Abstract
CRISPR/Cas9 is a powerful technology widely used for genome editing, with the potential to be used for correcting a wide variety of deleterious disease-causing mutations. However, the technique tends to generate more indels (insertions and deletions) than precise modifications at the target sites, which might not resolve the mutation and could instead exacerbate the initial genetic disruption. We sought to develop an improved protocol for CRISPR/Cas9 that would correct mutations without unintended consequences. As a case study, we focused on achondroplasia, a common genetic form of dwarfism defined by missense mutation in the Fgfr3 gene that results in glycine to arginine substitution at position 374 in mice in fibroblast growth factor receptor 3 (Fgfr3-G374R), which corresponds to G380R in humans. First, we designed a GFP reporter system that can evaluate the cutting efficiency and specificity of single guide RNAs (sgRNAs). Using the sgRNA selected based on our GFP reporter system, we conducted targeted therapy of achondroplasia in mice. We found that we achieved higher frequency of precise correction of the Fgfr3-G374R mutation using Cas9 protein rather than Cas9 mRNA. We further demonstrated that targeting oligos of 100 and 200 nucleotides precisely corrected the mutation at equal efficiency. We showed that our strategy completely suppressed phenotypes of achondroplasia and whole genome sequencing detected no off-target effects. These data indicate that improved protocols can enable the precise CRISPR/Cas9-mediated correction of individual mutations with high fidelity.
Keywords: fibroblast growth factor receptor (FGFR), CRISPR/Cas, gene therapy, genetic disease, bone
Introduction
CRISPR/Cas9 is a RNA-guided genome editing tool developed from a microbial adaptive immune defense system (1–3). Because of its high efficiency and easy-to-use nature, the CRISPR/Cas9 system is revolutionizing many research fields, including gene functional study, medical research, and genomic editing. Since its introduction into mammalian cells in 2013, many important progresses have been made in various scientific fields (4–6). In particular for gene therapy, several studies have demonstrated the power of CRISPR/Cas9 technology in mouse models for the correction of human hereditary genetic diseases, such as, cataract disorder (7), tyrosinemia (8), thalassemia (9), Duchenne muscular dystrophy (10–12), liver diseases (13, 14) and so on. These approaches are potentially translatable for human clinical therapy. Meanwhile studies also revealed some technical difficulties that CRISPR/Cas9 system encounters, such as it preferably generates more small insertions and deletions (Indels) than precise modifications, and off-target effect, i.e. binding and cutting at sites with shared homology (3, 15–17).
Achondroplasia is the most common genetic form of dwarfism inherited as an autosomal dominant disorder (18). People with achondroplasia have impaired ability in the longitudinal growth of long bone from endochondral ossification at the epiphyseal growth plate, leading to the short status. Homozygous achondroplasia predisposes its carriers to neonatal lethal condition, whereas heterozygous achondroplastic adults can be as short as 62.8 cm (24.7 in). More than 96% of patients with achondroplasia have a Gly to Arg transition at position 1138 (G1138A) of the fibroblast growth factor receptor 3 (FGFR3) gene, resulting in the Gly to Arg substitution at position 380 of the FGFR3 protein (19, 20), which corresponds to the Gly to Arg at 374 (G1120A for DNA) in mouse Fgfr3 (21). Although human growth hormone has been used to aid growth in other dwarfism, it does not help people with achondroplasia. The only way is the controversial surgery of limb-lengthening, which will lead to patients suffering huge pain. Therefore, developing a radical treatment is desired.
Using our previously generated mouse Fgfr3-G374R (corresponds to Fgfr3G1120A) achondroplasia model (21), here we show that high frequency of targeted correction of the G374R can be achieved by co-injection into zygotes of Cas9 protein and a single guide RNA (sgRNA)4 optimized for specifically targeting the mutant allele. The correction occurs via homology-directed repair (HDR) based on an exogenously supplied single strand donor DNA without inducing nonspecific off targeting events. The resulting mice showed normal body size, were fertile, and able to transmit the corrected allele to their progeny. Thus, our study provides proof for using the CRISPR/Cas9 system to correct this dominate genetic disease.
Results
Evaluation of genome editing efficiency and specificity of sgRNAs against Fgfr3-G374R by sgRNA testers
Three overlapping sgRNAs of 20 nt were designed to spanning the Gly to Arg mutation in Fgfr3 gene (Fig. S1A), the top 10 predicted off-target locus with NGG PAM (protospacer adjacent motif) sequence were listed for further analysis (Fig. S1, B–D). The highest homology for these sgRNAs is WT Fgfr3 that contains 1 mismatched base at the “Gly to Arg” mutation site. This is followed by 1 mismatches of the Cnnm2 gene to sgRNA1; 2 mismatches of noncoding region locus Chr2: 10643681 to sgRNA2, and 2 mismatches of Extl3 genes to sgRNA3, respectively. Increasing mismatches to these sgRNAs up to 4 bases in some other genes of varying homology are shown in Fig. S1, B–D.
We then tested the cutting efficiency of the sgRNAs using a GFP reporter plasmid (pCAG-EGx-xFP-BbsI), which carries tandemly duplicated defective GFPs with a mutation in their 3′ and 5′, respectively. A dual BbsI cutting site was also introduced between two mutated GFPs for cloning of sgDNA+NGG to be tested (Fig. 1A). For each sgRNA, we cloned the mutant form and the corresponding WT form, which carries one base mismatch with the mutant form, into the tester construct (Fig. 1B) to generate 3 pairs of tester reporters. Then, each of the reporters alone or together with their corresponding CRISPR/Cas9 construct were transfected into 293 cells to test their cutting efficiency and specificity.
