Abstract
The addition of a single β-d-GlcNAc sugar (O-GlcNAc) by O-GlcNAc-transferase (OGT) and O-GlcNAc removal by O-GlcNAcase (OGA) maintain homeostatic O-GlcNAc levels on cellular proteins. Changes in protein O-GlcNAcylation regulate cellular differentiation and cell fate decisions, but how these changes affect erythropoiesis, an essential process in blood cell formation, remains unclear. Here, we investigated the role of O-GlcNAcylation in erythropoiesis by using G1E-ER4 cells, which carry the erythroid-specific transcription factor GATA-binding protein 1 (GATA-1) fused to the estrogen receptor (GATA-1–ER) and therefore undergo erythropoiesis after β-estradiol (E2) addition. We observed that during G1E-ER4 differentiation, overall O-GlcNAc levels decrease, and physical interactions of GATA-1 with both OGT and OGA increase. RNA-Seq–based transcriptome analysis of G1E-ER4 cells differentiated in the presence of the OGA inhibitor Thiamet-G (TMG) revealed changes in expression of 433 GATA-1 target genes. ChIP results indicated that the TMG treatment decreases the occupancy of GATA-1, OGT, and OGA at the GATA-binding site of the lysosomal protein transmembrane 5 (Laptm5) gene promoter. TMG also reduced the expression of genes involved in differentiation of NB4 and HL60 human myeloid leukemia cells, suggesting that O-GlcNAcylation is involved in the regulation of hematopoietic differentiation. Sustained treatment of G1E-ER4 cells with TMG before differentiation reduced hemoglobin-positive cells and increased stem/progenitor cell surface markers. Our results show that alterations in O-GlcNAcylation disrupt transcriptional programs controlling erythropoietic lineage commitment, suggesting a role for O-GlcNAcylation in regulating hematopoietic cell fate.
Keywords: O-GlcNAcylation, O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), hematopoiesis, GATA transcription factor, gene regulation, differentiation, post-translational modification, transcription, erythropoiesis, myelopoiesis, G1E-ER4, FOG-1
Introduction
Hematopoiesis begins early in embryonic development and continues throughout adulthood to produce and replenish the various cell types comprising the blood system. Distinct cellular lineages are derived from hematopoietic stem cells via the activation of cell-specific gene expression programs, spatiotemporally controlled by lineage-specific transcription factors that activate or inactivate downstream genes (1) The molecular mechanisms by which lineage-specific transcriptional programs are established and maintained in different hematopoietic cell types are broadly understood, but the role of post-translational modifications (PTMs)5 in these transcriptional cascades is not well defined. PTMs have many fundamental biological functions and present an exciting new avenue to explore in the context of hematopoiesis. Understanding how extracellular signals and the cellular environment regulate hematopoiesis through PTMs has significant implications regarding the modulation of lineage-specific gene expression and offers the potential for identification and development of novel therapies to treat blood-related disorders.
Erythropoiesis is the process by which hematopoietic stem cells and progenitors proliferate and differentiate into mature erythrocytes. Erythroid differentiation is regulated by lineage-specific transcription factors that orchestrate proper cellular differentiation. GATA-1, a dual zinc finger DNA-binding protein that recognizes WGATAR motifs, is a key erythroid-specific transcription factor that coordinates erythropoietic gene programs during maturation (2–5). At the onset of erythroid differentiation and as it proceeds, many GATA-1 target genes are activated or repressed (6–9). The co-occupancy of DNA-binding sites by GATA-1 and other erythroid-specific or ubiquitous transcription factors, including coactivators and corepressors, determines the gene transcription status to some extent (7, 10–12). Epigenetic modification of histones and variation of WGATAR-adjacent DNA-binding motifs play a role (7, 8, 13–15), but these alone do not adequately explain how GATA-1–containing protein complexes are modulated to positively or negatively regulate gene expression.
Post-translational modification of transcription complexes during hematopoiesis might regulate lineage-specific activity by altering protein–protein interactions within these complexes or by modifying nearby chromatin structure (16–19). Recently, we demonstrated that O-GlcNAcylation modulates GATA-1–mediated repression of the Aγ-globin promoter (20). O-GlcNAcylation is the modification of serine and threonine residues by β-d-GlcNAc (O-GlcNAc). This monomeric sugar moiety is added and removed by the O-GlcNAc–processing enzymes, O-GlcNAc-transferase (OGT) and O-GlcNAcase (OGA), respectively (21). Dynamic O-GlcNAc cycling is critical for the proper regulation of gene transcription. O-GlcNAc is one of the histone code marks (22, 23), and it modulates other epigenetic changes on chromatin, such as DNA methylation (24, 25), histone acetylation (26), and histone methylation (25, 27). TET (ten-eleven translocation) proteins and host cell factor 1 colocalize with OGT at gene promoters associated with activating histone marks to promote target gene transcription (23–25, 28). OGT also interacts with mSin3A or components of the polycomb repressive complexes 1 and 2 to mediate gene silencing by forming stable repressor complexes (26, 27, 29). In addition, OGT and OGA regulate preinitiation complex formation and RNA polymerase II activity (30–32).
OGT and OGA interact with the GATA-1–FOG-1–Mi2β (NuRD) repressor complex at the Aγ-globin promoter, and O-GlcNAcylation of the NuRD chromatin-remodeling complex is necessary for GATA-1–FOG-1–Mi2β repressor formation (20). Other studies have demonstrated that O-GlcNAc cycling on chromatin modifiers is critical for modulation of gene transcription and normal development (33, 34). Thus, O-GlcNAc might regulate hematopoietic cell differentiation and maturation. During embryonic stem cell differentiation into neurons, cellular O-GlcNAc levels decrease (35) as well as those in epidermal keratinocytes (36), cardiomyocytes (37), and myoblasts (38). O-GlcNAc also decreases in K562 erythroleukemia cells following treatments that induce terminal differentiation (20). However, the overall O-GlcNAc level increases during differentiation of mouse chondrocyte (39) and osteoblast cells (40). These data suggest the hypothesis that O-GlcNAc cycling contributes to the regulation of gene expression programs during hematopoietic cell differentiation.
To test this hypothesis, we used well-established erythroid and myeloid cell systems that can be induced to undergo terminal differentiation. For an erythroid model, we used the G1E-ER4 cell line, an erythroid progenitor cell line derived from murine GATA-1− erythroid (G1E) cells that stably express GATA-1–ER, a fusion product of GATA-1 and human estrogen receptor ligand-binding domain (6). GATA-1 is the master regulatory transcription factor for erythropoiesis. When GATA-1 activity is restored following the addition of β-estradiol (E2), G1E-ER4 cells undergo erythroid differentiation (6, 41). For a myeloid cell model, we used the NB4 and HL60 myeloid leukemia cell lines, which differentiate into neutrophil-like cells following addition of all-trans-retinoic acid (42). In this work, we demonstrate that 1) erythropoietic cell differentiation resulted in a marked reduction of total O-GlcNAc levels; 2) interaction among GATA-1–ER, OGT, and OGA was enhanced after E2-induced differentiation; 3) perturbation of O-GlcNAc cycling affected 1,173 differentially expressed genes, including 47 erythroid-specific GATA-1 target genes; 4) OGA inhibition by Thiamet-G (TMG) led to an amplification of GATA-1–mediated activation or repression of GATA-1 target genes; 5) TMG treatment impaired myeloid cell differentiation into neutrophil-like cells; and 6) prolonged TMG treatment disrupted erythroid differentiation in G1E-ER4 cells. We conclude that O-GlcNAcylation modulates lineage-specific transcriptional cascades, thus contributing to the control of hematopoietic differentiation.
Results
O-GlcNAc levels decrease during erythropoiesis
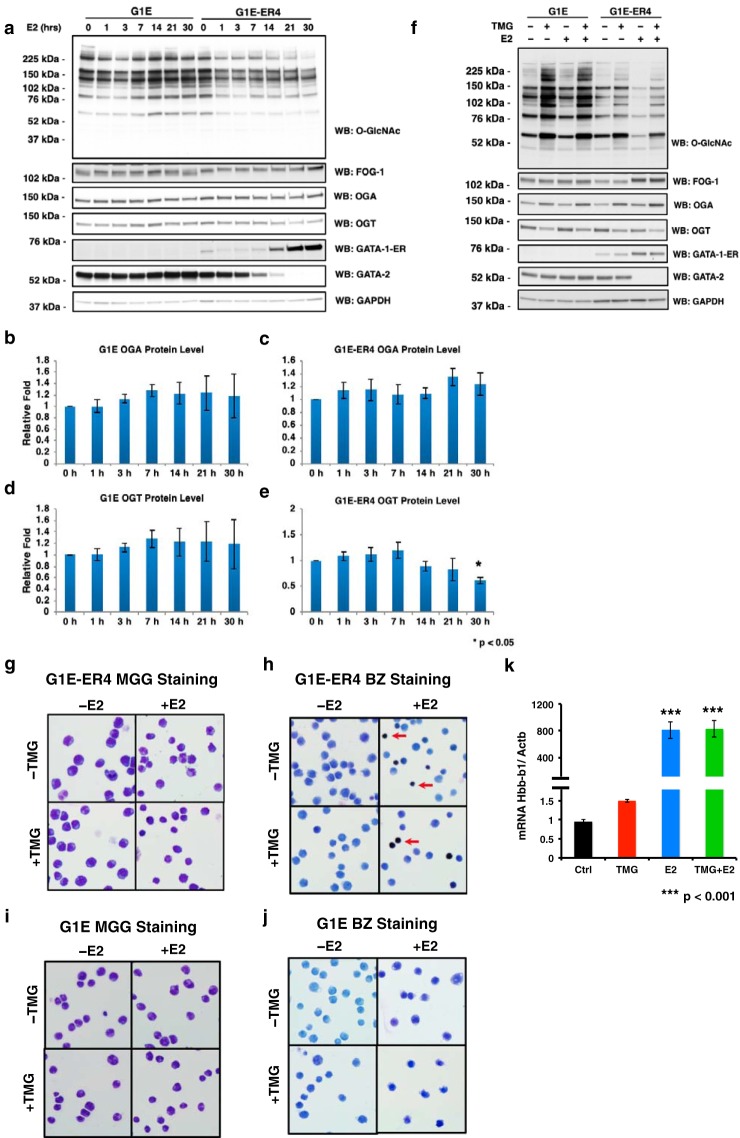
To determine whether O-GlcNAcylation is involved in erythroid differentiation, we treated GATA-1–ER cells with E2 to trigger erythroid differentiation. Expression of GATA-1–ER increased as reported previously (43, 44); Gata2 transcription was repressed by GATA-1, resulting in the decrease of GATA-2 protein level (15). Expression of friend of GATA-1 (FOG-1) also increased as reported previously (6) (Fig. 1a). Interestingly, the total cellular O-GlcNAc level was dramatically decreased after E2 treatment. We found that the level continued to decrease during differentiation, reaching its lowest level at 30 h post-E2 administration. The decreased O-GlcNAc level was associated with a reduction of OGT protein levels and an increase of OGA protein levels (Fig. 1, a, c, and e). However, the large decrease in cellular O-GlcNAcylation after E2 treatment suggests that the targeting of OGT and OGA to specific substrates might also be altered during differentiation. The O-GlcNAc level, as well as OGT and OGA expression, did not change over time in G1E negative control cells (Fig. 1, a, b, and d), which, unlike G1E-ER4 cells, lack inducible GATA-1 function (Fig. 1a). These data indicate that the O-GlcNAc level decreases during erythropoiesis and that the decline was a direct result of GATA-1 activity.
