proliferator–activated receptor (PPAR) signaling and arachidonic acid pathways were altered in liver; extracellular matrix–related processes were enriched among hypothalamic DEGs; and core histone genes were upregulated in adipose DEGs (Fig. 2C–2E). In addition, TG biosynthesis and glucose metabolism pathways were also moderately enriched among adipose DEGs, whereas few changes were seen for genes involved in adipocyte differentiation (57).
Next, we compared the enriched pathways from our DEGs with those identified from independent GEO datasets as described above to evaluate whether distinct study-specific signatures could converge onto similar biological processes. The replicated pathways across studies include steroid hormone biosynthesis, retinol metabolism, and fatty acid metabolism, suggesting that these processes were consistently influenced by BPA under varying exposure windows and dosages (Fig. 4D). At FDR <5%, 56.1% of the significant pathways in our study were replicated in one or more independent studies (P < 1e-4 compared with random expectation via a permutation test; Fig. 4E). Pairwise comparison revealed relatively higher overlap ratios between our study and individual independent studies than between the previous studies, despite the greater similarity in the study design among the previous studies (Fig. 4E).
Prenatal BPA exposure induces tissue-specific epigenetic alterations in male weaning offspring
Consistent with the observed gene expression disruptions at the transcriptomic level, we observed numerous methylomic alterations using RRBS, which characterizes DNA methylation states of millions of potential epigenetic sites at single-base resolution. At FDRs <5%, 5136, 104, and 476 DMCs were found in adipose, hypothalamus, and liver tissues, respectively (57). The DMCs show distinct expression patterns between the control and BPA groups, and samples within each group generally agree with one another on the upregulation or downregulation (Fig. 5A). When comparing our adipose methylation signatures with a previous study (40), we were able to replicate five out of seven peak hypomethylated genes and six out of nine peak hypermethylated genes. Interestingly, BPA induced local methylation changes in Gm26917 and Yam1, two lncRNAs with no previously known link to BPA, consistently across three tissues (Fig. 5B). The majority of the DMCs are located in intergenic regions (32% to 38%), followed by introns (31% to 37%) and exons (13% to 15%), but there is a paucity of DMCs in the promoter region (3% to 5%) (57). Contrary to predictions that promoter regions may be more prone to epigenetic changes, we found that within-gene and intergenic methylation alterations in DNA methylation are more prevalent, a pattern consistently observed in previous epigenomic studies (50, 87). In addition, 5.0%, 8.6%, and 8.1% DMCs overlap with repetitive DNA elements in adipose, hypothalamus, and liver, respectively, recapitulating a previous report of the interaction between BPA and repetitive DNA (88).
Figure 5.
Prenatal BPA exposure induced methylomic level alteration in adipose, hypothalamus, and liver. (A) Heat map of methylation level changes for the DMCs. Color indicates change in methylation ratio, with red and blue indicating upregulation and downregulation by BPA, respectively. (B) Venn diagram of genes with local DMCs between tissues shows tissue-specific and shared genes mapped to DMCs. (C) Significantly enriched pathways that satisfied FDR <1% across DMCs from adipose, hypothalamus, and liver tissues. Enrichment P value is determined by MSEA. (D) Fold enrichment for positive correlations (red bars) or negative correlations (blue bars) between DMCs and local DEGs, assessed by different gene regions. *P < 0.05; **P < 0.01; ***P < 0.0001; enrichment P values were determined using Fisher exact test. Ctrl, control; NGF, nerve growth factor; UTR, untranslated region.
For DMCs that are located within or adjacent to genes, we further tested whether the local genes adjacent to those DMCs show enrichment for known functional categories. Unlike DEGs, top processes enriched for DMCs concentrated on intracellular and extracellular communication and signaling-related pathways such as axon guidance, extracellular matrix organization, and nerve growth factor signaling (Fig. 5C; full results in an online repository) (57). The affected genes in these processes are related to cellular structure, cell adhesion, and cell migration, indicating that these functions may be particularly vulnerable to BPA-induced epigenetic modulation.
Potential regulatory role of DMCs in transcriptional regulation of BPA-induced DEGs
To explore the role of DMCs in regulating DEGs, we evaluated the connection between transcriptome and methylome by correlating the expression level of DEGs with the methylation ratio of their local DMCs. For the DEGs in adipose, hypothalamus, and liver tissue, we identified 42, 36, and 278 local DMCs for which methylation ratios were significantly correlated with gene expression. At a global level, compared with non-DEGs, DEGs are more likely to contain local correlated DMCs (57). A closer look into the expression-methylation correlation by different chromosomal regions further revealed a context-dependent correlation pattern (Fig. 5D). In adipose and liver, the 3% to 5% of DMCs in promoter regions tend to show significant enrichment for negative correlation with DEGs, whereas gene body methylations for DEGs are more likely to show signficant enrichment for positive correlation with gene expression. In hypothalamus, however, positive correlations between DEGs and DMCs are more prevalent across different gene regions. In addition, liver DMCs within lncRNAs were uniquely enriched for negative correlation with lncRNA expression, although the lack of a reliable mouse lncRNA target database prevented us from further investigating whether downstream targets of the lncRNAs were enriched in the DEGs. Specific examples of DEGs showing significant correlation with local DMCs include adipose DEG Slc25a1 (solute carrier family 25 member 1; involved in TG biosynthesis), hypothalamic DEG Mvk (mevalonate kinase; involved in cholesterol biosynthesis), and liver DEG Gm20319 (an lncRNA with unknown function) (57). These results support a role of BPA-induced differential methylation in altering the expression levels of adjacent genes.
Pervasive influence of prenatal BPA exposure on the liver TF network
BPA is known to bind to diverse types of nuclear receptors such as estrogen receptors and PPARs that function as TFs, thus influencing the action of downstream genes (89, 90). PPARg in particular has been shown to be a target of BPA in mouse and human and mechanistically linking BPA exposure with its associated effect on weight gain and increased adipogenesis (91–93). To explore the TF regulatory landscape underlying BPA exposure based on our genome-wide data, we leveraged tissue-specific TF regulatory networks from the FANTOM5 project (72) and integrated them with our BPA transcriptome profiling data. No TF was found to be differentially expressed in adipose tissue, whereas 1 TF (Pou3f1) and 14 TFs (such as Esrra, Hnf1a, Pparg, Tcf21, and Srebf1) were found to be differentially expressed in hypothalamus and liver, respectively. Due to the temporal nature of TF action, changes in TF levels may precede the downstream target genes and not be reflected in the transcriptomic profiles measured at the time of euthanization. Therefore, we further curated the target genes of TFs from FANTOM5 networks and tested the enrichment for the target genes of each TF among our tissue-specific DEGs (57). This analysis confirmed that BPA perturbs the activity of the downstream targets for estrogen receptors Esrrg (P = 1.4e-3; FDR 1.9%) and Esrra (P = 0.03; FDR 13%) in liver, as well as Esr1 in both adipose (P = 7.2e-3; FDR 10.6%) and liver (P = 7.2e-3; FDR 4.7%). Targets of Pparg were also perturbed in liver (P = 4.1e-3; FDR 3.8%). Therefore, we demonstrated that our data-driven network modeling is able to not only recapitulate results from previous in vitro and in vivo studies showing that BPA influences estrogen signaling and PPAR signaling (90), but also uniquely point to the tissue specificity of these BPA target TFs.
In addition to these expected TFs, we identified 14 adipose TFs and 61 liver TFs for which target genes were significantly enriched for BPA DEGs at FDR <5%. Many of these TFs showed much stronger enrichment for BPA DEGs among their downstream targets than the estrogen receptors (57). The adipose TFs include nuclear TF Y subunit α (Nfya) and fatty acid synthase (Fasn), both implicated in adipocyte energy metabolism (94). The liver TFs include multiple genes from the hepatocyte nuclear factors family and the CCAAT-enhancer-binding proteins family, which are critical for liver development and function, suggesting a pervasive influence of BPA on liver TF regulation.