Figure 1.
Cutting efficiency and off-target assay for sgRNAs. A, map for pCAG-EGx-xFP-BbsI tester, a dual BbsI cutting site was flanked by a 3′-mutated GFP and a 5′-mutated GFP. sgRNA oligos plus NGG PAM sequence are inserted into the pCAG-EGxxFP-BbsI plasmid through the dual BbsI cutting site to generate the tester reporter for each single target sgRNA. B, sequence of sgRNA oligos used for targeting vector (PX330) and tester vector cloning. C, tester assay for the Fgfr3 mutant sgRNA targeting constructs. PX330-sgRNA2 and its corresponding mutant tester or WT tester are co-transfected into 293FT cells, 24 h later, the cells were examined for GFP intensity to estimate the cutting efficiency. D, quantification of GFP positive cells and its intensity related to testers construct generated by sgRNA2.
Our data indicated that transfection of these testers alone did not generate obvious GFP signaling, which is consistent with the fact that tandem duplicated GFPs are defective in the pCAG-EGx-xFP-BbsI construct. On the other hand, co-transfection of all three pairs of testers with their corresponding sgRNA-CRISPR/Cas9 (PX330-sgRNA) constructs generated GFP positive cells. This is because the mutant GFPs can be repaired due to homologous recombination stimulated by CRISPR/Cas9-mediated double strand break at the target site, generating GFP+ cells (Fig. 1C). The GFP expression level should reflect the cutting efficiency of sgRNA-guided CRISPR/Cas9 to the targeted sequence. The data indicated that sgRNA1 had the highest cutting effect for the Fgfr3 mutant sequence, however, it also cut the corresponding WT sequence (one base mismatch to the MT sequence) with high efficiency. sgRNA3 cut much more efficiently than sgRNA1, however, sgRNA3 also cut the WT sequence and generated obvious GFP positive signaling (Fig. S2, A–C). Thus, sgRNA2 had moderate efficiency for cutting the Fgfr3 mutant allele among the three sgRNAs tested, whereas no cutting at its corresponding WT allele (Fig. 1, C and D), reflecting its high specificity to the mutant allele. Because avoiding the off-target effect is most crucial, we selected sgRNA2 for our subsequent in vivo experiments.
Targeted correction of dwarf phenotype associated with Fgfr3-G374R mutation in mice
We used the mouse model of dominant achondroplasia caused by a defined Gly to Arg mutation in exon 10 of the Fgfr3 gene (21). As achondroplasia mice grow poorly throughout development and have reduced fertility, a Loxp-neo-Loxp (lnl) cassette was inserted into intron 10 to block the expression of the mutant allele (Fig. 2A). Heterozygous mice for the mutation (Fgfr3G1120A−lnl/+) were normal, whereas homozygous mice (Fgfr3G1120A-lnl/G1120A-lnl) survived to adulthood and were fertile, although exhibited skeleton abnormalities similar to Fgfr3−/− mice generated by gene targeting (22). Deletion of the lnl cassette from mutant mice by breeding with mice carrying the Ella-Cre transgene will allow the expression of Fgfr3G1120A allele, generating mice with a dwarf phenotype (Fig. 2A). Our data indicated that the cross between homozygous male Fgfr3G1120A-lnl/G1120A-lnl mice with homozygous female Ella-Cre mice could generate dwarf phenotypes in all offspring (Fgfr3G1120A/+; Ella-Cre, or Fgfr3G1120A), thus the fertilized zygotes from this breeding were collected for pronuclear injection.
Figure 2.
Targeted correction of Fgfr3-G374R mutation by CRISPR/Cas9 system. A, flow chat of the breeding procedure. Male mice homozygous for Fgfr3G1120A-ln/lG1120A-lnl, which phenotypically mimic Fgfr3−/− mice, were crossed with homozygous Ella-Cre transgenic female mice to cut off the ploxPneo gene to activate the Fgfr3 point-mutated gene. All offspring were heterozygous for the activated Fgfr3 mutation (Fgfr3G1120A/+) and exhibited achondroplasia phenotypes (an example was shown in the lower right panel). Oocytes were isolated from this cross and Cas9 protein or mRNA, sgRNA, and ssDO were injected into the pronuclear to correction the mutation. B, the sequence of mutant Fgfr3 gene. Fgfr3 gene harbors a G1120A point mutation in exon 10 causing G374R transition. G to A point mutation generated a SfcI restriction site (CTACAG) that can be identified by restriction enzyme SfcI. Primers P1 and P3 were used for SfcI cutting assay, amplicon from primers P1 and P2 were used for the Sanger sequence. C, PCR amplicon (374 bp) was cut with SfcI to identify the indel generation. Without any change, the fragment can be cut into 2 fragments of 198 and 176 bp each. 1–6, pups from injected embryos; 7, parental EIIa-Cre mouse; 8, parental Fgfr3G1120A-lnl/G1120A-lnl. D, sequence around the target from 6 pups revealed by Sanger sequencing. E, further Sanger sequence was conducted to verify the gene correction.
Cas9 mRNA or protein, sgRNA2 RNA, and single strand donor WT DNA were co-injected into the pronuclear of Fgfr3G1120A zygotes, which were then implanted into pseudopregnant female mice for further development. Of a total of 63 live pups born, 42 mice were phenotypically normal (Table 1), suggesting the mutant allele was either disrupted or corrected.
