Abstract
Objective:
To measure consumption and absorption of human milk oligosaccharides (HMOs) in a cohort of premature infants treated with probiotic Bifidobacterium breve
Methods:
Twenty-nine premature infants (median gestational age 28 weeks, range 23–32 weeks) cared for in the neonatal intensive care unit of the King Edward and Princess Margaret Hospital in Perth, Australia were treated with Bifidobacterium breve at a dose of 1.66 billion organisms per day. Samples of feces, urine, and milk were obtained at initiation of the probiotic and again three weeks later. 16S ribosomal RNA from the feces was analyzed by next generation sequencing. Quantitation of HMO content of the milk, urine, and feces was performed using nano-high performance liquid chromatography-chip/time-of-flight mass spectrometry.
Results:
There was heterogeneity in colonization with bifidobacteria. “Responders” received milk with higher percentages of fucosylated HMOs and had higher percentages of bifidobacteria and lower percentages of Enterobacteriaceae in their feces than “non-responders.” Several individual HMOs in the milk were associated with changes in fecal bifidobacteria over time. Changes over time in milk, fecal and urine HMOs suggested heterogeneity among HMO structures in consumption by microbes in the gut lumen and absorption from the intestine.
Conclusions:
Colonization of the premature infant intestinal tract with probiotic B. breve is influenced by prebiotic HMOs. B. breve is a selective consumer of HMOs in the premature infant.
Keywords: probiotic, prebiotic, necrotizing enterocolitis, glycan, intestinal microbiota
Introduction
Necrotizing enterocolitis (NEC) is a common and devastating disease that predominantly affects premature infants. Dysbiosis is an alteration in the composition of the microbiota of a given anatomic niche associated with disease. The evidence for a central role of intestinal dysbiosis in the pathogenesis of NEC is compelling and includes data from animal1–3 and human studies.4–7 Probiotics are dietary supplements containing live commensal microbes. Multiple randomized clinical trials of probiotics have demonstrated decreased risk of NEC and death in premature infants,8 though the mechanisms by which probiotics provide protection and the preferred probiotic microbes for this vulnerable population are unclear.9
Human milk oligosaccharides (HMOs) are abundant in volume and in structural variety in human milk and yet the infant does not produce glycosidases capable of digesting these glycans raising the question of why the mother expends significant energy to create these structures. A limited number of commensal intestinal microbes produce glycosidases and transport molecules and are thus able to consume HMOs suggesting that the primary function of these glycans is to nourish and shape the infant’s intestinal microbiota. In addition, sialylated HMOs have been shown in animal models to promote growth and impact metabolic pathways in the liver, muscle and brain.10 One proposed mechanism by which some Bifidobacterium species are able to out-compete other gut microbes in the infant intestine is the degree to which these microbes express bacterial transport proteins and glycosidases to ingest and digest HMOs as a food source.11 Only Bifidobacterium longum subsp. infantis is able to consume all of the HMOs present in human milk, while Bifidobacterium bifidum digests some HMOs and transports select portions of the HMOs into its cytoplasm for consumption.12,13 Significant variability among strains of Bifidobacterium breve in the capacity to utilize HMOs as an energy source has been demonstrated.14 Analysis of HMOs that pass undigested through the gastrointestinal (GI) tract and appear in the feces and of HMOs that are absorbed from the GI tract and filtered by the kidneys have been proposed as potential markers of dysbiosis.15
Fucosyltransferase 2 is encoded by the gene FUT2 on chromosome 19. 20% of the population of many parts of the world are homozygous for mutations in FUT2 that render them unable to create α(1,2) fucosyl linkages in glycans on cell surfaces and in secretions (including human milk). These individuals have historically been referred to as “non-secretors,” and appear to have increased risk for some gastrointestinal diseases (e.g. Crohn’s disease)16 and decreased risk for others (e.g. Norwalk virus infection).17 Evidence of association between the most common mutation in the FUT2 gene and NEC has been mixed,18,19 raising the possibility that the secretor status of the mother may be as important in influencing the intestinal microbiota as the secretor status of the infant. Our group has previously demonstrated that term infants breast-fed by “secretor” mothers have higher numbers of fecal bifidobacteria than infants of “non-secretor” mothers and that B. infantis is the dominant Bifidobacterium in the feces of infants of “secretor” mothers while B. breve was found in the feces of infants of both “secretor” and “non-secretor” mothers.20
In this study we sought to characterize the impact of administration of B. breve on both the fecal microbiota and the HMO composition in the urine and stool of a cohort of premature infants. As B. breve is a “selective” consumer of HMOs (consumes some HMO structures but not others with strain to strain variation), this cohort provides an opportunity to study differences in colonization of the premature gut based on HMO consumption. We also sought to determine differences in response to B. breve based on maternal secretor status.
Methods
Informed consent for participation in the cohort study was obtained from the parents of each infant. The study was approved by the women and newborn health service human research ethics committee which is registered with the Australian health ethics committee. 29 premature infants were enrolled and received B. breve strain M16-V (Morinaga Milk Industries, Japan) at a dose of 1.66 billion organisms once daily as soon as enteral feedings were tolerated. The frequency was increased to twice daily (approximately 3.33 billion organisms per day) once feed volume was > 50 ml/kg/d. Samples of infant feces and urine and maternal milk were collected close to the time of initiation of the probiotic and again 3 weeks later. All samples were frozen at the time of collection and were shipped to the U.S. on dry ice.
