Abstract
N-methyl-D-aspartate receptors (NMDARs) are widely distributed in the brain with high concentrations in the telencephalon where they modulate synaptic plasticity, working memory, and other functions. While the actions of the predominate GluN2 NMDAR subunits, GluN2A and GluN2B are relatively well understood, the function of GluN2C and GluN2D subunits in the telencephalon is largely unknown. To better understand the possible role of GluN2C subunits, we used fluorescence in situ hybridization (FISH) together with multiple cell markers to define the distribution and type of cells expressing GluN2C mRNA. Using a GluN2C-KO mouse as a negative control, GluN2C mRNA expression was only found in non-neuronal cells (NeuN-negative cells) in the hippocampus, striatum, amygdala, and cerebral cortex. For these regions, a significant fraction of GFAP-positive cells also expressed GluN2C mRNA. Overall, for the telencephalon, the globus pallidus and olfactory bulb were the only regions where GluN2C was expressed in neurons. In contrast to GluN2C, GluN2D subunit mRNA colocalized with neuronal and not astrocyte markers or GluN2C mRNA in the telencephalon (except for the globus pallidus). GluN2C mRNA did, however, colocalize with GluN2D in the thalamus where neuronal GluN2C expression is found. These findings strongly suggest that GluN2C has a very distinct function in the telencephalon compared to its role in other brain regions and compared to other GluN2-containing NMDARs. NMDARs containing GluN2C may have a specific role in regulating L-glutamate or D-serine release from astrocytes in response to L-glutamate spillover from synaptic activity.
Keywords: N-methyl-D-aspartate receptor, L-glutamate, astrocyte, cortex, hippocampus, mRNA, GluN2C, GluN2D
Introduction
N-methyl-D-aspartate receptors (NMDARs) are a family of ligand-gated ion channels activated by the primary excitatory neurotransmitter of the central nervous system, L-glutamate. NMDARs have important roles in cognition, working memory, and synaptic plasticity but they are also involved in a number of neurological/psychiatric disorders such as schizophrenia, traumatic brain injury, Alzheimer disease, epilepsy, and depression [1,2]. These complex actions of NMDARs are generated by a diverse family of NMDAR subunits which display distinct biochemical and physiological properties.
NMDARs are tetrameric assemblies containing two GluN1 subunits and two subunits from the GluN2 (GluN2A-GluN2D) and/or GluN3 (GluN3A, GluN3B) subunit families. Receptors with different GluN2 subunits have distinct functional properties in terms of voltage-dependency, agonist affinity, and deactivation kinetics [2–6]. These properties, together with the varied distributions of the GluN2 subunits, can account for much of the functional diversity of NMDARs. To date, most studies of NMDAR function in the telencephalon have focused upon GluN2A and GluN2B subunits because these are the predominant GluN2 subunits in the forebrain. GluN2C-containing NMDARs are important for cerebellar function/motor coordination [7,8] and thalamic reticular nucleus modulation of delta oscillations [9], but relatively little is known about the role of GluN2C-containing NMDARs in the telencephalon.
While the distribution of GluN2C subunit expression has been well characterized, an important question remains as to their localization in the telencephalon. GluN2C subunits are highly expressed in cerebellar granular cells, moderately expressed in various lateral thalamic nuclei and in the mitral cells and glomerular layer of the olfactory bulb, and weakly expressed in the telencephalon (cortex, hippocampus, amygdala, and striatum) [4,10]. However, the identity of the cells expressing GluN2C in the telencephalon has been unclear. In the rodent cortex, hippocampus, and striatum, GluN2C (and GluN2D) have a diffuse, non-laminar distribution pattern that is consistent with both interneurons and glial cells [4,10]. Noting that the GluN2C-labelled cells were also found in the white matter and had small nuclei that stain well for hematoxylin, Watanabe and colleagues proposed that the cortical GluN2C-labelled cells were glia [10]; see also [11]. However, RT-PCR studies reported that parvalbumin positive cells display significant levels of GluN2C mRNA and no detectible GluN2D mRNA [12]. Additionally, other recent studies have used two different transgenic animals which express ß-galactosidase as an indicator for cells expressing the GluN2C gene. These studies reported ß-gal staining in neurons of the cerebral cortex [13,14]. This localization is also consistent with layer IV neuronal labeling by GluN2C probes in human and monkey cortex [15,16].
The purpose of this study was to take advantage of a GluN2C knockout mouse as a negative control and to use highly sensitive fluorescence in situ hybridization (FISH) together with multiple cell-specific markers to further define the distribution and identity of cells that express GluN2C subunit mRNA in the telencephalon and to compare this distribution to that found for GluN2D subunits. These results suggest that in the telencephalon, GluN2C mRNA is almost exclusively expressed by non-neuronal cells whereas GluN2D subunit mRNA is primarily expressed in neurons.
Methods
Animals and section preparation
Adult C57BL/6 wild-type mice and ß-galactosidase knock-in (GluN2C-KO) mice [13] on the same background, were used to identify GluN2C mRNA signal and confirm its specificity. Mice were sacrificed under deep isoflurane anesthesia and brains were removed and immediately frozen on powdered dry ice. Brains were cut at a thickness of 20 μm, thaw-mounted onto Superfrost Plus™ slides, and air-dried. Sections were fixed in ice-cold 4% paraformaldehyde, washed in phosphate-buffered saline (PBS), dehydrated in 70% ethanol, and stored in 95% ethanol at 4°C.
cRNA probe design
For GluN2C FISH, fluorescein- and digoxigenin-labeled cRNA probes were designed to detect GluN2C mRNA in wild-type mice but not in GluN2C-KO mice. A fragment of rat GluN2C cDNA (360–2402 nucleotide residues; GenBank accession number, NM_012575.3) that corresponds to the deleted fragment in the GluN2C-KO mouse genome [13] was subcloned into the pSPT 19 plasmid vector. For GluN2D ISH, complete coding sequence of rat GluN2D cDNA (Accession number: L31611.1) was used to synthesize digoxigenin- and fluorescein-labeled cRNA probes. GluN2C and GluN2D cRNA probes were prepared by in vitro transcription (DIG RNA Labeling Kit; from Roche), and fragmented by alkaline digestion as previously described [17].
Combined fluorescence in situ hybridization and immunohistochemistry
To combine FISH and immunohistochemistry (IHC) techniques in frozen sections, we used an adjusted protocol based on previous protocols [18,19]. Sections were rehydrated in 70% ethanol and TNT buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.05% Tween 20), acetylated in fresh solution of 0.25% acetic anhydride in 0.1 M triethanolamine-HCl, pH 8.0, for 10 min, and rinsed in TNT. Sections were prehybridized in hybridization buffer (50% formamide, 50 mM Tris-HCl, pH 7.5, 1× Denhardt’s solution, 600 mM NaCl, 200g/mL yeast tRNA, 1 mM ethylenediaminetetraacetic acid (EDTA), and 10% dextran sulfate) for 1 hour. Sections were then hybridized with GluN2C and/or GluN2D cRNA probes-containing hybridization buffer at 63–67°C overnight and then were washed stringently at 61–65°C with frequent gentle agitation, as follow: 5× SSC for 10 min, 4× SSC/ 50% formamide for 20 min, 2× SSC/ 50% formamide for 20 min, and 0.1× SSC for 10 min. After rinsing in TNT, sections were incubated in 2% H2O2 in TNT for 10 min, washed in TNT, and blocked with 5% bovine serum and normal goat serum/TNT for 1 h. Sections were incubated with peroxidase-conjugated anti-digoxigenin or anti-fluorescein antibody (Roche Diagnostics, 1:100–1:200) diluted in TNB for 1 h, and washed. Cy3-TSA or fluorescein-TSA (tyramide signal amplification plus kit, PerkinElmer) was used to develop the signal. For dual color FISH detection, sections were treated with 100 mM sodium azide to block residual peroxidase activity [20], and incubated with the antibody that detect the other mRNA target. Signal was developed using Cy3-TSA or fluorescein-TSA.
