Abstract
The human macrophage galactose-type lectin (MGL) is a C-type lectin characterized by a unique specificity for terminal GalNAc residues present in the tumor-associated Tn antigen (αGalNAc-Ser/Thr) and its sialylated form, the sialyl-Tn antigen. However, human MGL has multiple splice variants, and whether these variants have distinct ligand-binding properties is unknown. Here, using glycan microarrays, we compared the binding properties of the short MGL 6C (MGLshort) and the long MGL 6B (MGLlong) splice variants, as well as of a histidine-to-threonine mutant (MGLshort H259T). Although the MGLshort and MGLlong variants displayed similar binding properties on the glycan array, the MGLshort H259T mutant failed to interact with the sialyl-Tn epitope. As the MGLshort H259T variant could still bind a single GalNAc monosaccharide on this array, we next investigated its binding characteristics to Tn-containing glycopeptides derived from the MGL ligands mucin 1 (MUC1), MUC2, and CD45. Strikingly, in the glycopeptide microarray, the MGLshort H259T variant lost high-affinity binding toward Tn-containing glycopeptides, especially at low probing concentrations. Moreover, MGLshort H259T was unable to recognize cancer-associated Tn epitopes on tumor cell lines. Molecular dynamics simulations indicated that in WT MGLshort, His259 mediates H bonds directly or engages the Tn-glycopeptide backbone through water molecules. These bonds were lost in MGLshort H259T, thus explaining its lower binding affinity. Together, our results suggest that MGL not only connects to the Tn carbohydrate epitope, but also engages the underlying peptide via a secondary binding pocket within the MGL carbohydrate recognition domain containing the His259 residue.
Keywords: molecular modeling, microarray, peptide array, alternative splicing, mutant, C-type lectin, carbohydrate specificity, MGL, Tn antigen, splice variant, glycopeptide, glycan, carbohydrate recognition domain
Introduction
The C-type lectin family is a large family of soluble and transmembrane proteins that share the specific recognition of carbohydrate structures, which in most cases is Ca2+-dependent (1). Carbohydrate binding is facilitated via a common carbohydrate recognition domain (CRD).6 The primary binding site within this CRD is composed of a conserved three-amino acid motif, which largely determines the class of glycans the lectin is able to engage (e.g. the Gln-Pro-Asp (QPD) motif predicts binding to galactose or GalNAc) (2). In addition, secondary binding sites fine-tune the carbohydrate recognition, thereby creating enormous diversity in C-type lectin specificity and function.
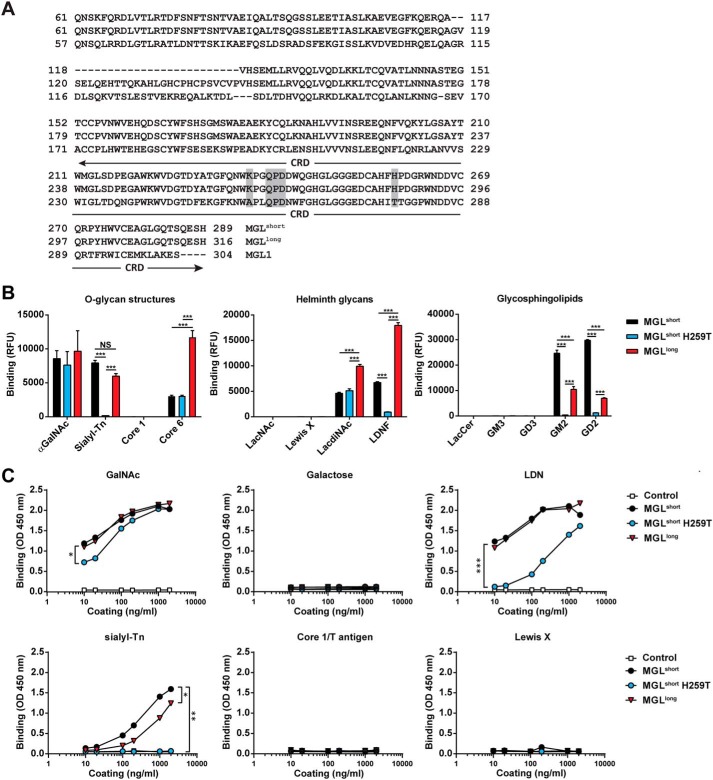
The human QPD-containing macrophage galactose-type lectin (MGL; CLEC10A or CD301) is exclusively expressed by macrophages and dendritic cells within the immune system (3). Triggering of MGL on these human dendritic cells conveys immune inhibitory signals, leading to production of the anti-inflammatory cytokines and the ability to block unwanted inflammatory responses (4–6). Humans harbor only one MGL gene, which is subject to extensive splicing, generating different long and short MGL isoforms. Alternative splicing mainly occurs around exon 6, encoding the terminal stalk region (7). The most abundantly expressed MGL subtype in dendritic cells is the short MGL 6C splice variant (Fig. 1A, from here on denoted as MGLshort) (7). Another relatively abundant variant is the MGL 6B variant, which compared with the MGLshort contains an additional 27 amino acids in its stalk region (Fig. 1A, MGLlong).