Figure 1.
O-GlcNAc levels decrease following the restoration of GATA-1 activity. G1E and G1E-ER4 cells were treated with TMG (OGA inhibitor) and/or E2 for 30 h. Cells were harvested following a time course (a) or at 30 h (f). Whole-cell lysates were subjected to immunoblotting. Relative OGA (b and c) and OGT (d and e) protein levels of G1E (b and d) and G1E-ER4 (c and e) were quantified by ImageJ 1.49v. * indicates p < 0.05 compared with 0 h. G1E-ER4 (g and h) and G1E (i and j) cells were subjected to May-Grünwald Giemsa (MGG) staining (g and i) for cell morphology and benzidine staining (BZ) (h and j) for hemoglobin content. The red arrow shows hemoglobin-positive cells. k, transcription level of the βmajor-globin gene (Hbb-b1) was measured by RT-qPCR, and β-actin (Actb) was used as an internal control. Boxes represent cropped blots. * indicates p < 0.05 compared with 0 h, and *** indicates p < 0.001 compared with control. Error bars represent S.E. WB, Western blotting.
We treated GATA-1–ER cells for 30 h with both E2 and TMG, a highly selective inhibitor of OGA that blocks removal of the O-GlcNAc moiety by the enzyme, to determine whether the reduction in total cellular O-GlcNAcylation is required for proper erythroid differentiation (45–47). Cells treated with TMG maintain the O-GlcNAc moiety on target proteins, leading to increased global levels of O-GlcNAcylation and altered O-GlcNAc cycling. TMG was added at the onset of E2-initiated differentiation (acute TMG treatment). Western blotting analyses showed an increased O-GlcNAc level following TMG treatment. FOG-1, GATA-1–ER, and GATA-2 protein levels were unaffected by TMG treatment (Fig. 1f). Morphologic maturation (Fig. 1g) and hemoglobin induction (Fig. 1h) were similar in E2-induced, ±acute TMG G1E-ER4 cells, and TMG did not affect βmajor-globin gene transcription (Fig. 1k). We did not observe changes in morphology (Fig. 1i) or hemoglobin induction in control G1E cells treated with E2 ± acute TMG (Fig. 1j), indicating that these cells did not undergo erythroid differentiation. We conclude that there were no significant observed differences in cell morphology, hemoglobin induction, or βmajor-globin gene transcription between E2-induced, ±acute TMG–treated G1E-ER4 cells within the first 30 h of induction. However, short-term TMG treatment did affect the overall O-GlcNAc levels in G1E-ER4 cells, suggesting the potential to disrupt cellular pathways involved in erythroid differentiation.
Prolonged OGA inhibition impairs erythropoiesis
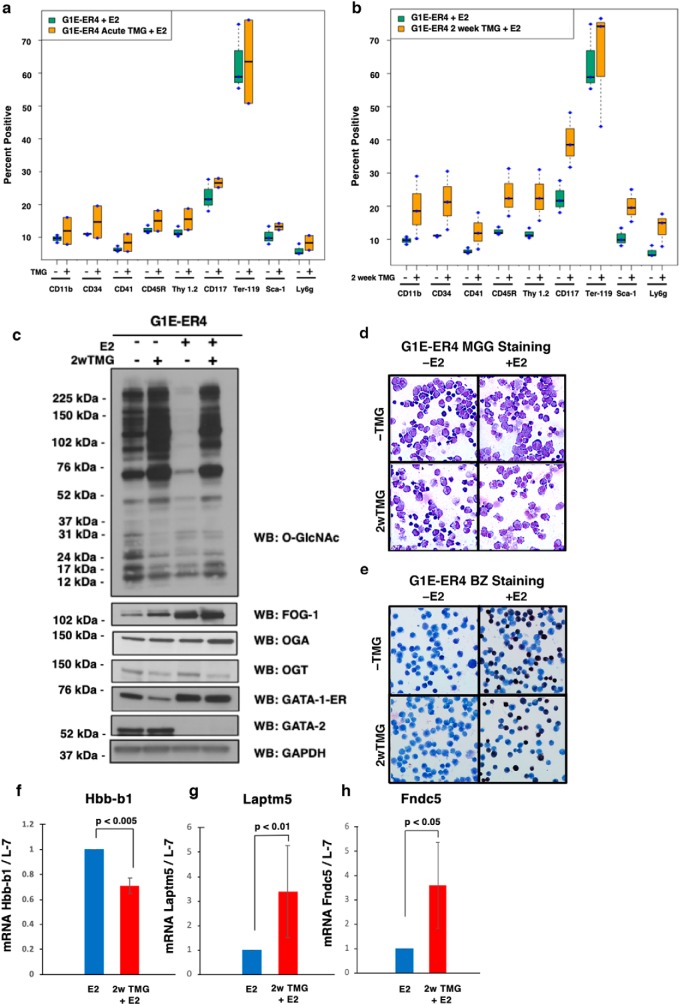
Previously, we demonstrated in cell lines and mouse tissue that sustained TMG treatment (14 days or longer) alters the metabolic demands of a cell and reprograms the transcriptome, leading to a variety of adaptive changes to TMG (48); therefore, we hypothesized that long-term TMG treatment would dramatically alter erythropoiesis compared with acute TMG treatment. G1E-ER4 cells were treated for 2 weeks with a daily dose of TMG and then differentiated with E2. To analyze how perturbation of O-GlcNAc cycling affects erythropoiesis, we performed flow cytometry on acute (TMG + E2) and long-term (2-week) TMG-treated G1E-ER4 cells. There was no significant difference after 30 h of E2 induction between treatment groups (data not shown). However, after 72 h of E2 induction, acute TMG treatment increased the expression of all cell surface markers (Fig. 2a). This increase was enhanced further in long-term TMG-treated G1E-ER4 cells (Fig. 2b). Overall O-GlcNAc levels and OGA expression increased during long-term TMG exposure, whereas OGT expression decreased (Fig. 2c). Long-term TMG exposure led to a slight decrease of GATA-1–ER pre-E2 induction and an increase in FOG-1 expression pre- and post-E2 induction (Fig. 2c). Sustained TMG treatment resulted in a 10% reduction of hemoglobin-positive cells compared with E2-only treatment (Fig. 2, d and e). Sustained TMG + E2 treatment reduced βmajor-globin expression by 34.5% compared with E2 treatment only (Fig. 2f). Laptm5 and Fndc5 gene expression increased 2.8- and 2.6-fold, respectively, with sustained TMG treatment and E2 (Fig. 2, g and h). These data demonstrate that sustained TMG treatment slows G1E-ER4 differentiation and alters the expression of erythroid genes.
Figure 2.
Sustained OGA inhibition modulates erythropoietic gene expression. TMG was added to G1E-ER4 cells at the same time as E2 induction. Treatment was either acute (72 h) or long-term (2 weeks). Flow cytometry analysis was performed on E2-only (data not shown), E2 + acute TMG (a), and long-term TMG treatment (2 weeks (2w)) + E2 (b). Whole cell lysates were prepared at 0 and 72 h for E2-only and long-term TMG treatment + E2 and subjected to immunoblotting (c). Cells were analyzed histologically by May-Grünwald Giemsa (MGG) staining (d) for cell morphology and benzidine staining (BZ) (e) for hemoglobin content. Transcription levels of the βmajor-globin gene (Hbb-b1) (f), Laptm5 (g), and Fndc5 (h) were measured by RT-qPCR; ribosome protein L-7 was used as an internal control. Boxes represent cropped blots. All experiments were performed with at least three biological replicates. Error bars represent S.D. WB, Western blotting.