For combined FISH/IHC experiments, after blocking sections in bovine serum and normal goat serum, sections were incubated with peroxidase-conjugated anti-digoxigenin or anti-fluorescein antibody (Roche Diagnostics, 1:100–1:200), mouse anti-NeuN antibody (Millipore, 1:100), and rabbit anti-glial fibrillary acidic protein (GFAP) antibody (Abcam, 1:1000) diluted in TNB at 4°C overnight. TNT-washed sections were then incubated with Cy3-TSA (tyramide signal amplification plus kit, PerkinElmer), and washed. Sections were then incubated with Alexa647-conjugated goat anti-mouse antibody (Invitrogen, 1:200) and Alexa488-conjugated goat anti-rabbit antibody (Invitrogen, 1:200) for 2 h. Sections were then rinsed in TNT and distilled water before being cover slipped using Fluoroshield™ with 4,6-diamidino-2-phenylindole (DAPI) medium (Sigma-Aldrich).
Quantification of signal and colocalization
Confocal fluorescent images were captured using a Zeiss confocal microscope. Z-series stack images were used for quantification and determination of colocalization. For quantitative analysis, cell or puncta counting was performed using 40× magnification. GluN2C mRNA puncta, GluN2D mRNA puncta, NeuN-positive cells, GFAP-positive cells, and DAPI-positive nuclei were identified and counted manually one-by-one in a single channel display mode. Colocalization of GluN2C mRNA, GluN2D mRNA, NeuN-positive cells, GFAP-positive cells, and GFAP/NeuN-negative cells were determined based on coincidence of these markers in the same cell as revealed by DAPI staining. Images shown here were projected using maximum intensity projection of Z-series stacks, and image brightness was adjusted using ImageJ program. Results were expressed as mean ± SEM.
Results
Specificity of antisense RNA probe to GluN2C mRNA
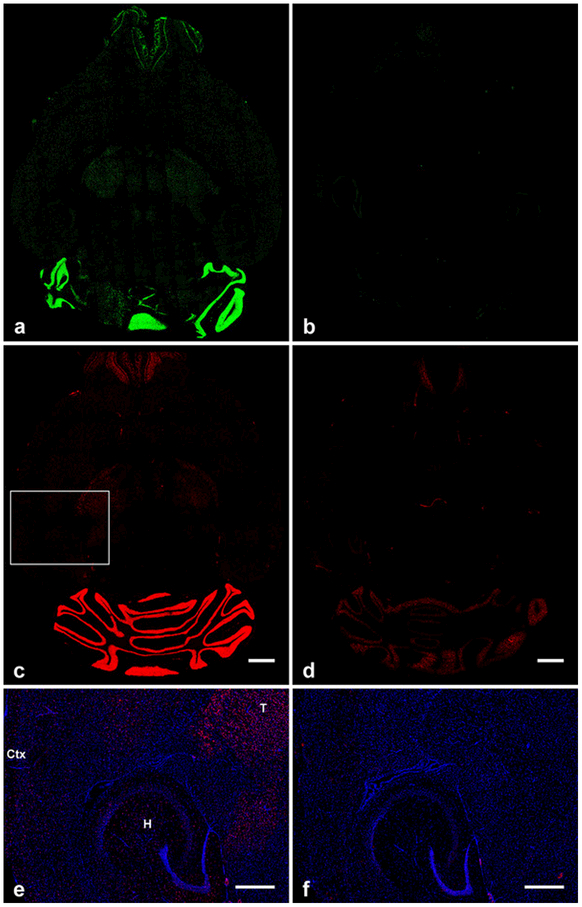
GluN2C mRNA distribution was studied in C57BL/6 wild type mice and GluN2C knockout mice in which most of the GluN2C coding region was replaced by ß-galactosidase. Since the GluN2C-KO mice have a fragment of the GluN2C gene left in their genome, we designed the antisense RNA probe so that its sequence only complements the deleted GluN2C exons in the GluN2C-KO mice. Non-isotopic in situ hybridization was performed using fluorescein-labeled and digoxygenin-labeled cRNA probes for GluN2C mRNA to determine GluN2C mRNA distribution in the brain. The probes displayed a signal for GluN2C mRNA that matches the previously described general GluN2C distribution in brain [3,6,10,21,22]. GluN2C mRNA signal was the strongest in the granular cell layer of the cerebellum and moderately strong in mitral cell and glomerular layers of the olfactory bulb. In the lateral thalamus, GluN2C mRNA signal level was low but higher than in striatum, hippocampus, and cortex. Importantly, cRNA probe signal discussed here was not observed in GluN2C-KO mice brain sections that were processed in parallel using the same FISH/IHC procedures and the same imaging conditions that detected GluN2C signal in wild-type brain (Figure 1).
Fig. 1.
Overall distribution and specificity of fluorescein-tagged (a, b), and digoxigenin-tagged (c-f) antisense RNA probe binding in horizontal mouse brain sections. (a, c) GluN2C mRNA signal in wild-type mouse brain showing the highest expression levels in the granular layer of the cerebellum. b, d, GluN2C-KO mouse brain sections showing very low (b) and low (d) background staining in cerebellum and olfactory bulb that was independent of the tag used. (e, f) Enlarged images of the cortex, hippocampus, and thalamus which are highlighted by the box in (c). Scattered GluN2C signal is displayed in these regions of wild-type mouse (e) but not in GluN2C-KO mouse (f). T, thalamus; Ctx, cortex; H, hippocampus. Scale bars: c, d, 1 mm; e, f, 500 μm
GluN2C mRNA distribution and cell-type identification in telencephalon and thalamus
Overall, GluN2C mRNA signal showed a greater density of labeled cells in the lateral thalamic structures in contrast to the signal in the telencephalon. In the cortex, the scattered, non-laminated GluN2C mRNA distribution was consistent with the distributions of cortical interneurons and glial cells. In preliminary experiments we evaluated GluN2C colocalization with the vesicular glutamate transporters 1 and 2 (VGluT1/2) and glutamate decarboxylase 67 (GAD67) as markers for L-glutamate and gamma-aminobutyric acid (GABA)-using neurons, respectively. We found no colocalization with either of these signals whereas GluN2D signal colocalized with GAD67 (data not shown). Thus, we focused further studies using a combination of FISH and IHC to characterize GluN2C colocalization with NeuN and GFAP as markers for neurons and astrocytes, respectively. As expected, signal for GFAP and NeuN did not colocalize with each other under our conditions. In representative brain regions, we determined the percentage of NeuN-positive cells, GFAP-positive astrocytes, and GFAP/NeuN-negative cells that expressed GluN2C mRNA.
Thalamus.
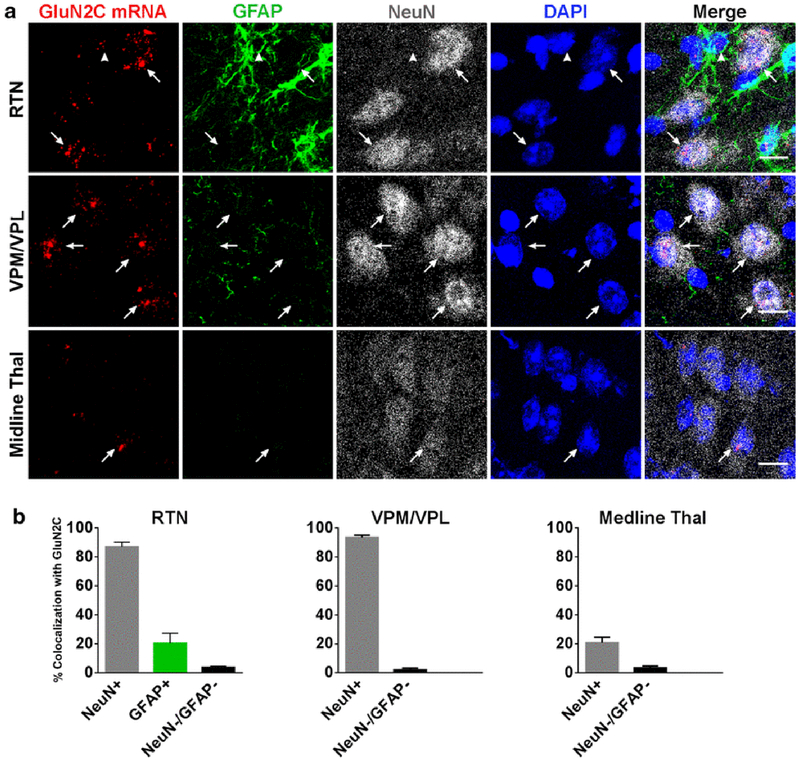
As previously reported, GluN2C mRNA was expressed by many thalamic nuclei [10]. Signal for GluN2C mRNA was displayed in the mediodorsal nucleus (nuc.), the ventral posteromedial nuc. (VPM), the ventral posterolateral nuc. (VPL), the posterior thalamic nuc., and the reticular thalamic nuc. (RTN). GluN2C mRNA signal was lower in the midline thalamic nuclei, and there was no signal in the medial and lateral habenular nuc. In the VPM/VPL and RTN, most NeuN-positive cells expressed GluN2C mRNA. GFAP-labeling was only strong in RTN, thus GFAP/GluN2C colocalization was only determined for this thalamic nucleus. Most of the RTN GFAP-positive cells were negative for GluN2C mRNA with only 20 ± 7 % of the GFAP-labeled cells expressing GluN2C mRNA. The percentage of NeuN-positive cells that were also positive for GluN2C in the VPM/VPL, RTN, and midline thalamus were 94 ± 1.6 %, 87 ± 3.2 %, and 21 ± 3.9 %, respectively (Figure 2).