Table 1.
CRISPR/Cas9-mediated phenotype correction
| Experiment group | Cas9 mRNA/protein | sgRNA | ssDO (200 nt) | No. of embryos injected | No. of pups born | No. of normal pups | Targeted knockin |
|---|---|---|---|---|---|---|---|
| ng/μl | |||||||
| I | mRNA (200 ng/μl) | 1 | 2.5 | 184 | 13 | 8 (61.6%) | 0/8 |
| II | mRNA (5 ng/μl) | 2.5 | 5 | 113 | 15 | 0 | NAa |
| III | Protein (30 ng/μl) | 2.5 | 10 | 395 | 35 | 34 (97.1%) | 9/34 (26.5%) |
a NA, not applicable.
We injected Cas9 mRNA at two concentrations, 200 and 5 ng/μl, which generated 13 and 15 pups, respectively. Of 13 pups generated by injection of 200 ng/μl mRNA, 8 (61.6%) where morphologically normal (Table 1). In contrast, none of the 15 mice generated by injection of 5 ng/μl of mRNA were normal, despite the increased concentrations of sgRNA and single strand DNA oligo (ssDO) by 2.5- and 2-fold, respectively, to increase genomic cutting and homologous recombination at the targeting site. This observation indicates that Cas9 mRNA at 5 ng/μl was too low to achieve effective genomic editing to the Fgfr3G1120A allele. Of note, 34 of 35 (97.1%) pups generated by injection of the Cas9 protein exhibited normal phenotype and 9 of them carry precision correction of the point mutation. Collectively, these data suggest that injection of Cas9 protein has a much higher efficiency in genome editing, although more conclusive data may be provided in future studies.
High frequency of targeted correction of the Fgfr3G1120A point mutation by the injection of Cas9 protein in mice
The correction of achondroplasia phenotype suggests that the Fgfr3G1120A allele in these mice was edited (either through targeted disruption or point mutation correction). Because the G to A transition at position 1120 (leading to G374R mutation) generates a SfcI restriction site (CTACAG) (Fig. 2B), thus change at the genomic level at this mutation site can be monitored by SfcI digestion. Therefore, to provide a quick analysis of the status of the mutation site, we used a pair of PCR primers flanking the G1120A mutation site to amplify a 374-bp fragment followed by restriction digestion with SfcI, which should cut this fragment into 176 and 198 bp if the CTACAG site is intact, whereas the loss of this site should render the SfcI resistance.
Of 13 samples isolated from phenotype-corrected mice generated by the injection of Cas9 mRNA, 8 were SfcI resistant (pup 1 in Fig. 2, B and C), indicating the loss of cutting site; and 5 were sensitive (pups 2 and 3, Fig. 2, B and C), suggesting the G1120A mutation was not corrected. To understand the reason why these animals were normal while keeping the mutation, we conducted Sanger's sequencing and the data indicated that pup 1 carried a 13-bp deletion, including the SfcI site, pup 2 had a deletion of “G,” and pup 3 had an insertion of “T,” respectively, in front of the SfcI site (Fig. 2D). In all three cases, the Fgfr3G1120A allele was effectively mutated due to the frameshift caused by indels, leading to the loss of the mutant phenotype.
In 34 DNA samples isolated from injection of the Cas9 protein, 25 (73.5%) carried indels that disrupted the Fgfr3G374R allele and the animal appeared normal. In the remaining 9 mice (26.5%), the “A” was replaced by G (pups 5 and 6, Fig. 2, C–E) directed by ssDO template (Correction), all of which destroyed the SfcI site. These data indicate that injection of the Cas9 protein elicits high genome editing efficiency with a significant fraction generated by precise correction. Similar high efficiency was observed when a loxP site was introduced into the EGFP locus through homologous recombination (Fig. 3).
Figure 3.
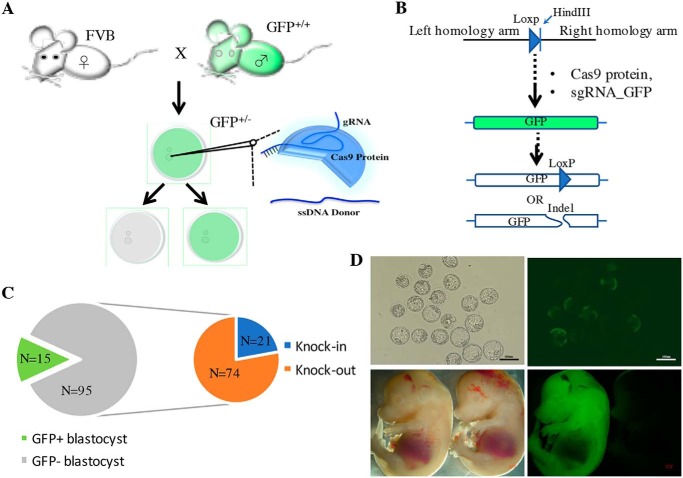
Knock-in a loxP site into GFP transgene through pronuclear injection of CRISPR/Cas9 system. A, WT FVB mice were mated with homozygous GFP male mice to get pronuclear stage GFP-positive embryos for microinjection. B, Cas9 protein, sgRNA (CAACTACAAGACCCGCGCCG) targeting GFP gene, and a single strand donor DNA, which contains a loxp sequence and HindIII sequence flanked by two 100-nt homology arms for both sides were co-injected. C, after injection, we have checked 110 blastocysts for the fluoresce and knock-in event by using HindIII restriction enzyme to cut the insertion site after PCR amplification. There are 95 (86.4%) embryos that lost the GFP signal, whereas the remaining 15 (13.6%) still expressed the GFP gene. Among 95 GFP negative blastocysts, there are only 21 blastocyst containing a HindIII cutting site that indicates the ssDNA donor gets knocked-in successfully. D, represent picture of GFP negative blastocysts and fetus at embryonic day 14.