Figure 1.
Differential GalNAc binding of MGL variants. A, amino acid sequence of the human MGL splice variants and the mouse MGL1. The stalk region and CRD are depicted, showing the 27-amino acid insertion in the stalk region of the MGLlong variant. Boxed in gray are the primary binding site (QPD) of MGL and the localization of the K237A and H259T mutations. B, binding of MGL variants determined by glycan array. Depicted are the binding to selected O-glycan core structures (left), Helminth-associated glycans (middle), and glycosphingolipids (right). Full glycan array results can be found in Table S1. Significance was calculated using two-way analysis of variance and Bonferroni's multiple-comparison test. Data represent mean ± S.E. LacCer, lactosylceramide. C, binding of the MGL variants to PAA-coated glycoconjugates was determined using an ELISA-based assay. Human IgG1 was used as a negative control. One representative experiment of three is shown. Significance was calculated by nonlinear regression followed by one-way analysis of variance and Bonferroni's multiple-comparison test. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
The carbohydrate specificity of the MGLshort variant has been widely studied through surface plasmon resonance, glycan microarrays, and cellular binding assays, revealing its exclusive specificity for terminal GalNAc moieties, such as the Tn antigen (αGalNAc-Ser/Thr), the LacdiNAc structure (LDN; GalNAcβ1–4GlcNAc), and the GalNAc-Tyr moiety (8–12). In QPD-containing C-type lectins, the equatorial/axial configuration of the 3-OH and 4-OH groups in the GalNAc are crucial for glycan binding and chelation of the Ca2+ ion (13). Indeed, from glycan microarray analysis, GalNAc structures extended at the 3- or 4-OH position fail to bind MGL (8). It is noteworthy that MGL tolerates substitutions at the 6-OH position and therefore recognizes the sialylated Tn antigen (Neu5Acα2–6GalNAcα-Ser/Thr) (10, 12).
Mice contain two functional copies of the MGL gene, termed MGL1 (CD301a) and MGL2 (CD301b) (14). Despite their high homology and the shared QPD motif, MGL1 and MGL2 differ substantially in their carbohydrate specificities. MGL1 mainly interacts with Lewis X (Galβ1–4(Fucα1–3)GlcNAc) and Lewis A (Galβ1–3(Fucα1–4)GlcNAc) (15), whereas MGL2 recognizes terminal α-/β-GalNAc ((sialyl)Tn antigen, LDN), as well as the terminal galactose in the core 1/T antigen and core 2 O-glycan structures (15). These disparities in ligand recognition have been further elucidated by Sakakura et al. (16), showing that whereas the galactose moiety of Lewis X is engaged by the QPD motif in the MGL1 CRD, the fucose residue is bound by a secondary binding site composed of Ala89 and Thr111 (corresponding to Ala256 and Thr278 in the full-length MGL1; Fig. 1A). These amino acids are replaced by arginine and serine in MGL2, respectively. Although no crystal structure of human MGL is available, glycan accommodation by MGLlong has been studied by saturation transfer-difference NMR measurements and molecular modeling, revealing fine-structural insights into the interaction of Gal/GalNAc and MUC1-Tn-glycopeptides with MGL (17). These NMR binding studies have verified that galactose is a much weaker ligand than GalNAc. The NHAc group of GalNAc participates in additional H-bond and CH-π interactions, which are absent in the case of galactose. Moreover, an increased number of H bonds were formed when MGL engaged a Tn-glycopeptide; however, this was not reflected by an improved affinity, probably due to the subtle enthalpy–entropy equilibrium of any ligand–protein recognition event.