Fig. 2.
Expression and colocalization of GluN2C mRNA in thalamus. (a) GluN2C mRNA colocalizes with NeuN-positive cells in RTN, VPM/VPL, and midline thal (arrows). GluN2C mRNA is expressed by a GFAP-positive cell in the RTN (arrowhead). (b) Percent of NeuN-positive, GFAP-positive, or NeuN-negative/GFAP-negative cells that were also labeled by GluN2C probe in the three thalamic regions. Scale bar = 10μm
Cerebral cortex.
Previous studies have shown that GluN2C mRNA hybridization signal is faint in the cerebral cortex and found in all cortical layers [10]. Consistent with those findings, in the present study, specific GluN2C signal was seen for scattered cells without evidence of a layered distribution. Thus, the GluN2C signal is not as would be expected for pyramidal or stellate neurons in the cerebral cortex which have a layer specific distribution. We quantified GluN2C mRNA distribution in the retrosplenial cortex and somatosensory cortex as examples of distinct cortical areas.
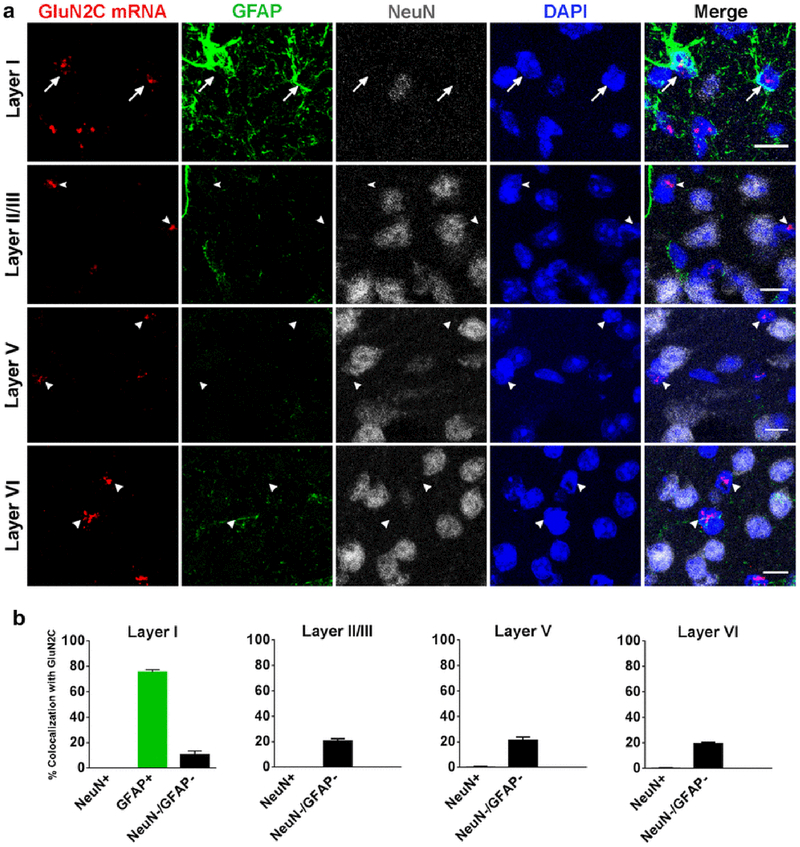
GluN2C mRNA signal was distributed throughout the cortical layers of the retrosplenial cortex and was not found in NeuN-positive cells. However, GluN2C mRNA was expressed in GFAP-positive cells and NeuN/GFAP-negative cells. GFAP-labeling was most strongly expressed in layer 1 of the retrosplenial cortex. Within this layer, 76 ± 1.8 % of the GFAP-positive astrocytes expressed GluN2C mRNA. In contrast, GFAP-labeling was weak and showed a very low number of GFAP-positive astrocytes in the other layers. Thus, GluN2C/GFAP mRNA colocalization was not analyzed in these layers. However, the few GFAP-positive cells in these layers were usually also positive for GluN2C signal (Figure 3). Cells that were either negative or positive for NeuN were quantified in all layers; only NeuN-negative cells colocalized with GluN2C signal (Figure 3).
Fig. 3.
GluN2C mRNA expression in multiple layers of retrosplenial cortex. (a) GluN2C mRNA is expressed in GFAP-positive cells in layer I (arrows). GluN2C mRNA (arrows/arrowheads) does not colocalize with NeuN-positive cells in any layer of the RSC. GluN2C mRNA colocalizes with NeuN-negative/GFAP-negative cells in the deep layers of the RSC. (b) Percent of NeuN-positive, GFAP-positive, or NeuN-negative/GFAP-negative cells that were also labeled by GluN2C probe in different layers of RSC. Scale bar = 10μm
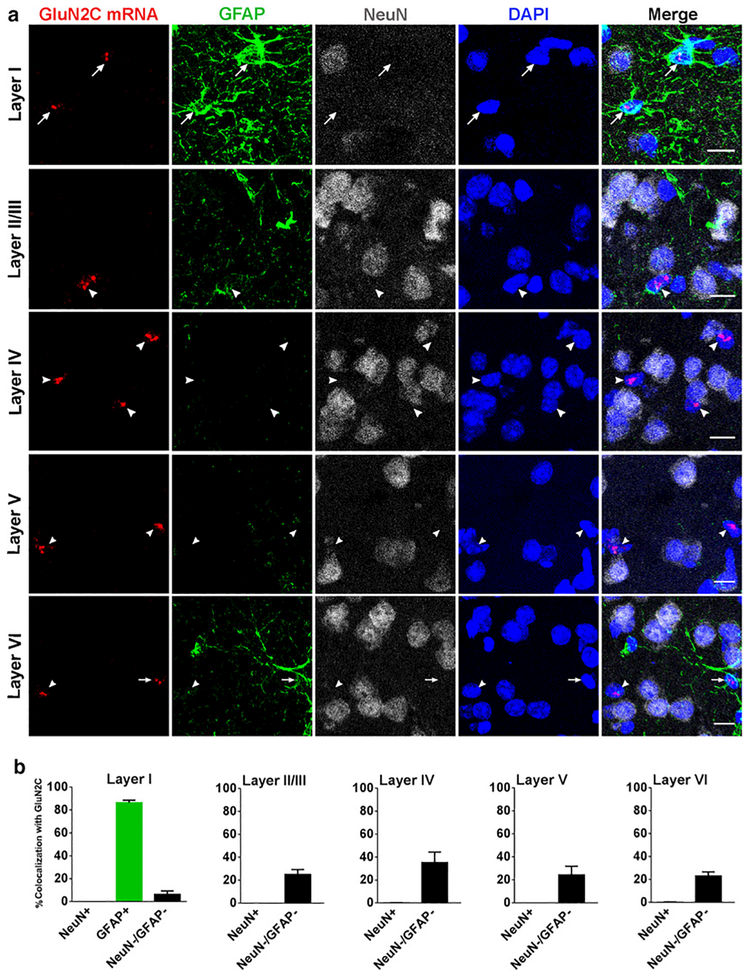
As found for retrosplenial cortex, expression of GluN2C mRNA in the somatosensory cortex was found in GFAP-positive cells and GFAP-negative/NeuN-negative cells but not in NeuN-positive cells. GFAP-labeling for the somatosensory cortex displayed the same pattern as found for the retrosplanial cortex, only cells in the superficial layer were labeled strongly with GFAP. We found that 86 ± 2.2 % of the GFAP-positive astrocytes expressed GluN2C mRNA. GFAP-labeling was again weak in the deeper cortical layers and thus GluN2C/GFAP colocalization was not quantified in these layers. However, when GFAP was present, GluN2C was frequently co-expressed. Most GluN2C-positive cells in the deeper layers were GFAP-negative/NeuN-negative cells (Figure 4).