Targeting single strand DNA oligos of 100 and 200 nt corrected Fgfr3G1120A mutation at equal efficiency in the achondroplasia mice
The correction of the point mutation is primarily mediated by targeting ssDOs, which provide a template for repairing the double strand break created by the cutting of CRISPR/Cas9. The length of the ssDO used in the above experiment is 200 nt. Next, we investigated whether supplying a shorter ssDO would still maintain comparable efficiency of HDR-mediated precise genome repair. Our data indicated that the injection of a ssDO of 101 nt yielded a phenotype correction efficiency of 97.5% (40/41), which is comparable with 97.1% (34/35) obtained by using the ssDO of 200 nt. DNA sequencing revealed that 31 (77.5%) mice carried indels, whereas the remaining 9 (22.5%) mice had their G to A mutation precisely corrected (Table 2).
Table 2.
Comparison of ssDO length for CRISPR/Cas9-mediated genomic editing
| ssDO | No. of embryos injected | No. of pups born | No. of normal pups | No. of pups (with indels) | No. of pups (correction) | |
|---|---|---|---|---|---|---|
| 200 nt | 395 | 35 | 34 | 25 | 9 (26.5%) | p = 0.86 |
| 101 nt | 371 | 41 | 40 | 31 | 9 (22.5%) |
Taken together, our data suggest that the Cas9 protein have higher efficiency to promote HDR-based genetic repairing, which was directed by exogenous oligos. 101 nt homologous ssDO keep the same efficiency for HDR compared with 200-nt ssDO.
CRISPR/Cas9-mediated correction or disruption of Fgfr3G374R mutation completely suppresses phenotypes of achondroplasia mice
The most significant features of achondroplasia mice include reduced body weight, dome-shaped head, short status, reduced lengths of long bone, most promptly in femurs and humerus (21) (Fig. 4A). We carefully examined all 18 mice that carried the corrected point mutation allele using these features of achondroplasia, as measured by bone length and body weight, and the data indicated that all these were absent in the phenotype-corrected mice and they were indistinguishable with WT mice (Fig. 4, A–D). One major concern of the achondroplasia therapy is whether the dwarf phenotype can be inherited to the pups. We therefore mated them with WT mice and then analyzed the pups. Progeny obtained from corrected Fgfr3 point mutation mice were all normal. DNA sequencing using primer against the mutant allele showed that the pups carried the repaired Fgfr3 allele originating from their parents, indicating that the corrected allele could be successfully transmitted to the next generation through the germline.
Figure 4.
Observation of gene corrected and mutant mice. A, whole mount skeleton staining of Fgfr3G1120A/+ dwarf (left) and corrected mice (right) at 1 month of age. B, X-ray images and its statistical analysis (C) showing the length of femur of dwarf and corrected mice. D, comparison of body weight between dwarf mice, corrected mice, and knockout mice.
The remaining 56 (25 from using of 200 nt ssDO and 31 from using 101-nt ssDO) mice that carry non-homologous end joining-mediated insertions or deletions were also phenotypically normal. Analysis of the nucleotide sequences of the Fgfr3 locus revealed that the mutant Fgfr3G374R allele was disrupted by indels at or near the G1120A mutation site in all these mice, whereas the remaining WT Fgfr3 allele is intact. This is consistent with our previous observation that mice heterozygous for the Fgfr3 knockout mutation is phenotypically normal (22).
Correction of G1120A mutation without off-targeting events
The CRISPR/Cas9 system sometimes suffers an off-target effect by cutting at sites with shared homology (15–17). To investigate this, we conducted whole-genome DNA sequencing for 7 mice delivered by one surrogate mother, including 3 mice with targeted mutation correction, 3 mice with indels (these 6 mice are phenotypically normal), and 1 mouse with un-corrected G1120A mutation (achondroplasia phenotype) revealed by SfcI digestion. As shown in Fig. 5, A and B, #1083 still contains the G to A mutation, whereas showing the dwarf phenotype. #1081, #1084, and #1089 only contained WT allele, suggesting the G1120A mutation was corrected. The remaining 3 mice, #1082, #1085, and #1086, exhibited mixed ratios of WT allele and indels. In the #1082 and #1086 mice, the WT allele was presented at 60 and 42%, respectively, which is considered the normal range of heterozygosity. Meanwhile, the #1082 mouse also contained a deletion of 17 bp and an insertion of a T, presented at 16 and 23%, respectively, suggesting this animal carries these two types of mutations in different portions of cells, presumably due to two distinct indels at 2- or 4-cell staged embryos. A similar situation was observed in #1086, which also carried insertion of a C and a T in different cells besides the WT allele. Mouse #1085 contained WT allele at 81.4% and an allele of 27 bp deletion at 18.6%, suggesting a precise correction of G1120A, and a deletion of 27 bp occurred in different cells at early 2-cell or 4-cell staged embryos. All these events (precision correction and indels) are mediated by CRISPR/Cas9 and effectively disrupted the Fgfr3G1120A mutation, resulting in the correction of achondroplasia phenotype of the mutant mice.
