Abstract
Lysine methylation is an important post-translational modification that is also present on mitochondrial proteins, but the mitochondrial lysine-specific methyltransferases (KMTs) responsible for modification are in most cases unknown. Here, we set out to determine the function of human family with sequence similarity 173 member B (FAM173B), a mitochondrial methyltransferase (MTase) reported to promote chronic pain. Using bioinformatics analyses and biochemical assays, we found that FAM173B contains an atypical, noncleavable mitochondrial targeting sequence responsible for its localization to mitochondria. Interestingly, CRISPR/Cas9-mediated KO of FAM173B in mammalian cells abrogated trimethylation of Lys-43 in ATP synthase c-subunit (ATPSc), a modification previously reported as ubiquitous among metazoans. ATPSc methylation was restored by complementing the KO cells with enzymatically active human FAM173B or with a putative FAM173B orthologue from the nematode Caenorhabditis elegans. Interestingly, lack of Lys-43 methylation caused aberrant incorporation of ATPSc into the ATP synthase complex and resulted in decreased ATP-generating ability of the complex, as well as decreased mitochondrial respiration. In summary, we have identified FAM173B as the long-sought KMT responsible for methylation of ATPSc, a key protein in cellular ATP production, and have demonstrated functional significance of ATPSc methylation. We suggest renaming FAM173B to ATPSc-KMT (gene name ATPSCKMT).
Keywords: ATP synthase, post-translational modification (PTM), metabolic regulation, mitochondrial respiratory chain complex, mitochondria, protein methylation, F1F0-ATPase, ATP synthase c-subunit, FAM173B, methyltransferase, oxidative phosphorylation, protein lysine methylation, KMT, ATPSc-KMT
Introduction
Many proteins are subject to post-translational methylation, and this can occur on several amino acid residues, including glutamine, histidine, arginine, and lysine (1–4). Methylation is catalyzed by specific methyltransferases (MTases)4 that use S-adenosylmethionine (AdoMet) as methyl donor (5, 6). Lysine (K)-specific MTases (KMTs) can transfer up to three methyl groups to the ϵ-amino group of a lysine, thus enabling generation of four potential variants of this residue (i.e. unmethylated and mono-, di-, and trimethylated lysine), which differ in hydrophobic properties and hydrogen-bonding capability but retain the positive charge (3, 7).
Lysine methylation and its functional importance has been most intensively studied in the case of histone methylation by SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain-containing KMTs (8). Here, methylation of specific lysines in the N-terminal histone tails are strong determinants of chromatin state and gene expression (9). However, lysine methylation is also frequently found on nonhistone proteins, and many of the responsible KMTs belong to a different MTase class, the so-called seven-β-strand (7BS) MTases (7, 10). For example, the human KMTs targeting eukaryotic elongation factors (eEF1A and eEF2) and chaperones, (VCP and Hsp70 proteins) were recently identified as 7BS KMTs (11–21).
Several mitochondrial proteins have been shown to be lysine-methylated (22), but only two mitochondrial KMTs have been identified so far. Both of these belong to the 7BS MTase family and target proteins located in the mitochondrial matrix (23–26). We and others recently showed that ETFβ-KMT (also known as METTL20) targets Lys-200 and Lys-203 of the β-subunit of electron transfer flavoprotein (ETFβ), whereas CS-KMT (also known as METTL12) targets Lys-395 of mitochondrial citrate synthase (CS) (23–26). Another prominent example of a lysine-methylated mammalian mitochondrial protein is the ATP synthase c-subunit (ATPSc), which is methylated on Lys-43, as reported in several independent studies (27–29). Also, ATPSc methylation was investigated in a wide range of metazoans, and Lys-43 was found to be invariably trimethylated (29).
The mitochondrial ATP synthase (ATPS) is responsible for the synthesis of ATP from ADP and phosphate in the process of oxidative phosphorylation (OxPhos), which utilizes the proton-motive force, generated during flow of electrons through the mitochondrial electron transport chain (ETC) to molecular oxygen (30). ATPS is a ∼600-kDa multiprotein complex (Complex V), and consists of two major parts: the oligomycin-binding domain (FO), which resides in the inner membrane, and the catalytic domain (F1), which faces the mitochondrial matrix. In metazoans, the central part of FO consists of a membrane-embedded ring of eight c-subunits that, together with proteins of the so-called “central stalk,” form the rotary element of the ATPS (31). During the return of protons from the intermembrane space back to the matrix, this rotary element is propelled, inducing recurrent conformational changes within the stationary F1 domain, enabling ATP synthesis (32). ATPSc is highly conserved throughout evolution, and humans have three distinct ATPSc-encoding genes. These encode an identical, 75-amino-acid-long, mature ATPSc protein but differ with respect to the encoded N-terminal presequence, which is cleaved off during mitochondrial import (33, 34).
During our efforts to identify novel human KMTs, we became interested in family with sequence similarity 173 member B (FAM173B), which, together with its close paralogue FAM173A, represent the closest human homologues of a broad-specificity KMT found in some archaea (denoted aKMT) (7, 35, 36). A genome-wide association study implicated FAM173B in chronic pain in humans (37), and indeed, we recently showed that FAM173B promoted chronic pain in a mouse model, in a fashion dependent on its MTase activity (38). We also found that FAM173B localized to mitochondria and that the recombinant enzyme displayed weak, nonspecific KMT activity on a lysine homopolymer (38). However, the actual biochemical function of FAM173B has remained obscure.
In the present study, we set out to identify the in vivo substrate(s) of FAM173B. Using FAM173B KO cells, we identified Lys-43 in ATPSc as target of FAM173B, and a FAM173B homologue from Caenorhabditis elegans showed the same activity. Moreover, lack of Lys-43 methylation decreased the efficiency of assembly of the ATPS complex and reduced its ATP-synthesizing activity.
Results
Phylogeny analysis of FAM173B
In addition to the previously mentioned FAM173A and aKMT proteins, homologues of human FAM173B are found in nonvertebrate animals, such as insects and nematodes, as well as in some bacteria (Table S1). To investigate the sequence similarity between FAM173B and these related proteins, we generated a sequence alignment of representative proteins, and a phylogram was constructed from the alignment (Fig. 1A). Clearly, the five categories of proteins (vertebrate FAM173A, vertebrate FAM173B, nonvertebrate FAM173-like, archaeal aKMT, and bacterial aKMT-like) clustered as five distinct groups in the phylogram. The archaeal and bacterial proteins located closely together at one end of the phylogram, whereas the FAM173B, FAM173A, and nonvertebrate FAM173-like proteins were found at the other end, with the nonvertebrate FAM173-like proteins localized closer to the FAM173B group than to the FAM173A group. This indicates that the nonvertebrate FAM173-like proteins may represent functional orthologues of FAM173B.
Figure 1.
Human FAM173B is an evolutionarily conserved mitochondrial protein. A, phylogram of FAM173-like proteins. The tree encompasses FAM173A (blue) and FAM173B (red) proteins from the vertebrates Homo sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), Salmo salar (Ss), Danio rerio (Dr), and Gallus gallus (Gg), as well as FAM173-like proteins from the nonvertebrate animals (green) Capitella teleta (Cte), C. elegans (Cel), Tribolium castaneum (Tca), Daphnia pulex (Dpu), Apis mellifera (Ame), Drosophila melanogaster (Dme), Aedes aegypti (Aae), and Hydra vulgaris (Hvu). Also included are various crenarchaeal homologues (purple), including the previously characterized aKMT enzyme from Sulfolobus islandicus (Sis) and corresponding homologues from Pyrolobus fumarii (Pfu), Hyperthermus butylicus (Hbu), Ignicoccus islandicus (Iis), Staphylothermus hellenicus (She), and Methanobacterium formicicum (Mfo), as well as the bacterial aKMT-like enzymes (brown) from Pseudomonas stutzeri (Pst), Salinibacter ruber (Sru), Rhodopseudomonas palustris (Rpa), and Agrobacterium (multispecies) (Agr). Accession numbers of used sequences are given in Table S1. B, sequence elements of the N-terminal portion of FAM173B and alignment of the N-terminal part of putative FAM173 orthologues from organisms denoted as in A. Colors indicate the position of nonconserved NTS (gray), predicted TMD (yellow), conserved preMT (orange), and N-terminal fragment of the MTase domain (green). Hallmark motifs of the 7BS MTase domain are indicated by black boxes. The position of the conserved acidic residue in motif Post I (Glu-117 in H. sapiens FAM173B), crucial for AdoMet binding, is marked with an asterisk. C, subcellular localization of FAM173B-derived GFP fusion proteins. A schematic representation of the used FAM173B-derived sequences is given (top), with the various domains denoted as in B. Shown are confocal fluorescence microscopy images of HeLa cells, fixed 24 h after transient transfection with plasmids encoding FAM173B-GFP, (Δ55)-FAM173B-GFP, or (56–90)-FAM173B-GFP. Cells were counterstained with anti-COX IV primary antibody, followed by Alexa Fluor 568–conjugated secondary antibody, to visualize the mitochondria, and with DAPI, to visualize the nuclei. Data were acquired through green (GFP), red (COX IV), and blue (DAPI) channels and merged.