Fig. 4.
GluN2C mRNA expression in somatosensory cortex. (a) GluN2C mRNA is expressed by GFAP-positive cells in layer I and layer VI (arrows). GluN2C mRNA does not colocalize with NeuN-positive cells in any layers of the SSC (arrows/arrowheads). GluN2C mRNA colocalizes with NeuN-negative/GFAP-negative cells (arrowheads). (b) Percent of NeuN-positive, GFAP-positive, or NeuN-negative/GFAP-negative cells that were also labeled by GluN2C probe in each of the layers of SSC. Scale bar = 10μm
Hippocampus and dentate gyrus.
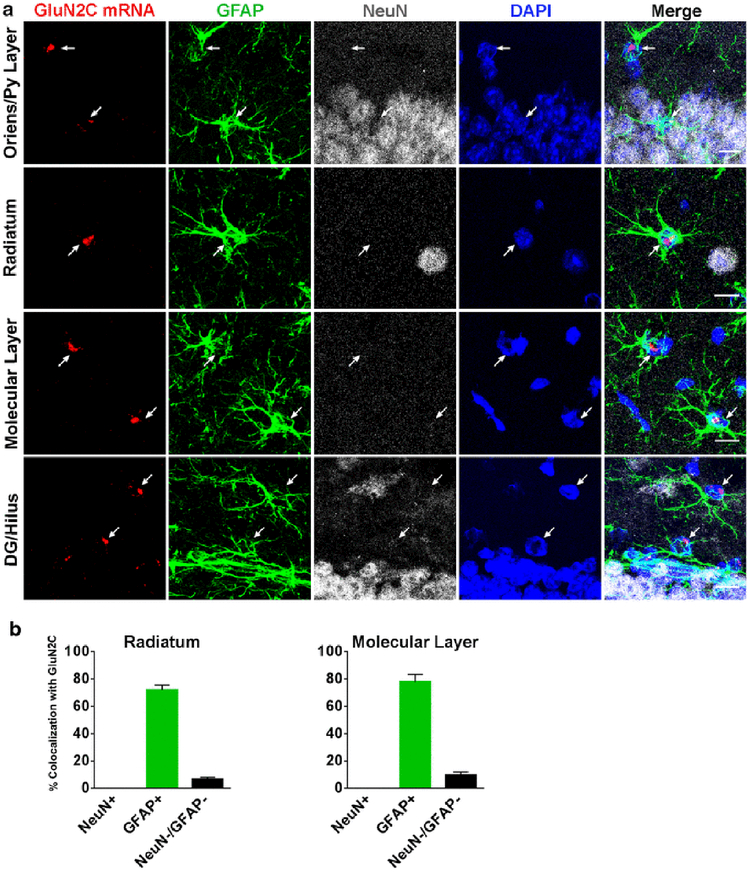
The GluN2C mRNA signal was distributed throughout the hippocampus and dentate gyrus. As previously reported by others, the distribution of cells labeled by GluN2C probe is inconsistent with GluN2C presence in pyramidal or granular cells. GluN2C-positive cells were found at low density in all hippocampal layers (oriens, pyramidale, radiatum, and lacunosum-moleculare) and in the dentate gyrus (granule cell layer, molecular layer, and hilus). NeuN-positive cells did not express GluN2C mRNA. GFAP-labeling was strong in both the hippocampus and dentate gyrus which allowed for quantification of GluN2C colocalization. GFAP was co-expressed with GluN2C mRNA by 72 ± 3.6 % in the stratum radiatum and by 78 ± 5.4 % in the molecular layer. Also, 7 ± 1.3 % and 10 ± 2 % of the NeuN/GFAP-negative cells expressed GluN2C mRNA in the radiatum and the molecular layer, respectively (Figure 5).
Fig. 5.
GluN2C mRNA expression in hippocampus. (a) GluN2C mRNA colocalizes with GFAP-positive cells in stratum oriens, the pyramidal cell layer, stratum radiatum, the molecular layer, the dentate gyrus-granular cell layer, and the hilus (arrows). GluN2C mRNA does not colocalize with NeuN-positive cells in these regions. GluN2C mRNA colocalizes with NeuN-negative/GFAP-negative cells. (b) Percent of NeuN-positive, GFAP-positive, or NeuN-negative/GFAP-negative cells that were also labeled by GluN2C probe in the stratum radiatum and the molecular layer. Scale bar = 10μm
Striatum, amygdala, and globus pallidus.
GluN2C mRNA signal was scattered throughout the striatum, amygdala, and globus pallidus at a low density. In the striatum and amygdala, NeuN-positive cells were not co-labeled with GluN2C mRNA signal. In contrast, GFAP-positive cells frequently co-expressed GluN2C mRNA. As found for the deep layers of the cerebral cortex, GFAP staining was weak in the basal ganglia, but when present was frequently co-expressed with GluN2C mRNA. GluN2C signal was also found in 20 ± 2.5 % and 24 ± 1.2 % of GFAP-negative/NeuN-negative cells in the striatum and amygdala, respectively. Unlike the striatum and the amygdala, GluN2C mRNA was expressed by NeuN-positive cells in the globus pallidus. 47 ± 7.8 % of the NeuN-positive cells and 2 ± 0.9 % of GFAP-negative/NeuN-negative cells were found to express GluN2C mRNA (Figure 6).
Fig. 6.
GluN2C mRNA expression in striatum, amygdala, and globus pallidus. (a) GluN2C mRNA colocalizes with GFAP-positive cells in the striatum and the amygdala (arrows). GluN2C mRNA colocalizes with NeuN-positive cells in the globus pallidus (arrow). GluN2C mRNA is also found in NeuN-negative/GFAP-negative cells (arrowhead). (b) Percent of NeuN-positive and NeuN-negative/GFAP-negative cells that were also labeled by GluN2C probe in the striatum, amygdala, and globus pallidus. Scale bar = 10μm
Comparison with GluN2D subunit distribution.
GluN2D subunits also display a low density, scattered distribution of mRNA expression in the hippocampus, striatum, and cerebral cortex [3,4,10,22–25]. Thus, in these areas, the GluN2C and GluN2D distributions appear similar and are difficult to distinguish from background staining levels. Using a full-length riboprobe to label GluN2D mRNA, we find the anatomically-specific pattern of staining with highest levels in the midline thalamus (paraventricular nucleus, central medial nucleus, nucleus reuniens, and rhomboid nucleus) and lower levels in lateral thalamus as others have reported for GluN2D mRNA [4,10] (Figure 7). This specific distribution also corresponds to L-[3H] glutamate binding sites that display a GluN2D-like pharmacological profile [3]. Using conditions that generate this specific GluN2D-like distribution, scattered cells throughout the telencephalon were labelled as has been reported. We find that these GluN2D-positive cells were also stained with NeuN and not with GFAP in the cerebral cortex, hippocampus, striatum, and thalamus (Figure 8).
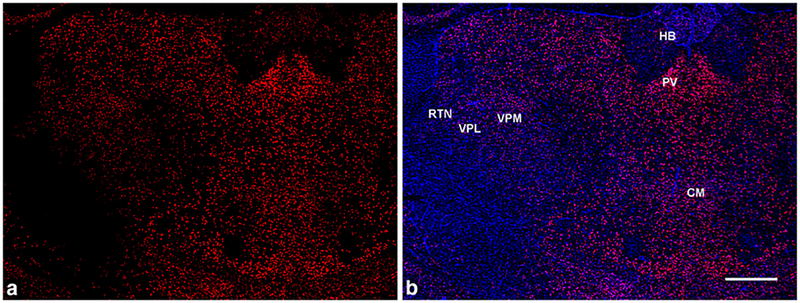
Fig. 7.