Figure 5.
Off-target analysis of sgRNA2 based on whole genome sequence. A, summary of genomic editing at the Fgfr3-G1120A site of 7 mice. #1083 still contains the Fgfr3-G1120A allele at about 40% suggesting no editing in the target region. #1081, #1084, and #1089 mice only contain WT allele, suggesting their Fgfr3-G1120A mutation was corrected. #1082 and #1086 mice contain WT allele with 60 and 42%, respectively, and also contain two different types of indels at lower percentages, suggesting these indels occurred at 2- or 4-cell staged embryos. #1085 mouse contain a WT allele at 81.4% and an allele of 27 bp deletion at 18.6%, suggesting the G1120A mutation may be corrected in some cells. B, types of mutations occurred in the Fgfr3-G1120A region of these 7 mice, including deletions of 17 or 27 bp, and two insertions of single nucleotides near the Fgfr3-G1120A mutation site, which effectively disrupted expression of the mutant allele.
Next, we examined the entire Fgfr3 locus spanning 15.4 kb and detected no alterations. We then studied all homologous loci (Fig. 1C) for sgRNA2 with 1–4 mismatched nucleotides and detected no indels generated. These analyses indicate that the G1120A mutation was corrected specifically without obvious off-targeting events.
Discussion
CRISPR/Cas9 is a powerful tool for genome editing that has been used in multiple organisms (1, 2, 4). However, CRISPR/Cas9, at its current stands, also suffers some limitations. For example, the CRISPR/Cas9 system frequently suffers an off-target effect by cutting at sites with shared homology (15–17). Furthermore, due to its high efficiency cutting at targeting sites, CRISPR/Cas9 preferably generates more indels than precise modifications, casting difficulty to generate the desired mutations. Using Fgfr3G374R achondroplasia mice as a model system, we have addressed some of the potential problems. Our optimized experiment conditions have achieved high specificity of targeted correction of achondroplasia with several navel features.
Unintended binding, modification, and cleavage of nucleic acids, so called the off-target effect, is a major challenge to the CRISPR/Cas9 system (3). To overcome this problem, many sgRNA designing tools have been developed to increase specificity of sgRNAs at the silicon level (23, 24). A number of methods have also been employed to avoid off-target cutting or identify undesired mutations, such as using single-based editors correct base-pairing to avoid inducing a dsDNA break, or replace Cas9 with Cas13a to target RNA editing (3). In our study, the off-target effect still occurred even if CRISPR design software was used to design our sgRNAs. Because of the newly designed GFP tester, we found two of three sgRNAs could cut the WT Fgfr3 sequence with a single mismatched base, whereas the other sgRNA cuts only at the MT Fgfr3 sequence. We used this sgRNA for further genome editing in mice and it indeed did not cut the corresponding WT Fgfr3 sequence with single mismatched base.
More recently, there are some discussions about whether or not the CRISPR/Cas9 system could generate unexpected mutations in vivo (25–29). To investigate this, we conducted whole genome sequencing, and detected no off-target events in the region near the targeting site, the entire Fgfr3 locus, and in all predicted potential off-target sites, which share varying degrees of homology with the target sequence. Altogether, we demonstrated that by using our optimized sgRNA, the CRISPR/Cas9-mediated targeted correction of the Fgfr3-G374R point mutation completely suppresses phenotypes of achondroplasia mice with high fidelity. This result demonstrates in principle the importance of using such a tester system to avoid potential off-target effects prior to the experiment. For example, if sgRNA 1 or 3 was used in our experiment, it might have generated off-target events. The GFP tester is user friendly, and reliable for providing functional evaluation of sgRNAs for their specificity and efficiency. From the long run, highly suspected sequences for off-target predicted by software could also be tested first to avoid any potential problems.
Both Cas9 mRNA and proteins have been widely used for achieving genome editing with variable frequencies (30, 31). Our data revealed significant higher frequency of CRRSPR/Cas9-mediated genome cutting at the target site and the precise correction of the G1120A point mutation by the Cas9 protein than Cas9 mRNA. For the underlying mechanism, we believe that there will be a gap between the time of injection of mRNA and producing the effective amount of protein by translation. However, this gap no longer exists by direct admission of Cas9 protein, as the protein can be immediately used for genome editing. We have also tested the knock-in efficiency by Cas9 protein to EGFP genes, and a comparable efficiency was achieved by introducing a 47-base loxP site into the EGFP gene. In conclusion, Cas9 protein achieves much higher efficiency than Cas9 mRNA of genome editing and introduction of knock-in mutation.
Conventional gene targeting by homologous recombination has high demand on the length of targeting homology to achieve ideal targeting efficiency (32). Because the CRISPR/Cas9 system generates double strand breaks at the target sites that greatly stimulates efficiency of homologous recombination, the long homologous arm of targeting constructs is less critical (32). Donor oligo or targeting constructs with less than 1000-bp homology arms are frequently used for genome editing in mouse zygotes (33). In our study, we first tested targeting ssDO with 200 nt (with around 100 nt on each side of the G to A mutation) and obtained genome editing frequency in 34/35 (97%) mice. In the 34 mice, 9 carry precise G1120A mutation correction (26.5%). To further test the effect of the length of homology, we injected a 101-nt ssDO and obtained comparable editing frequency and mutation correction rate. These data demonstrate that the targeting oligos of 101 and 200 nt precisely corrected the mutation at similar efficiency. As oligos of 101 nt can be generated much more economically, our finding is of great significance when designing targeting ssODs.