FAM173B contains an atypical mitochondrial targeting sequence
In a previous study, we found that mouse Fam173b is localized to mitochondria (38). Proteins that are targeted to mitochondria commonly contain a classic N-terminal mitochondrial targeting sequence (MTS), which is amphiphilic and α-helical (39). These classic MTS-containing proteins utilize a specific import machinery, and the MTS is cleaved off during the import process. However, more than half of all proteins targeted to mitochondria do not contain a classic MTS but use alternative mechanisms (39). To elucidate the targeting of FAM173B, we set out to identify sequence determinants required for its mitochondrial import. In addition to the C-terminal MTase domain, human FAM173B contains ∼90 residues of N-terminal sequence, but no typical MTS was detected by algorithms such as MitoProt (40). However, various algorithms for prediction of α-helical hydrophobic transmembrane domains (TMDs) (e.g. TMpred, Phobius, and TMHMM, available at www.expasy.org), predicted the presence of a TMD in the N-terminal portion of FAM173 homologues from various animals (Fig. 1B and Table S2). This TMD was located between a stretch of nonconserved N-terminal sequence (NTS) and a segment of conserved sequence that precedes the annotated MTase domain (preMT) (Fig. 1B).
We set out to investigate the role of the conserved TMD and preMT domains in targeting FAM173B to mitochondria. As previously observed (38), a fusion protein between FAM173B and GFP was efficiently targeted to mitochondria (Fig. 1C), as shown by extensive co-localization with cytochrome c oxidase subunit IV (COX IV). Interestingly, deletion of the first 55 amino acids, encompassing the NTS and TMD, did not abrogate mitochondrial targeting, suggesting that targeting may be provided by the preMT domain. Indeed, a fusion protein between the preMT domain and GFP was efficiently targeted to the mitochondria (Fig. 1C), indicating that the preMT domain can act as an autonomous MTS and likely is responsible for mitochondrial targeting of FAM173B. Moreover, Western blot analysis of mitoplasts from cells expressing C-terminally FLAG-tagged FAM173B (FAM173B-FLAG) revealed a single species of FLAG-tagged protein, with an apparent molecular mass of ∼30 kDa (Fig. S1), corresponding to the predicted mass of FAM173B-FLAG (29.2 kDa). Thus, our results indicate that FAM173B is not processed during mitochondrial import and that the preMT domain of FAM173B encompasses an atypical MTS, which is retained in the mature protein.
FAM173B is responsible for methylation of the ATP synthase c-subunit at Lys-43
To functionally characterize human FAM173B, the corresponding gene was disrupted in the haploid cell line HAP1 using CRISPR/Cas9 technology, with guide RNAs designed to target a sequence located upstream of motif “Post I,” which contains a catalytically critical acidic residue (i.e. Glu-117 in FAM173B) required for AdoMet binding to 7BS MTases (17, 18, 23, 24, 41). Through sequencing of genomic DNA, a clone of FAM173B KO cells was identified that contained a frameshift mutation in exon 2, encoding a truncated version of the FAM173B protein devoid of motif Post I, as well as the downstream portion of the MTase domain, and therefore predicted to be enzymatically inactive (for details, see “Experimental procedures”).
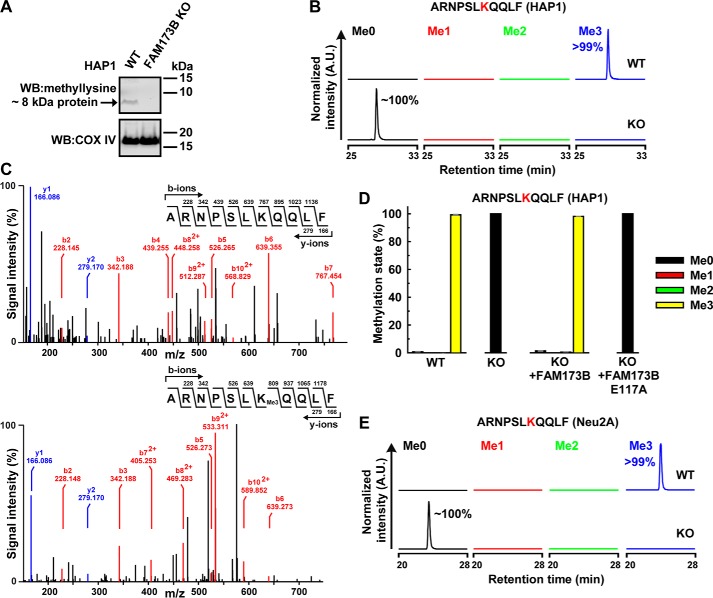
Given the previously observed mitochondrial localization and weak KMT activity of FAM173B, we analyzed cell extracts from HAP1-derived cells enriched in mitochondrial membranes for the presence of lysine-methylated proteins, using Western blotting and a previously characterized anti-methyllysine antibody (26). Several bands representing methylated proteins were detected in such extracts, but intriguingly, one band with an apparent mass of ∼8 kDa was present in unmodified HAP1 WT cells but absent in FAM173B KO cells (Fig. 2A). To identify the corresponding protein, cell extracts were resolved by SDS-PAGE, and the relevant (∼8-kDa) portion was subjected to chymotrypsin treatment, followed by MS analysis. A methylated peptide, ARNPSLKQQLF, corresponding to residues 37–47 of human ATPSc, was identified (amino acid numbering according to the mature ATPSc sequence). This peptide was nearly fully (>99%) trimethylated in HAP1 WT cells but was completely unmethylated in FAM173B KO cells (Fig. 2B). MS/MS fragmentation analysis of this peptide demonstrated the methylation site to be Lys-43 (Fig. 2C), which was previously shown to be trimethylated in all metazoans (29).
Figure 2.
Human FAM173B mediates methylation of ATPSc at Lys-43 in cells. A, a methylated protein of ∼8 kDa is detected in unmodified HAP1 WT cells, but not in FAM173B KO cells. Extracts enriched in mitochondrial membrane proteins were prepared from HAP1 WT and FAM173B KO cells and resolved by SDS-PAGE. Proteins were transferred by Western blotting to a membrane, which was probed with antibody raised against methylated lysine residues. A second membrane, containing parallel samples, was probed with anti-COX IV antibody, to demonstrate equal loading. B, FAM173B KO abrogates ATPSc methylation in HAP1 cells. Cell extracts, prepared as in A, were resolved by SDS-PAGE, and the portion of gel corresponding to the 7–12-kDa region was chymotrypsin-digested and analyzed by MS. Shown are representative, normalized extracted ion chromatograms, gated for different methylation states of ATPSc-derived, chymotrypsin-generated peptide, encompassing residues 37–47 of human mature ATPSc present in HAP1 WT or FAM173B KO cells, with Lys-43 marked in red. Percentages indicate the area under each peak, relative to the total area of all peaks. A.U., arbitrary units. C, MS/MS fragmentation spectra show the absence of ATPSc methylation at Lys-43 in FAM173B KO cells (top) and the presence of trimethylation in HAP1 WT cells (bottom). D, complementation of FAM173B KO cells with FAM173B restores the methylation of ATPSc at Lys-43. Extracts were prepared from HAP1 WT, FAM173B KO, or KO cells expressing FLAG-tagged FAM173B, either nonmutated or E117A-mutated (see Fig. S1), and analyzed by MS as in B. Shown are the mean relative intensities of MS signals, gated for different methylation states of the indicated, ATPSc-derived peptide, with Lys-43 marked in red. Error bars, range of values from three independent analyses of each cell line. E, similar to B, but for mouse ATPSc from neuroblastoma-derived (Neu2A) cells.
To confirm that FAM173B is directly responsible for methylation of ATPSc at Lys-43, we complemented FAM173B KO cells with an ectopically expressed gene encoding FAM173B-FLAG or with a corresponding gene encoding a predicted enzymatically inactive mutant, E117A (Fig. S1). We then investigated the methylation status of Lys-43 in the complemented cells and found that complementation with WT FAM173B restored the methylation back to the fully trimethylated state observed in WT cells (Fig. 2D). In contrast, complementation with the gene encoding the E117A mutant failed to restore methylation. To provide additional evidence for FAM173B-dependent methylation of ATPSc, the Fam173b gene was knocked out in mouse neuroblastoma (Neu2A)-derived cells (for details, see “Experimental procedures”), and the methylation status of Lys-43 was assessed. Reassuringly, Lys-43 was found to be fully trimethylated in the unmodified WT Neu2A cells, whereas it was unmethylated in the corresponding Fam173b KO cells (Fig. 2E). We also analyzed a panel of rat organs and found in all cases that ATPSc was fully trimethylated at Lys-43 (Fig. S2), indicating that the modification is constitutive. In summary, these results demonstrate that the enzymatic activity of FAM173B is required for methylation of ATPSc at Lys-43 in cells.