GluN2D mRNA distribution in the thalamus in coronal brain sections. (a, b) GluN2D signal (red) is higher in the midline thalamic nuclei (including paraventricular (PV) and central medial (CM) nuclei) compared to the habenula (HB) and the lateral thalamus - ventrobasal complex (VPM/VPL) and RTN. (b) GluN2D mRNA probe (red) co-stained with DAPI (blue). Scale bar = 500μm
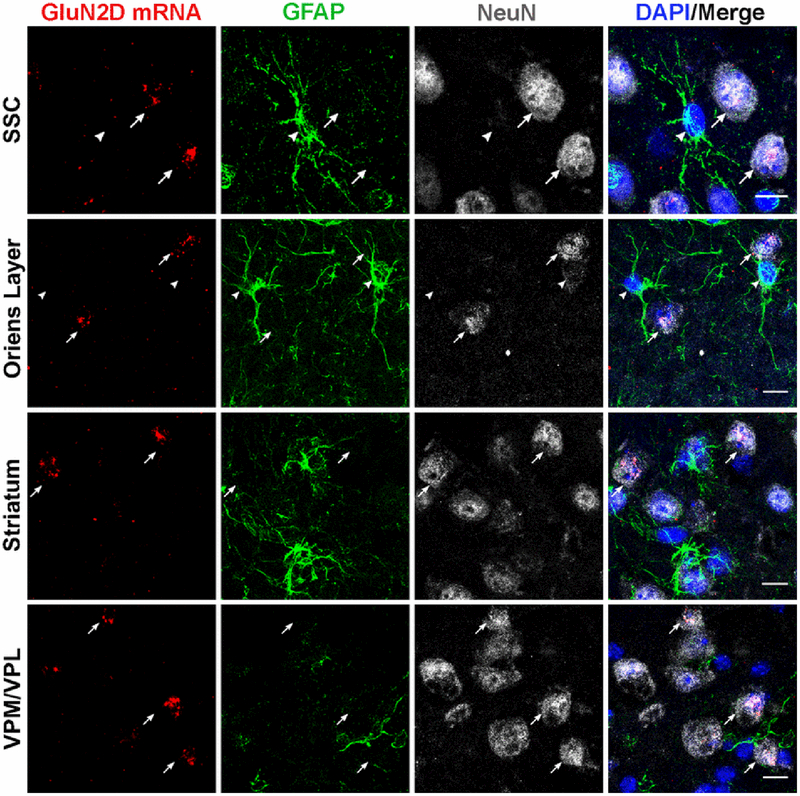
Fig. 8.
GluN2D mRNA expression colocalizes with NeuN in the somatosensory cortex, hippocampus (e.g. stratum oriens), striatum and the thalamic VPM/VPL nuclei (arrows). GluN2D mRNA did not colocalize with GFAP-positive cells (arrowheads). Scale bar = 10μm
Thus, GluN2C colocalized with GFAP in the telencephalon whereas GluN2D riboprobes did not. Conversely, GluN2D colocalized with NeuN, but GluN2C did not colocalize with this neuronal marker in the telencephalon other than in the globus pallidus. The idea that GluN2C and GluN2D are expressed in distinct cell populations in the telencephalon, was further supported by the absence of colocalization of these two probes in the cortex, hippocampus, and striatum (Figures 8, 9C). These two probes, did however, colocalize in the thalamus (reticular nucleus and ventral posterior nucleus), a region where GluN2C is also expressed in neurons. Importantly, of the mouse genome, the GRIN2C (GluN2C) and GRIN2D (GluN2D) genes are more closely related to each other than any other gene. Consequently, non-specific probe labeling of other mRNAs would most likely cause a false positive of GluN2C/GluN2D colocalization and thus would not account for the dissimilar distributions seen for the GluN2C and GluN2D probes. Since a GluN2D knockout that does not express some GluN2D mRNA was not available, we were not able to define background staining levels of the GluN2D probe and so we did not attempt a quantitative colocalization analysis.
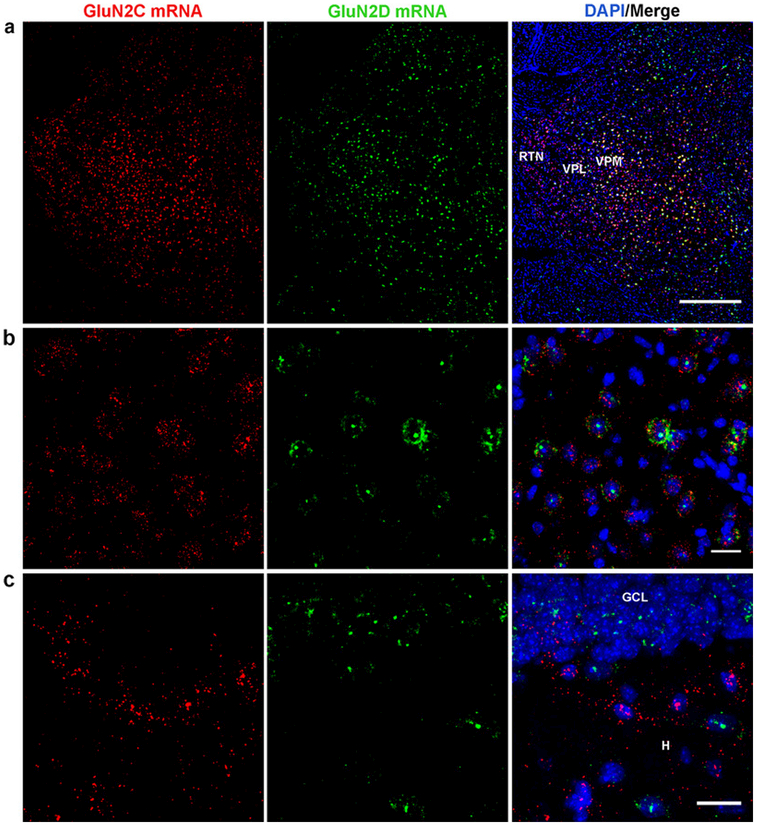
Fig. 9.
Localization of GluN2C and GluN2D mRNA in hippocampus and thalamus. GluN2C and GluN2D mRNA display partial colocalization in the lateral thalamus (VPM/VPL,RTN) low magnification (a) and high magnification (VPM/VPL) (b). (c) GluN2C and GluN2D mRNA did not colocalize in the dentate gyrus granule cell layer (GCL) or hilus (H). (c). Scale bars: a, 500 μm; b, c, 20 μm
Discussion
Studies have consistently shown that GluN2C mRNA is found in cerebellar granule cells, lateral nuclei of the thalamus, and in the mitral cells and glomerular layer of the olfactory bulb. However, the identity of cells expressing GluN2C in the telencephalon is less clear and the literature is inconsistent. In the present study, we find that essentially all of the GluN2C signal in the telencephalon is expressed in glial cells and colocalized well with GFAP when GFAP was present. The only evidence that we found for a neuronal localization of GluN2C in the telencephalon was for the globus pallidus and the olfactory bulb. These results are consistent with the original characterizations of GluN2C mRNA distribution [4,10] which reported GluN2C in cells sparsely scattered throughout each region of the telencephalon and that these cells are likely to be glial cells based on their small size and dark staining by hematoxylin [10]. In the same tissue sections, GluN2C mRNA was expressed in neurons of the thalamus and cerebellum, thus both the neuronal marker and GluN2C probe were staining as expected in these sections.
In contrast to GluN2C, the GluN2D probe consistently colocalized with NeuN and not with GluN2C. Given the lack of a layered distribution for GluN2D signal in the cortex and hippocampus, the GluN2D-positive neurons are likely to be interneurons. This result is consistent with demonstrations that GluN2D expression in GABAergic cells in the cerebral cortex and hippocampus [23,26,27].