In summary, we have demonstrated that the CRISPR/Cas9 system can be used to cure achondroplasia in mouse by directly correcting the genetic defect through homology-mediated gene editing. Our GFP reporter can be used to select sgRNAs with high specificity and reasonable cutting efficiency to avoid off-target effects. Microinjection of Cas9 protein greatly improves genome editing frequency. Targeting oligo of 101 nt, which is more easily affordable than longer oligos, can be reliably used for the editing. The combination of these factors greatly facilitates the introduction of the desired genome modification with high fidelity.
Experimental procedures
Animal model
As previously described to bypass the lethal effect of Fgfr3G1120A mutation, a lnl cassette is inserted between exons 10 and 11 to knockout the mutant Fgfr3 gene (21). Mice harboring homozygous Fgfr3G1120A-lnl/G1120A-lnl exhibit phenotypes similar to Fgfr3−/− mice (22) and are maintained to serve as stud mice. Homozygote of EIIa-Cre (JAX, ME) female mice were superovulated by injection with pregnant mare's serum gonadotropin followed by human chorionic gonadotropin, and then crossed with Fgfr3G1120A-lnl/G1120A-lnl stud mice to delete the lnl cassette and obtain Fgfr3 point mutant embryos for pronuclear injection. All animal procedures were performed under the ethical guidelines of the University of Macau (animal protocol number: UMAEC-037-2015).
sgRNA reporter plasmid construction
sgRNA reporter plasmid (pCAG-EGx-xFP-BbsI) was constructed by modifying pCAG-EGxxFP (Addgene). Briefly, a short dual BbsI cassette was amplified from plasmid PX330 (1) and inserted into the pCAG-EGxxFP digested with EcoRI to obtain pCAG-EGxxFP-BbsI (Fig. 1A). sgRNA tester sequence was designed based on the sgRNA sequence including the NGG PAM sequence, then the adapter was added for further cloning. To clone the target sequence into the reporter plasmid, paired oligos with adaptor were synthesized (Fig. 1A). Each pair of oligos was phosphorylated and annealed followed by ligation with a BbsI pre-digested reporter plasmid.
Microinjection
sgRNA(s) were designed by using Optimized CRISPR Design online tools (34), then ligation with PX330 plasmid. sgRNAs with a T7 promoter were amplified by PCR and in vitro transcribed using a MEGAshortscript T7 kit (Thermo Fisher Scientific, MA). After transcription, the sgRNAs were purified with a MEGAclear kit (Thermo Fisher Scientific, MA) according to the manufacturer's instructions. The sequences used for preparation of template for in vitro transcription of sgRNA2 was amplified from PX330-sgRNA2 using primers (TTAATACGACTCACTATAGGCGCAGGCGTCCTCAGCTAC and AAAAGCACCGACTCGGTGCC).
Two single strand donor DNA oligo (ssDO) of the total length of 200 and 101 nt were designed based on WT Fgfr3 genomic DNA. The ssDO-200 nt contains 99 and 100 nt at 5′ and 3′ to the G to A point mutation, respectively; and the ssODN-101 nt contains 50 nt on each side flanking the G to A point mutation as shown; Wt-200nt, AAGGCTGGATGAGGCCCCAAAATTTGTATCTTTGCAGCTGAGGAGGAGCTGATGGAAACTGATGAGGCTGGCAGCGTGTACGCAGGCGTCCTCAGCTACGGGGTGGTCTTCTTCCTCTTCATCCTGGTGGTGGCAGCTGTGATACTCTGCCGCCTGCGCAGTCCCCCAAAGAAGGGCTTGGGCTCGCCCACCGTGCACAA; and Wt-101nt: TGATGGAAACTGATGAGGCTGGCAGCGTGTACGCAGGCGTCCTCAGCTACGGGGTGGTCTTCTTCCTCTTCATCCTGGTGGTGGCAGCTGTGATACTCTGC.
EIIa-Cre female mice were superovulated and mated with Fgfr3G1120A-lnl/G1120A-lnl males. Pronuclear stage embryos were collected followed by microinjection punched by FemtoJet device (Eppendorf). Cas9 mRNA or proteins were co-injected with sgRNA and ssDO. For Cas9 mRNA injection, different concentrations of Cas9 mRNAs (5 or 200 ng/μl) and sgRNA (2.5 or 1 ng/μl) were mixed with ssDO (5 or 2.5 ng/μl) injected into the embryos. The injected embryos were transferred into pseudopregnant female mice.
Genotyping, DNA sequencing analysis, and Sanger sequencing
The target site (sequence around Fgfr3G1120A mutation site) was amplified from genomic DNA of the genetic modified mice by PCR with primers (Fgfr3-s and Fgfr3-a) for SfcI digestion diagnosis, and flanking primers (Fgfr3-s and Loxp-a) for Sanger sequence; Fgfr3-s (P1), CTCTTCTCCAAGTATCCCAGGTCC; Fgfr3-a (P2), CCTGCTGGGACTCTAGGAGACAC; and Loxp-a (P3), CGAAGTTATCTAGAGTCGACCATCG.