Fecal Next Generation Sequencing
Bacterial DNA was extracted from the stool specimens and DNA library construction was carried out as previously described21,22 and submitted to the UC Davis Genome Center DNA Technologies Core for sequencing on an Illumina MiSeq instrument (Illumina, San Diego CA). QIIME software package (University of Colorado. Boulder CO, version 1.7.0) was used for quality filtering and demultiplexing the resulting sequencing data.23 OTUs were assigned using UCLUST (drive5.com, Tiburon, CA) based on 97% pairwise identity24 and taxonomic classification was based on the Ribosomal Database Project classifier (Michigan State University, East Lansing, MI) against a representative subset of the Greengenes 16S rRNA database (Second Genome, South San Francisco, CA, gg_otus_12_10 release).25,26 Taxonomic relative abundance data from the QIIME data analysis were used to calculate correlations between HMO consumption and fecal bacteria abundance.
HMO Extraction
Free HMOs were extracted from whole breast milk, fecal, and urine samples from 29 mother-infant pairs following previously reported methods for oligosaccharide extraction.27–30 Briefly, 25 μL of each milk sample was aliquoted onto a 96-well plate, diluted, and defatted via centrifugation. Proteins were then removed by ethanol precipitation at −80°C for 1.5 hours. The resulting glycans were reduced with 1.0 M NaBH4 at 65°C for 1.5 hours, then purified and desalted on solid phase extraction (SPE) graphitized carbon cartridges (GCC). The solvent was evaporated from the eluent fractions, and after reconstitution, the samples were diluted for analysis. Each fecal sample was weighed out and diluted to 0.1 mg/mL in deionized water and shaken overnight at 4°C. Following centrifugation, 200 μL of supernatant was aliquoted onto a 96-well plate, and the same steps for protein precipitation, reduction, and SPE were followed as for milk extraction. Urine (200 μL) was also aliquoted onto a 96-well plate, again with the same protein removal, reduction, and SPE purification steps. An extra SPE C8 step in fecal and urine clean-up was used prior to GCC purification.29
Nano-High Performance Liquid Chromatography-Chip/Time-of-Flight Mass Spectrometry
Analysis of the extracted oligosaccharides was performed on a nano-high performance liquid chromatography (HPLC)-chip/time-of-flight (TOF) mass spectrometer (MS). The HPLC system used was an Agilent 1200 series, which incorporates a dual pump system and a microfluidic chip packed with porous graphitized carbon (PGC). The capillary pump flows at a rate of 4.0 μL/min and a two μL injection volume was used to load each sample onto the 40 nL enrichment column of the chip. The nano pump achieves separation on the 75 μL × 43 mm analytical column of the chip with a binary gradient of aqueous solvent A (3% acetonitrile:water (v/v) in 0.1% formic acid) and organic solvent B (90% acetonitrile:water (v/v) in 0.1% formic acid). This system is coupled to an Agilent 6220 series TOF MS via chip cube interface and run in the positive mode following the method developed and optimized for HMO separation.28,31 A dual nebulizer electrospray source was used to calibrate the instrument, with calibrant ions ranging from m/z 118.086 to 2721.895.
Oligosaccharide Analysis
Data were collected using the Agilent MassHunter Workstation Data Acquisition version B.02.01, and analyzed with the Agilent MassHunter Qualitative Analysis B.03.01 software. Oligosaccharides were identified using the Find Compounds by Molecular Feature function, with an in-house program for peak alignment. Individual HMOs were assigned by matching retention time and exact mass within 20 ppm error to previously developed HMO libraries.28,31 The glycans were divided into different classes based on monosaccharide composition (fucosylated, structures with fucose but not sialic acid; sialylated, structures with sialic acid but not fucose; total fucosylation, all structures with fucose; total sialylation: all structures with sialic acid; fucosylated and sialylated, structures with both fucose and sialic acid; and undecorated, structures with neither fucose nor sialic acid), and to determine relative abundances of each class, abundances were divided by the total oligosaccharide count for each mother. To determine relative abundances of individual compounds, the same calculation was performed as for HMO class. Fecal samples were run in separate batches, so absolute ion counts were normalized to an external HMO pool to account for instrument variations and batch effects. Individual compounds that were identified in greater than 50% of samples were included, and for samples in which that compound was either not identified or extracted by the software, a value of half the limit of detection was given.
Secretor Status Determination
Phenotypic secretor status was determined for each mother by analyzing the relative abundance of certain HMOs in her milk. Absolute abundances of compounds with known α(1,2) fucosylation were summed and normalized to each mother’s total oligosaccharide abundance. Mothers with a relative α(1,2) fucosylated abundance >6% were determined to be phenotypic secretors, those lower were considered non-secretors. This method was modified from a previously developed method for determining secretor status.32 The structures identified with an α(1,2) fucose linkage and used for this determination were 2’-fucosyllactose (2’FL), lactodifucotetraose (LDFT), trifucosyllacto-N-hexaose (TFLNH), and difucosyllacto-N-hexaose a (DFLNHa).