A putative FAM173B homologue from C. elegans methylates ATPSc at Lys-43
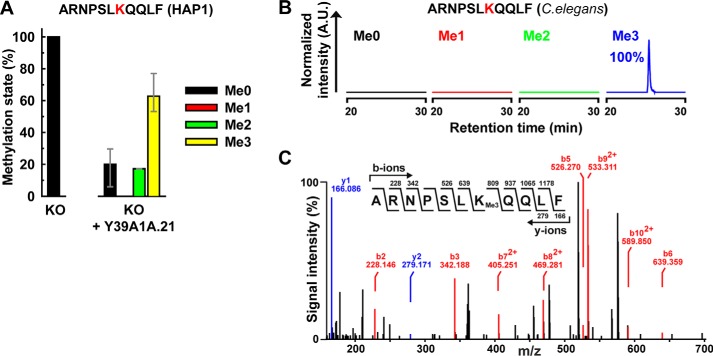
Our generated phylogram (Fig. 1A) suggested that the FAM173-like proteins present in nonvertebrate animals may be functional orthologues of vertebrate FAM173B. To further investigate this, HAP1-derived FAM173B KO cells were complemented with the gene encoding the FAM173-like protein from C. elegans, Y39A1A.21 (Fig. S3), and the methylation status of Lys-43 in ATPSc was assessed. Indeed, complementation of KO cells with Y39A1A.21 largely restored trimethylation of ATPSc at Lys-43 (Fig. 3A), indicating that Y39A1A.21 is capable of methylating human ATPSc. This further indicates that the Y39A1A.21 protein methylates C. elegans ATPSc in vivo, and, indeed, Lys-43 of the mature nematode ATPSc (NP_001022966.1) was found to be present exclusively in the trimethylated state (Fig. 3, B and C). (Note that the relevant ATPSc-derived peptide is identical in humans and worms and gives similar MS/MS spectra). Taken together, the above results strongly indicate that protein Y39A1A.21 is the KMT responsible for trimethylation of Lys-43 of ATPSc in C. elegans and that FAM173-like proteins from other nonvertebrate animals have the same biochemical function.
Figure 3.
The putative FAM173B orthologue from C. elegans methylates Lys-43 in ATPSc. A, complementation of FAM173B KO cells with the putative FAM173B orthologue from C. elegans (Y39A1A.21) restores the methylation of ATPSc at Lys-43. The methylation status of ATPSc from HAP1 FAM173B KO, or KO cells expressing FLAG-tagged Y39A1A.21 protein was analyzed as in Fig. 2D. Error bars, range of values from three independent analyzes of KO cells and three different clones of complemented cells, as indicated in Fig. S3. B, in vivo methylation of ATPSc from C. elegans. Extracts enriched in mitochondrial membrane proteins were prepared from worms and resolved by SDS-PAGE. The portion of the gel corresponding to ATPSc was chymotrypsin-digested and analyzed by MS. Shown are representative, normalized extracted ion chromatograms gated for different methylation states of the indicated chymotrypsin-generated peptide from C. elegans ATPSc, with Lys-43 marked in red. Percentages indicate the area under each peak, relative to the total area of all peaks. A.U., arbitrary units. C, MS/MS fragmentation spectrum demonstrating trimethylation of Lys-43 in ATPSc from C. elegans.
Lack of FAM173B-mediated ATPSc methylation affects the assembly of the ATP synthase complex
To investigate the functional consequences of FAM173B-mediated methylation of ATPSc, we set out to compare various features between cells that have Lys-43 trimethylation (i.e. HAP1 WT and FAM173B KO cells complemented with FAM173B) and cells that lack this modification (i.e. FAM173B KO and KO cells complemented with E117A-mutated FAM173B).
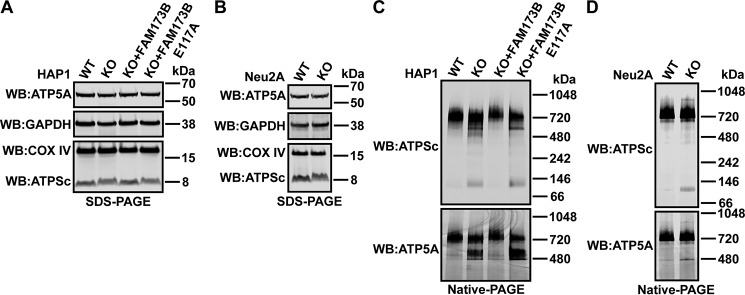
First, we observed that the relative amount of COX IV (loading control for mitochondria) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; loading control for cytosol) was not significantly changed in various HAP1- and Neu2A-derived cells (Fig. 4, A and B), indicating that the lack of ATPSc methylation does not affect the mitochondrial content of cells.
Figure 4.
Methylation at Lys-43 affects incorporation of ATPSc into the ATP synthase complex. A and B, denaturing gel (SDS-PAGE) and Western blot analysis of proteins present in cell extracts. A, unmodified HAP1 (WT), FAM173B KO, or KO cells complemented with FLAG-tagged FAM173B, either nonmutated or E117A-mutated, were lysed in 1% Triton X-100–containing lysis buffer. 50 μg of protein from lysates was resolved by SDS-PAGE and transferred by Western blotting (WB) to a membrane, which was probed with anti-COX IV antibody (loading control for mitochondria) and reprobed with anti-GAPDH antibody (loading control for cytosol). The same membrane was reprobed with anti-ATP5A and anti-ATPSc antibodies. Shown are images from a representative experiment. B, similar to A, but for lysates prepared from Neu2A-derived mouse cells. C and D, nondenaturing gel (native PAGE) and Western blot analysis of ATP synthase subunits present in mitoplast extracts. C, 4 μg of protein from mitoplast extracts prepared from HAP1-derived cells, as in A, was resolved by native PAGE and transferred by Western blotting to a membrane, which was probed with anti-ATPSc antibody and reprobed with anti-ATP5A antibody. Shown are images from a representative experiment. D, similar to C, but for mitoplast extracts prepared from Neu2A-derived mouse cells.
Next, because ATPSc is part of the multimeric ATPS complex, we set out to test whether methylation of ATPSc affects its ability to form high-molecular weight complexes, using native gel electrophoresis and Western blotting. We observed that in all four HAP1-derived cell lines, the majority of ATPSc was present as part of a protein complex of ∼720 kDa in size, which also contained the ATPS F1 α-subunit (ATP5A) (Fig. 4C) and is predicted to represent the full ATPS F1FO complex (42, 43). However, in FAM173B-deficient HAP1 cells, several other ATPSc-containing bands were markedly increased, including bands located between the 480 and 720 kDa markers and a prominent band located between the 66 and 146 kDa markers, the latter one likely corresponding to the ATPSc octamer, possibly in association with some additional subunits of ATPS FO complex. The latter, smaller ATPSc-containing band was also observed in Neu2A cells and was found to be substantially stronger in the Fam173b KO cells, relative to the WT cells (Fig. 4D). Interestingly, in the ATPSc methylation-proficient HAP1 cells, the ATPS α-subunit (ATP5A) was primarily found in the ∼720 kDa band corresponding to the full F1FO complex, but in the methylation-deficient cells, a strong band of ∼480 kDa was also observed (Fig. 4C). A similar band was also observed in the Neu2A cells and was found to be more prominent in the Fam173b KO cells, relative to the WT cells (Fig. 4D). Moreover, we found that whereas the total amount of ATPSc was similar regardless of its methylation status, its apparent mobility in (denaturing) SDS-PAGE was different between the methylated and unmethylated forms (Fig. 4, A and B), and the effect was even more pronounced when mitoplast extracts (rather than whole-cell extracts) were analyzed (Figs. S4 and S5). When trimethylated, the ATPSc protein migrated as a focused band with an apparent mass of ∼8 kDa, whereas the unmethylated ATPSc was observed as a more diffuse band with a somewhat slower migration rate (Fig. 4 (A and B) and Figs. S4 and S5). Taken together, the above results demonstrate that the lack of ATPSc methylation leads to accumulation of low-molecular weight assembly intermediates and indicate that ATPSc methylation plays an important role in ensuring optimal assembly and/or stability of the human F1FO ATPS complex.
FAM173B-mediated ATPSc methylation promotes the activity of the ATPS complex
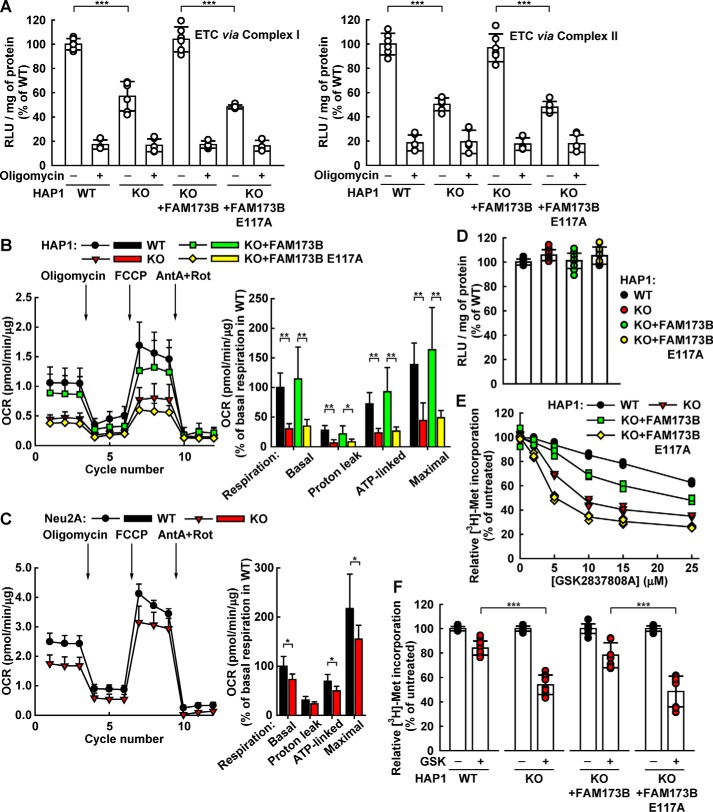
We also set out to investigate how the lack of Lys-43 methylation in ATPSc may affect the ATP-synthesizing ability of the ATPS complex. To this end, we assessed the ability of digitonin-permeabilized cells to generate ATP from ADP and Pi, when supplied with glutamate and malate as a source of electrons for Complex I of the ETC. Alternatively, digitonin-permeabilized cells were supplied with succinate, in the presence of rotenone (inhibitor of Complex I), as a source of electrons for Complex II of the ETC. We found that digitonin-permeabilized HAP1 WT cells were able to generate ATP when electrons were supplied to the ETC via either Complex I or Complex II and that oligomycin (inhibitor of mitochondrial ATPS), as expected, efficiently inhibited ATP formation (Fig. 5A). Interestingly, FAM173B KO cells showed ∼50% reduction in the amount of generated ATP, compared with WT cells, indicating that methylation of ATPSc at Lys-43 influences the efficiency of OxPhos-driven ATP synthesis. Most importantly, complementation of KO cells with enzymatically active FAM173B, but not with enzymatically inactive (E117A-mutated) enzyme, restored ATP generation to the levels observed in WT cells (Fig. 5A). Thus, lack of ATPSc methylation in HAP1 cells appears to cause reduced ATP synthesis through OxPhos.