Myeloid to neutrophil-like differentiation is impaired with TMG treatment
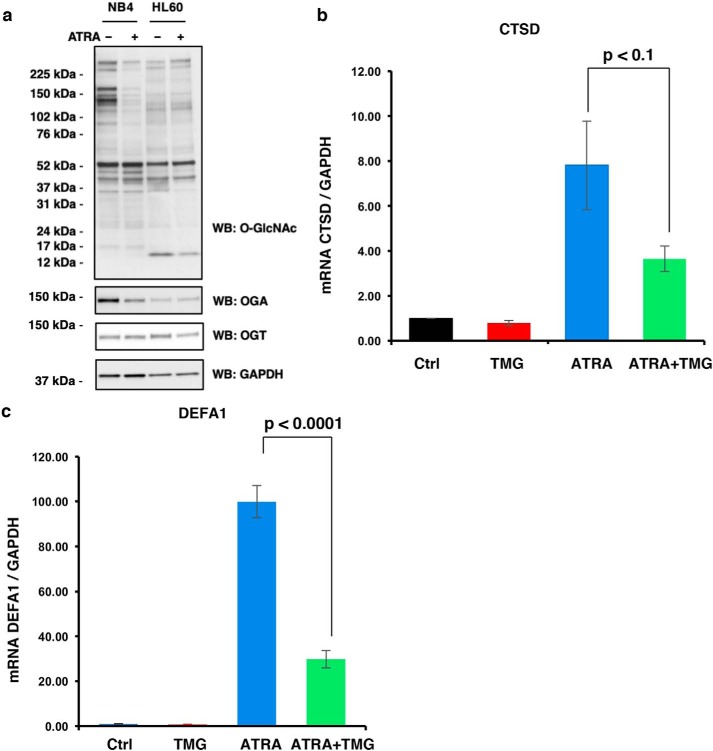
Based on our G1E-ER4 cell data, we hypothesized that O-GlcNAcylation might be involved in the differentiation of other hematopoietic lineages. Human NB4 and HL60 myeloid leukemia cells differentiate into neutrophil-like cells following the addition of all-trans-retinoic acid (ATRA) (42). Therefore, we treated cells with ATRA for 48 h prior to harvesting and analysis. In both cell lines, we measured a decline in O-GlcNAc levels. The O-GlcNAcylation change in the NB4 cells affected a large number of O-GlcNAc–modified proteins, whereas in HL60 cells proteins at 76, 44, 37, and 12 kDa showed declines in O-GlcNAcylation (Fig. 3a). Similar to G1E-ER4 cells, OGT levels did not decline during differentiation, suggesting that OGT substrate–targeting proteins change after ATRA induction. Next, we treated NB4 cells with TMG concomitantly with ATRA induction and measured transcription of the cathepsin D (CTSD) and defensin α1 (DEFA1) genes, which increase during neutrophil-like differentiation (49, 50). ATRA treatment led to a robust increase in CTSD (Fig. 3b) and DEFA1 (Fig. 3c) mRNA expression. TMG treatment decreased CTSD and DEFA1 gene expression following ATRA induction. These data phenocopy gene expression changes during G1E-ER4 cell differentiation and suggest that 1) O-GlcNAc levels decline as hematopoietic cells differentiate, 2) substrate targeting of OGT and OGA changes during differentiation, and 3) disruption in O-GlcNAc homeostasis impairs differentiation.
Figure 3.
Neutrophil-like differentiation is altered with TMG treatment. NB40 or HL60 cells were induced to differentiate with ATRA ± TMG. Cells were harvested at 48 h, and whole-cell lysates were subjected to immunoblotting (a). Transcription levels of genes CTSD (b) and DEFA1 (c) induced by ATRA treatment were analyzed by qPCR. The experiments were repeated three times. GAPDH was used as an internal control. Error bars represent S.E. Ctrl, control.
GATA-1 interacts with OGT and OGA in G1E-ER4 cells
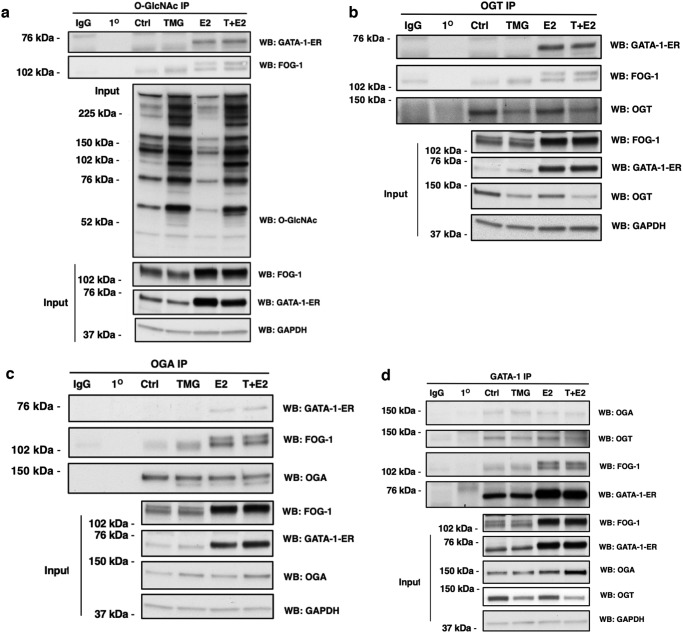
Our previous study in human erythroleukemia K562 cells or murine chemical inducer of dimerization–dependent human β-globin locus yeast artificial chromosome bone marrow cells demonstrated that OGT and OGA interact with the NuRD repressor complex at the −566 GATA site of the Aγ-globin gene promoter and that the Mi2β–OGT–OGA interactions change when γ-globin transcription is altered (20). Thus, we assessed whether OGT and OGA interact with GATA-1–ER in G1E-ER4 cell lysates treated with TMG and/or E2 via coimmunoprecipitation experiments using antibodies against O-GlcNAc (RL2), OGT, OGA, and GATA-1. We found that GATA-1–ER was poorly coimmunoprecipitated by O-GlcNAc, OGT, and OGA antibodies prior to E2 treatment. Following E2 treatment, O-GlcNAc, OGT, and OGA robustly coimmunoprecipitated GATA-1–ER (Fig. 4, a–c). O-GlcNAc, OGT, and OGA coimmunoprecipitated FOG-1 before and after E2 treatment, although E2 treatment increased the amount of coimmunoprecipitated FOG-1 with a molecular weight shift suggesting an increase in FOG-1 phosphorylation (51, 52) (Fig. 4, a–c). GATA-1 antibody coimmunoprecipitated OGT and OGA before and after E2 treatment (Fig. 4d). These data demonstrate that the interaction among GATA-1–ER, OGT, and OGA, as well as the O-GlcNAcylation of GATA-1–ER-interacting proteins, increases during G1E-ER4 cell differentiation.
Figure 4.
GATA-1 interacts with OGT and OGA in G1E-ER4 cells. G1E-ER4 cells were treated with TMG (T) and/or E2 for 30 h. Cells were harvested and subjected to IP. O-GlcNAcylated protein, OGT, OGA, and GATA-1–ER were immunoprecipitated with a specific O-GlcNAc antibody (RL2) (a) and OGT (b), OGA (c), and GATA-1 (d) antibodies, respectively. Blots were probed with RL2, GATA-1, FOG-1, OGT, OGA, and GAPDH, respectively. Isotype (normal rabbit or mouse IgG) IP and antibody-alone precipitation (1°) were used as negative controls. Forty micrograms of total lysates were used as input. All experiments were performed with at least three biological replicates. Boxes represent cropped blots. WB, Western blotting; Ctrl, control.
Inhibition of OGA with TMG changes erythroid gene transcription networks
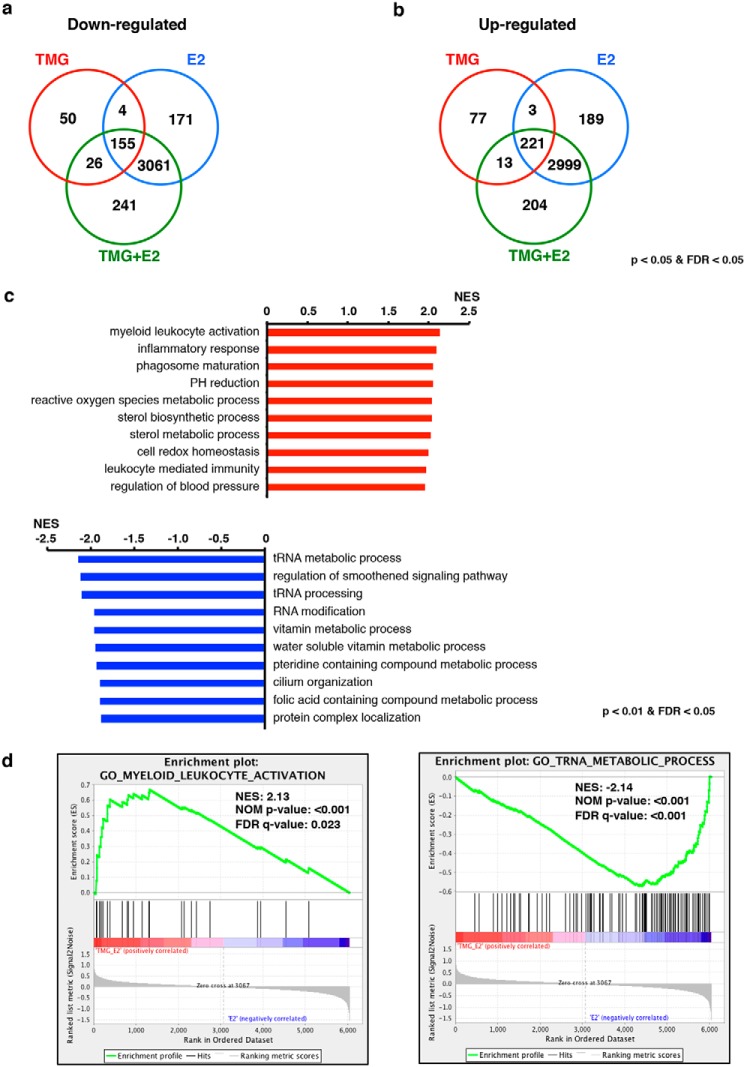
Based on our previous work (20) and the current study, we have observed that OGT and OGA interact with GATA-1 in multiple erythroid cell lines; next, we tested whether disruptions of O-GlcNAc homeostasis altered the erythroid gene transcription network during G1E-ER4 differentiation. We performed next generation RNA-sequence (RNA-Seq) analysis on G1E-ER4 cells without treatment, with acute TMG or E2 treatment only, and with E2 + acute TMG treatment for 30 h because this time point corresponds to previously published GATA-1 next generation chromatin immunoprecipitation (ChIP)-sequencing data (7). 8,271 transcripts (Table S1) were included in the RNA-Seq analysis from the different treatments, and the shared genes that were down-regulated (Fig. 5a) or up-regulated (Fig. 5b) are shown. Our RNA-Seq data suggest that inhibition of OGA by TMG changes transcription levels of numerous genes during erythropoiesis. Gene set enrichment analysis (GSEA) of the 8,271 transcripts was performed to characterize the biological processes altered by inhibition of OGA (53, 54). The top 10 up-regulated (top panel) and down-regulated (bottom panel) biological processes are listed comparing acute TMG + E2 with E2 treatment only (Fig. 5c). Examples of enrichment analysis plots are shown in Fig. 5d. Interestingly, biological processes of myeloid-leukocyte activation and inflammatory response were up-regulated. To further evaluate our RNA-Seq data, we compared the transcription profile of G1E-ER4 cells treated with E2 + acute TMG with that of E2 only–treated cells (Fig. 6a). 1,173 transcripts (Table S2) were differentially expressed in E2 + acute TMG compared with E2 treatment only; 530 genes were up-regulated, and 643 were down-regulated (Fig. 6, b and c), including 47 erythroid-specific genes (Table S3) (55). Together, these results indicate that transcriptional networks were changed when OGA was inhibited with TMG during erythropoiesis and that alteration of O-GlcNAc cycling in GATA-1–ER cells modulates terminal differentiation of erythroid gene programs.