Other studies have suggested that GluN2C is expressed in cortical neurons. Using a transgenic approach to express the ß-gal reporter under control of the endogenous GluN2C promotor, neurons of layer 6 of the retrosplenial cortex were found to express ß-gal [13]. Similarly, in a different mouse construct, putting ß-gal under control of a large portion of the GluN2C promotor resulted in ß-gal labelled neurons in layer 4 of the somatosensory cortex [14]. This latter study also provided evidence for functional GluN2C-containing NMDAR in neurons by showing single channel properties indicative of GluN2C-containing receptors. Using the GluN2C-KO/ ß-gal mouse [13] (the GluN2C knockout control in this study), we find ß-gal expression in both layer IV of somatosensory cortex and layer VI of the retrosplenial cortex.
The possible expression of GluN2C in cortical layer IV is consistent with studies of monkey brain in which riboprobes for GluN2C specifically labelled layer IV of somatosensory cortex area 3b [16] and human brain studies where riboprobes labelled the granular layer of prefrontal cortex and visual cortex [15]. As has been suggested, neuronal layer IV GluN2C expression may be specific to primates [15]. In rodents, there is no evidence of layer IV-specific staining; GluN2C probe binding is uniformly scattered throughout the cortex [3,10,22,24,28]. Interestingly, stellate layer IV cortical neurons in a mouse model of tuberous sclerosis complex display thalamocortical synaptic responses with GluN2C-like pharmacological properties whereas wildtype mice do not [29]. Thus, GluN2C expression in rodent stellate cells may normally be suppressed, but can be expressed under pathological conditions. Perhaps GluN2C promoter driven beta-gal expression escapes this regulatory suppression.
In this study, we used the GluN2C knockout as a negative control and a probe targeting the RNA sequence missing in the knockout to label GluN2C mRNA in wildtype mice. By using identical hybridization / imaging conditions for parallel experiments in wildtype and knockout mice, we were able to distinguish low levels of background staining from positive staining. This approach should greatly reduce any false positives due to the probe binding to non-GluN2C RNA. Furthermore, since GluN2D is the most closely related gene to GluN2C and the GluN2D probe did not colocalize with GluN2C in the telencephalon, neither probe appears to be labelling the other’s mRNA. However, since our GluN2C probe does have some complementary homology to GluN2A, B, D mRNA, defining conditions that eliminated this binding could conceivably have reduced the levels of probe binding to GluN2C mRNA. Thus, we cannot rule out that in some cases the absence of staining is a false negative, especially if the cell had very low levels of GluN2C mRNA. Using these conditions, probe binding displayed punctate staining near the nucleus as has been reported for GluN2D probes [23] and thus we do not appear to be detecting mRNA in dendrites or astrocyte processes.
GluN2C mRNA colocalized with GFAP in the telencephalon when GFAP was present. For telencephalic regions that poorly stain for GFAP (e.g. deep cortical layers), GluN2C signal was found predominately in NeuN-negative/GFAP-negative cells, which are likely to be astrocytes that are not expressing GFAP. NeuN staining was robust in these regions, but did not colocalize with GluN2C. Relatively few neuronal types are not stained by NeuN and these are predominately expressed outside of telencephalon [30–32]. It is also possible that some of the GluN2C+/NeuN-/GFAP- cells include oligodendrocytes or other glial cell populations. Overall, these results are consistent with the findings of RNA-seq transcriptomes for the cerebral cortex [33,34]. From these studies, GluN2C mRNA from the cerebral cortex was much more highly expressed in astrocytes than in oligodendrocytes, microglia, and neurons. Immunohistochemical studies indicate the presence of GluN1 and GluN2A and/or GluN2B in astrocyte processes [35] while evaluation of RNA from isolated astrocytes indicates the presence of mRNA for GluN1, GluN2B, and GluN2C [36], or find mRNA for all NMDAR subunits but with relatively high levels of GluN2C and low levels of GluN2D expression [37].
Electrophysiological studies in cortical and hippocampal astrocytes have demonstrated functional NMDARs in astrocytes, and these receptors can mediate neuron-to-glia communication [38–40]. The astrocytic endfeet contain NMDARs and AMPARs, and enwrap the synaptic clefts. By this mechanism, astrocytes can sense the glutamate concentration in the synapse. Stimulation of NMDARs in astrocytes can increase intracellular calcium levels [40,41]. In turn, this increase in intracellular calcium may potentially cause or enhance the calcium-dependent release of gliotransmitters such as ATP, D-serine, and L-glutamate into the perisynaptic region. D-serine/L-glutamate released from astrocytes appears to be released at sites directly opposing neuronal extrasynaptic NMDA receptors [42] and thereby causing slow inward NMDA receptor-mediated currents in neurons [43–47].
Pharmacological studies support a role for functional NMDARs containing GluN2C and/or GluN2D subunits in astrocytes [48]. Agonist-evoked NMDAR currents in cortical astrocytes display low sensitivity to magnesium blockade and are especially sensitive to the partially-selective GluN2C/D antagonist UBP141 [48,49]. Similarly, cortical glial cell transmembrane NMDA receptor currents activated by stimulating cortical neuronal afferents are likewise preferentially blocked by UBP141 and not by GluN2B-selective antagonists [48]. Among GluN subunits, GluN2C, GluN1, and GluN3A display the strongest expression in cortical astrocytes as reported in RNA-sequencing / transcriptome databases [33,50]. Thus, it is possible that astrocytes express a heterotrimeric GluN1/GluN2C/GluN3A receptor, as suggested by [48] which would also impart low magnesium sensitivity or GluN2C and GluN3A subunits may be in separate NMDAR complexes in the same or different astrocytes.
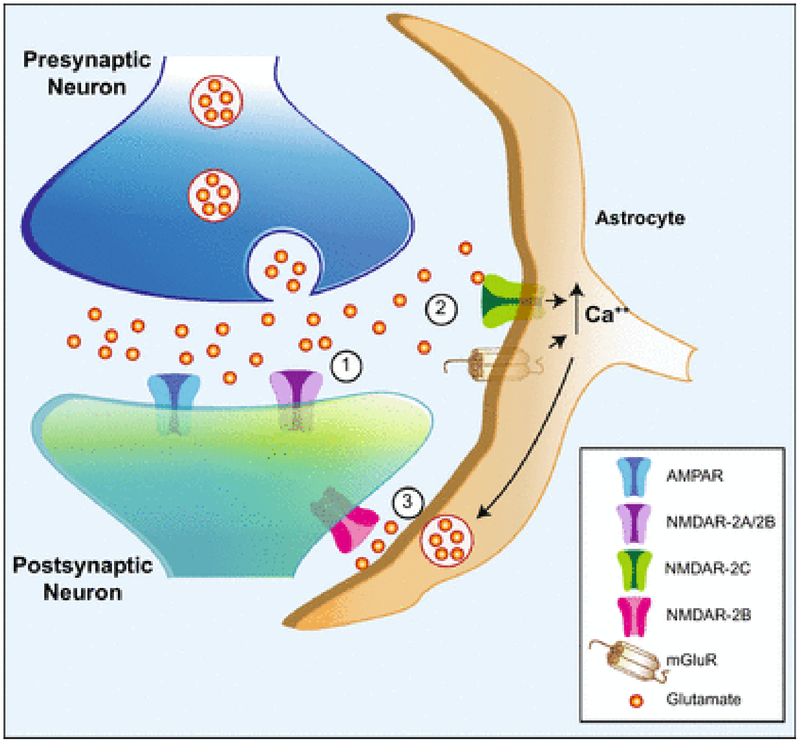
The ability of astrocyte NMDARs to elicit glutamate release and a subsequent extrasynaptic NMDAR neuronal current might account for extrasynaptic NMDAR currents that are seen upon short burst stimulation in the hippocampus [51,52]. These long-lasting neuronal NMDAR currents are activated by glutamate spillover during repetitive synaptic stimulation in the CA3-CA1 hippocampal synapse. This response is especially sensitive to PPDA, UBP141, and ifenprodil, suggesting an involvement of GluN2C/D and GluN2B subunits. The present study suggests the possibility that GluN2C in astrocytes could be responding to the glutamate spillover to cause glutamate release from astrocytes which in turn may activate neuronal extrasynaptic GluN2B-containing receptors (Fig. 10).
Fig. 10.