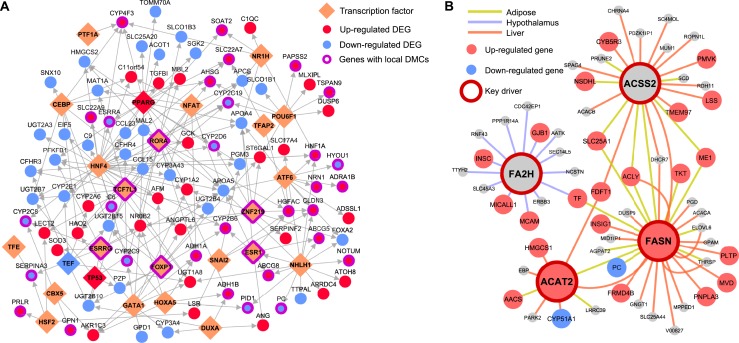
We further extracted the subnetwork containing 89 unique downstream targets of the significant liver TFs that are also liver DEGs. This subnetwork showed significant enrichment for genes involved in metabolic pathways such as steroid hormone biosynthesis and fatty acid metabolism. The regulatory subnetwork for the top liver TFs (FDR <5%) revealed a highly interconnected TF subnetwork that potentially senses BPA exposure and in turn governs the expression levels of their targets (Fig. 6A), with Pparg and Hnf4 among the core TFs. Some of the TFs in this network, including Esr1, Esrrg, Foxp1, and Tcf7l1, also had local DMCs identified in our study, indicating that BPA may perturb this liver TF subnetwork via local modification of DNA methylation of key TFs.
Figure 6.
TFs and KDs orchestrate BPA-induced gene expression level changes. (A) Liver TF regulatory networks for the top ranked TFs (FDR <5%) based on enrichment of liver DEGs among TF downstream targets. Network topology was based on FANTOM5. For TFs with >20% overlapping downstream targets, only the TF with the lowest FDR is shown. (B) Gene-gene regulatory subnetworks (BNs) for cross-tissue KDs. Network topology was based on BN modeling of each tissue using genetic and transcriptome datasets from mouse and human populations (57). For each tissue, if two or more datasets were available for a given tissue, a network for each dataset was constructed, and a consensus network was derived by keeping only the high-confidence network edges between genes (edges appearing in two or more studies).
Identification of potential non-TF regulators governing BPA induced molecular perturbations
To further identify regulatory genes that mediate the action of BPA on downstream targets through non-TF mechanisms, we leveraged data-driven tissue-specific BNs generated from multiple independent human and mouse studies (57). These data-driven networks are complementary to the TF networks used previously and have proven valuable for accurately predicting gene-gene regulatory relationships and novel KDs that were experimentally validated (49–52, 95). KDs were defined as network nodes for which surrounding subnetworks are significantly enriched for BPA exposure–related DEGs. At FDR <1%, we identified 21, 1, and 100 KDs in adipose, hypothalamus, and liver, respectively (57). The top KDs in adipose (top 5 KDs Acss2, Pc, Agpat2, Slc25a1, and Acly), hypothalamus (Fa2h), and liver (top 5 KDs Dhcr7, Aldh3a2, Fdft1, Mtmr11, and Hmgcr) were involved in cholesterol, fatty acid, and glucose metabolism processes. In addition, three KDs—Acss2 (acetyl-coenzyme A synthetase 2), Acat2 (acetyl-coenzyme A acetyltransferase 2), and Fasn (fatty acid synthase)—were involved in the upregulation of DEGs in both adipose and liver, despite the fact that few DEG signatures overlap across tissues (Fig. 6B). These KDs are consistent with the observed increased expression of several genes implicated in lipogenesis, including Fasn, and help explain the liver accumulation of TGs when mice are exposed to BPA (75). Together, these results indicate that BPA may engage certain common regulators that have tissue-specific targets. The distinct upregulatory pattern within the subnetworks of individual KDs supports the potential functional importance of KDs in orchestrating the action of downstream genes. These KDs, along with the TFs from the previous analysis, may represent regulatory targets that transmit the in vivo biological effects of BPA.
BPA transcriptomic and methylomic signatures are related to metabolic traits in mice
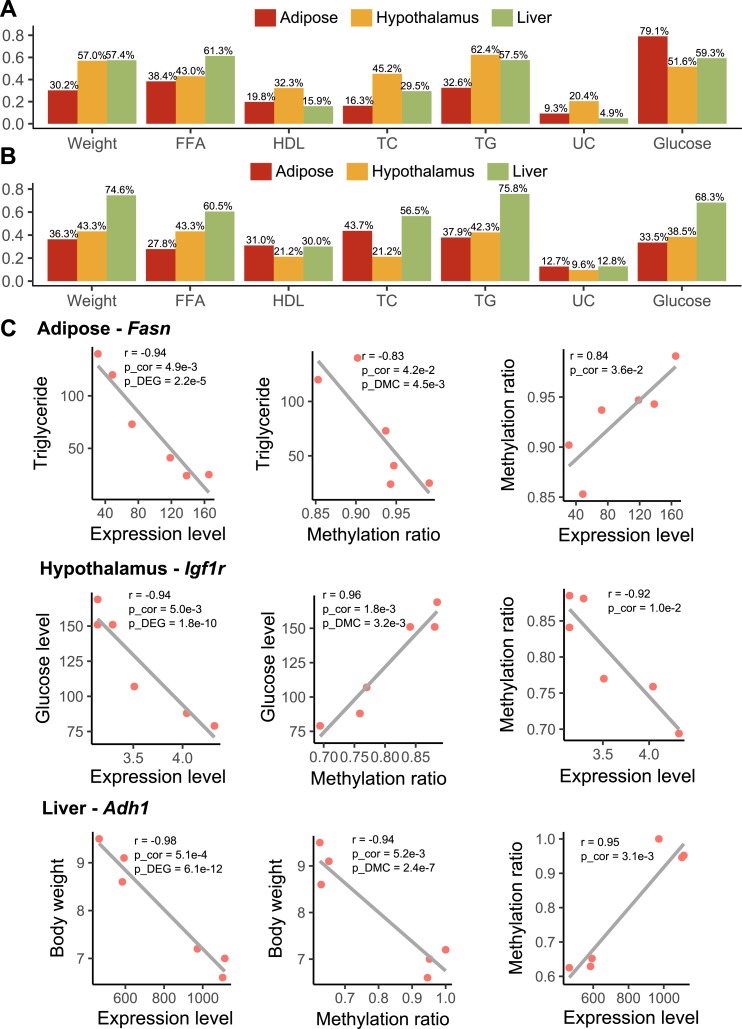
To assess the relationship between the BPA molecular signatures and metabolic traits in the mouse model, the DEGs and DMCs from individual tissues were tested for correlation with the measured metabolic traits: body weight, FFAs, total cholesterol, HDL cholesterol, TGs, and blood glucose. At P < 0.05, over two-thirds of tissue-specific DEGs and >60% of DMCs were identified to be correlated with at least one metabolic trait (Fig. 7A and 7B). Notably, liver DEGs exhibited stronger correlation with FFAs and TGs, whereas adipose DEGs were uniquely associated with glucose level, which is consistent with the pathway annotation results for these tissues. In contrast, liver DMCs showed stronger correlations with metabolic traits than those from adipose and hypothalamus tissues.
Figure 7.
Correlation among gene expression, DNA methylation, and metabolic traits. (A) Percentage of tissue-specific DEGs that are correlated with metabolic traits (P < 0.05). (B) Percentage of tissue-specific DMCs that are correlated with metabolic traits (P < 0.05). (A and B) P values were determined using Pearson correlation test. (C) Pairwise correlation among expression level, methylation ratio, and metabolic profiles (TGs, glucose level, and body weight) for Fasn, Igf1r, and Adh1. p_cor, P value was determined using Pearson correction test; p_DEG was determined using differential expression test; p_DMC was determined using differential methylation test. Each dot represents a mouse. TC, total cholesterol; UC, unesterified cholesterol.
Cross-examination of correlation across gene expression, DNA methylation, and metabolic traits revealed 35 consistent DEG-DMC-trait associations (3 in adipose, 4 in hypothalamus, and 28 in liver) (57). For example, in adipose tissue, Fasn (also a perturbed TF hot spot in adipose and a shared KD in adipose and liver) was correlated with its exonic DMC at chr11:120816457, and both were correlated with TG level; in hypothalamus, Igf1r (insulin-like growth factor 1 receptor) was correlated with its intronic DMC at chr7:68072768, and both were correlated with blood glucose level; in liver, Adh1 (alcohol dehydrogenase 1A) was correlated with its intronic DMC at chr3:138287690, and both were correlated with body weight (Fig. 7C). These results suggest that BPA alters local DMCs of certain genes to regulate gene expression, which may in turn regulate distinct metabolic traits.