Figure 5.
Inhibition of OGA changes gene transcription network during erythropoiesis. Venn diagrams of down- (a) and up-regulated (b) transcripts in TMG, E2, and E2 + acute TMG treatments compared with control, respectively, are shown. Transcripts with a p value <0.05 and FDR <0.05 were included in the analysis (c). The top 10 up- (top) and down-regulated (bottom) biological processes analyzed by GSEA are ranked by normalized enrichment score (NES) for TMG + E2– and E2 only–treated G1E-ER4 cells (d). Enrichment plots for E2 + acute TMG– and E2 only–treated G1E-ER4 cells show up-regulated biological process myeloid leukocyte activation (left) and down-regulated biological process tRNA metabolic process (right). A nominal (NOM) p value <5% and an FDR q value <25% were used for biological process analysis.
Figure 6.
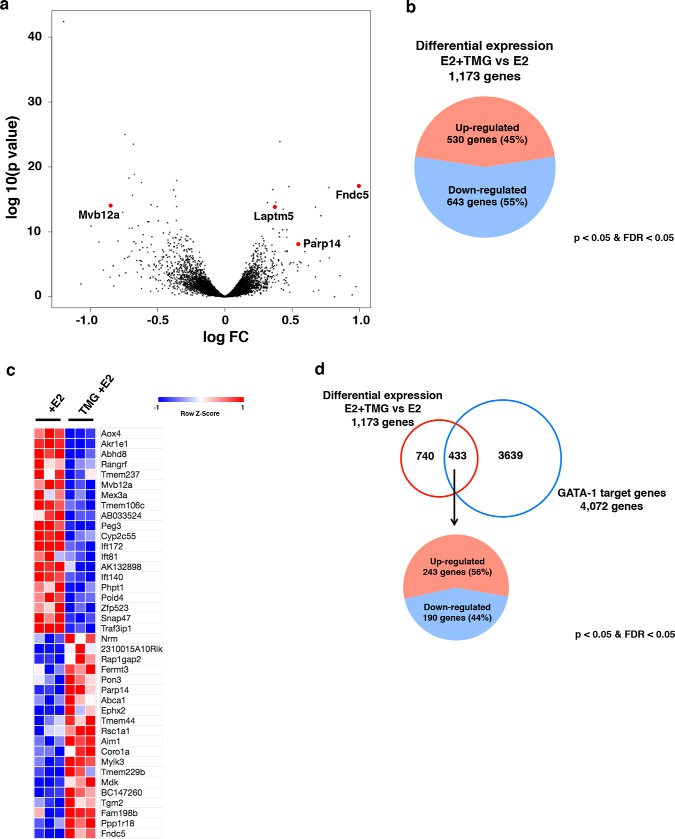
O-GlcNAc regulates GATA-1 target gene transcription during erythropoiesis. The volcano plot shows differentially expressed genes in E2 + acute TMG treatment compared with E2-only treatment (a). Each black dot represents a single gene from RNA-Seq analysis with the negative log of p value plotted on the y axis, and the log of the -fold change (FC) plotted on the x axis. 1,173 genes were differentially expressed in E2 + acute TMG treatment compared with E2 only treatment, and four genes (Laptm5, Fndc5, Parp14, and Mvb12a) of interest are highlighted in red. The pie chart shows the distribution of up- (45%) and down-regulated (55%) genes differentially expressed (b). Transcripts with a p value <0.05 and an FDR <0.05 were included in the analysis. The heat map shows the top 20 differentially expressed genes (c). The Venn diagram shows differential expression of 1,173 genes (E2 + acute TMG treatment versus E2-only treatment) and 4,072 GATA-1 target genes from the database. Of 433 differentially expressed GATA-1 target genes, 243 genes were up-regulated, and 190 genes down-regulated (d).
O-GlcNAc cycling modulates GATA-1–ER target gene transcription during erythropoiesis
To evaluate how O-GlcNAc cycling modulates GATA-1–ER target gene transcription, we used an online database, Harmonizome (56), to search for GATA-1 target genes present in G1E-ER4 cells after E2 treatment. A total of 4,072 GATA-1 target genes were selected from the database by combining nine sets of mouse GATA-1–binding site profiles in G1E-ER4 cells from the ENCODE database (56, 57). 433 of these genes (Table S4) were differentially expressed in E2 + acute TMG–treated cells compared with E2 only–treated cells (Fig. 6d). Of the 433 genes, 243 were up-regulated, and 190 were down-regulated (Fig. 6d). GSEA was performed on these 433 genes. Immune response and defense response biological processes, which are characteristic of leukocytes, were observed to be up-regulated in E2 + acute TMG–treated cells compared with E2 only–treated cells (data not shown).
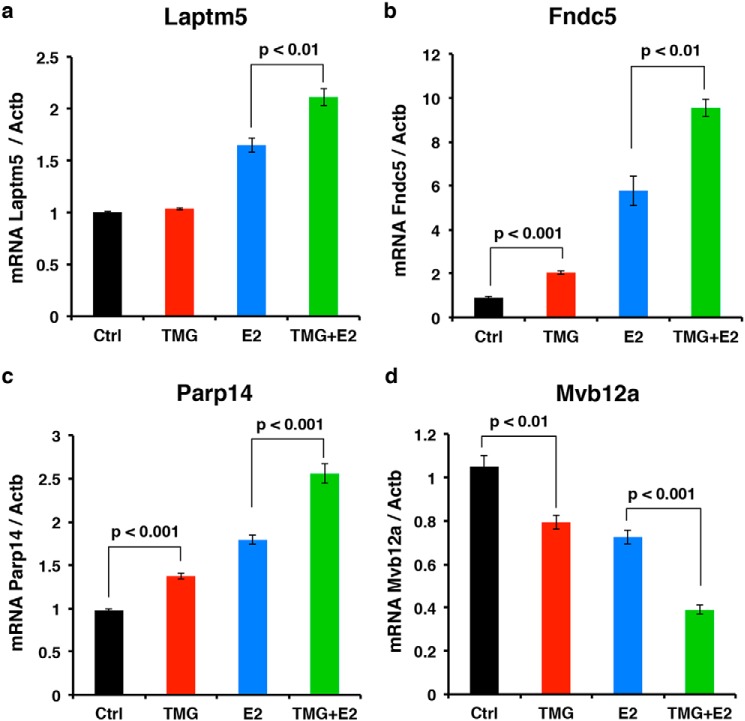
Next, four genes were selected for reverse transcription–quantitative polymerase chain reaction (RT-qPCR) analysis to assess the quality of our RNA-Seq data. Laptm5, Fndc5, Parp14, and Mvb12a are all GATA-1 target genes that showed robust transcription level changes following E2 + acute TMG treatment compared with E2-only treatment in our RNA-Seq data analysis. Following E2-only treatment for 30 h, Laptm5, Fndc5, and Parp14 transcription levels were increased compared with control (Fig. 7, a–c), whereas the Mvb12a transcription level decreased (Fig. 7d). When cells were treated with E2 + acute TMG, Laptm5, Fndc5, and Parp14 transcription levels were increased further compared with E2-only treatment (Fig. 7, a–c), whereas Mvb12a transcription decreased further (Fig. 7d). These results were consistent with the -fold changes from our RNA-Seq data. We repeated identical measurements on the expression of these genes in G1E cells (negative control); no change was detected in the expression of these genes with TMG, E2, or E2 + acute TMG treatment (data not shown). These data confirm that subsets of GATA-1 target genes are affected by alterations in O-GlcNAc homeostasis during erythropoiesis.
Figure 7.
Validation of O-GlcNAc–regulated GATA-1 target gene by RT-qPCR. Transcription levels of GATA-1 target genes Laptm5 (a), Fndc5 (b), Parp14 (c), and Mvb12a (d) were analyzed by RT-qPCR after cells were treated with E2 + acute TMG or E2 only. The experiments were repeated four times. β-Actin (Actb) was used as an internal control. Error bars represent S.D. Ctrl, control.