Schematic diagram showing a potential role for GluN2C-containing NMDARs in the tripartite synapse. (1) L-Glutamate released from presynaptic nerve endings activates AMPA receptors and GluN2A-and/or GluN2B-containing NMDARs (NMDAR-2A/2B) on the postsynaptic dendrite. (2) With sufficient synaptic activation, glutamate spillover activates GluN2C-containing NMDARs (NMDAR-2C) and metabotropic glutamate receptors (mGluR) on the astrocyte, leading to increased cytoplasmic calcium levels from extracellular and intracellular sources, respectively. (3) In response to elevated intracellullar calcium from intracellular stores, and possibly from GluN2C currents, astrocytes release glutamate (and other gliotransmitters) which acts on extrasynaptic GluN2B-containing NMDARs (NMDAR-2B) on the postsynaptic structure on the same neuron, and potentially additional neurons, to modulate excitability and synaptic plasticity.
GluN2C-containing NMDARs have 3 properties that make them ideal as detectors of synaptic L-glutamate spillover. They display a high affinity for L-glutamate and glycine/D-serine [4,6,53,54] so they can detect low, extrasynaptic concentrations of L-glutamate. They are also weakly inhibited by Mg++ so their channel activity does not require coincident depolarization as do GluN2A or GluN2B-containing NMDARs [4,6,21,53,54]. Thirdly, GluN2C-containing NMDARs do not desensitize [4,55], so these receptors would stay responsive to slowly changing or tonic extracellular L-glutamate. Interestingly, GluN2D-containing receptors also display these properties, so they may display a similar role in the detection of low concentrations of extrasynaptic L-glutamate in order to tonically activate inhibitory interneurons [56] in addition to their activation by synaptic activity [26,27].
Taken together, these results suggest that astrocytes with GluN2C-containing NMDA receptors can modulate the excitability of neuronal NMDA receptors throughout the telencephalon. Since schizophrenia is thought to reflect NMDA receptor hypofunction, it is interesting that the expression of GluN2C mRNA in the dorsolateral prefrontal cortex was reduced in people with schizophrenia [57] and that GluN2C knockout mice display a phenotype resembling schizophrenia [58]. Therefore, the modulation of GluN2C function may represent a new target to treat schizophrenia.
Acknowledgments:
This study was funded by the National Institutes of Health grants MH60252 and GM110768 and the Edna Ittner Pediatric Research Foundation. The authors thank Dr. Andres Buonanno and colleagues for having provided the GluN2C-KO mouse line, Dr. Emily Harrison for technical advice, Dr. Masahiko Watanabe for helpful discussions and Robin Taylor for expert graphical assistance.
References
- 1.Yamamoto H, Hagino Y, Kasai S and Ikeda K, (2015) Specific Roles of NMDA Receptor Subunits in Mental Disorders. Curr. Mol. Med 15, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ and Dingledine R, (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev 62, 405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buller AL, Larson HC, Schneider BE, Beaton JA, Morrisett RA and Monaghan DT, (1994) The molecular basis of NMDA receptor subtypes: native receptor diversity is predicted by subunit composition. J. Neurosci 14, 5471–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monyer H, Burnashev N, Laurie DJ, Sakmann B and Seeburg PH, (1994) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 12, 529–540. [DOI] [PubMed] [Google Scholar]
- 5.Mishina M, Mori H, Araki K, Kushiya E, Meguro H, Kutsuwada T, Kashiwabuchi N, Ikeda K, Nagasawa M and Yamazaki M, (1993) Molecular and functional diversity of the NMDA receptor channel. Ann. N. Y. Acad. Sci 707, 136–152. [DOI] [PubMed] [Google Scholar]
- 6.Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N and Masu M, (1993) Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J. Biol. Chem 268, 2836–2843. [PubMed] [Google Scholar]
- 7.Ebralidze AK, Rossi DJ, Tonegawa S and Slater NT, (1996) Modification of NMDA receptor channels and synaptic transmission by targeted disruption of the NR2C gene. J. Neurosci 16, 5014–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadotani H, Hirano T, Masugi M, Nakamura K, Nakao K, Katsuki M and Nakanishi S, (1996) Motor discoordination results from combined gene disruption of the NMDA receptor NR2A and NR2C subunits, but not from single disruption of the NR2A or NR2C subunit. J. Neurosci 16, 7859–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Buonanno A, Vertes RP, Hoover WB and Lisman JE, (2012) NR2C in the thalamic reticular nucleus; effects of the NR2C knockout. PLoS One. 7, e41908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe M, Inoue Y, Sakimura K and Mishina M, (1993) Distinct distributions of five N-methyl-D-aspartate receptor channel subunit mRNAs in the forebrain. J. Comp. Neurol 338, 377–390. [DOI] [PubMed] [Google Scholar]
- 11.Landwehrmeyer GB, Standaert DG, Testa CM, Penney JB,Jr AB, (1995) NMDA receptor subunit mRNA expression by projection neurons and interneurons in rat striatum. J. Neurosci 15, 5297–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi D, Keeler B, Zhang W, Houle JD and Gao WJ, (2009) NMDA receptor subunit expression in GABAergic interneurons in the prefrontal cortex: application of laser microdissection technique. J. Neurosci. Methods 176, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karavanova I, Vasudevan K, Cheng J and Buonanno A, (2007) Novel regional and developmental NMDA receptor expression patterns uncovered in NR2C subunit-beta-galactosidase knock-in mice. Mol. Cell. Neurosci 34, 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binshtok AM, Fleidervish IA, Sprengel R and Gutnick MJ, (2006) NMDA receptors in layer 4 spiny stellate cells of the mouse barrel cortex contain the NR2C subunit. J. Neurosci 26, 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherzer CR, Landwehrmeyer GB, Kerner JA, Counihan TJ, Kosinski CM, Standaert DG, Daggett LP, Velicelebi G, Penney JB and Young AB, (1998) Expression of N-methyl-D-aspartate receptor subunit mRNAs in the human brain: hippocampus and cortex. J. Comp. Neurol 390, 75–90. [DOI] [PubMed] [Google Scholar]
- 16.Munoz A, Woods TM and Jones EG, (1999) Laminar and cellular distribution of AMPA, kainate, and NMDA receptor subunits in monkey sensory-motor cortex. J. Comp. Neurol 407, 472–490. [DOI] [PubMed] [Google Scholar]
- 17.Angerer LM and Angerer RC (1992) In situ hybridization to cellular RNA with radiolabeled RNA probes, in In Situ Hybridization: A Practical Approach, Wilkinson DG ed., IRL Press, Oxford University Press, Oxford, pp. 15–32. [Google Scholar]
- 18.Chaudhuri AD, Yelamanchili SV and Fox HS, (2013) Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Front. Cell. Neurosci 7, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasaki M, Matsui M and Watanabe M, (2010) Preferential localization of muscarinic M1 receptor on dendritic shaft and spine of cortical pyramidal cells and its anatomical evidence for volume transmission. J. Neurosci 30, 4408–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King RS and Newmark PA, (2013) In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol 13, 8-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B and Seeburg PH, (1992) Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 256, 1217–1221. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe M, Inoue Y, Sakimura K and Mishina M, (1992) Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 3, 1138–1140. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki M, Okada R, Takasaki C, Toki S, Fukaya M, Natsume R, Sakimura K, Mishina M, Shirakawa T and Watanabe M, (2014) Opposing role of NMDA receptor GluN2B and GluN2D in somatosensory development and maturation. J. Neurosci 34, 11534–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudolf GD, Cronin CA, Landwehrmeyer GB, Standaert DG, Penney JB Jr, and Young AB, (1996) Expression of N-methyl-D-aspartate glutamate receptor subunits in the prefrontal cortex of the rat. Neuroscience. 73, 417–427. [DOI] [PubMed] [Google Scholar]