Whole genome sequencing
Total DNA was extracted from mouse tail using DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany). Whole genome sequencing libraries were prepared using the DNA Library Prep kit (New England Biolabs) in accordance with the manufacturer's instructions. Briefly, 1 μg of DNA was sheared using a Covarias sonicator, then followed by end-repair, ligation, and amplification. Whole genome sequencing libraries were evaluated using BioAnalyzer (Agilent, CA) and quantitative PCR. Libraries were sequenced on an Illumina Hiseq ×10 sequencer using 2 × 150 bp cycles to meet coverage of 30 times for further analysis.
Bioinformatics analyses
Sequenced reads were mapped to the mouse (Mus musculus) genome (mm10) using Burrows-Wheeler Aligner (version 0.7.15). Unique mapped reads were used for the further analysis. GATK best practices (version 4.0.3) workflows were used to identify SNV(s) and Indel(s). Variants were further filtered to exclude variants in dbSNPv150. Variant annotations were done via SnpEff (version 4.3T) The top potential off-target sites were predicted by Optimized CRISPR Design online tools (34).
X-ray and whole mount skeletal preparation
For X-ray imaging, animals were euthanatized with CO2. After removing the skin, photographs were taken by using an X-ray machine (Bruker, MA). For skeletal preparation, the carcasses were eviscerated, fixed in 95% ethanol, stained with Alizarin red S and Alcian blue, cleared by KOH treatment, and then stored in glycerol as previously described (22).
Author contributions
K. M., X. Z., J. Z., X. X., and C.-X. D. resources; K. M., X. Z., J. Z., X. X., and C.-X. D. data curation; K. M., X. Z., and J. Z. software; K. M., X. X., and C.-X. D. funding acquisition; K. M., X. Z., and S. M. S. validation; K. M., X. Z., S. M. S., Z. H., U. I. C., X. X., and C.-X. D. investigation; K. M., X. Z., and S. M. S. visualization; K. M., X. Z., S. M. S., J. Z., Z. H., U. I.C., X. X., and C.-X. D. methodology; K. M., X. X., and C.-X. D. writing-original draft; K. M. and C.-X. D. writing-review and editing; X. Z., J. Z., X. X., and C.-X. D. formal analysis; X. X. and C.-X. D. conceptualization; X. X. and C.-X. D. supervision; X. X. and C.-X. D. project administration.
Supplementary Material
Acknowledgments
We thank members of the C. Deng and X. Xu laboratories for helpful advice and discussion.
This work was supported by a Chair Professor Grant (to C. D.) and multi-year research Grant (MYRG) 2016-00088-FHS (to X. X.) by the University of Macau, Macau SAR, China, Macao Science and Technology Development Fund (FDCT) Grants 065/2015/A2, 094/2015/A3 (to C. D.), 027/2015/A1 (to X. X.), and 111/2017/A (to K. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- sgRNA
- single guide RNA
- HDR
- homology-directed repair
- nt
- nucleotide(s)
- PAM
- protospacer adjacent motif
- lnl
- Loxp-neo-Loxp
- oligo
- oligonucleotide
- ssDO
- single strand donor DNA oligo
- EGFP
- enhanced green fluorescent protein.
References
- 1. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., and Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knott G. J., and Doudna J. A. (2018) CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S., Sun H., Miao K., and Deng C. X. (2016) CRISPR-Cas9: from genome editing to cancer res. Int. J. Biol. Sci. 12, 1427–1436 10.7150/ijbs.17421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luther D. C., Lee Y. W., Nagaraj H., Scaletti F., and Rotello V. M. (2018) Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin. Drug Deliv. 15, 905–913 10.1080/17425247.2018.1517746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen S., Sanjana N. E., Zheng K., Shalem O., Lee K., Shi X., Scott D. A., Song J., Pan J. Q., Weissleder R., Lee H., Zhang F., and Sharp P. A. (2015) Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 10.1016/j.cell.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., and Li J. (2013) Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13, 659–662 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 8. Yin H., Xue W., Chen S., Bogorad R. L., Benedetti E., Grompe M., Koteliansky V., Sharp P. A., Jacks T., and Anderson D. G. (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 10.1038/nbt.2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ou Z., Niu X., He W., Chen Y., Song B., Xian Y., Fan D., Tang D., and Sun X. (2016) The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Sci. Rep. 6, 32463 10.1038/srep32463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tabebordbar M., Zhu K., Cheng J. K. W., Chew W. L., Widrick J. J., Yan W. X., Maesner C., Wu E. Y., Xiao R., Ran F. A., Cong L., Zhang F., Vandenberghe L. H., Church G. M., and Wagers A. J. (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411 10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nelson C. E., Hakim C. H., Ousterout D. G., Thakore P. I., Moreb E. A., Castellanos Rivera R. M., Madhavan S., Pan X., Ran F. A., Yan W. X., Asokan A., Zhang F., Duan D., and Gersbach C. A. (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Long C., Amoasii L., Mireault A. A., McAnally J. R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J. M., Bassel-Duby R., and Olson E. N. (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 10.1126/science.aad5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K., Batshaw M. L., and Wilson J. M. (2016) A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 34, 334–338 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin H., Song C. Q., Dorkin J. R., Zhu L. J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A., Gupta A., Bolukbasi M. F., Walsh S., Bogorad R. L., Gao G., Weng Z., Dong Y., Koteliansky V., Wolfe S. A., Langer R., Xue W., and Anderson D. G. (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333 10.1038/nbt.