Inhibition of OGA decreases the occupancy of the Laptm5 intronic GATA-binding site by GATA-1, O-GlcNAc, OGT, and OGA
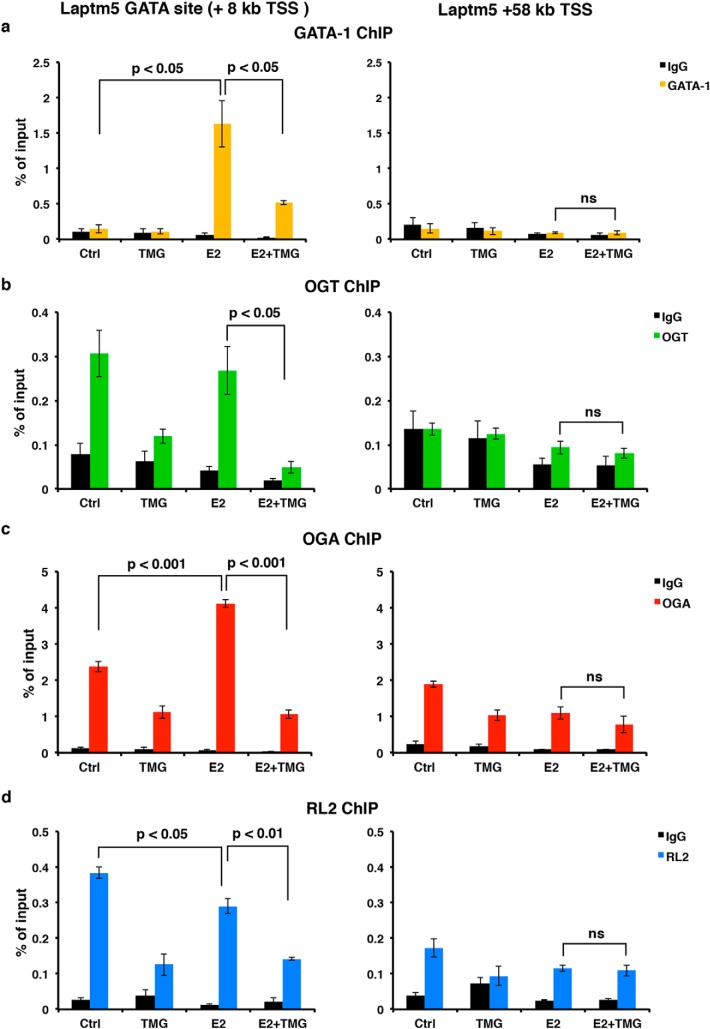
Based on our RNA-Seq and GSEA analysis, we observed that inhibition of OGA by TMG altered a subset of GATA-1 target genes during erythropoiesis, especially those associated with immune response and defense response genetic pathways. Laptm5, a target of GATA-1–ER activation, is specifically expressed in hematopoietic cells and is preferentially expressed in immune cells (58, 59). LAPTM5 positively modulates inflammatory signaling pathways and cytokine secretion in macrophages (59) and is up-regulated in G1E-ER4 after E2 treatment for 30 h. We performed ChIP assays at the GATA-binding site located in the first intron of Laptm5, ∼8 kb downstream from the transcription start site (TSS) (7). Using antibodies against GATA-1, OGT, OGA, and O-GlcNAc to assess the occupancy of this site by GATA-1–ER, OGT, OGA, and O-GlcNAc, we observed in untreated control G1E-ER4 cells an enrichment in the occupancy of OGT, OGA, and an O-GlcNAcylation (Fig. 8, b–d), suggesting that O-GlcNAcylation at the Laptm5 GATA-binding site might be essential for maintaining the basal level of Laptm5 transcription. Following GATA-1–ER restoration by E2, GATA-1–ER occupancy dramatically increased; however, E2 + acute TMG treatment reduced the amount of GATA-1–ER occupancy at this site (Fig. 8a, left panel). OGT occupancy did not change, and OGA occupancy increased after E2 treatment compared with the untreated control (Fig. 8, b and c, left panel). We hypothesize that this contributes to the decrease in overall O-GlcNAc levels at this GATA-binding site (Fig. 8d, left panel).
Figure 8.
Inhibition of OGA decreases the occupancy of GATA-1–ER, OGT, OGA, and the O-GlcNAc level at Laptm5 GATA-binding site. GATA-1 (a), OGT (b), OGA (c), and O-GlcNAc (RL2) (d) ChIP assays were performed using G1E-ER4 cells treated with E2 + acute TMG or E2 only. ChIP DNA was analyzed by qPCR using a set of primers targeting the Laptm5 GATA-binding site (+8 kb TSS) and +58 kb TSS, respectively. Normal rabbit IgG served as a negative control. All experiments were performed with at least three biological replicates. ns, not significant. Error bars represent S.D. Ctrl, control.
Interestingly, the occupancy of OGT and OGA and the overall O-GlcNAc level at this GATA-binding site decreased after E2 + acute TMG treatment compared with E2-only treatment (Fig. 8, b–d, left panel). A similar pattern was observed when acute TMG–only treatment was compared with the untreated control (Fig. 8, b–d, left panel). To show that these changes were specific to the GATA-binding site, we used a non-GATA-1–binding DNA region 50 kb downstream of the GATA-binding site as a negative control. We did not observe any changes when comparing E2 + acute TMG treatment with E2-only treatment (Fig. 8, a–d, right panel). These data suggest that O-GlcNAc homeostasis plays a role in GATA-1 regulation of the Laptm5 gene and that acute TMG treatment alters the occupancy by GATA-1–ER, OGT, and OGA at this binding site.
Discussion
In this study, we utilized a well-established cell model of erythropoiesis, G1E-ER4 cells, to investigate the role of O-GlcNAc during GATA-1–mediated erythroid differentiation. After the restoration of GATA-1 activity, overall O-GlcNAc levels dramatically decreased with an increase of OGA and a decrease of OGT protein levels. The decrease in O-GlcNAcylation during differentiation was also seen in two myeloid leukemia cell lines after induction into more neutrophil-like cells. In addition, GATA-1–ER restoration promoted the interaction of GATA–1-ER with OGT and OGA. RNA-Seq analysis of G1E-ER4 cells treated with E2 + acute TMG demonstrated a change in transcription level of 1,173 genes compared with cells treated with E2 only, including 433 GATA-1 target genes. The transcription level changes were orthogonally validated on selected genes by RT-qPCR. One of these genes, Laptm5, was subjected to further proof-of-principle analysis. ChIP data demonstrated that the occupancy of the Laptm5 GATA-binding site by GATA-1 and OGA was increased and that the overall O-GlcNAc level was decreased following GATA-1 activation; inhibition of OGA with TMG diminished these changes. Acute TMG treatment impaired expression of neutrophil genes during ATRA-mediated differentiation of myeloid cells. Interestingly, sustained TMG treatment in G1E-ER4 reduced βmajor-globin gene expression but increased expression of Fndc5 and Laptm5. Together, these data suggest that O-GlcNAc plays a role in regulating a subset of GATA-1–controlled erythroid genes and that disruption of O-GlcNAc homeostasis impairs erythropoietic cell differentiation, potentially maintaining the cells at a more progenitor-like stage.
We previously demonstrated that GATA-1 interacts with OGT and OGA in mouse erythroleukemia BirA cells (20), and our current results validate these findings. O-GlcNAcylation of GATA-1 was not detected, but GATA-1–interacting proteins are likely modified by O-GlcNAc because following differentiation, the O-GlcNAc antibody pulled down GATA-1. The interaction of OGT and OGA with GATA-1–ER and O-GlcNAcylation of GATA-1–ER-interacting proteins during erythropoiesis could change protein–protein interactions within GATA-1–containing transcriptional complexes, thereby contributing to GATA-1–mediated gene activation or repression. Our RNA-Seq analysis suggests that inhibition of OGA by TMG alters gene transcription during erythropoiesis and that these changes may reflect either reprogramming of the cell to a more myeloid-leukocyte–like lineage or delayed progenitor stage differentiation. Thus, O-GlcNAc may play a role in determining hematopoietic cell fate at an earlier developmental stage. Inhibition of OGA by TMG in hematopoietic stem cells prior to lineage commitment may change the O-GlcNAcylation pattern and skew hematopoietic stem cell differentiation toward a particular lineage. Previous reports show that elevated O-GlcNAc in mouse embryonic stem cells delays the onset of neuronal differentiation (35), whereas increased O-GlcNAcylation skews human pluripotent stem cell differentiation toward adipose mesoderm markers and away from ectoderm markers (29). Because myeloid leukemia cells also show reduced O-GlcNAcylation during differentiation, OGA inhibition might reduce hematopoietic differentiation in general or shift commitment toward another hematopoietic lineage.
Of the 1,173 differentially expressed genes (E2 + acute TMG compared with E2 only), we determined that 47 of these genes are erythroid-specific genes, including Gfi1b (61), Alas2 (62), and Gata2 (3), which are critical for normal erythroid differentiation. The Gfi1b gene encodes for a transcription factor important in promoting erythrocyte differentiation, Gata2 expression inhibits erythrocyte differentiation, and the Alas2 gene is critical in porphyrin biosynthesis. We also found that 433 of the differentially expressed genes were under control of GATA-1 with 243 genes up-regulated and 190 down-regulated. Notably, 84% of activated GATA-1 target genes became more activated in the presence of TMG, whereas 79% of repressed GATA-1 target genes became more repressed, indicating that inhibition of OGA amplifies GATA-1–mediated transcriptional effects. One of the GATA-1 target genes is Laptm5, which is expressed in hematopoietic cells (58). The Laptm5 gene encodes a transmembrane receptor that is associated with lysosomes and has been shown to positively modulate inflammatory signaling pathways and cytokine secretion in macrophages (59). Transcription of Laptm5 was activated following GATA-1 restoration in G1E-ER4 cells, and the activation was enhanced in the presence of TMG. Strikingly, the occupancy of the Laptm5 GATA-binding site by GATA-1–ER, OGT, and OGA after inhibition of OGA decreased as did the O-GlcNAc level. Even though GATA-1–ER was decreased following E2 + acute TMG treatment, TMG amplified the increase of Laptm5 transcription compared with E2 treatment only. Thus, TMG treatment and subsequent retention of O-GlcNAc–modified proteins may stabilize the interaction of GATA-1 with other transcription factors, thereby reducing the recruitment or turnover of these proteins. Hence, even with reduced occupancy by GATA-1 at the GATA-binding site, the remaining GATA-1 complex may be more stable or more active in promoting transcription. These data further argue for the importance of the rate of O-GlcNAc cycling at GATA-binding sites. Reduction of O-GlcNAc cycling at the Laptm5 gene with E2 + acute TMG treatment may lead to the recruitment of other transcriptional coactivators or stabilize the GATA-1–containing activator complex at the Laptm5 GATA-binding site, resulting in increased transcription.