Relevance of BPA signature to human complex traits/diseases
Human observational studies have associated developmental BPA exposure with a wide variety of human diseases ranging from cardiometabolic diseases to neuropsychiatric disorders (15, 16, 96). Large-scale human GWAS offer an unbiased view of the genetic architecture for various human traits/diseases, and intersections of the molecular footprints of BPA in our mouse study with human disease risk genes can help infer the potential disease-causing properties of BPA in humans. From the GWAS Catalog (74), we collected associated genes for 161 human traits/diseases (traits with <50 associated genes were excluded) and evaluated the enrichment for the trait-associated genes among DEG and DMC signatures. At FDR <5%, no trait was found to be significantly enriched for BPA DEGs. Surprisingly, despite the differences among tissue-specific DMCs (Fig. 5B), 19 out of the 161 traits showed consistently strong enrichment for DMCs across all 3 tissues at FDR <1%. The top traits include body mass index (BMI) and type 2 diabetes (Table 1). As DNA methylation status is known to determine long-term gene expression pattern instead of immediate dynamic gene regulation, the BMI and diabetes-associated genes may be under long-term programming by BPA-induced differential methylation, thereby affecting later disease risks.
Table 1.
Top 5 Human Traits for Which Associated Genes in GWAS Are Enriched for DMCs Across Adipose, Hypothalamus, and Liver at FDR <1% in MSEA
| Human Trait | Adipose |
Hypothalamus |
Liver |
|||
|---|---|---|---|---|---|---|
| P | FDR, % | P | FDR, % | P | FDR, % | |
| Obesity-related traits | 1.28E-16 | 0.00 | 3.03E-15 | 0.00 | 2.71E-19 | 0.00 |
| BMI | 1.30E-13 | 0.00 | 3.74E-07 | 0.00 | 9.66E-12 | 0.00 |
| Postbronchodilator FEV1/FVC ratio | 8.17E-09 | 0.00 | 1.45E-08 | 0.00 | 3.67E-07 | 0.00 |
| Type 2 diabetes | 1.21E-05 | 0.03 | 8.97E-09 | 0.00 | 0.001243 | 0.92 |
| Platelet distribution width | 8.16E-08 | 0.00 | 7.62E-05 | 0.16 | 5.20E-05 | 0.12 |
Abbreviations: FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity.
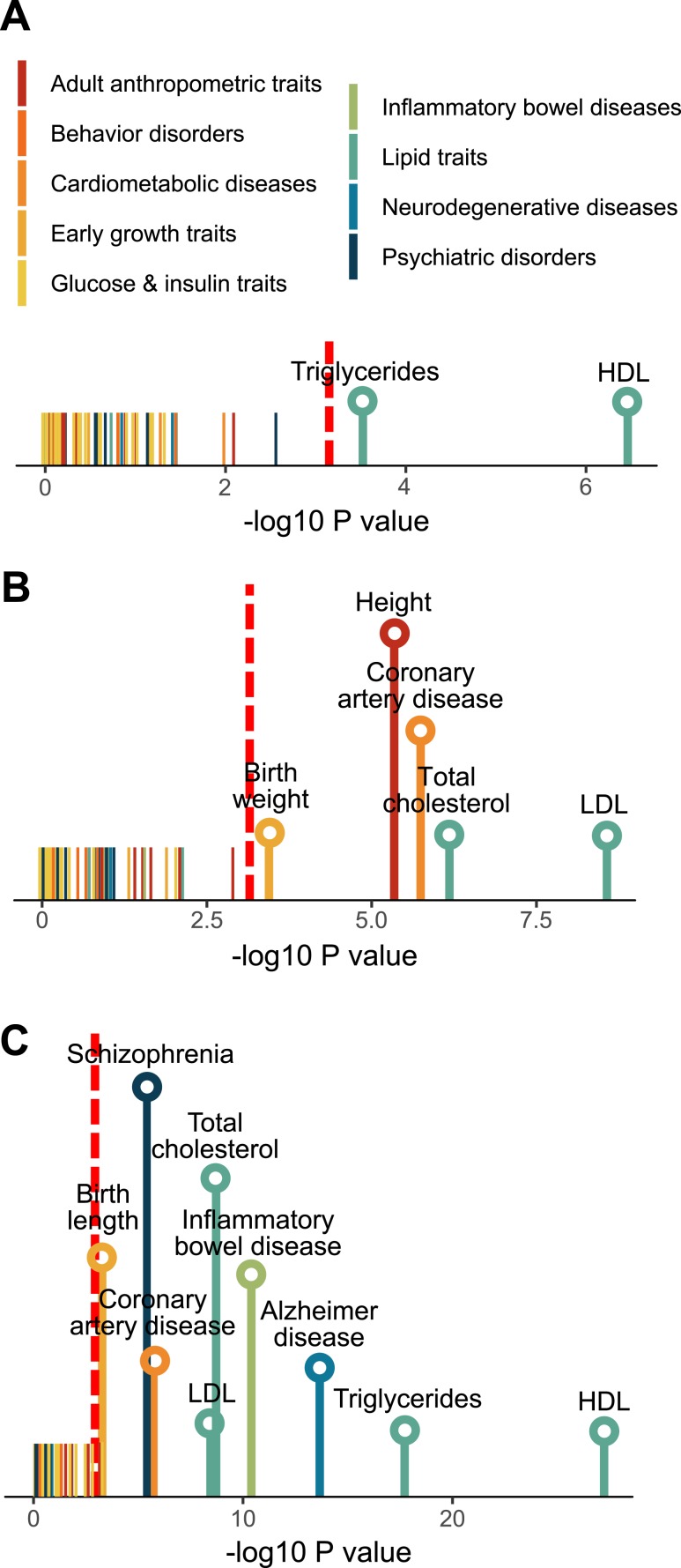
The previous analysis involving the GWAS catalog focused only on small sets of the top candidate genes for various diseases and may have limited statistical power. To improve the statistical power, we curated the full summary statistics from 61 human GWAS that are publicly available (covering millions of SNP-trait associations in each GWAS), which enabled us to extend the assessment of disease association by considering additional human disease genes with moderate to low effect sizes (see “Materials and Methods”). This analysis showed that DEGs from all three tissues exhibited consistent enrichment for genes associated with lipid traits such as TGs, low-density lipoprotein cholesterol, and HDL cholesterol (Fig. 8A–8C). Interestingly, enrichment for birth weight and birth length was also observed for hypothalamus and liver signatures, respectively. Liver DEGs were also significantly associated with coronary artery disease, inflammatory bowel disease, Alzheimer’s disease, and schizophrenia. Top DEGs driving the inflammatory bowel disease association involve immune and inflammatory response genes (PSMB9, TAP1, and TNF), whereas association with Alzheimer’s disease and schizophrenia involves genes related to cholesterol homeostasis (APOA4, ABCG8, and SOAT2) and mitochondrial function (GCDH, PDPR, and SHMT2), respectively. These results suggest that tissue-specific targets of BPA are connected to diverse human complex diseases through both the central nervous system and peripheral tissues.
Figure 8.
Association of differential expression signatures from (A) adipose, (B) hypothalamus, and (C) liver with 61 human traits/diseases, color-coded into nine primary disease categories. P values are determined using MSEA. Red dashed line indicates the cutoff for Bonferroni-corrected P = 0.05. Names of traits/diseases for which P values did not pass Bonferroni-corrected cutoff are not shown. LDL, low-density lipoprotein.
Discussion
This multitissue, multiomics integrative study represents a systems biology investigation of prenatal BPA exposure. By integrating systematic profiling of the transcriptome and methylome of multiple metabolic tissues with phenotypic trait measurements, large-scale human association datasets, and network analysis, we uncovered insights into the molecular regulatory mechanisms underlying the health effects of prenatal BPA exposure. Specifically, we identified tens to thousands of tissue-specific DEGs and DMCs involved in diverse biological functions such as metabolic pathways (oxidative phosphorylation/tricarboxylic acid cycle, fatty acid, cholesterol, glucose metabolism, and PPAR signaling), extracellular matrix, focal adhesion, and inflammation (arachidonic acid) with DMCs partially explaining the regulation of DEGs. Network analysis helped reveal potential regulatory circuits post-BPA exposure and pinpointed both tissue-specific and cross-tissue regulators of BPA activities, including TFs such as estrogen receptors, Pparg, Srebf1, and Hnf1a, and non-TF KDs such as Fasn. We also identified previously under-studied targets such as Cyp51 and lncRNAs across tissues, Fa2h in hypothalamus, and Nfya in adipose tissue. Furthermore, the BPA gene signatures and the predicted regulators were found to be linked to a wide spectrum of disease-related traits in both mouse and human.