Statistics
Descriptive statistics for the cohort, t-tests comparing percentages of fecal bacteria and comparing percentages of HMOs, and simple linear regression of fecal bacterial families with fecal HMOs were calculated using STATA version 12.1. Logarithmic transformation was performed to facilitate determination of changes in percentage bifidobacteria, changes in milk, fecal, and urine HMOs and changes in the ratios of fecal HMOs:milk HMOs and urine HMOs:milk HMOs over time, using the mathematical relationship log( Y / X ) = log( Y ) – log(X). Spearman correlation coefficients were calculated using SAS version 9.4. Fold changes in the ratios of fecal HMOs:milk HMOs and urine HMOs:milk HMOs over time were calculated by exponentiating the point estimates and 95% confidence intervals for the mean differences in log-transformed quantities.
Results
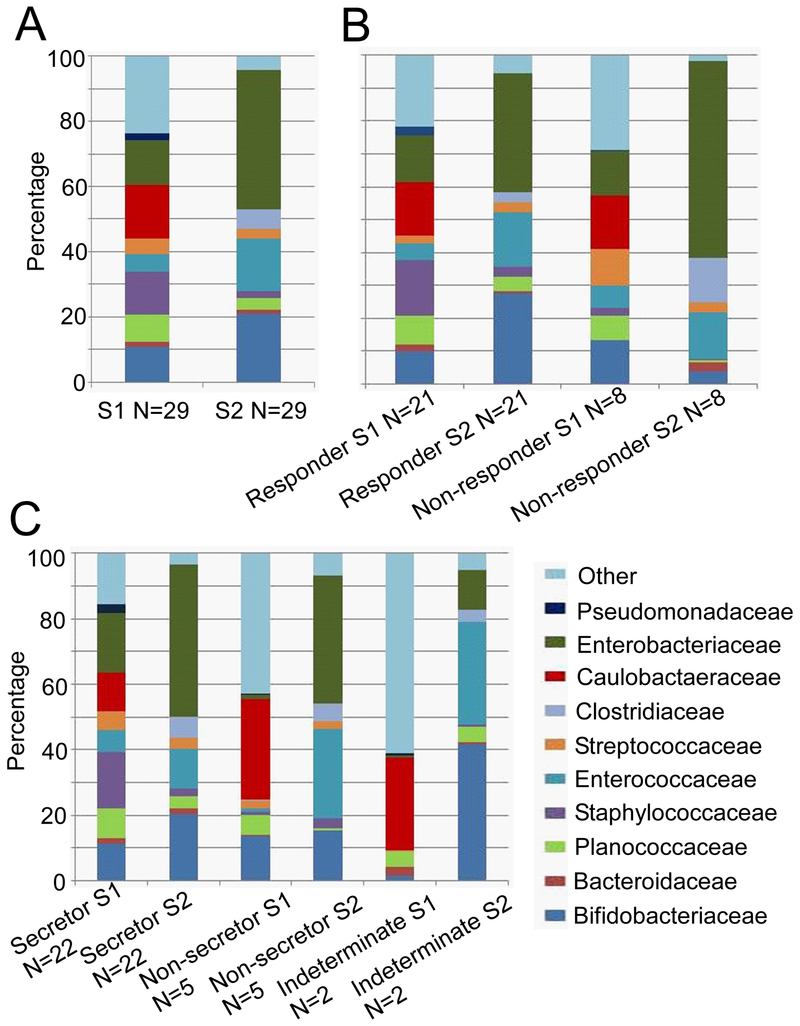
Table 1 summarizes demographic details and outcomes in the cohort. The results of NGS of the fecal microbiota are summarized in Figure 1. As expected, there were marked differences in microbial composition over time (panel A). An increase in Enterobacteriaceae over time is common in premature infants33 and was pronounced in this cohort. There was heterogeneity in colonization with bifidobacteria among the infants. To characterize this, we divided the cohort into “responders” and “non-responders” with the latter group defined as having < 6% bifidobacteria in the second stool sample. Non-responders had significantly higher percentages of Enterobacteriaceae (p=0.01) and Clostrideaceae (p=0.02) in the second fecal sample than responders (panel B). Infants with secretor mothers, delivery type, and antibiotic treatment did not differ between responder and non-responder infants. We chose 6% as an initial cut-off between responders and non-responders based on observations that premature infants generally have very low levels of bifidobacteria even with feeding of human milk (0–20%).22,33,34 We re-defined “non-responders” twice, first as infants with < 10% bifidobacteria in the second stool sample and second as infants with < 20% bifidobacteria and found similar results (supplemental digital content Figure 1A and B)
Table 1.
Description of the cohort
| Demographics | |
| Gestational age at birth, completed weeks* | 29 (27, 31) |
| Apgar scores at 1 min* | 6 (4, 7) |
| Apgar scores at 5 min* | 8 (7, 9) |
| Gender (M, F) | 12, 17 |
| Delivery type (caesarean, vaginal) | 19, 10 |
| Age at first probiotic dose, days* | 5 (3, 5) |
| Diet (EBM, PDHM) | 29, 14 |
| Full enteral feeding, days* | 8 (7, 11) |
| Antibiotic days between sample 1 and sample 2* | 0 (0, 0) |
| Outcomes | |
| NEC (stage 2, stage 3) | 2, 0 |
| CLD | 12 |
| Home on oxygen | 0 |
| ROP | 6 |
| ROP laser | 2 |
EBM = expressed breast milk, PDHM = pasteurized donor human milk,
median (1st and 3rd quartiles)
Figure 1:
Next generation sequencing of the fecal microbiota at the family level. (A) All 29 enrolled infants, S1 = sample 1, S2 = sample 2. (B) Responders (more than 6% bifidobacteria in S2) vs non-responders (less than 6% bifidobacteria in S2). (C) Secretors vs non-secretors.