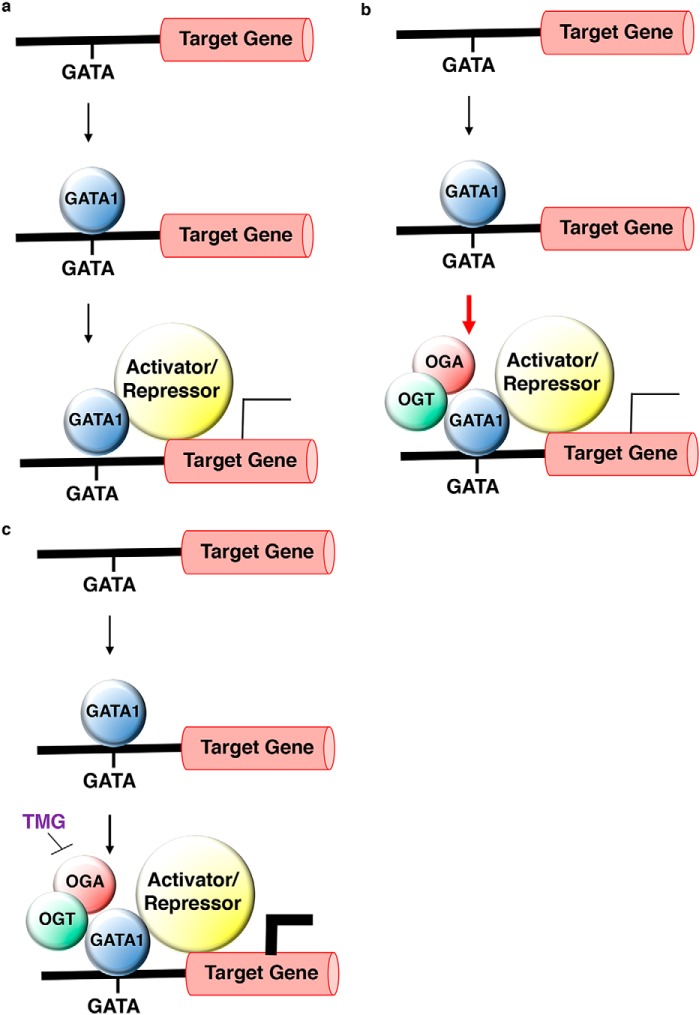
Based on the Laptm5 data, we propose the following general model for OGT, OGA, and GATA-1 regulation of transcription during erythropoiesis. Following GATA-1 restoration by E2, GATA-1 transcriptional activator complexes are recruited to GATA-binding sites located in noncoding regions of genes, such as promoters and introns (7, 57), resulting in increased gene expression (Fig. 9a). OGT interacts with and modifies a component or components of these complexes, thereby promoting the recruitment of other transcriptional coactivators. OGA then removes O-GlcNAc from one or more of the proteins in the complexes, leading to complete recruitment of the promoter (Fig. 9b). When OGA is inhibited by TMG, O-GlcNAc cycling at the activator complexes is disrupted, leading to decreased cycling of OGT, OGA, and GATA-1 at the GATA-binding sites; however, a stable population of activator or repressor complex stays bound to the promoter, amplifying the regulation on the gene (Fig. 9c). In addition, it is possible that the components of the activator complex might also change in the absence of OGA function. Regardless, we hypothesize that this alternate activator/repressor complex is likely to be more stable and active due to the changes in protein–protein interactions within the complexes or alterations of the nearby chromatin structure, resulting in enhanced transcription or repression of the target genes.
Figure 9.
Model of OGT/OGA-mediated activation/repression of GATA-1 target genes during erythropoiesis. Following GATA-1 restoration by E2, a GATA-1 transcriptional activator/repressor complex (yellow circle) is recruited to GATA sites located in cis-regulatory regions of genes, resulting in their expression (general model for GATA-1 gene activation/repression) (a). A subset of GATA-1 target genes utilize OGT/OGA as components of the transcriptional complex to activate/repress GATA-1 target genes under normal phycological conditions. The red arrow indicates an increased amount of OGT, OGA, and GATA-1 at the GATA-1–binding site (b). When OGA is inhibited by TMG, O-GlcNAc cycling at the activator/repressor complex is reduced (small black arrow). Even though there is reduced occupancy of the OGT, OGA, and GATA-1 complex at the GATA-1–binding site, the remaining complex does appear more stable, leading to enhanced GATA-1–mediated gene activation/repression (c).
We hypothesize that O-GlcNAcylation is required for proper hematopoietic cell differentiation. Our data are supported by other published studies of differentiation (20, 35–38) that also show a decrease of global O-GlcNAcylation level, suggesting that in most cells differentiation requires this reduction. The relationship of reduced O-GlcNAc levels to differentiation deserves further study. Many cells enter the cell cycle before differentiation, suggesting that a decrease in O-GlcNAc might enhance cell cycle progression (63). Alternately, cells might redistribute metabolites away from the hexosamine biosynthetic pathway that makes UDP-GlcNAc, the metabolic substrate for OGT, to other metabolic pathways needed for growth (64). Because O-GlcNAcylation is a regulator of transcriptional programs involved with lineage-specific differentiation and O-GlcNAcylation regulates transcription factors, RNA polymerase II, and chromatin-remodeling functions (65), alterations to O-GlcNAc homeostasis may have a dramatic effect on gene expression programs. Thus, disruption of O-GlcNAc homeostasis caused by disease or nutrient excess may result in altered hematopoietic cell differentiation.
Materials and methods
Antibodies
Primary antibodies and secondary antibodies for immunoblotting were used at 0.5 μg/ml and 1:10,000 dilution, respectively. Antibodies for immunoprecipitation (IP) and ChIP assay were used at 2 μg/reaction. Anti-OGT (AL-34) and -OGA (345) were gracious gifts from Dr. Gerald Hart at The Johns Hopkins University School of Medicine. Anti-O-linked GlcNAc antibody (RL2) (12440061) and rabbit anti-goat IgG HRP (31402) were purchased from Thermo Fisher Scientific. Anti-GAPDH antibody (ab9484) and anti-GATA-2 antibody (ab109241) were purchased from Abcam. Anti-chicken IgY HRP (A9046) was purchased from Sigma. Goat anti-rabbit IgG HRP (170-6515) and goat anti-mouse IgG HRP (170-6516) were purchased from Bio-Rad. Goat anti-rat IgG HRP (NA935V) was purchased from GE Healthcare. GATA-1 antibody (sc-265 X) and FOG antibody (sc-9361 X) were purchased from Santa Cruz Biotechnology. Antibodies used exclusively for IP, normal rabbit IgG (sc-2027), normal mouse IgG (sc-2025), and rat IgG (sc-2026), were purchased from Santa Cruz Biotechnology. Anti-OGT (DM-17) antibody (O6264) and anti-OGA antibody (SAB4200267) were purchased from Sigma. Antibodies used exclusively for ChIP, rabbit control IgG (ab46540) and anti-GATA-1 antibody (ab11852), were purchased from Abcam. ChIP grade mouse (G3A1) mAb IgG1 isotype control (5415) was purchased from Cell Signaling Technology. OGT antibody (61355) was purchased from Active Motif. Flow cytometry antibodies PE anti-mouse CD11b (557397), PE anti-mouse CD34 (551387), PE anti-mouse CD41 (558040), PE anti-mouse CD45R/B220 (553089), PE anti-mouse CD90.2 (553005), PE anti-mouse CD117 (553869) PE anti-mouse TER-119/erythroid cells (553673), PE anti-mouse Ly-6A/E (553336), and PE anti-mouse Ly6G and Ly-6C (553128) were purchased from BD Biosciences. Normal rat IgG (sc-2026) was purchased from Santa Cruz Biotechnology.
Cell culture
G1E and G1E-ER4 cells were kind gifts from Dr. Soumen Paul at the University of Kansas Medical Center. These lines were cultured as described previously (6, 41). Cells were incubated at 37 °C in 5% CO2 in a 95% humidified incubator. Cells were treated with 10 μm TMG (S.D. Specialty Chemicals) and/or 1 μm E2 (E2758, Sigma) (41). After 30 h, cells were harvested for subsequent analysis. For sustained TMG treatment, TMG was added to cells every day (10 μm) for 2 weeks prior to E2 induction. NB4 and HL60 cells were grown as described previously (66). ATRA (R2625, Sigma) was used at 1 μm in DMSO.
Western blotting and IP
Cells were lysed on ice as described previously (20). Whole-cell lysates were used for electrophoresis and subjected to Western blotting as described previously (20). All Western blotting results were repeated in three independent experiments, and the representative images are shown. IP was performed as described previously (20). We used 2 mg of cell lysates and 2 μg of antibody for each reaction. OGA and OGT relative protein levels were measured by analyzing the band densities using ImageJ 1.49 (67) and then normalized to the density of GAPDH.
Benzidine staining and May-Grünwald/Giemsa stain
Benzidine staining and May-Grünwald/Giemsa stain were performed as described previously (6) to assess the hemoglobin content and morphology changes, respectively. Cell stain images were visualized and photographed using a Nikon Eclipse 80i digital microscope (Nikon Instruments Inc., Melville, NY).
Total RNA isolation and RT-PCR
Total RNA was isolated from 5 × 106 cells using TRI Reagent (T9424, Sigma) according to the manufacturer's instructions. For the RT reactions, 0.5 μg of total RNA was reverse transcribed to cDNA using iScript Reverse Transcription Supermix (170-8841, Bio-Rad) following the manufacturer's instructions. Reactions were incubated in a thermal cycler (Model 2720, Applied Biosystems) using the following protocol: priming for 5 min at 25 °C, RT for 20 min at 46 °C, RT inactivation for 1 min at 95 °C, and hold at 4 °C.
RNA sequencing
RNA was extracted using TRI Reagent according to manufacturer's instructions, and RNA-Seq libraries were prepared using the Illumina TruSeq Stranded mRNA Library Preparation kit Set A (RS-122-2101, Illumina) according to the manufacturer's instructions. RNA libraries were sequenced using an Illumina HiSeq 2500 Sequencing System.
RNA-Seq data analysis
FastQC (0.11.2) and RSEM (1.2.22) software were used to assess the quality of the RNA-Seq results, align the reads to the mouse genome reference GRCm38/mm10, and calculate the gene expression values. R (3.2.2) and EdgeR (3.14.0) software were used to normalize the expression values using the TMM method (weighted trimmed mean of M-values) followed by differential expression analysis. To reduce the burden of multiple testing in differential gene expression analyses, a filter was initially applied to reduce the number of genes. Genes were removed if they did not present a meaningful gene expression across all samples; only genes with counts per million of >10 in at least two samples were considered in differential expression analyses. The Benjamini and Hochberg procedure was used to control the false discovery rate (FDR). The following R packages were utilized for calculations and visualizations: EdgeR and Gplot.