Figure 5.
Lack of ATPSc methylation leads to reduced mitochondrial ATP synthesis and respiration. A, ablation of ATPSc methylation reduces the efficiency of ATP synthesis by ATPS. Unmodified HAP1 (WT), FAM173B KO, or KO cells complemented with FLAG-tagged FAM173B, either nonmutated or E117A-mutated, were permeabilized with digitonin, and their potential to generate ATP was tested by incubating with ADP and either with glutamate and malate to supply electrons to the ETC via Complex I (left) or with succinate and rotenone to supply electrons to the ETC via Complex II (right). The ATP synthesis experiment was performed either in the absence or presence of oligomycin. Generated ATP was detected using a luminescence-based assay, and the results are expressed as RLU generated per mg of protein. Shown are results from three independent experiments, run in duplicates, and the values have been normalized to those obtained for WT cells in the absence of oligomycin. Error bars, S.D. (n = 6). B, lack of ATPSc methylation reduces mitochondrial respiration in HAP1-derived cells. OCRs of cells indicated as in A were measured by a Seahorse analyzer under basal conditions and after the sequential addition of oligomycin, FCCP, and a mixture of antimycin A and rotenone (AntA + Rot). Typical OCR traces from a representative experiment are presented (left), with arrows indicating the time of addition of the indicated compounds. Error bars, S.D. (n = 5). Mitochondrial respiration dissected into individual components (i.e. basal respiration, respiration due to proton leak, ATP synthesis-linked respiration, and maximal respiration). Shown are the average values from two independent experiments. Error bars, S.D. (n = 10). C, similar to B, but for mitochondrial respiration of neuroblastoma (Neu2A)-derived mouse cells. D, comparison of steady-state ATP levels in HAP1-derived cells. The indicated cells were grown overnight, and then the cellular ATP levels were detected and reported as in A. Shown are results from four independent experiments, run in replicates, normalized to values obtained for WT cells. Error bars, S.D. (n = 11). E and F, differential sensitivity of HAP1-derived cells to inhibition of protein synthesis by the lactate dehydrogenase A inhibitor GSK2837808A (GSK). The indicated cells were treated with GSK for 24 h, with [3H]methionine present during the last 4 h of incubation. Cellular protein was precipitated with 10% TCA, and incorporated [3H]methionine was assayed by scintillation counting. E, titration of the inhibitory effect of GSK on protein synthesis in HAP1-derived cells. F, inhibition of protein synthesis by 10 μm GSK. The experiment is identical to that in E, except that only a single concentration (10 μm) was tested, but with more replicates. Shown are results from five independent experiments, run in replicates, normalized to values obtained in untreated cells. Error bars, S.D. (n = 12). *, p < 0.01; **, p < 0.001; ***, p < 0.0001.
Mitochondrial ATP synthesis is driven by the respiratory chain, which consumes molecular oxygen. One may therefore predict that the reduced ATP synthesis observed in FAM173B-deficient cells would be accompanied by a reduction in oxygen consumption. We measured cellular oxygen consumption in the FAM173B-proficient and -deficient cell lines using a Seahorse analyzer. In these experiments, the oxygen consumption rate is measured under basal conditions (OCRbasal) and after sequential addition of (a) oligomycin (OCRoligomycin), (b) FCCP (uncoupling protonophore that dissipates mitochondrial membrane potential; OCRFCCP), and (c) a mixture of antimycin A (inhibitor of Complex III) + rotenone (OCRAntA+Rot). This allows dissection of the mitochondrial respiration into separate components (i.e. basal respiration (OCRbasal − OCRAntA+Rot), respiration due to proton leak (OCRoligomycin − OCRAntA+Rot), ATP synthesis-linked respiration (OCRbasal − OCRoligomycin), and maximal respiration (OCRFCCP − OCRAntA+Rot)). We observed that in HAP1-derived FAM173B-deficient cells, the basal respiration and the ATP synthesis–linked respiration were both reduced by more than 50% compared with the FAM173B-proficient cells (Fig. 5B). Moreover, the proton leak and the maximal respiration also showed over 50% decrease in FAM173B-deficient cells compared with FAM173B-proficient cells. In line with these observations, Fam173b-deficient Neu2A cells also displayed decreased respiration (Fig. 5C). In conclusion, the observed reduction in ATP synthesis in FAM173B-deficient cells (Fig. 5A) is mirrored by reduction in oxygen consumption (Fig. 5, B and C).
We also compared the steady-state levels of ATP between the various ATPSc methylation-deficient and -proficient HAP1-derived cells and found these levels to be similar (Fig. 5D). This is not surprising, as cellular ATP levels are generally under tight regulation (44). Thus, the reduction in OxPhos-dependent ATP synthesis caused by lack of ATPSc methylation may be compensated for by down-regulation of processes that consume large quantities of ATP, such as protein synthesis. Moreover, it is well-established that cultured cell lines often generate ATP not primarily through OxPhos, but rather by anaerobic glycolysis, a phenomenon known as the “Warburg effect.” Thus, effects of deficient ATPSc methylation on cellular ATP production may be masked and/or compensated for by anaerobic, glycolytic ATP production. To elucidate potential effects of deficient ATPSc methylation on cellular energy metabolism in the absence of glycolysis, we used GSK2837808A, an inhibitor of lactate dehydrogenase A (45), and the energy-demanding process of protein synthesis as a readout of cellular ability to generate ATP. Clearly, treatment of HAP1 cells with GSK2837808A caused a reduction in protein synthesis, but the effect was much stronger in FAM173B KO cells than in WT cells, and complementation of the KO cells with enzymatically active FAM173B partly abrogated the GSK2837808A sensitivity (Fig. 5, E and F). Thus, the observed sensitivity of the FAM173B-deficient cells to protein synthesis inhibition by GSK2837808A (Fig. 5, E and F), further supports that these cells are less proficient in using OxPhos (Fig. 5, A and B) to compensate for inhibition of anaerobic glycolysis.
Discussion
In the present study, we have unraveled the biochemical function of the largely uncharacterized human mitochondrial MTase FAM173B and showed that this enzyme is the long-sought KMT responsible for methylation of Lys-43 in ATPSc. Moreover, we demonstrated that lack of FAM173B-mediated methylation both affected the assembly of the ATPS complex and modulated its ability to mediate ATP synthesis.
We observed increased accumulation of low-molecular weight intermediates of ATPS complex in cells lacking ATPSc methylation, which indicates that the assembly and/or the stability of Complex V is affected. Inhibition of cellular protein synthesis with cycloheximide for several hours did not reduce the amount of fully assembled ATPS or its individual subunits, regardless of the ATPSc methylation status.5 Thus, the assembled Complex V is stable, and the observed low-molecular weight intermediates in KO cells likely result from aberrant assembly of the ATPS complex, rather than from decreased stability.
We showed that ATP synthesis through OxPhos was reduced in permeabilized cells lacking ATPSc methylation, and these results were further corroborated by measurements of oxygen consumption, which revealed decreased ATP synthesis–linked mitochondrial respiration. Somewhat unexpectedly, we found that also the maximal mitochondrial respiration was reduced in FAM173B-deficient cells (Fig. 5, B and C), whereas the mitochondrial content of cells and the relative amount of ATPS subunits and COX IV remained unaffected (Fig. 4 (A and B) and Figs. S4 and S5). Therefore, the reduction of maximal respiration in FAM173B-deficient cells may possibly reflect an adaptation of the maximum respiratory capacity to match the activity of the ATP synthase. An alternative explanation is that FAM173B, in addition to targeting ATPSc, also methylates other components of the mitochondrial respiratory system and enhances their performance.