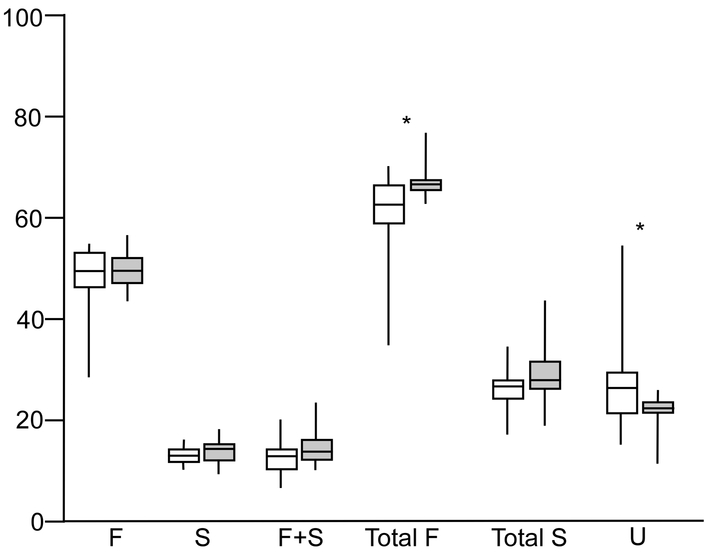
To address our primary hypothesis, that HMO composition in the milk fed to these infants influenced colonization with bifidobacteria, we performed several analyses. First, we compared milk HMO classs between responders and non-responders and found higher percentages of total fucosylated HMOs (those with one or more fucose plus those with fucose plus sialic acid) and lower percentages of undecorated HMOs (those lacking both fucose and sialic acid) in the milk fed to non-responder babies (Figure 2). The same patterns were consistent with the two alternate definitions of responder and non-responder (supplemental digital content Figure 2A and B). Second, we calculated Spearman correlation coefficients for the change in log percentage of fecal bifidobacteria from sample one to sample two with three different sample two measurements: a) the percentage of individual HMOs in milk sample two (5 of 44 HMO structures analyzed demonstrated significant correlations (supplemental digital content Table 1)); b) the ratio of fecal HMOs to milk HMOs in sample two, a surrogate marker for HMO consumption or absorption that is essentially normalized to control for changes in milk consumption, (one HMO structure demonstrated a significant correlation (IFLNH I); c) the ratio of urine HMOs to milk HMOs in sample two, a surrogate marker for absorption normalized for milk consumption, (one HMO structure demonstrated a significant correlation (IFLNH III). Third, we calculated Spearman correlation coefficients for the change in log percentage of fecal bifidobacteria from sample one to sample two with the change from sample one to sample two in two different ratios: a) the ratio of fecal HMOs to milk HMOs (significant correlations presented in Table 2) and b) the ratio of urine HMOs to milk HMOs (no significant correlations). Finally, we compared fecal samples from infants with secretor mothers and infants with non-secretor mothers and found no significant differences (Figure 1, panel C). To confirm this fourth method, we compared the mean change in log percentage of bifidobacteria from fecal sample one to fecal sample two between infants with secretor mothers and infants of non-secretor mothers and found no difference.
Figure 2:
(A) HMO classes in the mothers’ milk of responders and non-responders (n= 2 from each dyad) F=fucosylated only, S=sialylated only, F+S=HMOs with both fucose and sialic acid, Total F=F+(F+S), Total S=S+(F+S), U=undecorated (no fucose or sialic acid). Shaded boxes are non-responders (n=8), Open boxes are responders (n=21). Boxes are the 25th and 75th centiles with the median, whiskers are the range. *= P<0.05 (t-test assuming non-equal variance).
Table 2:
Correlations between changes in bifidobacteria from sample one to sample two with changes in the fecal HMO: milk HMO ratio from sample one to sample two.
| HMO class | Spearman’s rho | P value | |
| Fucosylated* | 0.47 | 0.02 | |
| Total Fucosylated* | 0.48 | 0.02 | |
| HMO structure | HMO class | Spearman’s rho | P value |
| pLNH | Undecorated | 0.50 | 0.01 |
| MFpLNH IV | Fucosylated | 0.60 | 0.002 |
| IFLNH I | Fucosylated | 0.52 | 0.01 |
| 5230b** | Fucosylated | 0.45 | 0.03 |
| LSTc+LSTb | Sialylated | 0.79 | 0.04 |
Fucosylated HMOs are structures with fucose but not sialic acid, total fucosylated HMOs include all structures with fucose.
Four number designations refer to the number of monosaccharides in this order: Hex_Fuc_HexNAc_Sialic acid
HMO composition in the milk changes over time and is highly variable in women delivering prematurely.35 Supplemental digital content Table 2 summarizes changes in class of HMOs and individual HMOs over time in milk, feces, and urine. For most classes of HMOs, the changes over time in the milk were paralleled by similar changes over time in the urine and feces. There were two notable exceptions. First, the class of HMOs containing both fucose and sialic acid decreased as a percentage of total HMOs in milk over time, but increased in both absolute numbers and percentages over time in the feces (suggesting minimal consumption by the gut microbes) and increased in absolute numbers in the urine (suggesting absorption from the gut). Second, the class of undecorated HMOs increased in milk over time both in absolute quantity and in percentage of total HMOs, but decreased in percentage in the feces suggesting that these structures were consumed by the gut microbes and/or absorbed from the gut (consistent with the increase in absolute numbers in the urine). Several individual HMO structures appeared to be consumed in the gut (present in milk but decreased over time or absent in the feces, e.g. DFLNO I and 6’SL) or readily absorbed from the gut and filtered into the urine (present in milk and in urine, e.g. LNT + LNnT and MFLNH I + III).