Although our multitissue, multiomics design limits the number of biological replicates we could have for each group, the large-scale disruption we observed in the transcriptome and methylome was consistent with previous reports (37, 40, 78, 97), with a number of differential genes and methylation signals replicating previous findings. Our qPCR results further strengthen the validity of our data. Additionally, we focused our analyses on evaluating the aggregated behavior of BPA signatures using both pathway analysis and network modeling to reduce the potential noise and false positives at an individual gene level, because the random chance to have multiple genes in the same pathway to be false positives is much lower. Indeed, we found a generally higher replication rate of the biological pathways between our study and the other studies than replication between the previous studies. Moreover, our unique study design of examining multiomics in multiple tissues in parallel yields higher comparability when integrating the results between data types and across tissues, as they were from the same set of animals and were profiled in the same conditions. For instance, the much larger numbers of DEGs and more coherent network perturbations revealed in the liver tissue compared with the other two tissues derived from the same set of animals suggest that liver might be a more sensitive target tissue for BPA than the other tissues, although adipose and hypothalamus also appear to be important targets.
Across all three tissues at the transcriptome level, we found that lipid metabolism– and energy homeostasis–related processes were consistently perturbed, with the scale of perturbation being strongest in liver. This aligns well with the signficant changes in the plasma lipid profiles we observed in the offspring, the reported perturbation of lipid metabolism in fetal murine liver (78), and the reported susceptibility for nonalcoholic fatty liver diseases following BPA exposure (79, 98, 99). The only shared gene across tissues, Cyp51, encodes a protein that catalyzes metabolic reactions including cholesterol and steroid biosynthesis and biological oxidation (100) and is a critical regulator for testicular spermatogenesis (101). The consistent alteration of Cyp51 across tissues suggests that this gene is a general target of BPA, with the potential to alter functions related to cholesterol, hormone, and energy metabolism. The liver signature replicated across our and previous studies (75, 76) and a top ranked TF regulator in our TF analysis, Srebf1, is a main regulator of lipid homeostasis, again supporting that metabolism is a central target of BPA (75, 76). We also revealed an intriguing link between BPA and lncRNAs across tissues, for which functional importance in developmental processes, disease progression, and response to BPA exposure was increasingly recognized yet underexplored (102). Our molecular data provide intriguing lncRNA candidates such as Gm20319, Gm26917, and Yam1 for future in-depth functional analyses.
For adipose tissue, clusters of genes responsible for core histones were found to be uniquely altered. Along with the strong adipose-specific differential methylation status, our results revealed gonadal adipose tissues as an especially vulnerable site for BPA-induced epigenetic reprogramming. Besides, developmental BPA exposure has been previously suggested to influence white adipocyte differentiation (86, 103, 104). However, the adipocyte differentiation pathway was not significantly enriched in our study. This is consistent with the report by Angle et al. (104), in which increased adipocyte number is only found in mouse offspring with prenatal BPA exposure at 5 μg/kg/d and 500 μg/kg/d, but not 5 mg/kg/d. Additionally, we found significant enrichment for TG biosynthesis and glucose metabolism genes at the differential methylation sites, suggesting that prenatal BPA exposure may affect fat storage and glucose homeostasis in the adipose tissue. Although in this study we mainly investigate gonadal adipose tissue as a surrogate for abdominal fat in the context of metabolic disorders, the information may be useful for exploring the relationship between this fat depot and the gonad.
Concerning the hypothalamus, our study uses next-generation sequencing technology to simultaneously investigate the effect of BPA on the transcriptome and DNA methylome. Hypothalamus is an essential brain region that regulates the endocrine system, peripheral metabolism, and numerous brain functions. We identified BPA-induced DEGs and DMCs that were enriched for extracellular matrix–related processes such as axon guidance, focal adhesion, and various metabolic processes. These hypothalamic pathways have been previously associated with metabolic (50, 51) and neurodegenerative diseases (50, 105), and they could underlie the reported disruption of hypothalamic melanocortin circuitry after BPA exposure (106). Our study highlights the hypothalamus as a critical target for BPA. However, our study used the whole hypothalamus containing heterogeneous nuclei, and future studies to examine individual hypothalamic nuclei such as the arcuate nucleus in the mediobasal hypothalamus will offer better resolution of the specific nuclei and cell types that may be targets of BPA.
By interrogating both the transcriptome and DNA methylome in matching tissues, we were able to directly assess both global and specific correlative relationships between DEGs and DMCs. Specifically, we found that DEGs are more likely to have correlated DMCs in the matching tissue, a trend that persists in nonpromoter regions. Our results corroborate previous findings regarding the importance of gene body methylation in disease etiology (107, 108). Given that >90% of DMCs were found in nonpromoter regions, closer investigation of the regulatory circuits involving these regions may unveil new insights into BPA response (87). Due to the severe multiple testing penalty that limits the statistical power to assess all pairwise correlations, our analysis was restricted to analyzing local relationships between the two molecular scales. For DMCs with no marked correlation to local gene expression, the underlying reasons could be: (i) the long-range gene regulation by DMCs through three-dimensional organization of nucleus, and (ii) the long-term impact on expression changes by DMCs, which is likely missed in our analysis in which DNA methylation and gene expression are measured at the same time point.
Known as an endocrine-disrupting chemical, BPA has been speculated to exert its primary biological action by modifying the activity of hormone receptors, including estrogen receptors, PPARg, and glucocorticoid receptors (90). Indeed, the activity for the downstream targets of Pparg and three estrogen and estrogen-related receptors were found to be disrupted in the liver by prenatal BPA exposure. More importantly, our unbiased data-driven analysis revealed many additional TFs and non-TF regulatory genes that also likely mediate BPA effects. In fact, many of the identified TF targets of BPA, such as Fasn, Srebf1, and several hepatic nuclear factors, showed much higher ranking in our regulator prediction analyses. In liver, a tightly interconnected TF subnetwork was highly concentrated with BPA-affected genes involved in metabolic processes such as cytochrome P450 system (Cyp3a25, Cyp2a12, and Cyp1a2), lipid (Apoa4, Abcg5, and Soat2), and glucose (Hnf1a, Adra1b, and Gck) regulation, with extensive footprints of altered methylation status in the TFs and other subnetwork genes. Therefore, our results support a widespread impact of BPA on liver transcriptional regulation, and the convergence of differential methylation and gene expression in this TF subnetwork implies that BPA perturbs this subnetwork via epigenetic regulation of the TFs, which in turn trigger transcriptomic alterations in downstream genes. In hypothalamus, we identified Fa2h as the strongest KD. This enzyme is highly expressed in the brain and is important for the production of sphingolipids containing 2-hyroxylated fatty acids, the most abundant lipid components of the myelin sheath. Mice lacking Fa2h have impaired myelin maintenance (109), and mutations in human FA2H have been associated with neurodegeneration (110), hereditary spastic paraplegia (111), and autism (112). In adipose, we discovered a regulatory axis governed by Nfya and Fasn that are known regulators of fatty acid metabolism and adipogenesis. NF-YA is a histone-fold domain protein that binds to the inverted CCAAT element in the Fasn promoter (94, 113), and both Nfya and Fasn were found to be significantly perturbed by BPA in our study. Moreover, Fasn also serves as a cross-tissue KD, governing distinct groups of upregulated lipid metabolism genes in adipose and liver post-BPA exposure, supporting its role in mediating the BPA-induced lipid dysregulation at the systemic level. A previous study has also shown BPA-induced effects on Fasn methylation after perinatal exposure (97). The significant correlation of gene expression and methylation for Fasn with TG level furthers implicates its role as a network-level regulator and biomarker for BPA-induced lipid dysregulation. Our observation of Fasn is consistent with evidence suggesting its susceptibility to methylation perturbation under obesogenic feeding (114) and its causal functional importance for fatty liver diseases (52, 115). The causal regulatory role of these genes in BPA activities warrants future testing via genetic manipulation studies, such as knocking down or overexpressing the predicted regulators to examine their ability to modulate BPA activities.