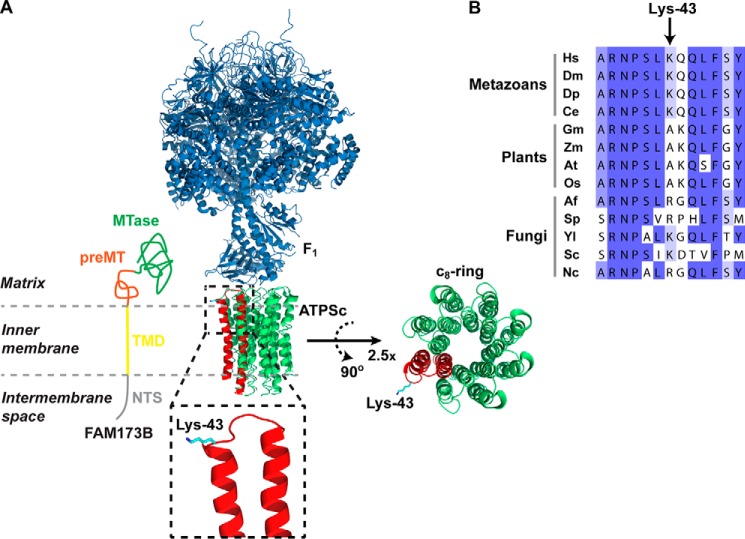
We found that human FAM173B contains, in the N-terminal part, a putative TMD domain, followed by atypical, noncleavable MTS. In mammalian cells, eight ATPSc monomers form a ring structure that traverses the inner mitochondrial membrane, and Lys-43 is localized on the matrix side of the membrane (Fig. 6A). Thus, we favor a model where FAM173B is localized in the mitochondrial matrix, with the TMD inserted into the inner membrane, thereby bringing the enzyme in proximity to its membrane-embedded substrate ATPSc (Fig. 6A).
Figure 6.
Localization of Lys-43 in the ATPS complex structure and in the ATPSc sequence. A, structural representation of the complex between the F1 and ATPSc moieties (c8-ring) of the bovine ATPS complex, generated from a previously published structure (31) (Protein Data Bank code 2XND) and its topology in the inner mitochondrial membrane. Rotation and magnification (×2.5) of the c8-ring was performed to better visualize the ring structure. F1 is shown in blue, and ATPSc is shown in green, except for one of the eight protomers of the c8-ring, which is shown in red, and Lys-43 is here indicated in cyan. Also, a model for the submitochondrial localization of FAM173B is presented, with annotations and coloring as in Fig. 1. B, alignment of ATPSc-derived sequences from various organisms. Shown is the sequence surrounding the methylation site, Lys-43, in human ATPSc, aligned with the corresponding sequences of ATPSc from various organisms: H. sapiens (Hs), D. melanogaster (Dm), D. pulex (Dp), C. elegans (Ce), Glycine max (Gm), Zea mays (Zm), Arabidopsis thaliana (At), Oryza sativa (Os), Aspergillus fumigatus (Af), Schizosaccharomyces pombe (Sp), Yarrowia lipolytica (Yl), Saccharomyces cerevisiae (Sc), and Neurospora crassa (Nc). Sequence accession numbers are given in Table S3.
Vertebrate genomes encode, in addition to FAM173B, the related MTase FAM173A, and we consider it highly likely that also FAM173A is a KMT, due to its high sequence similarity to FAM173B (Fig. 1A). Interestingly, FAM173A has sequence homology to FAM173B also in the TMD/preMT region (i.e. in the portion responsible for targeting to mitochondria) (Fig. 1B). We therefore predict that also FAM173A is found in mitochondria, with the same submitochondrial localization as FAM173B, likely modifying an inner mitochondrial membrane protein on a Lys residue that faces the matrix side.
We have, in recent years, unraveled the function of various human KMTs, and we were generally able to demonstrate the enzymatic activity of the recombinant KMT on the relevant substrate in vitro, using either a recombinant substrate or cell extracts (typically from KMT KO cells) (12, 16–18, 20, 23, 24). Unlike ATPSc, however, none of these substrates are membrane proteins, and we have not been able to demonstrate robust in vitro activity of recombinant FAM173B on ATPSc, neither on the recombinant protein nor in mitochondrial extracts. In a previous study, we found, using [3H]AdoMet and fluorography, that recombinant FAM173B mediated methylation of undefined high-molecular weight material in extracts from HEK293 cells (38), but we have not been able to reproduce these results with the HAP1 cells used here. Also, we found, in several of our previous studies, that the structural context and state of the target protein strongly influenced its ability to become methylated. For example, the chaperone protein VCP forms, similarly to ATPSc, a ring-shaped oligomeric structure with a central pore, but VCP-KMT–mediated methylation of VCP on Lys-315, which is lining the pore, can only occur on monomeric VCP, prior to assembly of the VCP hexamer (18). In conclusion, our inability to observe in vitro activity of FAM173B on ATPSc may have several explanations related to the accessibility and oligomeric state of ATPSc. However, the observed requirement for enzymatically active FAM173B for ATPSc methylation in cells, taken together with the previously reported mitochondrial localization and observed KMT activity of FAM173B, does, in our view, provide convincing evidence that this MTase directly catalyzes ATPSc methylation on Lys-43. Taken together with the apparent presence of FAM173B orthologues in all metazoans, our observations that both human and nematode FAM173B target ATPSc further strengthen the notion that Lys-43 trimethylation by FAM173B is ubiquitous among metazoans.
Interestingly, Lys-43 in ATPSc has been demonstrated, by molecular dynamics simulations, to play an important role in binding the anionic lipid cardiolipin, which is considered an essential component of the ATPS complex (46). Simulations showed that the native, methylated ATPSc bound less cardiolipin than its demethylated counterpart, and it was suggested that interaction of Lys-43 with cardiolipin may help in efficient propelling of the c8-ring rotor by protons returning from the intermembrane space to the matrix (46). Thus, one may speculate that FAM173B-mediated methylation of ATPSc has evolved to functionally optimize its interaction with cardiolipin.
In vertebrate ATP synthases, ATPSc forms a ring consisting of eight monomers, the so-called c8-ring, but interestingly, different types of rings are formed in other species. For example, ATPSc from yeast mitochondria forms c10-rings (47), whereas c14-rings are found in plant chloroplasts (48). Although ATPSc shows strong sequence conservation between different species, Lys-43 and surrounding residues are not particularly well conserved across the tree of life (Fig. 6B). Thus, one may speculate that small variations in this region may contribute to the observed variations in c-ring size and that also trimethylation of Lys-43 may play a role in modulating or optimizing ring formation.
During the last few years, several human 7BS MTases have been established as KMTs, and these are highly specific enzymes that, in most cases, target single lysine residue in one protein. Therefore, a naming nomenclature has been established for these enzymes, which is based on the substrate specificity. In particular, the mitochondrial MTases METTL20 and METTL12 were found to target ETFβ and CS, respectively, and were consequently redubbed ETFβ-KMT and CS-KMT (gene names EFTBKMT and CSKMT, respectively) (23, 24). Thus, we suggest that FAM173B be renamed ATPSc-KMT (gene name ATPSCKMT) in keeping with this prevailing nomenclature.
Experimental procedures
Gene cloning and mutagenesis
Plasmid constructs and the strategy used to generate them are described in detail in Table S4. In brief, ORFs were amplified by PCR and cloned into the indicated vectors by standard ligation-dependent cloning or by ligation-independent cloning using the In-Fusion® HD Cloning Plus kit (Takara). Mutations within ORFs were introduced by site-directed mutagenesis using PCR splicing by overhang extension (SOEing; for details, see supporting information). Sanger sequencing was used to verify the identity and the integrity of all cloned constructs.
Bioinformatics analysis
NCBI Basic Local Alignment Search Tool (BLAST) (49) was used to identify protein sequences homologous to human FAM173B and ATPSc, found in Tables S1 and S3, respectively. Multiple-sequence alignments and secondary structure predictions were generated using algorithms embedded in the JalView interface (http://www.jalview.org/)6 (50). A phylogenetic tree was generated from the alignment using the PhyML program embedded within the www.phylogeny.fr6 package (51, 52). The tree was visualized using the FigTree program (https://github.com/rambaut/figtree/tree/master/release/common).6
Cell cultures
HeLa cells were grown in RPMI 1640 GlutaMAX medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 100 units/ml penicillin and 0.1 mg/ml streptomycin (P/S). Human HAP1-derived cells were grown in IMDM GlutaMAX medium supplemented with 10% FBS and P/S. Mouse Neu2A-derived cells were grown in DMEM GlutaMAX (high-glucose, pyruvate) medium supplemented with 10% FBS and P/S.
Generation of stable cell lines
HAP1 FAM173B KO cells were generated as a (nonexclusive) custom project by Horizon Genomics (Austria). The relevant gene was disrupted in haploid HAP1 parental cells using CRISPR-Cas9, with guide RNAs designed to target part of exon 2 of the human FAM173B gene located upstream of motif Post I, which is required for enzymatic activity of 7BS MTases. Individual clones were selected by limiting dilution, and frame-shifting events within the FAM173B gene were determined by sequencing of genomic DNA. The FAM173B-deficient cell line contains a 2-bp deletion within exon 2, resulting in generation of a truncated version of the FAM173B protein, consisting of the initial 56 amino acids of FAM173B, followed by 21 residues of out-of-frame sequence, and this cell line is commercially available (Horizon Discovery HZGHC000533c006). Complementation of HAP1 FAM173B KO cells was performed similarly as described previously (24), by transfection with a p3xFLAG-CMV-14-derived plasmid encoding either unmodified or E117A-mutated human FAM173B or Y39A1A.21, the FAM173 homologue from C. elegans, all bearing a C-terminal 3xFLAG tag. Transfected cells were selected with 1 mg/ml Geneticin (Gibco) and expanded in medium containing Geneticin. Individual clones of complemented cells were screened by Western blotting for the presence of FLAG tag and COX IV (as loading control), using appropriate antibodies (see “Western blot analysis”).