To confirm these observations, we used paired t-tests to identify differences in the logarithm of the two ratios between the first and second samples. Significant fold changes in the log fecal HMO:milk HMO ratio are presented in Table 3. Consistent with the observations above, the class of undecorated HMOs and four individual undecorated HMOs showed significant decreases in the log fecal HMO:milk HMO ratio over time (best explained by either consumption by intestinal microbes, absorption from the intestinal tract, and/or increase over time in milk intake and/or milk HMO content). The observation that the class of fucosylated HMOs and one individual HMO (MFpLNH IV) showed significant increases in the log fecal HMO:milk HMO ratio over time (best explained by either limited consumption by intestinal microbes, minimal absorption from the intestinal tract, and/or decrease over time in milk HMO content) while other individual fucosylated HMOs showed the opposite pattern underscores the heterogeneity in the interactions between the consumed HMOs, the intestinal microbiota and host gut permeability. In this cohort, these differences are likely due in large part to differences in consumption of specific fucosylated HMOs by the probiotic B. breve. Significant fold changes in the log urine HMO:milk HMO ratio are presented in Supplemental digital content Table 3. It is noteworthy that as a class fucosylated HMOs show a marked increase over time suggesting high levels of absorption from the gut and excretion by the kidneys while individual fucosylated HMOs show either a significant increase or decrease (further evidence of heterogeneity among HMOs).
Table 3.
Fold changes in the log fecal HMO:milk HMO ratio from sample one to sample two
| HMO Group | Fold change (95% CIs) | P value | |
|---|---|---|---|
| Fucosylated | 6.6 (3.6–12) | <0.0001 | |
| Fucosylated and sialylated | 7.6 (2.0–29) | 0.004 | |
| Total fucosylated | 7.9 (4.4–14) | <0.0001 | |
| Undecorated | 0.021 (0.0092–0.047) | <0.0001 | |
| HMO structure | HMO group | ||
| 3FL | Fucosylated | 0.056 (0.011–0.28) | 0.001 |
| LNT | Undecorated | 0.0052 (0.00047–0.058) | 0.0003 |
| LNT+LNnT | Undecorated | 0.018 (0.0031–0.10) | <0.0001 |
| LNFP II | Fucosylated | 0.046 (0.0067–0.32) | 0.003 |
| LNH | Undecorated | 0.077 (0.011–0.53) | 0.01 |
| LNnH | Undecorated | 0.046 (0.0066–0.33) | 0.004 |
| MFpLNH IV | Fucosylated | 16 (3.1–80) | 0.002 |
| 4120a | Fucosylated | 0.11 (0.014–0.84) | 0.03 |
| DFLNHc | Fucosylated | 0.033 (0.0086–0.13) | <0.0001 |
| 5130b | Fucosylated | 0.14 (0.024–0.81) | 0.03 |
| DFLNO I/DFLNO II | Fucosylated | 0.12 (0.057–0.26) | <0.0001 |
| 6’SL | Sialylated | 0.083 (0.01–0.69) | 0.02 |
Numbers less than one represent fold changes in the negative direction suggesting disappearance from the intestinal lumen over time consistent with either consumption of the HMO by gut microbes or absorption of the HMO from the gut. Numbers greater than one suggest increases in the gut lumen over time consistent with minimal absorption from the gut or decreased consumption by gut microbes.
Simple linear regression analysis comparing the fecal microbial classes other than bifidobacteria to the fecal composition of HMO classes yielded few significant associations. Among the second stool samples, for Enterobacteriaceae no correlations were noted, while for Bacteroides there was a weak positive correlation with percentage fecal undecorated HMOs (supplemental digital content figure 3).
Discussion
In term infants who receive predominantly their mother’s own milk, bifidobacteria are common colonizers of the infant gut accounting for up to 90% of total intestinal bacteria in developing countries.36 In western countries, the percentages of bifidobacteria are lower, even in healthy breast-fed infants who were born vaginally and have not been exposed to antibiotics.37,38 These differences likely play a role in the many compelling observations supporting the hygiene or “old friends” hypothesis, including the increased incidences of type 1 diabetes and food allergies seen in developed countries.38 In premature infants, bifidobacteria are conspicuously absent even when the diet is exclusively human milk.33 Differing species of bifidobacteria vary in their capacity to colonize the premature infant, based in large part on their capacity to utilize HMOs as a nutrient source.22 Differences in bifidobacteria at the subspecies and strain level have been characterized.39,40 Probiotics have been shown in several cohort studies and placebo-controlled randomized clinical trials to decrease the risk of NEC with several Bifidobacterium species demonstrating efficacy.41 We have previously demonstrated colonization and a decreased incidence of NEC in a cohort study of the B. breve strain administered to this cohort.42,43 The largest clinical trial to date of probiotics for the prevention of NEC in premature infants found no difference between B. breve and placebo.44 In that trial, the administered strain differed from the one studied herein, the dose administered was low compared to other similar trials, cross-contamination between the treatment and placebo groups was common, and the infants who were colonized with the probiotic (as opposed to the infants that received the probiotic) had lower rates of NEC, late-onset sepsis and death.45
The dominance of Enterobacteriaceae in this cohort is consistent with other reports33 and is important given the compelling evidence that members of this family of bacteria include Klebsiella pneumonia and Escherichia coli, both of which have been demonstrated to predispose to NEC. In a previous probiotic trial in premature infants we found a decrease in fecal Enterobacteriaceae with administration of B. longum subsp. infantis but not with B. animalis subsp. lactis.22 In this cohort, the percentage of Enterobacteriaceae increased over time in spite of B. breve administration.