ChIP assay
The ChIP assay was performed using a method published previously (20, 60). Briefly, chromatin was prepared and sheared as described (20). For each reaction, sheared chromatin was diluted with IP buffer (150 mm NaCl, 5 mm EDTA, 0.5% (v/v) Nonidet P-40, 1% (v/v) Triton X-100, 40 mm GlcNAc, and 50 mm Tris-HCl, pH 7.5) supplemented with protease inhibitor and incubated with 2 μg of control IgG or specific antibody, respectively, at 4 °C overnight. Protein G Dynabeads (10004D, Thermo Fisher Scientific) were added to the mixture according to the manufacturer's instructions followed by rotation at 4 °C for 2 h. Dynabeads were washed with 1 ml of cold IP buffer six times and then mixed with 10% (w/v) Chelex 100 slurry (1421253, Bio-Rad); for input samples, 10% of sheared chromatin was used and processed at the same time as the ChIP samples. Samples were boiled at 95 °C for 10 min followed by treatment with RNase A and proteinase K. Samples were boiled again for 10 min to inactivate proteinase K and centrifuged at 12,000 × g for 1 min at 4 °C. The supernatants were transferred to fresh tubes and stored at −20 °C for subsequent qPCR analysis.
RT-qPCR
RT-qPCR was performed as described previously (20). The primer sequences for measuring target gene transcription levels are listed in Table S5. The reactions were run in a CFX96 Touch Real-Time PCR Detection System (185-5195, Bio-Rad).
Flow cytometry
Cell surface markers were characterized using flow cytometry analysis. Cells were first counted with a hemocytometer to ensure that 5 × 105 cells were used per surface marker. Cells were centrifuged, the supernatant was discarded, and the pellets were washed twice in ice-cold PBS and 0.1% BSA (w/v). Pellets were then resuspended in 100 μl of ice-cold PBS and 0.1% BSA (w/v) per surface marker test and then blocked with 1% (v/v) normal rat serum for 30 min. After washing twice in 1 ml of ice-cold PBS and 0.1% BSA (w/v), cells were again resuspended in 100 μl of ice-cold PBS and 0.1% BSA (w/v) per surface marker test and then incubated with 0.5 μg of antibody in the dark for 40 min. Cells were then washed twice in 1 ml of ice-cold PBS and 0.1% BSA (w/v) and resuspended in 350 μl of PBS and 0.1% BSA (w/v) for flow cytometry analysis. 10,000 cells were analyzed per surface marker using an LSR II BD Biosciences flow cytometer (FACSDiva v.8 software). Dead cells were gated from the analysis.
qPCR data analysis
Quantification cycle (Cq) values were calculated by CFX ManagerTM software (Bio-Rad). For cDNA RT-qPCR data, the dynamic ranges of RT and amplification efficiency were evaluated before applying the ΔΔCq method to calculate relative gene expression change. The transcription level of the target gene was normalized to the internal control as -fold change. For ChIP DNA qPCR data, the Cq value was normalized as a percentage of input. Data generated in at least three independent experiments are presented as mean ± S.E.; the two-tailed Student t test statistic was applied with p < 0.05 considered to be a significant difference.
GSEA
GSEA was performed according to the GSEA user guide (53, 54). For gene ontology analysis, biological process gene sets from the Molecular Signature Database (MsigDB) were used. Pathways with nominal p value <5% and FDR q value <25% were included in the analysis.
Author contributions
Z. Z. and S. G. data curation; Z. Z. and S. G. software; Z. Z. and S. G. formal analysis; Z. Z., M. P. P., L. V. N., and J. D. F. validation; Z. Z., M. P. P., S. G., L. V. N., H. F., and J. D. F. investigation; Z. Z., M. P. P., and S. G. visualization; Z. Z., M. P. P., S. G., L. V. N., and K. R. P. methodology; Z. Z., M. P. P., L. V. N., J. D. F., K. R. P., and C. S. writing-original draft; Z. Z., M. P. P., L. V. N., J. D. F., D. C. K., K. R. P., and C. S. writing-review and editing; D. C. K., K. R. P., and C. S. resources; D. C. K., K. R. P., and C. S. supervision; K. R. P. and C. S. conceptualization; K. R. P. and C. S. funding acquisition; C. S. project administration.
Supplementary Material
Acknowledgment
We thank Diana Kalinowska from University of Kansas Medical Center (KUMC) Department of Pharmacology, Toxicology and Therapeutics for assistance with the ChIP assay.
This work was supported by NIDDK, National Institutes of Health Grants R01DK100595 (to C. S. and K. R. P.) and R01HL111264 (to K. R. P.) and a University of Kansas Medical Center (KUMC) Biomedical Research Training Program grant (to Z. Z.). Partial support was provided by NIGMS, National Institutes of Health Grants P20GM103549, P30GM118247, and P20GM103418. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S5.
- PTM
- post-translational modification
- OGT
- O-GlcNAc-transferase
- O-OGA
- O-GlcNAcase
- GATA-1
- GATA-binding protein 1
- ER
- estrogen receptor
- E2
- β-estradiol
- TMG
- Thiamet-G
- LAPTM5
- lysosomal protein transmembrane 5
- NuRD
- GATA-1–FOG-1–Mi2β
- G1E
- GATA-1− erythroid
- FOG-1
- friend of GATA-1
- ATRA
- all-trans-retinoic acid
- CTSD
- cathepsin D
- DEFA1
- defensin α1
- RT-qPCR
- reverse transcription–quantitative polymerase chain reaction
- TSS
- transcription start site
- IP
- immunoprecipitation
- HRP
- horseradish peroxidase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PE
- phycoerythrin
- FDR
- false discovery rate
- Cq
- quantification cycle.
References
- 1. Goode D. K., Obier N., Vijayabaskar M. S., Lie-A-Ling M., Lilly A. J., Hannah R., Lichtinger M., Batta K., Florkowska M., Patel R., Challinor M., Wallace K., Gilmour J., Assi S. A., Cauchy P., et al. (2016) Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell 36, 572–587 10.1016/j.devcel.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pevny L., Simon M. C., Robertson E., Klein W. H., Tsai S. F., D'Agati V., Orkin S. H., and Costantini F. (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349, 257–260 10.1038/349257a0 [DOI] [PubMed] [Google Scholar]
- 3. Orkin S. H. (1992) GATA-binding transcription factors in hematopoietic cells. Blood 80, 575–581 [PubMed] [Google Scholar]
- 4. Fujiwara Y., Browne C. P., Cunniff K., Goff S. C., and Orkin S. H. (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 93, 12355–12358 10.1073/pnas.93.22.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., and Engel J. D. (1990) Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 4, 1650–1662 10.1101/gad.4.10.1650 [DOI] [PubMed] [Google Scholar]
- 6. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., and Weiss M. J. (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147 10.1182/blood-2004-04-1603 [DOI] [PubMed] [Google Scholar]
- 7. Cheng Y., Wu W., Kumar S. A., Yu D., Deng W., Tripic T., King D. C., Chen K. B., Zhang Y., Drautz D., Giardine B., Schuster S. C., Miller W., Chiaromonte F., Zhang Y., et al. (2009) Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 19, 2172–2184 10.1101/gr.098921.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu W., Cheng Y., Keller C. A., Ernst J., Kumar S. A., Mishra T., Morrissey C., Dorman C. M., Chen K. B., Drautz D., Giardine B., Shibata Y., Song L., Pimkin M., Crawford G. E., et al. (2011) Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 21, 1659–1671 10.1101/gr.125088.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kingsley P. D., Greenfest-Allen E., Frame J. M., Bushnell T. P., Malik J., McGrath K. E., Stoeckert C. J., and Palis J. (2013) Ontogeny of erythroid gene expression. Blood 121, e5–e13 10.1182/blood-2012-04-422394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C. R., Gregory G. D., Hardison R. C., and Blobel G. A. (2009) SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113, 2191–2201 10.1182/blood-2008-07-169417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu M., Riva L., Xie H., Schindler Y., Moran T. B., Cheng Y., Yu D., Hardison R., Weiss M. J., Orkin S. H., Bernstein B. E., Fraenkel E., and Cantor A. B. (2009) Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36, 682–695 10.1016/j.molcel.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soler E., Andrieu-Soler C., de Boer E., Bryne J. C., Thongjuea S., Stadhouders R., Palstra R. J., Stevens M., Kockx C., van Ijcken W., Hou J., Steinhoff C., Rijkers E., Lenhard B., and Grosveld F. (2010) The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 24, 277–289 10.1101/gad.551810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fujiwara T., O'Geen H., Keles S., Blahnik K., Linnemann A. K., Kang Y. A., Choi K., Farnham P. J., and Bresnick E. H. (2009) Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681 10.1016/j.molcel.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., Wang Z., Zhang J., and Lim S. H. (2009) Core promoter sequence of SEMG I spans between the two putative GATA-1 binding domains and is responsive to IL-4 and IL-6 in myeloma cells. Leuk. Res. 33, 166–169 10.1016/j.leukres.2008.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bresnick E. H., Lee H. Y., Fujiwara T., Johnson K. D., and Keles S. (2010) GATA switches as developmental drivers. J. Biol. Chem. 285, 31087–31093 10.1074/jbc.R110.159079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boyes J., Byfield P., Nakatani Y., and Ogryzko V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396, 594–598 10.1038/25166 [DOI] [PubMed] [Google Scholar]
- 17. Lamonica J. M., Vakoc C. R., and Blobel G. A. (2006) Acetylation of GATA-1 is required for chromatin occupancy. Blood 108, 3736–3738 10.1182/blood-2006-07-032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee H. Y., Johnson K. D., Fujiwara T., Boyer M. E., Kim S. I., and Bresnick E. H. (2009) Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol. Cell 36, 984–995 10.1016/j.molcel.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kadri Z., Lefevre C., Goupille O., Penglong T., Granger-Locatelli M., Fucharoen S., Maouche-Chretien L., Leboulch P., and Chretien S. (2015) Erythropoietin and IGF-1 signaling synchronize cell proliferation and maturation during erythropoiesis. Genes Dev. 29, 2603–2616 10.1101/gad.267633.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Z., Costa F. C., Tan E. P., Bushue N., DiTacchio L., Costello C. E., McComb M. E., Whelan S. A., Peterson K. R., and Slawson C. (2016) O-Linked N-acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase interact with Mi2β protein at the Aγ-globin promoter. J. Biol. Chem. 291, 15628–15640 10.1074/jbc.M116.721928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hart G. W. (2014) Three decades of research on O-GlcNAcylation—a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front. Endocrinol. 5, 183 10.3389/fendo.2014.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakabe K., Wang Z., and Hart G. W. (2010) β-N-Acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 10.1073/pnas.1009023107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gambetta M. C., and Müller J. (2015) A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma 124, 429–442 10.1007/s00412-015-0513-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Q., Chen Y., Bian C., Fujiki R., and Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 10.1038/nature11742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., et al. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 10.1038/emboj.2012.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X., Zhang F., and Kudlow J. E. (2002) Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 10.1016/S0092-8674(02)00810-3 [DOI] [PubMed] [Google Scholar]