Mouse Neu2A Fam173b KO cells were generated in-house, using CRISPR-Cas9, by transfecting with pSpCas9(BB)-2A-GFP (PX458) plasmid (53) containing single guide RNA (caccGCGTGCCTGCAACTTCGAAGC) designed to target part of exon 2 of the mouse Fam173b gene located upstream of motif Post I. GFP-positive cells were sorted by FACS, individual clones were selected by limiting dilution, and frame-shifting events within the Fam173B gene were determined by sequencing of genomic DNA using the following primers: forward, GATGGAGCGAGTAGGAACG; reverse, TCATCTGGGGCACACCAAAA. The Fam173b-deficient Neu2A cell line contains a mixture of two Fam173b gene alleles, with either a 1- or 2-bp deletion within exon 2, both resulting in generation of truncated versions of the protein, consisting of the initial 70 amino acids of mouse Fam173b, followed by either 27 or 2 residues of out-of-frame sequence, respectively.
Transient transfection and fluorescence microscopy
HeLa cells were transiently transfected with pEGFP-N1–derived plasmids (Clontech), encoding human full-length FAM173B or two truncated versions thereof (i.e. a deletion mutant where the initial N-terminal 55 amino acids were deleted (Δ55) or a mutant consisting only of amino acids 56–90 of FAM173B), both fused to the N terminus of enhanced GFP. 24 h after transfection, cells were fixed in cold acetone for 15 min and then incubated overnight with anti-COX IV antibody, followed by an anti-rabbit secondary antibody coupled to Alexa Fluor 568 (Molecular Probes). 1 μg/ml DAPI was used as nuclear counterstaining. Cell staining was then analyzed using an Olympus FluoView 1000 (Ix81) confocal fluorescence microscopy system with a PlanApo ×100, numerical aperture 1.1 oil objective (Olympus). The different fluorophores were excited at 405 nm (DAPI), 488 nm (GFP), and 559 nm (Alexa Fluor 568). A Kalman filter was used to record multichannel images.
Cell lysis and preparation of mitoplast extracts enriched for mitochondrial inner membrane proteins
Cells were grown in ∅15-cm plates until 50–80% confluent, washed once with PBS, and harvested by scraping in 2 ml of PBS. Cells were centrifuged (3 min, 750 × g), PBS was removed, and cell pellets were frozen at −20 °C until needed. Cells were lysed for 5 min at 4 °C with Lysis Buffer 1 (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5% glycerol, 1% Triton X-100), supplemented with protease inhibitor mix (P8340, Sigma-Aldrich), and cleared by centrifugation (11,000 × g, 5 min, 4 °C) to obtain clear lysate.
Preparation of cell extracts enriched for mitochondrial inner membrane proteins was performed at 4 °C, based on a protocol described previously (54), with important modifications. Frozen cell pellets were thawed on ice, resuspended in 600 μl of PBS containing 2 mg/ml digitonin and protease inhibitor mix (P8340), and incubated for 5 min on ice. The suspension was centrifuged (11,000 × g, 10 min), the supernatant was discarded, and the pellet was resuspended in 300 μl of PBS containing 0.5 mg/ml digitonin and protease inhibitors. The suspension was again centrifuged (11,000 × g, 5 min), and the mitoplast pellet, enriched for mitochondrial inner membranes, was resuspended by vigorous vortexing in 70 μl of Extraction Buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1% n-dodecyl-β-d-maltoside, 5% glycerol, and protease inhibitors), and incubated on ice for 5 min. The suspension was centrifuged (16,100 × g, 5 min), and n-dodecyl-β-d-maltoside–extracted proteins were recovered in the supernatant. Protein concentration in extracts was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
Frozen pellets of the nematode C. elegans or frozen fragments of rat organs were thawed on ice, resuspended in PBS containing 2 mg/ml digitonin and protease inhibitor mix (P8340), and mechanically fragmented by vortexing with sharp glass beads. Non-solubilized material and glass beads were removed by centrifugation (500 × g, 2 min). The supernatant was then used to prepare mitoplast extracts, similarly as described above.
Western blot analysis
Proteins present in clear lysate (50 μg) or mitoplast extracts (30 μg) were prepared as described above, resolved by SDS-PAGE, and transferred to polyvinylidene difluoride Immobilon®-FL transfer membrane (Merck), which was stained with Ponceau S and blocked using Odyssey® Blocking Buffer (TBS) (LI-COR) diluted 1:1 (v/v) in TBS. The membrane was then incubated with primary antibodies: mouse anti-FLAG (F1804, Sigma-Aldrich), rabbit anti-COX IV (ab16056, Abcam), rabbit anti-methylated lysine (ab23366, Abcam), mouse anti-ANT2 (H00000292-B01P, Abnova), mouse anti-ATP5A (ab14748, Abcam), rabbit anti-GAPDH (ab9485), or rabbit anti-ATPSc (ab181243, Abcam), diluted in Odyssey® Blocking Buffer mixed 1:1 (v/v) with TBS, containing 0.05% Tween 20. The primary antibodies were detected with LI-COR secondary antibodies coupled with IR fluorescent dyes (either goat anti-mouse IRDye® 680RD or goat anti-rabbit IRDye® 800CW) according to the manufacturer's instructions and visualized using the LI-COR Odyssey CLx imaging system. When needed, the same membrane was reprobed with different primary and secondary antibodies, without stripping. Precision Plus protein dual color standards (Bio-Rad) or Cameleon Duo prestained protein ladder (LI-COR) were used to evaluate the size of polypeptides visualized by Western blotting. The signal intensities detected for various proteins were integrated using the software analysis tool embedded in the basic software package of the Odyssey imaging system. The signal ratio between ATPSc and COX IV, detected in various HAP1-derived cell lines, was normalized to that observed in unmodified (WT) cells.
Analysis of protein complexes under nondenaturing conditions
For analysis of protein complexes under nondenaturing conditions, 4 μg of protein from mitoplast extracts was resolved using 4–16% gradient native PAGE BisTris gels (Thermo Fisher Scientific), according to the manufacturer's instructions. Proteins were then transferred by Western blotting to a polyvinylidene difluoride membrane and analyzed as described above. NativeMark unstained protein standard (Thermo Fisher Scientific) was used to evaluate the size of protein complexes.
Mass spectrometry analysis
Proteins present in mitoplast extracts were resolved by SDS-PAGE, stained with Coomassie, and the portion of gel containing the protein of interest was excised and subjected to in-gel chymotrypsin (Roche Applied Science) digestion. The resulting proteolytic fragments were analyzed by LC-MS, similarly as described previously (20).
MS data were analyzed using in-house maintained human, rat, mouse, and C. elegans protein sequence databases using SEQUESTTM and Proteome DiscovererTM (Thermo Fisher Scientific). The mass tolerances of a fragment ion and a parent ion were set as 0.5 Da and 10 ppm, respectively. Methionine oxidation, cysteine carbamido-methylation, and lysine and arginine methylation were selected as variable modifications. MS/MS spectra of peptides corresponding to methylated ATPSc were manually searched by Qual Browser (version 2.0.7).
Measurement of ATP levels in cells
Cells were seeded in a 96-well plate format, at a density of 3 × 104 cells/well, and grown overnight. Cells were removed from the incubator and processed immediately, by assaying the intracellular ATP level using the Luminescent ATP Detection Assay Kit (ab113849, Abcam), according to the manufacturer's instructions. This assay is based on the emission of light (luminescence) during ATP-dependent oxidation of d-luciferin by firefly luciferase, and the intensity of luminesce is proportional to the concentration of ATP present in the sample. Samples were transferred to white 96-well plates, and the luminescence was recorded using a Wallac Victor2 1420 multilabel plate reader (PerkinElmer Life Sciences). The individual luminescence intensities were normalized to cellular protein content and reported as relative luminescence units (RLU)/mg of protein. The protein content was assayed in parallel wells, as follows. After removing the medium, the cells were gently washed with PBS and lysed in 25 μl of Lysis Buffer 2 (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1% Triton X-100), and the protein content was determined using the Pierce BCA Protein Assay Kit. To compare results obtained for different cell lines during various independent experiments, the data within the individual experiments were normalized to values obtained for HAP1 WT cells and are reported as percentage of signal (in RLU/mg protein) observed in WT cells.