One unique aspect of this study is the linking of the molecular landscape of prenatal BPA exposure to traits/diseases in both mouse and human. In our mouse study, the observed changes in body weight, lipid profiles, and glucose level are highly concordant with the functions of the molecular targets. For instance, prenatal BPA exposure perturbs both the expression levels and local DNA methylation status of Fasn, Igf1r, and Adh1. These DEGs and their local DMCs also significantly correlate with phenotypic outcomes, thus serving as examples of how DNA methylation and gene regulation bridge the gap between BPA exposure and phenotypic manifestation. To further enhance the translatability of our findings from mouse to human, we searched for human diseases linked to the BPA-affected genes. An intriguing discovery is the prominent overrepresentation of differential methylation signals in adipose, hypothalamus, and liver within known genes related to obesity and type 2 diabetes, supporting that BPA may affect obesity and diabetes risk through systemic reprogramming of DNA methylation. More sophisticated analyses incorporating the BPA differential gene expression and the full statistics of human GWAS corroborated the observed connection between prenatal BPA exposure and lipid homeostasis (116), birth weight (117), and coronary artery disease (15) reported in observational studies. Moreover, our findings suggest the involvement of prenatal BPA exposure in the development of inflammatory bowel syndrome, schizophrenia, and Alzheimer’s disease. These associations warrant future investigations.
Designed as the discovery phase of a comprehensive investigation of in vivo BPA activities, our study opens numerous future lines of investigation. First, our current molecular studies focused on male tissues because of the stronger phenotypes observed in males. Our phenotypic examination and qPCR experiments on females support much subtler changes in females, and future studies will require larger sample sizes to uncover female-specific biology. Secondly, our study design does not address whether the observed BPA genomic effects are direct or indirect, and radiotracing or substrate-binding experiments are needed to elucidate this question. Thirdly, the causal link between the genomic effects observed and the phenotypes that result from BPA exposure is not established, and genetic perturbation experiments are required to test the causal roles of the predicted regulators of BPA actions. Gene annotation accuracy may also affect the results and interpretation. Lastly, we tested in utero BPA exposure at one dose via oral gavage, which can cause prenatal stress and confound the results, and examined phenotypes and molecular profiles only at weaning age as a proof-of-concept for our systems biology framework. Considering that the effects of early-life exposure to BPA are highly variable and dependent on factors such as the dose, window, route (e.g., using food as an alternative), and frequency of exposure as well as genetic background, age, and sex (14), future studies testing these additional variables using large sample sizes are necessary to generate a comprehensive understandings of BPA risks under various exposure conditions.
In summary, our study represents a multitissue, multiomics integrative investigation of prenatal BPA exposure. The systems biology framework we applied revealed how BPA triggers cascades of regulatory circuits involving numerous TFs and non-TF regulators that coordinate diverse molecular processes within and across core metabolic tissues, thereby highlighting that BPA exerts its biological functions via much more diverse targets than previously thought. As such, our findings offer a comprehensive systems-level understanding of tissue sensitivity and molecular perturbations elicited by prenatal BPA exposure and offer promising candidates for targeted mechanistic investigation as well as much-needed network-level biomarkers of prior BPA exposure. The strong influence of BPA on metabolic pathways and cardiometabolic phenotypes merits its characterization as a general metabolic disruptor posing systemic health risks.
Acknowledgments
We thank Zhe Ying for assistance in collecting mice hypothalamus tissue and Dr. Guanglin Zhang for assistance in the RRBS experiments.
Financial Support: L.S. is supported by a University of California, Los Angeles, Dissertation Year Fellowship, Eureka Scholarship, Hyde Scholarship, Burroughs Wellcome Fund Inter-School Program in Metabolic Diseases Fellowship, and the China Scholarship Council. G.D. is supported by NIEHS/National Institutes of Health Grant T32ES015457. P.A. is supported by National Institutes of Health/NIEHS Grant R01-ES02748701 and the Burroughs Wellcome Foundation. X.Y. is supported by National Institutes of Health Grant DK104363 and the Leducq Foundation.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- BMI
body mass index
- BN
Bayesian network
- BPA
bisphenol A
- DEG
differentially expressed gene
- DMC
differentially methylated CpG
- EDC
endocrine-disrupting chemical
- FDR
false discovery rate
- FFA
free fatty acid
- GEO
Gene Expression Omnibus
- GWAS
genome-wide association studies
- KD
key driver
- lncRNA
long noncoding RNA
- MetS
metabolic syndrome
- MSEA
Marker Set Enrichment Analysis
- NIEHS
National Institute for Environmental Health Sciences
- PPAR
peroxisome proliferator–activated receptor
- qPCR
quantitative RT-PCR
- RNA-seq
RNA-sequencing
- RRBS
reduced representation bisulfite sequencing
- SNP
single nucleotide polymorphism
- TF
transcription factor
- TG
triglyceride
- wKDA
Weighted Key Driver Analysis
References
- 1. Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. Developmental origins of non-communicable disease: implications for research and public health. Environ Health. 2012;11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boekelheide K, Blumberg B, Chapin RE, Cote I, Graziano JH, Janesick A, Lane R, Lillycrop K, Myatt L, States JC, Thayer KA, Waalkes MP, Rogers JM. Predicting later-life outcomes of early-life exposures. Environ Health Perspect. 2012;120(10):1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heindel JJ, Vandenberg LN. Developmental origins of health and disease: a paradigm for understanding disease cause and prevention. Curr Opin Pediatr. 2015;27(2):248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect. 2008;116(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haugen AC, Schug TT, Collman G, Heindel JJ. Evolution of DOHaD: the impact of environmental health sciences. J Dev Orig Health Dis. 2015;6(2):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai WT. Human health risk on environmental exposure to bisphenol-A: a review. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2006;24(2):225–255. [DOI] [PubMed] [Google Scholar]
- 7. Sun C, Leong LP, Barlow PJ, Chan SH, Bloodworth BC. Single laboratory validation of a method for the determination of Bisphenol A, Bisphenol A diglycidyl ether and its derivatives in canned foods by reversed-phase liquid chromatography. J Chromatogr A. 2006;1129(1):145–148. [DOI] [PubMed] [Google Scholar]
- 8. Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA). Reprod Toxicol. 2007;24(2):139–177. [DOI] [PubMed] [Google Scholar]
- 9. Rubin BS, Paranjpe M, DaFonte T, Schaeberle C, Soto AM, Obin M, Greenberg AS. Perinatal BPA exposure alters body weight and composition in a dose specific and sex specific manner: The addition of peripubertal exposure exacerbates adverse effects in female mice. Reprod Toxicol. 2017;68:130–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hao M, Ding L, Xuan L, Wang T, Li M, Zhao Z, Lu J, Xu Y, Chen Y, Wang W, Bi Y, Xu M, Ning G. Urinary bisphenol A concentration and the risk of central obesity in Chinese adults: A prospective study. J Diabetes. 2018;10(6):442–448. [DOI] [PubMed] [Google Scholar]