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin F., Sánchez-Hernandez S., Gutiérrez-Guerrero A., Pinedo-Gomez J., and Benabdellah K. (2016) Biased and unbiased methods for the detection of off-target cleavage by CRISPR/Cas9: an overview. Int. J. Mol. Sci. 17, e1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rastogi A., Murik O., Bowler C., and Tirichine L. (2016) PhytoCRISP-Ex: a web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing. BMC Bioinformatics 17, 261 10.1186/s12859-016-1143-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J. B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J. S., and Concordet J. P. (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ornitz D. M., and Legeai-Mallet L. (2017) Achondroplasia: development, pathogenesis, and therapy. Dev. Dyn. 246, 291–309 10.1002/dvdy.24479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bellus G. A., Hefferon T. W., Ortiz de Luna R. I., Hecht J. T., Horton W. A., Machado M., Kaitila I., McIntosh I., and Francomano C. A. (1995) Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 56, 368–373 [PMC free article] [PubMed] [Google Scholar]
- 20. Stoilov I., Kilpatrick M. W., and Tsipouras P. (1995) A common FGFR3 gene mutation is present in achondroplasia but not in hypochondroplasia. Am. J. Med. Genet. 55, 127–133 10.1002/ajmg.1320550132 [DOI] [PubMed] [Google Scholar]
- 21. Wang J. M., Du. X. L., Li C. L., Yin L. J., Chen B., Sun J., Su N., Zhao L., Song R. H., Song W. W., Chen L., and Deng C. X. (2004) Gly374Arg mutation in Fgfr3 causes achondroplasia in mice. Chinese J. Med. Genet. 21, 537 [PubMed] [Google Scholar]
- 22. Deng C., Wynshaw-Boris A., Zhou F., Kuo A., and Leder P. (1996) Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84, 911–921 10.1016/S0092-8674(00)81069-7 [DOI] [PubMed] [Google Scholar]
- 23. Cui Y., Xu J., Cheng M., Liao X., and Peng S. (2018) Review of CRISPR/Cas9 sgRNA design tools. Interdiscip. Sci. 10, 455–465 10.1007/s12539-018-0298-z [DOI] [PubMed] [Google Scholar]
- 24. Listgarten J., Weinstein M., Kleinstiver B. P., Sousa A. A., Joung J. K., Crawford J., Gao K., Hoang L., Elibol M., Doench J. G., and Fusi N. (2018) Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng. 2, 38–47 10.1038/s41551-017-0178-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lareau C. A., Clement K., Hsu J. Y., Pattanayak V., Joung J. K., Aryee M. J., and Pinello L. (2018) Response to “unexpected mutations after CRISPR-Cas9 editing in vivo.” Nat. Methods 15, 238–239 10.1038/nmeth.4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nutter L. M. J., Heaney J. D., Lloyd K. C. K., Murray S. A., Seavitt J. R., Skarnes W. C., Teboul L., Brown S. D. M., and Moore M. (2018) Response to “unexpected mutations after CRISPR-Cas9 editing in vivo.” Nat. Methods 15, 235–236 10.1038/nmeth.4559 [DOI] [PubMed] [Google Scholar]
- 27. Kim S. T., Park J., Kim D., Kim K., Bae S., Schlesner M., and Kim J. S. (2018) Response to “unexpected mutations after CRISPR-Cas9 editing in vivo.” Nat. Methods 15, 239–240 10.1038/nmeth.4554 [DOI] [PubMed] [Google Scholar]
- 28. Wilson C. J., Fennell T., Bothmer A., Maeder M. L., Reyon D., Cotta-Ramusino C., Fernandez C. A., Marco E., Barrera L. A., Jayaram H., Albright C. F., Cox G. F., Church G. M., and Myer V. E. (2018) Response to “unexpected mutations after CRISPR-Cas9 editing in vivo.” Nat. Methods 15, 236–237 10.1038/nmeth.4552 [DOI] [PubMed] [Google Scholar]
- 29. Lescarbeau R. M., Murray B., Barnes T. M., and Bermingham N. (2018) Response to “unexpected mutations after CRISPR-Cas9 editing in vivo.” Nat. Methods 15, 237 10.1038/nmeth.4553 [DOI] [PubMed] [Google Scholar]
- 30. Chu V. T., Weber T., Graf R., Sommermann T., Petsch K., Sack U., Volchkov P., Rajewsky K., and Kühn R. (2016) Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16, 4 10.1186/s12896-016-0234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L., Shao Y., Guan Y., Li L., Wu L., Chen F., Liu M., Chen H., Ma Y., Ma X., Liu M., and Li D. (2015) Large genomic fragment deletion and functional gene cassette knock-in via Cas9 protein mediated genome editing in one-cell rodent embryos. Sci. Rep. 5, 17517 10.1038/srep17517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng C., and Capecchi M. R. (1992) Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 12, 3365–3371 10.1128/MCB.12.8.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yao X., Zhang M., Wang X., Ying W., Hu X., Dai P., Meng F., Shi L., Sun Y., Yao N., Zhong W., Li Y., Wu K. L., Li W. P., Chen Z., and Yang H. (2018) Tild-CRISPR allows for efficient and precise gene knockin in mouse and human cells. Dev. Cell 45, 526–536 10.1016/j.devcel.2018.04.021 [DOI] [PubMed] [Google Scholar]
- 34. Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O., Cradick T. J., Marraffini L. A., Bao G., and Zhang F. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.