- 27. Chu C. S., Lo P. W., Yeh Y. H., Hsu P. H., Peng S. H., Teng Y. C., Kang M. L., Wong C. H., and Juan L. J. (2014) O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U.S.A. 111, 1355–1360 10.1073/pnas.1323226111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., et al. (2012) Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 10.1126/science.1221711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maury J. J., El Farran C. A., Ng D., Loh Y. H., Bi X., Bardor M., and Choo A. B. (2015) RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 15, 182–189 10.1016/j.scr.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 30. Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., and Lewis B. A. (2012) Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287, 23549–23561 10.1074/jbc.M111.330910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewis B. A., Burlingame A. L., and Myers S. A. (2016) Human RNA polymerase II promoter recruitment in vitro is regulated by O-linked N-acetylglucosaminyltransferase (OGT). J. Biol. Chem. 291, 14056–14061 10.1074/jbc.M115.684365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Resto M., Kim B. H., Fernandez A. G., Abraham B. J., Zhao K., and Lewis B. A. (2016) O-GlcNAcase is an RNA polymerase II elongation factor coupled to pausing factors SPT5 and TIF1β. J. Biol. Chem. 291, 22703–22713 10.1074/jbc.M116.751420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olivier-Van Stichelen S., and Hanover J. A. (2015) You are what you eat: O-linked N-acetylglucosamine in disease, development and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 18, 339–345 10.1097/MCO.0000000000000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Howerton C. L., Morgan C. P., Fischer D. B., and Bale T. L. (2013) O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. U.S.A. 110, 5169–5174 10.1073/pnas.1300065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Speakman C. M., Domke T. C., Wongpaiboonwattana W., Sanders K., Mudaliar M., van Aalten D. M., Barton G. J., and Stavridis M. P. (2014) Elevated O-GlcNAc levels activate epigenetically repressed genes and delay mouse ESC differentiation without affecting naive to primed cell transition. Stem Cells 32, 2605–2615 10.1002/stem.1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sohn K. C., Lee E. J., Shin J. M., Lim E. H., No Y., Lee J. Y., Yoon T. Y., Lee Y. H., Im M., Lee Y., Seo Y. J., Lee J. H., and Kim C. D. (2014) Regulation of keratinocyte differentiation by O-GlcNAcylation. J. Dermatol. Sci. 75, 10–15 10.1016/j.jdermsci.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 37. Kim H. S., Park S. Y., Choi Y. R., Kang J. G., Joo H. J., Moon W. K., and Cho J. W. (2009) Excessive O-GlcNAcylation of proteins suppresses spontaneous cardiogenesis in ES cells. FEBS Lett. 583, 2474–2478 10.1016/j.febslet.2009.06.052 [DOI] [PubMed] [Google Scholar]
- 38. Ogawa M., Mizofuchi H., Kobayashi Y., Tsuzuki G., Yamamoto M., Wada S., and Kamemura K. (2012) Terminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta 1820, 24–32 10.1016/j.bbagen.2011.10.011 [DOI] [PubMed] [Google Scholar]
- 39. Andrés-Bergós J., Tardio L., Larranaga-Vera A., Gómez R., Herrero-Beaumont G., and Largo R. (2012) The increase in O-linked N-acetylglucosamine protein modification stimulates chondrogenic differentiation both in vitro and in vivo. J. Biol. Chem. 287, 33615–33628 10.1074/jbc.M112.354241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koyama T., and Kamemura K. (2015) Global increase in O-linked N-acetylglucosamine modification promotes osteoblast differentiation. Exp. Cell Res. 338, 194–202 10.1016/j.yexcr.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 41. Grass J. A., Jing H., Kim S. I., Martowicz M. L., Pal S., Blobel G. A., and Bresnick E. H. (2006) Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 26, 7056–7067 10.1128/MCB.01033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drach J., Lopez-Berestein G., McQueen T., Andreeff M., and Mehta K. (1993) Induction of differentiation in myeloid leukemia cell lines and acute promyelocytic leukemia cells by liposomal all-trans-retinoic acid. Cancer Res. 53, 2100–2104 [PubMed] [Google Scholar]
- 43. Lamonica J. M., Deng W., Kadauke S., Campbell A. E., Gamsjaeger R., Wang H., Cheng Y., Billin A. N., Hardison R. C., Mackay J. P., and Blobel G. A. (2011) Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc. Natl. Acad. Sci. U.S.A. 108, E159–E168 10.1073/pnas.1102140108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manavathi B., Lo D., Bugide S., Dey O., Imren S., Weiss M. J., and Humphries R. K. (2012) Functional regulation of pre-B-cell leukemia homeobox interacting protein 1 (PBXIP1/HPIP) in erythroid differentiation. J. Biol. Chem. 287, 5600–5614 10.1074/jbc.M111.289843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 10.1038/nchembio.96 [DOI] [PubMed] [Google Scholar]
- 46. Yuzwa S. A., Shan X., Macauley M. S., Clark T., Skorobogatko Y., Vosseller K., and Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 8, 393–399 10.1038/nchembio.797 [DOI] [PubMed] [Google Scholar]
- 47. Hastings N. B., Wang X., Song L., Butts B. D., Grotz D., Hargreaves R., Fred Hess J., Hong K. K., Huang C. R., Hyde L., Laverty M., Lee J., Levitan D., Lu S. X., Maguire M., et al. (2017) Inhibition of O-GlcNAcase leads to elevation of O-GlcNAc tau and reduction of tauopathy and cerebrospinal fluid tau in rTg4510 mice. Mol. Neurodegener. 12, 39 10.1186/s13024-017-0181-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan E. P., McGreal S. R., Graw S., Tessman R., Koppel S. J., Dhakal P., Zhang Z., Machacek M., Zachara N. E., Koestler D. C., Peterson K. R., Thyfault J. P., Swerdlow R. H., Krishnamurthy P., DiTacchio L., et al. (2017) Sustained O-GlcNAcylation reprograms mitochondrial function to regulate energy metabolism. J. Biol. Chem. 292, 14940–14962 10.1074/jbc.M117.797944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baines K. J., Simpson J. L., Wood L. G., Scott R. J., and Gibson P. G. (2011) Systemic upregulation of neutrophil alpha-defensins and serine proteases in neutrophilic asthma. Thorax 66, 942–947 10.1136/thx.2010.157719 [DOI] [PubMed] [Google Scholar]
- 50. Yoon S. G., Cheong H. J., Kim S. J., Kim K. H., Lee S. C., Lee N., Park H. S., and Won J. H. (2013) Src family kinase inhibitor PP2 has different effects on all-trans-retinoic acid or arsenic trioxide-induced differentiation of an acute promyelocytic leukemia cell line. Cancer Res. Treat. 45, 126–133 10.4143/crt.2013.45.2.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Villén J., Beausoleil S. A., Gerber S. A., and Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 10.1073/pnas.0609836104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., et al. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 54. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li L., Freudenberg J., Cui K., Dale R., Song S. H., Dean A., Zhao K., Jothi R., and Love P. E. (2013) Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood 121, 4575–4585 10.1182/blood-2013-01-479451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rouillard A. D., Gundersen G. W., Fernandez N. F., Wang Z., Monteiro C. D., McDermott M. G., and Ma'ayan A. (2016) The Harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. ENCODE Project Consortium (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 10.1126/science.1105136 [DOI] [PubMed] [Google Scholar]
- 58. Adra C. N., Zhu S., Ko J. L., Guillemot J. C., Cuervo A. M., Kobayashi H., Horiuchi T., Lelias J. M., Rowley J. D., and Lim B. (1996) LAPTM5: a novel lysosomal-associated multispanning membrane protein preferentially expressed in hematopoietic cells. Genomics 35, 328–337 10.1006/geno.1996.0364 [DOI] [PubMed] [Google Scholar]
- 59. Glowacka W. K., Alberts P., Ouchida R., Wang J.-Y., and Rotin D. (2012) LAPTM5 protein is a positive regulator of proinflammatory signaling pathways in macrophages. J. Biol. Chem. 287, 27691–27702 10.1074/jbc.M112.355917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nelson J. D., Denisenko O., and Bomsztyk K. (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 10.1038/nprot.2006.27 [DOI] [PubMed] [Google Scholar]
- 61. Saleque S., Cameron S., and Orkin S. H. (2002) The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16, 301–306 10.1101/gad.959102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nakajima O., Takahashi S., Harigae H., Furuyama K., Hayashi N., Sassa S., and Yamamoto M. (1999) Heme deficiency in erythroid lineage causes differentiation arrest and cytoplasmic iron overload. EMBO J. 18, 6282–6289 10.1093/emboj/18.22.6282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., and Hart G. W. (2005) Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 280, 32944–32956 10.1074/jbc.M503396200 [DOI] [PubMed] [Google Scholar]
- 64. Ferrer C. M., Sodi V. L., and Reginato M. J. (2016) O-GlcNAcylation in cancer biology: linking metabolism and signaling. J. Mol. Biol. 428, 3282–3294 10.1016/j.jmb.2016.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hardivillé S., and Hart G. W. (2014) Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213 10.1016/j.cmet.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Z., Cao L., Kang R., Yang M., Liu L., Zhao Y., Yu Y., Xie M., Yin X., Livesey K. M., and Tang D. (2011) Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy 7, 401–411 10.4161/auto.7.4.14397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.