- 11. Beydoun HA, Khanal S, Zonderman AB, Beydoun MA. Sex differences in the association of urinary bisphenol-A concentration with selected indices of glucose homeostasis among U.S. adults. Ann Epidemiol. 2014;24(2):90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teppala S, Madhavan S, Shankar A. Bisphenol A and metabolic syndrome: Results from NHANES. Int J Endocrinol. 2012;2012:598180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mouneimne Y, Nasrallah M, Khoueiry-Zgheib N, Nasreddine L, Nakhoul N, Ismail H, Abiad M, Koleilat L, Tamim H. Bisphenol A urinary level, its correlates, and association with cardiometabolic risks in Lebanese urban adults. Environ Monit Assess. 2017;189(10):517. [DOI] [PubMed] [Google Scholar]
- 14. Wassenaar PNH, Trasande L, Legler J. Systematic review and meta-analysis of early-life exposure to bisphenol A and obesity-related outcomes in rodents. Environ Health Perspect. 2017;125(10):106001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han C, Hong YC. Bisphenol A, hypertension, and cardiovascular diseases: epidemiological, laboratory, and clinical trial evidence. Curr Hypertens Rep. 2016;18(2):11. [DOI] [PubMed] [Google Scholar]
- 16. Ranciere F, Lyons JG, Loh VH, Botton J, Galloway T, Wang T, Shaw JE, Magliano DJ. Bisphenol A and the risk of cardiometabolic disorders: a systematic review with meta-analysis of the epidemiological evidence. Environ Health. 2015;14:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stahlhut RW, Myers JP, Taylor JA, Nadal A, Dyer JA, vom Saal FS. Experimental BPA exposure and glucose-stimulated insulin response in adult men and women. J Endocrine Soc. 2018;2(10):1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J, Yu P, Qian W, Li Y, Zhao J, Huan F, Wang J, Xiao H. Perinatal bisphenol A exposure and adult glucose homeostasis: identifying critical windows of exposure. PLoS One. 2013;8(5):e64143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryan KK, Haller AM, Sorrell JE, Woods SC, Jandacek RJ, Seeley RJ. Perinatal exposure to bisphenol-a and the development of metabolic syndrome in CD-1 mice. Endocrinology. 2010;151(6):2603–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyawaki J, Sakayama K, Kato H, Yamamoto H, Masuno H. Perinatal and postnatal exposure to bisphenol A increases adipose tissue mass and serum cholesterol level in mice. J Atheroscler Thromb. 2007;14(5):245–252. [DOI] [PubMed] [Google Scholar]
- 21. Rubin BS, Soto AM. Bisphenol A: Perinatal exposure and body weight. Mol Cell Endocrinol. 2009;304(1-2):55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. García-Arevalo M, Alonso-Magdalena P, Rebelo Dos Santos J, Quesada I, Carneiro EM, Nadal A. Exposure to bisphenol-A during pregnancy partially mimics the effects of a high-fat diet altering glucose homeostasis and gene expression in adult male mice. PLoS One. 2014;9(6):e100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 2013;8(1):e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Susiarjo M, Xin F, Bansal A, Stefaniak M, Li C, Simmons RA, Bartolomei MS. Bisphenol A exposure disrupts metabolic health across multiple generations in the mouse. Endocrinology. 2015;156(6):2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bansal A, Rashid C, Xin F, Li C, Polyak E, Duemler A, van der Meer T, Stefaniak M, Wajid S, Doliba N, Bartolomei MS, Simmons RA. Sex- and dose-specific effects of maternal bisphenol A exposure on pancreatic islets of first- and second-generation adult mice offspring. Environ Health Perspect. 2017;125(9):097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Camacho J, Truong L, Kurt Z, Chen YW, Morselli M, Gutierrez G, Pellegrini M, Yang X, Allard P. The memory of environmental chemical exposure in C. elegans is dependent on the Jumonji demethylases jmjd-2 and jmjd-3/utx-1. Cell Reports. 2018;23(8):2392–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baillie-Hamilton PF. Chemical toxins: a hypothesis to explain the global obesity epidemic. J Altern Complement Med. 2002;8(2):185–192. [DOI] [PubMed] [Google Scholar]
- 28. Heindel JJ. Endocrine disruptors and the obesity epidemic. Toxicol Sci. 2003;76(2):247–249. [DOI] [PubMed] [Google Scholar]
- 29. Newbold RR, Padilla-Banks E, Jefferson WN, Heindel JJ. Effects of endocrine disruptors on obesity. Int J Androl. 2008;31(2):201–208. [DOI] [PubMed] [Google Scholar]
- 30. European Food Safety Authority Scientific opinion on the risks to public health related to the presence of bisphenol A (BPA) in foodstuffs. EFSA J. 2015;13(1):3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bisphenol A. (BPA) Joint Emerging Science Working Group; Department of Health and Human Services. 2014 updated review of literature and data on bisphenol A (CAS RN 80-05-7). https://www.fda.gov/downloads/food/ingredientspackaginglabeling/foodadditivesingredients/ucm424071.pdf. Published 6 June 2014. Accessed 8 October 2017.
- 32. National Toxicology Program NTP research report on the CLARITY-BPA core study: a perinatal and chronic extended-dose-range study of bisphenol A in rats. NTP RR 9. 10.22427/NTP-RR-9. Published September 2018. Accessed 16 November 2018. [DOI] [PubMed]
- 33. Beronius A, Johansson N, Rudén C, Hanberg A. The influence of study design and sex-differences on results from developmental neurotoxicity studies of bisphenol A: implications for toxicity testing. Toxicology. 2013;311(1-2):13–26. [DOI] [PubMed] [Google Scholar]
- 34. Ariemma F, D’Esposito V, Liguoro D, Oriente F, Cabaro S, Liotti A, Cimmino I, Longo M, Beguinot F, Formisano P, Valentino R. Low-dose bisphenol-A impairs adipogenesis and generates dysfunctional 3T3-L1 adipocytes. PLoS One. 2016;11(3):e0150762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ben-Jonathan N, Hugo ER, Brandebourg TD. Effects of bisphenol A on adipokine release from human adipose tissue: Implications for the metabolic syndrome. Mol Cell Endocrinol. 2009;304(1-2):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olsvik PA, Skjærven KH, Søfteland L. Metabolic signatures of bisphenol A and genistein in Atlantic salmon liver cells. Chemosphere. 2017;189:730–743. [DOI] [PubMed] [Google Scholar]
- 37. Lejonklou MH, Dunder L, Bladin E, Pettersson V, Rönn M, Lind L, Waldén TB, Lind PM. Effects of low-dose developmental bisphenol A exposure on metabolic parameters and gene expression in male and female Fischer 344 rat offspring. Environ Health Perspect. 2017;125(6):067018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anderson OS, Kim JH, Peterson KE, Sanchez BN, Sant KE, Sartor MA, Weinhouse C, Dolinoy DC. Novel epigenetic biomarkers mediating bisphenol A exposure and metabolic phenotypes in female mice. Endocrinology. 2017;158(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma Y, Xia W, Wang DQ, Wan YJ, Xu B, Chen X, Li YY, Xu SQ. Hepatic DNA methylation modifications in early development of rats resulting from perinatal BPA exposure contribute to insulin resistance in adulthood. Diabetologia. 2013;56(9):2059–2067. [DOI] [PubMed] [Google Scholar]
- 40. Taylor JA, Shioda K, Mitsunaga S, Yawata S, Angle BM, Nagel SC, Vom Saal FS, Shioda T. Prenatal exposure to bisphenol A disrupts naturally occurring bimodal DNA methylation at proximal promoter of fggy, an obesity-relevant gene encoding a carbohydrate kinase, in gonadal white adipose tissues of CD-1 mice. Endocrinology. 2018;159(2):779–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Faulk C, Kim JH, Jones TR, McEachin RC, Nahar MS, Dolinoy DC, Sartor MA. Bisphenol A-associated alterations in genome-wide DNA methylation and gene expression patterns reveal sequence-dependent and non-monotonic effects in human fetal liver. Environ Epigenet. 2015;1(1):dvv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nahar MS, Kim JH, Sartor MA, Dolinoy DC. Bisphenol A-associated alterations in the expression and epigenetic regulation of genes encoding xenobiotic metabolizing enzymes in human fetal liver. Environ Mol Mutagen. 2014;55(3):184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang T, Pehrsson EC, Purushotham D, Li D, Zhuo X, Zhang B, Lawson HA, Province MA, Krapp C, Lan Y, Coarfa C, Katz TA, Tang WY, Wang Z, Biswal S, Rajagopalan S, Colacino JA, Tsai ZT, Sartor MA, Neier K, Dolinoy DC, Pinto J, Hamanaka RB, Mutlu GM, Patisaul HB, Aylor DL, Crawford GE, Wiltshire T, Chadwick LH, Duncan CG, Garton AE, McAllister KA, Bartolomei MS, Walker CL, Tyson FL; TaRGET II Consortium . The NIEHS TaRGET II Consortium and environmental epigenomics. Nat Biotechnol. 2018;36(3):225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Messerlian C, Martinez RM, Hauser R, Baccarelli AA. ‘Omics’ and endocrine-disrupting chemicals - new paths forward. Nat Rev Endocrinol. 2017;13(12):740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. López M, Nogueiras R, Tena-Sempere M, Diéguez C. Hypothalamic AMPK: a canonical regulator of whole-body energy balance. Nat Rev Endocrinol. 2016;12(7):421–432. [DOI] [PubMed] [Google Scholar]