- 25.Standaert DG, Landwehrmeyer GB, Kerner JA, Penney JB Jr, and Young AB, (1996) Expression of NMDAR2D glutamate receptor subunit mRNA in neurochemically identified interneurons in the rat neostriatum, neocortex and hippocampus. Brain Res. Mol. Brain Res 42, 89–102. [DOI] [PubMed] [Google Scholar]
- 26.Perszyk RE, DiRaddo JO, Strong KL, Low CM, Ogden KK, Khatri A, Vargish GA, Pelkey KA, Tricoire L, Liotta DC, Smith Y, McBain CJ and Traynelis SF, (2016) GluN2D-Containing N-methyl-d-Aspartate Receptors Mediate Synaptic Transmission in Hippocampal Interneurons and Regulate Interneuron Activity. Mol. Pharmacol 90, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Engelhardt J, Bocklisch C, Tonges L, Herb A, Mishina M and Monyer H, (2015) GluN2D-containing NMDA receptors-mediate synaptic currents in hippocampal interneurons and pyramidal cells in juvenile mice. Front. Cell. Neurosci 9, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akazawa C, Shigemoto R, Bessho Y, Nakanishi S and Mizuno N, (1994) Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J. Comp. Neurol 347, 150–160. [DOI] [PubMed] [Google Scholar]
- 29.Lozovaya N, Gataullina S, Tsintsadze T, Tsintsadze V, Pallesi-Pocachard E, Minlebaev M, Goriounova NA, Buhler E, Watrin F, Shityakov S, Becker AJ, Bordey A, Milh M, Scavarda D, Bulteau C, Dorfmuller G, Delalande O, Represa A, Cardoso C, Dulac O, Ben-Ari Y and Burnashev N, (2014) Selective suppression of excessive GluN2C expression rescues early epilepsy in a tuberous sclerosis murine model. Nat. Commun 5, 4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weyer A and Schilling K, (2003) Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J. Neurosci. Res 73, 400–409. [DOI] [PubMed] [Google Scholar]
- 31.Sarnat HB, Nochlin D and Born DE, (1998) Neuronal nuclear antigen (NeuN): a marker of neuronal maturation in early human fetal nervous system. Brain Dev 20, 88–94. [DOI] [PubMed] [Google Scholar]
- 32.Mullen RJ, Buck CR and Smith AM, (1992) NeuN, a neuronal specific nuclear protein in vertebrates. Development. 116, 201–211. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA and Wu JQ, (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancarci BO, Toker L, Tripathy SJ, Li B, Rocco B, Sibille E and Pavlidis P, (2017) Cross-Laboratory Analysis of Brain Cell Type Transcriptomes with Applications to Interpretation of Bulk Tissue Data. eNeuro. 4, 10.1523/ENEURO.0212-17.2017. eCollection 2017 Nov-Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conti F, DeBiasi S, Minelli A and Melone M, (1996) Expression of NR1 and NR2A/B subunits of the NMDA receptor in cortical astrocytes. Glia. 17, 254–258. [DOI] [PubMed] [Google Scholar]
- 36.Schipke CG, Ohlemeyer C, Matyash M, Nolte C, Kettenmann H and Kirchhoff F, (2001) Astrocytes of the mouse neocortex express functional N-methyl-D-aspartate receptors. FASEB J 15, 1270–1272. [DOI] [PubMed] [Google Scholar]
- 37.Dzamba D, Honsa P, Valny M, Kriska J, Valihrach L, Novosadova V, Kubista M and Anderova M, (2015) Quantitative Analysis of Glutamate Receptors in Glial Cells from the Cortex of GFAP/EGFP Mice Following Ischemic Injury: Focus on NMDA Receptors. Cell. Mol. Neurobiol 35, 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lalo U, Pankratov Y, Kirchhoff F, North RA and Verkhratsky A, (2006) NMDA receptors mediate neuron-to-glia signaling in mouse cortical astrocytes. J. Neurosci 26, 2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letellier M, Park YK, Chater TE, Chipman PH, Gautam SG, Oshima-Takago T and Goda Y, (2016) Astrocytes regulate heterogeneity of presynaptic strengths in hippocampal networks. Proc. Natl. Acad. Sci. U. S. A 113, E2685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palygin O, Lalo U, Verkhratsky A and Pankratov Y, (2010) Ionotropic NMDA and P2X1/5 receptors mediate synaptically induced Ca2+ signalling in cortical astrocytes. Cell Calcium 48, 225–231. [DOI] [PubMed] [Google Scholar]
- 41.Verkhratsky A, Rodriguez JJ and Parpura V, (2012) Calcium signalling in astroglia. Mol. Cell. Endocrinol 353, 45–56. [DOI] [PubMed] [Google Scholar]
- 42.Bezzi P, Gundersen V, Galbete JL, Seifert G, Steinhauser C, Pilati E and Volterra A, (2004) Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nat. Neurosci 7, 613–620. [DOI] [PubMed] [Google Scholar]
- 43.Parri HR, Gould TM and Crunelli V, (2001) Spontaneous astrocytic Ca2+ oscillations in situ drive NMDAR-mediated neuronal excitation. Nat. Neurosci 4, 803–812. [DOI] [PubMed] [Google Scholar]
- 44.Angulo MC, Kozlov AS, Charpak S and Audinat E, (2004) Glutamate released from glial cells synchronizes neuronal activity in the hippocampus. J. Neurosci 24, 6920–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fellin T, Pascual O, Gobbo S, Pozzan T, Haydon PG and Carmignoto G, (2004) Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron. 43, 729–743. [DOI] [PubMed] [Google Scholar]
- 46.Perea G and Araque A, (2005) Properties of synaptically evoked astrocyte calcium signal reveal synaptic information processing by astrocytes. J. Neurosci 25, 2192–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee CJ, Mannaioni G, Yuan H, Woo DH, Gingrich MB and Traynelis SF, (2007) Astrocytic control of synaptic NMDA receptors. J. Physiol 581, 1057–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palygin O, Lalo U and Pankratov Y, (2011) Distinct pharmacological and functional properties of NMDA receptors in mouse cortical astrocytes. Br. J. Pharmacol 163, 1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morley RM, Tse HW, Feng B, Miller JC, Monaghan DT and Jane DE, (2005) Synthesis and pharmacology of N1-substituted piperazine-2,3-dicarboxylic acid derivatives acting as NMDA receptor antagonists. J. Med. Chem 48, 2627–2637. [DOI] [PubMed] [Google Scholar]
- 50.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ and Barres BA, (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lozovaya NA, Grebenyuk SE, Tsintsadze TS, Feng B, Monaghan DT and Krishtal OA, (2004) Extrasynaptic NR2B and NR2D subunits of NMDA receptors shape ‘superslow’ afterburst EPSC in rat hippocampus. J. Physiol 10.1113/jphysiol.2004.063792 Epub. 558, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa BM, Feng B, Tsintsadze TS, Morley RM, Irvine MW, Tsintsadze V, Lozovaya NA, Jane DE and Monaghan DT, (2009) N-methyl-D-aspartate (NMDA) receptor NR2 subunit selectivity of a series of novel piperazine-2,3-dicarboxylate derivatives: preferential blockade of extrasynaptic NMDA receptors in the rat hippocampal CA3-CA1 synapse. J. Pharmacol. Exp. Ther 331, 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T and Arakawa M, (1992) Molecular diversity of the NMDA receptor channel. Nature 358, 36–41. [DOI] [PubMed] [Google Scholar]
- 54.Ikeda K, Nagasawa M, Mori H, Araki K, Sakimura K, Watanabe M, Inoue Y and Mishina M, (1992) Cloning and expression of the epsilon 4 subunit of the NMDA receptor channel. FEBS Lett. 313, 34–38. [DOI] [PubMed] [Google Scholar]
- 55.Krupp JJ, Vissel B, Heinemann SF and Westbrook GL, (1996) Calcium-dependent inactivation of recombinant N-methyl-D-aspartate receptors is NR2 subunit specific. Mol. Pharmacol 50, 1680–1688. [PubMed] [Google Scholar]
- 56.Riebe I, Seth H, Culley G, Dosa Z, Radi S, Strand K, Frojd V and Hanse E, (2016) Tonically active NMDA receptors--a signalling mechanism critical for interneuronal excitability in the CA1 stratum radiatum. Eur. J. Neurosci 43, 169–178. [DOI] [PubMed] [Google Scholar]
- 57.Beneyto M and Meador-Woodruff JH, (2008) Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology.1 33, 2175–2186. [DOI] [PubMed] [Google Scholar]
- 58.Gupta SC, Ravikrishnan A, Liu J, Mao Z, Pavuluri R, Hillman BG, Gandhi PJ, Stairs DJ, Li M, Ugale RR, Monaghan DT and Dravid SM, (2016) The NMDA receptor GluN2C subunit controls cortical excitatory-inhibitory balance, neuronal oscillations and cognitive function. Sci. Rep 6, 38321. [DOI] [PMC free article] [PubMed] [Google Scholar]