In a previous study of a cohort of premature infants in the U.S., we demonstrated that the composition of HMOs in the mother’s milk influences the fecal microbiota and that there are significant differences among HMO structures in absorption from the gut, excretion in the urine, and consumption by gut microbes. In that study the infants had not received any probiotic or prebiotic supplements. There were correlations between the class of fucoslyated HMOs in the milk and decreased Enterobacteriales in the infant feces (significant individual HMO structures included 2’FL, LDFT, and LNFP V) and between the class of undecorated HMOs in the milk and increased Enterobacteriales in the infant feces (one significant HMO structure: LNnH). As expected the primary consumers of HMOs (bifidobacteria and Bacteroides) were present in very small amounts.46 The current study differs in that all of the infants received the probiotic B. breve with most of the infants showing colonization by the time of the second sample. Regardless of how we defined “responders” and “non-responders” we found similar associations with undecorated HMOs more abundant in the milk fed to responder babies and total fucosylated HMOs more abundant in the milk fed to non-responders. The most likely explanation is that the undecorated HMOs are indeed a significant part of the explanation as to why some premature infants become colonized by probiotic B breve while others do not. Likely HMO candidates include LNH, LNT, LNnT and LNnH (see supplemental table 1 and table 3) It is also likely that some structures that contain both fucose and sialic acid and some structures with fucose but not sialic acid are not able to be consumed by the bifidobacteria present in these babies (including the probiotic B. breve). A high abundance of these structures in the milk would mean fewer HMOs that are available as a food source for the bifidobacteria resulting in poor colonization with the probiotic. This is consistent with the data in table 3 where the classes of fucosylated, fucosylated + sialylated, and total fucosylated and the single HMO MFpLNH IV were all associated with an increase in the log feces:milk ratio over time. In our previous study, we saw trends towards differences in the infant fecal microbiota based on maternal secretor status, but this was not seen in the current larger cohort.
Over 100 HMO structures have been identified (75 of these structures are presented in Table 3 of a recent methods paper29). We selected 46 of the most abundant structures for this study. Several HMO structures are worthy of individual comment. First, 2’FL (2’-fucosyllactose), which is abundant in the milk of secretor mothers but essentially absent from non-secretor mothers, can be synthesized at a reasonable cost, and is now added to some infant formulas. This HMO is consumed in vitro by only 5 of 26 strains of B. breve.14 A recent study in the premature piglet model found no alteration in the intestinal microbiota, digestive function, or incidence/severity of NEC with addition of 2’FL to the diet,47 while in the rodent model 2’FL is protective against NEC and increases mesenteric perfusion.48,49 In the current cohort we found no changes in 2’FL in the milk, urine, or feces over time, no significant differences between fecal bifidobacteria based on maternal secretor status and no associations between maternal milk 2’FL content and the intestinal microbiota, all suggesting that the administered strain of B. breve is not a consumer of 2’FL. Second, 3FL is found in the milk of both secretors and non-secretors, was consumed by 10 of 26 strains of B. breve,14 increased in the milk over time, but was not found in significant quantities in many of the stool specimens (suggesting consumption by gut microbes) or urine specimens (suggesting a lack of absorption from the gut). This was confirmed by the almost 20 fold decrease in the log fecal:milk and urine:milk ratios over time (Table 3 and Supplemental digital content table 3), suggesting that this HMO is consumed by the administered B. breve strain. Third, DSLNT (disialyllacto-N-tetraose) has been demonstrated to decrease NEC incidence/severity in the rodent model.50 In the current cohort and in our previous cohort, DSLNT was in very low abundance in all specimens. Fourth, the abundant undecorated HMO LNT is aggressively consumed by all 26 tested B. breve strains,14 increased in the milk over time, decreased in the feces over time (suggesting consumption by gut microbes) and increased in the urine over time (suggesting absorption). Finally, B. breve strains in general are able to utilize large but not small sialylated HMOs.14 In the current study, the large HMOs, LSTb and LSTc decrease over time in the milk, are uncommon in feces (suggesting consumption), and increase over time in the urine (suggesting absorption), while the small HMO 3’SL decreases over time in the milk, but does not change over time in the feces or urine (suggesting a lack of both consumption and absorption). Interestingly 6’SL, also a small sialylated HMO, appears to be consumed by the gut microbes in this cohort.