- 46. Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4(1):177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choe SS, Huh JY, Hwang IJ, Kim JI, Kim JB. Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Front Endocrinol (Lausanne). 2016;7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci. 2013;9(2):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mäkinen VP, Civelek M, Meng Q, Zhang B, Zhu J, Levian C, Huan T, Segrè AV, Ghosh S, Vivar J, Nikpay M, Stewart AF, Nelson CP, Willenborg C, Erdmann J, Blakenberg S, O’Donnell CJ, März W, Laaksonen R, Epstein SE, Kathiresan S, Shah SH, Hazen SL, Reilly MP, Lusis AJ, Samani NJ, Schunkert H, Quertermous T, McPherson R, Yang X, Assimes TL; Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium . Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10(7):e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meng Q, Ying Z, Noble E, Zhao Y, Agrawal R, Mikhail A, Zhuang Y, Tyagi E, Zhang Q, Lee JH, Morselli M, Orozco L, Guo W, Kilts TM, Zhu J, Zhang B, Pellegrini M, Xiao X, Young MF, Gomez-Pinilla F, Yang X. Systems nutrigenomics reveals brain gene networks linking metabolic and brain disorders. EBioMedicine. 2016;7:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shu L, Chan KHK, Zhang G, Huan T, Kurt Z, Zhao Y, Codoni V, Trégouët DA, Yang J, Wilson JG, Luo X, Levy D, Lusis AJ, Liu S, Yang X; Cardiogenics Consortium . Shared genetic regulatory networks for cardiovascular disease and type 2 diabetes in multiple populations of diverse ethnicities in the United States. PLoS Genet. 2017;13(9):e1007040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chella Krishnan K, Kurt Z, Barrere-Cain R, Sabir S, Das A, Floyd R, Vergnes L, Zhao Y, Che N, Charugundla S, Qi H, Zhou Z, Meng Y, Pan C, Seldin MM, Norheim F, Hui S, Reue K, Lusis AJ, Yang X. Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst. 2018;6(1):103–115.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104(32):13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol A exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9(4):e1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24(7):2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kitchin KT, Ebron MT. Further development of rodent whole embryo culture: solvent toxicity and water insoluble compound delivery system. Toxicology. 1984;30(1):45–57. [DOI] [PubMed] [Google Scholar]
- 57. Shu L, Meng Q, Diamante G, Tsai B, Chen Y-W, Mikhail A, Luk H, Ritz B, Allard P, Yang X. Data from: Prenatal bisphenol A exposure in mice induces multitissue multiomics disruptions linking to cardiometabolic disorders. figshare 2018. Deposited 12 November 2018 10.6084/m9.figshare.7451069.v2. [DOI] [PMC free article] [PubMed]
- 58. SEQC/MAQC-III Consortium A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32(9):903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meng Q, Zhuang Y, Ying Z, Agrawal R, Yang X, Gomez-Pinilla F. Traumatic brain injury induces genome-wide transcriptomic, methylomic, and network Perturbations in brain and blood predicting neurological disorders. EBioMedicine. 2017;16:184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen Y, Shu L, Qiu Z, Lee DY, Settle SJ, Que Hee S, Telesca D, Yang X, Allard P. Exposure to the BPA-substitute Bisphenol S causes unique alterations of germline function. PLoS Genet. 2016;12(7):e1006223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30(3):301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews S. Fast QC: a quality control tool for high throughput sequence data. Available at: www.bioinformatics.babraham.ac.uk/projects. Accessed 10 December 2015.
- 63. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10(1):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun D, Xi Y, Rodriguez B, Park HJ, Tong P, Meong M, Goodell MA, Li W. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 2014;15(2):R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics. 2017;33(15):2381–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shu L, Zhao Y, Kurt Z, Byars SG, Tukiainen T, Kettunen J, Orozco LD, Pellegrini M, Lusis AJ, Ripatti S, Zhang B, Inouye M, Mäkinen VP, Yang X. Mergeomics: multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genomics. 2016;17(1):874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marbach D, Lamparter D, Quon G, Kellis M, Kutalik Z, Bergmann S. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016;13(4):366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Marmugi A, Ducheix S, Lasserre F, Polizzi A, Paris A, Priymenko N, Bertrand-Michel J, Pineau T, Guillou H, Martin PG, Mselli-Lakhal L. Low doses of bisphenol A induce gene expression related to lipid synthesis and trigger triglyceride accumulation in adult mouse liver. Hepatology. 2012;55(2):395–407. [DOI] [PubMed] [Google Scholar]
- 76. Melis JP, Derks KW, Pronk TE, Wackers P, Schaap MM, Zwart E, van Ijcken WF, Jonker MJ, Breit TM, Pothof J, van Steeg H, Luijten M. In vivo murine hepatic microRNA and mRNA expression signatures predicting the (non-)genotoxic carcinogenic potential of chemicals. Arch Toxicol. 2014;88(4):1023–1034. [DOI] [PubMed] [Google Scholar]
- 77. Meng Z, Wang D, Yan S, Li R, Yan J, Teng M, Zhou Z, Zhu W. Effects of perinatal exposure to BPA and its alternatives (BPS, BPF and BPAF) on hepatic lipid and glucose homeostasis in female mice adolescent offspring. Chemosphere. 2018;212:297–306. [DOI] [PubMed] [Google Scholar]
- 78. Ilagan Y, Mamillapalli R, Goetz LG, Kayani J, Taylor HS. Bisphenol-A exposure in utero programs a sexually dimorphic estrogenic state of hepatic metabolic gene expression. Reprod Toxicol. 2017;71:84–94. [DOI] [PubMed] [Google Scholar]
- 79. Shimpi PC, More VR, Paranjpe M, Donepudi AC, Goodrich JM, Dolinoy DC, Rubin B, Slitt AL. Hepatic lipid accumulation and Nrf2 expression following perinatal and peripubertal exposure to bisphenol A in a mouse model of nonalcoholic liver disease. Environ Health Perspect. 2017;125(8):087005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Susiarjo M, Xin F, Stefaniak M, Mesaros C, Simmons RA, Bartolomei MS. Bile acids and tryptophan metabolism are novel pathways involved in metabolic abnormalities in BPA-exposed pregnant mice and male offspring. Endocrinology. 2017;158(8):2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang D, Zhu W, Yan S, Meng Z, Yan J, Teng M, Jia M, Li R, Zhou Z. Impaired lipid and glucose homeostasis in male mice offspring after combined exposure to low-dose bisphenol A and arsenic during the second half of gestation. Chemosphere. 2018;210:998–1005. [DOI] [PubMed] [Google Scholar]
- 82. Nishizawa H, Imanishi S, Manabe N. Effects of exposure in utero to bisphenol A on the expression of aryl hydrocarbon receptor, related factors, and xenobiotic metabolizing enzymes in murine embryos. J Reprod Dev. 2005;51(5):593–605. [DOI] [PubMed] [Google Scholar]