ATP synthesis in digitonin-permeabilized cells
ATP synthesis in digitonin-permeabilized cells was performed based on a protocol described elsewhere (55), with important modifications. Cells were seeded in a 96-well plate format, at a density of 3 × 104 cells/well, and grown overnight. Medium was removed, and cells were permeabilized by incubating at room temperature, for 3 min, in PBS containing 50 μg/ml digitonin. Next, PBS was removed, and permeabilized cells were incubated at 37 °C, for 30 min, in Incubation Buffer (25 mm Tris-HCl, pH 7.4, 10 mm KCl, 10 mm KH2PO4, 0.1 mm MgCl2, 2 mm EDTA), supplemented with either (a) 5 mm malate, 5 mm glutamate, and 0.4 mm ADP, to stimulate mitochondrial ATP production by supplying ETC with electrons via Complex I, or (b) 10 mm succinate, 0.4 mm ADP, and 2 μg/ml rotenone (inhibitor of Complex I), to stimulate mitochondrial ATP production by supplying ETC with electrons only via Complex II. In each case, 2 μg/ml oligomycin was used as control, to assess ATP production in the absence of functional mitochondrial ATP synthase. Next, the level of produced ATP was assayed using the Luminescent ATP Detection Assay Kit (ab113849, Abcam), similarly as described above. The luminescence intensities were normalized by the cells' protein content. The data within an individual experiment were additionally normalized to the values obtained for HAP1 WT cells and are reported as percentage of signal (in RLU/mg protein) observed in WT cells in the absence of oligomycin.
Measurement of mitochondrial respiration by Seahorse analyzer
HAP1 cells were seeded on noncoated XF24 plates (Seahorse Bioscience), at a density of 2.5 × 104 cells/well, and N2A cells were seeded on poly-l-lysine–coated XF24 plates, at a density of 1.25 × 104 cells/well, and grown overnight. Cells were washed and placed in Seahorse XF-assay medium (pH 7.4) containing 25 mm glucose, 4 mm glutamine, and 1 mm pyruvate at 37 °C for 1 h. The Seahorse Bioscience XF24 Analyzer (Seahorse Bioscience) was used to measure OCRs under basal conditions, and after the sequential addition of oligomycin (2 μm), FCCP (2 μm), and a mixture of antimycin A and rotenone (AntA + Rot, 2 μm each), which were injected after cycles 3, 6, and 9, respectively. Each assay cycle consisted of 1.5 min of mixing, 2 min of waiting, and 3 min of OCR measurements. For each condition, three cycles were used to determine the average OCR under given conditions (i.e. OCRbasal, OCRoligomycin, OCRFCCP and OCRAntA+Rot). The measured OCR was normalized for protein content and expressed in pmol of O2 consumed per minute per μg of protein (pmol/min/μg). OCRs measured under different conditions were used to calculate the individual components of mitochondrial respiration (i.e. basal respiration (OCRbasal − OCRAntA+Rot), respiration due to proton leak (OCRoligomycin − OCRAntA+Rot), ATP synthesis-linked respiration (OCRbasal − OCRoligomycin), and maximal respiration (OCRFCCP − OCRAntA+Rot)). Two independent experiments were run, each consisting of five replicates of individual cell lines. The data within an individual experiment were normalized to basal respiration in WT cells, and combined results from two experiments are reported as percentage of basal respiration observed in WT.
Measurement of protein synthesis by [3H]methionine incorporation
Cells were seeded in a 24-well format, at a density of 2 × 105 cells/well, grown overnight, and then treated with the indicated concentrations of GSK2837808A or DMSO (as carrier) for 24 h. During the last 4 h of incubation, 10 μCi/ml [3H]methionine was present in the medium. Next, medium was removed, and cells were precipitated with 10% TCA, and TCA-insoluble material was subjected to scintillation counting. The amount of radioactivity incorporated in cells treated with GSK2837808A was normalized to values obtained for control (DMSO)-treated cells.
Statistical analysis
The independent two-sample Student's t test was used to evaluate the probability (p value) that the means of two populations are not different.
Author contributions
J. M. and P. Ø. F. conceived and planned the study. J. M. and P. Ø. F. planned experiments. J. M., R. P., A. Y. H., A. M., I. K., H. L. D. M. W., B. B., F. Z., and N. E. performed experiments: J. M., A. Y. H. and I. K. cloning and mutagenesis; R. P. and A. Y. H. cell microscopy; J. M. cell work, Western blotting, and preparation of samples; A. M. and J. M. mass spectrometry; H. L. D. M. W., B. B., F. Z., and N. E. generation of Neu2 cells with Fam173b KO and Seahorse analyzer experiments. J. M. and P. Ø. F. wrote the manuscript. All authors analyzed data and read and commented on the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Lars Eide (Oslo University Hospital) for insightful discussions and Dr. Erna Davydova (Department of Biosciences, University of Oslo) for critical reading of the manuscript. We thank Dr. Magnus Jakobsson for assistance during the initial phase of this project. We thank Dr. Marianne Fyhn (Department of Biosciences, University of Oslo) for providing rat organs. We thank Dr. Hilde Nilsen (Department of Clinical Molecular Biology, University of Oslo) for providing C. elegans. We thank the Oslo NorMIC Imaging Platform (Department of Biosciences, University of Oslo) for the use of cell-imaging equipment.
This work was supported by Research Council of Norway Grant FRIMEDBIO-240009 and Norwegian Cancer Society Grant 107744-PR-2007-0132. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4 and Figs. S1–S5.
J. M. Małecki and P. Ø. Falnes, unpublished data.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MTase
- methyltransferase
- 7BS
- seven β-strand
- AdoMet
- S-adenosylmethionine
- ANT
- adenine nucleotide translocator
- AntA
- antimycin A
- ATPS
- ATP synthase
- ATP5A
- ATP synthase α-subunit
- ATPSc
- ATP synthase c-subunit
- COX IV
- cytochrome c oxidase subunit IV
- CS
- citrate synthase
- ETC
- electron transport chain
- ETFβ
- electron transfer flavoprotein β-subunit
- FCCP
- carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- KMT
- lysine-specific methyltransferase
- MTS
- mitochondrial targeting sequence
- OxPhos
- oxidative phosphorylation
- RLU
- relative luminescence units
- Rot
- rotenone
- TMD
- transmembrane domain
- VCP
- valosine-containing protein
- preMT
- segment of conserved sequence that precedes the annotated MTase domain
- FAM173B
- family with sequence similarity 173 member B
- NTS
- N-terminal sequence
- OCR
- oxygen consumption rate
- FBS
- fetal bovine serum
- P/S
- penicillin/streptomycin
- DAPI
- 4′,6-diamidino-2-phenylindole
- BisTris
- bis(2-hydroxyethyl)aminotris(hydroxymethyl)methane.
References
- 1. Bedford M. T. (2007) Arginine methylation at a glance. J. Cell Sci. 120, 4243–4246 10.1242/jcs.019885 [DOI] [PubMed] [Google Scholar]
- 2. Figaro S., Scrima N., Buckingham R. H., and Heurgué-Hamard V. (2008) HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1 FEBS Lett. 582, 2352–2356 10.1016/j.febslet.2008.05.045 [DOI] [PubMed] [Google Scholar]
- 3. Moore K. E., and Gozani O. (2014) An unexpected journey: lysine methylation across the proteome. Biochim. Biophys. Acta 1839, 1395–1403 10.1016/j.bbagrm.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Webb K. J., Zurita-Lopez C. I., Al-Hadid Q., Laganowsky A., Young B. D., Lipson R. S., Souda P., Faull K. F., Whitelegge J. P., and Clarke S. G. (2010) A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J. Biol. Chem. 285, 37598–37606 10.1074/jbc.M110.170787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petrossian T. C., and Clarke S. G. (2011) Uncovering the human methyltransferasome. Mol. Cell Proteomics 10, M110.000976 10.1074/mcp.M110.000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schubert H. L., Blumenthal R. M., and Cheng X. (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 10.1016/S0968-0004(03)00090-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falnes P. Ø., Jakobsson M. E., Davydova E., Ho A. Y., and Małecki J. (2016) Protein lysine methylation by seven-β-strand methyltransferases. Biochem. J. 473, 1995–2009 10.1042/BCJ20160117 [DOI] [PubMed] [Google Scholar]
- 8. Herz H. M., Garruss A., and Shilatifard A. (2013) SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38, 621–639 10.1016/j.tibs.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greer E. L., and Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lanouette S., Mongeon V., Figeys D., and Couture J. F. (2014) The functional diversity of protein lysine methylation. Mol. Syst. Biol. 10, 724 10.1002/msb.134974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., and Coulombe B. (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 9, e1003210 10.1371/journal.pgen.1003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davydova E., Ho A. Y., Malecki J., Moen A., Enserink J. M., Jakobsson M. E., Loenarz C., and Falnes P. Ø. (2014) Identification and characterization of a novel evolutionarily conserved lysine-specific methyltransferase targeting eukaryotic translation elongation factor 2 (eEF2) J. Biol. Chem. 289, 30499–30510 10.1074/jbc.M114.601658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dzialo M. C., Travaglini K. J., Shen S., Loo J. A., and Clarke S. G. (2014) A new type of protein lysine methyltransferase trimethylates Lys-79 of elongation factor 1A. Biochem. Biophys. Res. Commun. 455, 382–389 10.1016/j.bbrc.2014.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamey J. J., Winter D. L., Yagoub D., Overall C. M., Hart-Smith G., and Wilkins M. R. (2016) Novel N-terminal and lysine methyltransferases that target translation elongation factor 1A in yeast and human. Mol. Cell Proteomics 15, 164–176 10.1074/mcp.M115.052449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamey J. J., Wienert B., Quinlan K. G. R., and Wilkins M. R. (2017) METTL21B is a novel human lysine methyltransferase of translation elongation factor 1A: discovery by CRISPR/Cas9 knockout. Mol. Cell Proteomics 16, 2229–2242 10.1074/mcp.M116.066308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., and Falnes P. Ø. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 function through lysine methylation. J. Biol. Chem. 288, 27752–27763 10.1074/jbc.M113.483248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakobsson M. E., Malecki J., Nilges B. S., Moen A., Leidel S. A., and Falnes P. Ø. (2017) Methylation of human eukaryotic elongation factor α (eEF1A) by a member of a novel protein lysine methyltransferase family modulates mRNA translation. Nucleic Acids Res. 45, 8239–8254 10.1093/nar/gkx432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Mælandsmo G. M., Egge-Jacobsen W., and Falnes P. Ø. (2012) Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038 10.1038/ncomms2041 [DOI] [PubMed] [Google Scholar]