Conclusions
HMOs shape the intestinal microbiota of the term breast-fed infant, but have a lesser impact in premature infants. The tremendous diversity of HMO structures suggests heterogeneity of function. As a class, fucosylated HMOs in the milk were associated with increased bifidobacteria in the feces of these infants, all of whom received probiotic B. breve, with several individual HMO structures in the milk associated with changes in bifidobacteria in the feces. However, there is diversity even among HMO classes as demonstrated by the marked ifferences among fucosylated structures in consumption, absorption and excretion in the urine. No single HMO structure is likely to be able to duplicate the complexity of human milk glycans. Maternal secretor status, as determined by abundance of α(1,2) fucosyl linkages in the milk was not a significant predictor of response to the administered probiotic.
Supplementary Material
Supplemental Table 1: Spearman correlation coefficients for the change in log percentage of fecal bifidobacteria from sample one to sample two with three outcomes
Supplemental Table 2: Changes in HMOs in milk, feces and urine over time.
Supplemental Table 3: Fold changes in the log urine HMO:milk HMO ratio from sample one to sample two.
Supplemental Figure 1: Next generation sequencing of the fecal microbiota at the family level A: Responders had >10% bifidobacteria in fecal sample 2; B: Responders had >20% bifidobacteria in fecal sample 2.
Supplemental Figure 2: HMO classes in the mothers’ milk of responders and non-responders (2 samples from each mother) F=fucosylated only, S=sialylated only, F+S=HMOs with both fucose and sialic acid, Total F=F+(F+S), Total S=S+(F+S), U=undecorated (no fucose or sialic acid). A: Responders had >10% bifidobacteria in fecal sample 2; B: Responders had >20% bifidobacteria in fecal sample 2. Shaded boxes are non-responders, Open boxes are responders. Boxes are the 25th and 75th centiles with the median, whiskers are the range. *= P<0.05 (t-test assuming non-equal variance).
Supplemental Figure 3: Correlation between percentage fecal Bacteroides and percentage fecal undecorated HMOs in the second fecal sample.
What is known:
Probiotic microbes decrease the risk of necrotizing enterocolitis and death in premature infants
Human milk oligosaccharides are selective prebiotics
B. breve M16-V has been shown to decrease necrotizing enterocolitis
B. breve strains are variable in their capacity to consume human milk oligosaccharides
What is new:
Colonization of the premature infant with B. breve M16-V is influenced by the composition of human milk oligosaccharides
B. breve M16-V is a selective consumer of human milk oligosaccharides (e.g. consumes 3FL and LNT but not 2’FL)
Secretor status of the mother is not a predictor of colonization with B. breve M16-V
Acknowledgements:
The authors express appreciation to Dr. Abe and Morinaga Milk Industry Co, Tokyo for provision of the probiotic B. breve.
Conflicts of interest and source of funding: D.A.M. and C.B.L are co-founders of Evolve Biosystems, a company focused on diet based manipulation of the gut microbiota. The other authors report no potential conflicts of interest. This work was funded in part by National Institutes of Health Awards AT007079, HD065122 and AT008759, and the Peter J. Shields Endowed Chair in Dairy Food Science.
References
- 1.Carlisle EM, Poroyko V, Caplan MS, Alverdy JA, Liu D. Gram negative bacteria are associated with the early stages of necrotizing enterocolitis. PloS one 2011;6:e18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cilieborg MS, Boye M, Molbak L, Thymann T, Sangild PT. Preterm birth and necrotizing enterocolitis alter gut colonization in pigs. Pediatric research 2011;69:10–6. [DOI] [PubMed] [Google Scholar]
- 3.Underwood MA, Arriola J, Gerber CW, et al. Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: alterations in inflammation, innate immune response, and the microbiota. Pediatric research 2014;76:326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. The ISME journal 2009;3:944–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mai V, Young CM, Ukhanova M, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PloS one 2011;6:e20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claud EC, Keegan KP, Brulc JM, et al. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 2013;1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner BB, Deych E, Zhou Y, et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 2016;387:1928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alfaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane database of systematic reviews (Online) 2014;4:CD005496. [DOI] [PubMed] [Google Scholar]
- 9.Patel RM, Denning PW. Therapeutic use of prebiotics, probiotics, and postbiotics to prevent necrotizing enterocolitis: what is the current evidence? Clinics in perinatology 2013;40:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charbonneau MR, O’Donnell D, Blanton LV, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 2016;164:859–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology (Reading, England) 2013;159:649–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asakuma S, Hatakeyama E, Urashima T, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. The Journal of biological chemistry 2011;286:34583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Scientific reports 2015;5:13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Moyano S, Totten SM, Garrido DA, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Applied and environmental microbiology 2013;79:6040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Li M, Wu S, et al. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. Journal of pediatric gastroenterology and nutrition 2015;60:825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGovern DP, Jones MR, Taylor KD, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Human molecular genetics 2010;19:3468–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, et al. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002;122:1967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrow AL, Meinzen-Derr J, Huang P, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. The Journal of pediatrics 2011;158:745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demmert M, Schaper A, Pagel J, et al. FUT 2 polymorphism and outcome in very-low-birth-weight infants. Pediatric research 2015;77:586–90. [DOI] [PubMed] [Google Scholar]