- 83. Johnson SA, Spollen WG, Manshack LK, Bivens NJ, Givan SA, Rosenfeld CS. Hypothalamic transcriptomic alterations in male and female California mice (Peromyscus californicus) developmentally exposed to bisphenol A or ethinyl estradiol. Physiol Rep. 2017;5(3):e13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Arambula SE, Belcher SM, Planchart A, Turner SD, Patisaul HB. Impact of low dose oral exposure to bisphenol A (BPA) on the neonatal rat hypothalamic and hippocampal transcriptome: a CLARITY-BPA Consortium study. Endocrinology. 2016;157(10):3856–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cheong A, Johnson SA, Howald EC, Ellersieck MR, Camacho L, Lewis SM, Vanlandingham MM, Ying J, Ho SM, Rosenfeld CS. Gene expression and DNA methylation changes in the hypothalamus and hippocampus of adult rats developmentally exposed to bisphenol A or ethinyl estradiol: a CLARITY-BPA consortium study. Epigenetics. 2018;13(7):704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Somm E, Schwitzgebel VM, Toulotte A, Cederroth CR, Combescure C, Nef S, Aubert ML, Hüppi PS. Perinatal exposure to bisphenol A alters early adipogenesis in the rat. Environ Health Perspect. 2009;117(10):1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lou S, Lee HM, Qin H, Li JW, Gao Z, Liu X, Chan LL, Kl Lam V, So WY, Wang Y, Lok S, Wang J, Ma RC, Tsui SK, Chan JC, Chan TF, Yip KY. Whole-genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol. 2014;15(7):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Faulk C, Kim JH, Anderson OS, Nahar MS, Jones TR, Sartor MA, Dolinoy DC. Detection of differential DNA methylation in repetitive DNA of mice and humans perinatally exposed to bisphenol A. Epigenetics. 2016;11(7):489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Acconcia F, Pallottini V, Marino M. Molecular mechanisms of action of BPA. Dose Response. 2015;13(4):1559325815610582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. MacKay H, Abizaid A. A plurality of molecular targets: The receptor ecosystem for bisphenol-A (BPA). Horm Behav. 2018;101:59–67. [DOI] [PubMed] [Google Scholar]
- 91. Wang J, Sun B, Hou M, Pan X, Li X. The environmental obesogen bisphenol A promotes adipogenesis by increasing the amount of 11β-hydroxysteroid dehydrogenase type 1 in the adipose tissue of children. Int J Obes. 2013;37(7):999–1005. [DOI] [PubMed] [Google Scholar]
- 92. Ahmed S, Atlas E. Bisphenol S- and bisphenol A-induced adipogenesis of murine preadipocytes occurs through direct peroxisome proliferator-activated receptor gamma activation. Int J Obes. 2016;40(10):1566–1573. [DOI] [PubMed] [Google Scholar]
- 93. Vafeiadi M, Roumeliotaki T, Myridakis A, Chalkiadaki G, Fthenou E, Dermitzaki E, Karachaliou M, Sarri K, Vassilaki M, Stephanou EG, Kogevinas M, Chatzi L. Association of early life exposure to bisphenol A with obesity and cardiometabolic traits in childhood. Environ Res. 2016;146:379–387. [DOI] [PubMed] [Google Scholar]
- 94. Nishi-Tatsumi M, Yahagi N, Takeuchi Y, Toya N, Takarada A, Murayama Y, Aita Y, Sawada Y, Piao X, Oya Y, Shikama A, Masuda Y, Kubota M, Izumida Y, Matsuzaka T, Nakagawa Y, Sekiya M, Iizuka Y, Kawakami Y, Kadowaki T, Yamada N, Shimano H. A key role of nuclear factor Y in the refeeding response of fatty acid synthase in adipocytes. FEBS Lett. 2017;591(7):965–978. [DOI] [PubMed] [Google Scholar]
- 95. Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153(3):707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Inadera H. Neurological effects of bisphenol A and its analogues. Int J Med Sci. 2015;12(12):926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim JH, Sartor MA, Rozek LS, Faulk C, Anderson OS, Jones TR, Nahar MS, Dolinoy DC. Perinatal bisphenol A exposure promotes dose-dependent alterations of the mouse methylome. BMC Genomics. 2014;15(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ke ZH, Pan JX, Jin LY, Xu HY, Yu TT, Ullah K, Rahman TU, Ren J, Cheng Y, Dong XY, Sheng JZ, Huang HF. Bisphenol A exposure may induce hepatic lipid accumulation via reprogramming the DNA methylation patterns of genes involved in lipid metabolism. Sci Rep. 2016;6(1):31331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yang S, Zhang A, Li T, Gao R, Peng C, Liu L, Cheng Q, Mei M, Song Y, Xiang X, Wu C, Xiao X, Li Q. Dysregulated autophagy in hepatocytes promotes bisphenol A-induced hepatic lipid accumulation in male mice. Endocrinology. 2017;158(9):2799–2812. [DOI] [PubMed] [Google Scholar]
- 100. Lewinska M, Juvan P, Perse M, Jeruc J, Kos S, Lorbek G, Urlep Z, Keber R, Horvat S, Rozman D. Hidden disease susceptibility and sexual dimorphism in the heterozygous knockout of Cyp51 from cholesterol synthesis. PLoS One. 2014;9(11):e112787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Keber R, Rozman D, Horvat S. Sterols in spermatogenesis and sperm maturation. J Lipid Res. 2013;54(1):20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Karlsson O, Baccarelli AA. Environmental health and long non-coding RNAs. Curr Environ Health Rep. 2016;3(3):178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Vom Saal FS, Nagel SC, Coe BL, Angle BM, Taylor JA. The estrogenic endocrine disrupting chemical bisphenol A (BPA) and obesity. Mol Cell Endocrinol. 2012;354(1-2):74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Angle BM, Do RP, Ponzi D, Stahlhut RW, Drury BE, Nagel SC, Welshons WV, Besch-Williford CL, Palanza P, Parmigiani S, vom Saal FS, Taylor JA. Metabolic disruption in male mice due to fetal exposure to low but not high doses of bisphenol A (BPA): evidence for effects on body weight, food intake, adipocytes, leptin, adiponectin, insulin and glucose regulation. Reprod Toxicol. 2013;42:256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vercruysse P, Vieau D, Blum D, Petersén Å, Dupuis L. Hypothalamic alterations in neurodegenerative diseases and their relation to abnormal energy metabolism. Front Mol Neurosci. 2018;11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. MacKay H, Patterson ZR, Abizaid A. Perinatal exposure to low-dose bisphenol-A disrupts the structural and functional development of the hypothalamic feeding circuitry. Endocrinology. 2017;158(4):768–777. [DOI] [PubMed] [Google Scholar]
- 107. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. [DOI] [PubMed] [Google Scholar]
- 108. Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9(6):823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zöller I, Meixner M, Hartmann D, Büssow H, Meyer R, Gieselmann V, Eckhardt M. Absence of 2-hydroxylated sphingolipids is compatible with normal neural development but causes late-onset axon and myelin sheath degeneration. J Neurosci. 2008;28(39):9741–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zaki MS, Selim L, Mansour L, Mahmoud IG, Fenstermaker AG, Gabriel SB, Gleeson JG. Mutations in FA2H in three Arab families with a clinical spectrum of neurodegeneration and hereditary spastic paraparesis. Clin Genet. 2015;88(1):95–97. [DOI] [PubMed] [Google Scholar]
- 111. Liao X, Luo Y, Zhan Z, Du J, Hu Z, Wang J, Guo J, Hu Z, Yan X, Pan Q, Xia K, Tang B, Shen L. SPG35 contributes to the second common subtype of AR-HSP in China: frequency analysis and functional characterization of FA2H gene mutations. Clin Genet. 2015;87(1):85–89. [DOI] [PubMed] [Google Scholar]
- 112. Scheid I, Maruani A, Huguet G, Leblond CS, Nygren G, Anckarsäter H, Beggiato A, Rastam M, Amsellem F, Gillberg IC, Elmaleh M, Leboyer M, Gillberg C, Betancur C, Coleman M, Hama H, Cook EH, Bourgeron T, Delorme R. Heterozygous FA2H mutations in autism spectrum disorders. BMC Med Genet. 2013;14(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, Jothi R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell. 2014;55(5):708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gracia A, Elcoroaristizabal X, Fernández-Quintela A, Miranda J, Bediaga NG, M de Pancorbo M, Rimando AM, Portillo MP. Fatty acid synthase methylation levels in adipose tissue: effects of an obesogenic diet and phenol compounds. Genes Nutr. 2014;9(4):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hui ST, Parks BW, Org E, Norheim F, Che N, Pan C, Castellani LW, Charugundla S, Dirks DL, Psychogios N, Neuhaus I, Gerszten RE, Kirchgessner T, Gargalovic PS, Lusis AJ. The genetic architecture of NAFLD among inbred strains of mice. eLife. 2015;4:e05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dallio M, Masarone M, Errico S, Gravina AG, Nicolucci C, Di Sarno R, Gionti L, Tuccillo C, Persico M, Stiuso P, Diano N, Loguercio C, Federico A. Role of bisphenol A as environmental factor in the promotion of non-alcoholic fatty liver disease: in vitro and clinical study. Aliment Pharmacol Ther. 2018;47(6):826–837. [DOI] [PubMed] [Google Scholar]
- 117. Veiga-Lopez A, Kannan K, Liao C, Ye W, Domino SE, Padmanabhan V. Gender-specific effects on gestational length and birth weight by early pregnancy BPA exposure. J Clin Endocrinol Metab. 2015;100(11):E1394–E1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental methods, figures, and tables are available at Figshare (doi.org/10.6084/m9.figshare.7451069.v2) (57). RNA-seq and RRBS data have been submitted to the Gene Expression Omnibus (GEO) under accession numbers GSE121603 (for RNA-seq) and GSE121604 (for RRBS).