- 19. Lipson R. S., Webb K. J., and Clarke S. G. (2010) Two novel methyltransferases acting upon eukaryotic elongation factor 1A in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 500, 137–143 10.1016/j.abb.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malecki J., Aileni V. K., Ho A. Y. Y., Schwarz J., Moen A., Sørensen V., Nilges B. S., Jakobsson M. E., Leidel S. A., and Falnes P. Ø. (2017) The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A) Nucleic Acids Res. 45, 4370–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shimazu T., Barjau J., Sohtome Y., Sodeoka M., and Shinkai Y. (2014) Selenium-based S-adenosylmethionine analog reveals the mammalian seven-β-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS One 9, e105394 10.1371/journal.pone.0105394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Małecki J., Ho A. Y., Moen A., Dahl H. A., and Falnes P. Ø. (2015) Human METTL20 is a mitochondrial lysine methyltransferase that targets the β subunit of electron transfer flavoprotein (ETFβ) and modulates its activity. J. Biol. Chem. 290, 423–434 10.1074/jbc.M114.614115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Małecki J., Jakobsson M. E., Ho A. Y. Y., Moen A., Rustan A. C., and Falnes P. Ø. (2017) Uncovering human METTL12 as a mitochondrial methyltransferase that modulates citrate synthase activity through metabolite-sensitive lysine methylation. J. Biol. Chem. 292, 17950–17962 10.1074/jbc.M117.808451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhein V. F., Carroll J., He J., Ding S., Fearnley I. M., and Walker J. E. (2014) Human METTL20 methylates lysine residues adjacent to the recognition loop of the electron transfer flavoprotein in mitochondria. J. Biol. Chem. 289, 24640–24651 10.1074/jbc.M114.580464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rhein V. F., Carroll J., Ding S., Fearnley I. M., and Walker J. E. (2017) Human METTL12 is a mitochondrial methyltransferase that modifies citrate synthase. FEBS Lett. 591, 1641–1652 10.1002/1873-3468.12649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen R., Fearnley I. M., Palmer D. N., and Walker J. E. (2004) Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with batten disease. J. Biol. Chem. 279, 21883–21887 10.1074/jbc.M402074200 [DOI] [PubMed] [Google Scholar]
- 28. Katz M. L., Christianson J. S., Norbury N. E., Gao C. L., Siakotos A. N., and Koppang N. (1994) Lysine methylation of mitochondrial ATP synthase subunit c stored in tissues of dogs with hereditary ceroid lipofuscinosis. J. Biol. Chem. 269, 9906–9911 [PubMed] [Google Scholar]
- 29. Walpole T. B., Palmer D. N., Jiang H., Ding S., Fearnley I. M., and Walker J. E. (2015) Conservation of complete trimethylation of lysine-43 in the rotor ring of c-subunits of metazoan adenosine triphosphate (ATP) synthases. Mol. Cell Proteomics 14, 828–840 10.1074/mcp.M114.047456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walker J. E. (2013) The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16 10.1042/BST20110773 [DOI] [PubMed] [Google Scholar]
- 31. Watt I. N., Montgomery M. G., Runswick M. J., Leslie A. G., and Walker J. E. (2010) Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 107, 16823–16827 10.1073/pnas.1011099107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abrahams J. P., Leslie A. G., Lutter R., and Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 10.1038/370621a0 [DOI] [PubMed] [Google Scholar]
- 33. Yan W. L., Lerner T. J., Haines J. L., and Gusella J. F. (1994) Sequence analysis and mapping of a novel human mitochondrial ATP synthase subunit 9 cDNA (ATP5G3). Genomics 24, 375–377 10.1006/geno.1994.1631 [DOI] [PubMed] [Google Scholar]
- 34. Gay N. J., and Walker J. E. (1985) Two genes encoding the bovine mitochondrial ATP synthase proteolipid specify precursors with different import sequences and are expressed in a tissue-specific manner. EMBO J. 4, 3519–3524 10.1002/j.1460-2075.1985.tb04111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu Y., Zhang Z., Wang Q., Luo Y., and Huang L. (2012) Identification and characterization of a highly conserved crenarchaeal protein lysine methyltransferase with broad substrate specificity. J. Bacteriol. 194, 6917–6926 10.1128/JB.01535-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niu Y., Xia Y., Wang S., Li J., Niu C., Li X., Zhao Y., Xiong H., Li Z., Lou H., and Cao Q. (2013) A prototypic lysine methyltransferase 4 from archaea with degenerate sequence specificity methylates chromatin proteins Sul7d and Cren7 in different patterns. J. Biol. Chem. 288, 13728–13740 10.1074/jbc.M113.452979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peters M. J., Broer L., Willemen H. L., Eiriksdottir G., Hocking L. J., Holliday K. L., Horan M. A., Meulenbelt I., Neogi T., Popham M., Schmidt C. O., Soni A., Valdes A. M., Amin N., Dennison E. M., et al. (2013) Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann. Rheum. Dis. 72, 427–436 10.1136/annrheumdis-2012-201742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Willemen H. L. D. M., Kavelaars A., Prado J., Maas M., Versteeg S., Nellissen L. J. J., Tromp J., Gonzalez Cano R., Zhou W., Jakobsson M. E., Małecki J., Posthuma G., Habib A. M., Heijnen C. J., Falnes P. Ø., and Eijkelkamp N. (2018) Identification of FAM173B as a protein methyltransferase promoting chronic pain. PLoS Biol. 16, e2003452 10.1371/journal.pbio.2003452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bolender N., Sickmann A., Wagner R., Meisinger C., and Pfanner N. (2008) Multiple pathways for sorting mitochondrial precursor proteins EMBO Rep. 9, 42–49 10.1038/sj.embor.7401126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Claros M. G., and Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 10.1111/j.1432-1033.1996.00779.x [DOI] [PubMed] [Google Scholar]
- 41. Małecki J., Dahl H. A., Moen A., Davydova E., and Falnes P. Ø. (2016) The METTL20 homologue from Agrobacterium tumefaciens is a dual-specificity protein-lysine methyltransferase that targets ribosomal protein L7/L12 and the β subunit of electron transfer flavoprotein (ETFβ). J. Biol. Chem. 291, 9581–9595 10.1074/jbc.M115.709261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu T., Pagadala V., and Mueller D. M. (2015) Understanding structure, function, and mutations in the mitochondrial ATP synthase. Microb. Cell 2, 105–125 10.15698/mic2015.04.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fujikawa M., Sugawara K., Tanabe T., and Yoshida M. (2015) Assembly of human mitochondrial ATP synthase through two separate intermediates, F1-c-ring and b-e-g complex. FEBS Lett. 589, 2707–2712 10.1016/j.febslet.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 44. Wang X., Zhang X., Wu D., Huang Z., Hou T., Jian C., Yu P., Lu F., Zhang R., Sun T., Li J., Qi W., Wang Y., Gao F., and Cheng H. (2017) Mitochondrial flashes regulate ATP homeostasis in the heart. Elife 6, e23908 10.7554/eLife.23908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Billiard J., Dennison J. B., Briand J., Annan R. S., Chai D., Colón M., Dodson C. S., Gilbert S. A., Greshock J., Jing J., Lu H., McSurdy-Freed J. E., Orband-Miller L. A., Mills G. B., Quinn C. J., et al. (2013) Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 1, 19 10.1186/2049-3002-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duncan A. L., Robinson A. J., and Walker J. E. (2016) Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc. Natl. Acad. Sci. U.S.A. 113, 8687–8692 10.1073/pnas.1608396113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stock D., Leslie A. G., and Walker J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 10.1126/science.286.5445.1700 [DOI] [PubMed] [Google Scholar]
- 48. Vollmar M., Schlieper D., Winn M., Büchner C., and Groth G. (2009) Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J. Biol. Chem. 284, 18228–18235 10.1074/jbc.M109.006916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGinnis S., and Madden T. L. (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25 10.1093/nar/gkh435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., and Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 52. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klement P., Nijtmans L. G., Van den Bogert C., and Houstěk J. (1995) Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal. Biochem. 231, 218–224 10.1006/abio.1995.1523 [DOI] [PubMed] [Google Scholar]
- 55. Van Bergen N. J., Blake R. E., Crowston J. G., and Trounce I. A. (2014) Oxidative phosphorylation measurement in cell lines and tissues. Mitochondrion 15, 24–33 10.1016/j.mito.2014.03.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.