- 20.Lewis ZT, Totten SM, Smilowitz JT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bokulich NA, Mills DA. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Applied and environmental microbiology 2013;79:5214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Underwood MA, Kalanetra KM, Bokulich NA, et al. A comparison of two probiotic strains of bifidobacteria in premature infants. The Journal of pediatrics 2013;163:1585–91 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 2011;108 Suppl 1:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninonuevo MR, Park Y, Yin H, et al. A strategy for annotating the human milk glycome. Journal of agricultural and food chemistry 2006;54:7471–80. [DOI] [PubMed] [Google Scholar]
- 28.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. Journal of proteome research 2010;9:4138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Leoz ML, Wu S, Strum JS, et al. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. Analytical and bioanalytical chemistry 2013;405:4089–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Totten SM, Wu LD, Parker EA, et al. Rapid-throughput glycomics applied to human milk oligosaccharide profiling for large human studies. Analytical and bioanalytical chemistry 2014;406:7925–35. [DOI] [PubMed] [Google Scholar]
- 31.Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. Journal of proteome research 2011;10:856–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Totten SM, Zivkovic AM, Wu S, et al. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. Journal of proteome research 2012;11:6124–33. [DOI] [PubMed] [Google Scholar]
- 33.La Rosa PS, Warner BB, Zhou Y, et al. Patterned progression of bacterial populations in the premature infant gut. Proceedings of the National Academy of Sciences of the United States of America 2014;111:12522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arboleya S, Sanchez B, Solis G, et al. Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: a functional inference study. International journal of molecular sciences 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Leoz ML, Gaerlan SC, Strum JS, et al. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. Journal of proteome research 2012;11:4662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huda MN, Lewis Z, Kalanetra KM, et al. Stool microbiota and vaccine responses of infants. Pediatrics 2014;134:E362–E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turroni F, Peano C, Pass DA, et al. Diversity of bifidobacteria within the infant gut microbiota. PloS one 2012;7:e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vatanen T, Kostic AD, d’Hennezel E, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016;165:842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugahara H, Odamaki T, Xiao J. Genotypic and phenotypic evaluation revealed the appropriateness of human-residential bifidobacteria for human use. Milk Science 2015;64:261–9. [Google Scholar]
- 40.Odamaki T, Horigome A, Sugahara H, et al. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. International journal of genomics 2015;2015:567809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vongbhavit K, Underwood MA. Prevention of necrotizing enterocolitis through manipulation of the intestinal microbiota of the premature infant. Clinical therapeutics 2016;38:716–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patole S, Keil AD, Chang A, et al. Effect of Bifidobacterium breve M-16V supplementation on fecal bifidobacteria in preterm neonates--a randomised double blind placebo controlled trial. PloS one 2014;9:e89511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patole SK, Rao SC, Keil AD, Nathan EA, Doherty DA, Simmer KN. Benefits of Bifidobacterium breve M-16V supplementation in preterm neonates - A Retrospective Cohort Study. PloS one 2016;11:e0150775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costeloe K, Hardy P, Juszczak E, Wilks M, Millar MR, Probiotics in Preterm Infants Study Collaborative G. Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial. Lancet 2016;387:649–60. [DOI] [PubMed] [Google Scholar]
- 45.Deshpande G, Rao S, Athalye-Jape G, Conway P, Patole S. Probiotics in very preterm infants: the PiPS trial. Lancet 2016;388:655. [DOI] [PubMed] [Google Scholar]
- 46.Underwood MA, Gaerlan S, De Leoz ML, et al. Human milk oligosaccharides in premature infants: absorption, excretion, and influence on the intestinal microbiota. Pediatric research 2015;78:670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cilieborg MS, Bering SB, Ostergaard MV, et al. Minimal short-term effect of dietary 2’-fucosyllactose on bacterial colonisation, intestinal function and necrotising enterocolitis in preterm pigs. The British journal of nutrition 2016;116:834–41. [DOI] [PubMed] [Google Scholar]
- 48.Autran CA, Schoterman MH, Jantscher-Krenn E, Kamerling JP, Bode L. Sialylated galacto-oligosaccharides and 2’-fucosyllactose reduce necrotising enterocolitis in neonatal rats. The British journal of nutrition 2016;116:294–9. [DOI] [PubMed] [Google Scholar]
- 49.Good M, Sodhi CP, Yamaguchi Y, et al. The human milk oligosaccharide 2’-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. The British journal of nutrition 2016;116:1175–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jantscher-Krenn E, Zherebtsov M, Nissan C, et al. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 2012;61:1417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Spearman correlation coefficients for the change in log percentage of fecal bifidobacteria from sample one to sample two with three outcomes
Supplemental Table 2: Changes in HMOs in milk, feces and urine over time.
Supplemental Table 3: Fold changes in the log urine HMO:milk HMO ratio from sample one to sample two.
Supplemental Figure 1: Next generation sequencing of the fecal microbiota at the family level A: Responders had >10% bifidobacteria in fecal sample 2; B: Responders had >20% bifidobacteria in fecal sample 2.
Supplemental Figure 2: HMO classes in the mothers’ milk of responders and non-responders (2 samples from each mother) F=fucosylated only, S=sialylated only, F+S=HMOs with both fucose and sialic acid, Total F=F+(F+S), Total S=S+(F+S), U=undecorated (no fucose or sialic acid). A: Responders had >10% bifidobacteria in fecal sample 2; B: Responders had >20% bifidobacteria in fecal sample 2. Shaded boxes are non-responders, Open boxes are responders. Boxes are the 25th and 75th centiles with the median, whiskers are the range. *= P<0.05 (t-test assuming non-equal variance).
Supplemental Figure 3: Correlation between percentage fecal Bacteroides and percentage fecal undecorated HMOs in the second fecal sample.