Our knowledge on MGL has substantially increased over the last decade; nevertheless, the carbohydrate fine specificities and ligand-binding properties of the different human MGL splice variants have never been directly compared and have been assumed to be identical. Here, we aimed to elucidate the molecular recognition features of human MGLshort and MGLlong isoforms by evaluating their carbohydrate specificities using glycan and Tn-peptide microarrays. In addition, we mutated the His259 in the CRD of human MGL, equivalent to Thr111 in MGL1, to Thr in the MGLshort variant (H259T) to elucidate whether this amino acid is also crucial for the exclusive GalNAc-binding properties of human MGL.
Results
Carbohydrate recognition profiles of MGL variants
The MGL 6B (MGLlong) and 6C (MGLshort) splice variants vary in a region of 27 amino acids present in the terminal stalk domain of MGLlong only (Fig. 1A). We constructed recombinant MGL-Fc proteins, consisting of the extracellular domains of MGLlong and MGLshort fused to the human IgG1-Fc domain for detection. Previously, Ala89 and Thr111 (Ala256 and Thr278 in the full-length MGL1) were shown to be crucial for the Lewis X preference of murine MGL1 (16). Based on these findings and our previous work using the MGLshort splice variant (8), we mutated human MGLshort at the corresponding amino acids (H259T and K237A) to elucidate whether these amino acids also contribute to the fine specificity of human MGL (Fig. 1A). In our initial screens, the MGLshort K237A/H259T double mutant showed identical binding characteristics as the MGLshort single H259T mutant (data not shown); therefore, we omitted the K237A/H259T mutant from further analyses. All MGL-Fc proteins displayed equal binding to three independent anti-MGL antibodies (Fig. S1B), confirming the correct folding and indicating that the H259T mutation did not induce significant structural changes.
We next screened the glycan array developed by the Consortium for Functional Glycomics (http://www.functionalglycomics.org),7 containing 610 glycans of natural and synthetic origin. As reported previously (8), MGLshort recognized the O-glycans αGalNAc, sialyl-Tn, and core 6 (GlcNAcβ1–6GalNAcα); the helminth glycans LDN and its fucosylated derivate (LDNF; GalNAcβ1–4(Fucα1–3)GlcNAc); and the glycosphingolipid structures GM2 (GalNAcβ1–4(Neu5Acα2–3)Galβ1–4Glc) and GD2 (GalNAcβ1–4(Neu5Acα2–8Neu5Acα2–3)Galβ1–4Glc) (Fig. 1B; full results in Table S1). Although differences were subtle, MGLlong displayed significantly higher binding to core 6, LDN, and LDNF than MGLshort and significantly lower binding to GM2 and GD2, as deduced by glycan microarrays. In contrast, the MGLshort H259T failed to interact with sialyl-Tn, LDNF, GM2, or GD2 on the array, whereas αGalNAc, the core 6 structure, and LDN were still bound (Fig. 1B). None of the MGL variants interacted with the core 1/T antigen, LacNAc, Lewis X, lactosylceramide, or GM3 structures on this array.
Next, we performed ELISA-based binding assays using polyacrylamide (PAA)-coupled glycoconjugates. All MGL variants, including the mutant, strongly bound the PAA-coupled GalNAc (Fig. 1C). Recognition of PAA-coupled sialyl-Tn and LDN by MGLshort and MGLlong was comparable, whereas MGLshort H259T showed diminished binding to both. Again, no binding could be detected to galactose, core 1/T antigen, or Lewis X. In contrast to the glycan microarray, no differential binding between MGLshort and MGLlong to the LDN moiety could be observed. The discrepancy with glycan microarrays might be explained by the multivalent nature of the PAA-glycoconjugate (20–25 glycans/molecule) versus the monovalent presentation on the array.
Differential glycan binding of the MGLshort H259T mutant
Overall, the carbohydrate recognition profiles of MGLshort and MGLlong appeared to be quite similar; therefore, we focused our further investigations on the differential recognition of MGLshort and the MGLshort H259T mutant. Although the Thr111 amino acid (corresponding to His259 in human MGL) has been implicated in the exclusive Lewis X specificity of mouse MGL1 (16), neither the MGLshort H259T nor the K237A/H259T double mutant recognized Lewis X on the glycan array or in the ELISA-based assay, indicating that these two amino acids are not sufficient to convey Lewis X recognition. Nevertheless, the His259 seems essential for binding extended GalNAc structures, such as the sialyl-Tn and glycosphingolipids, and might therefore be a critical component of a secondary binding site in the MGL CRD.
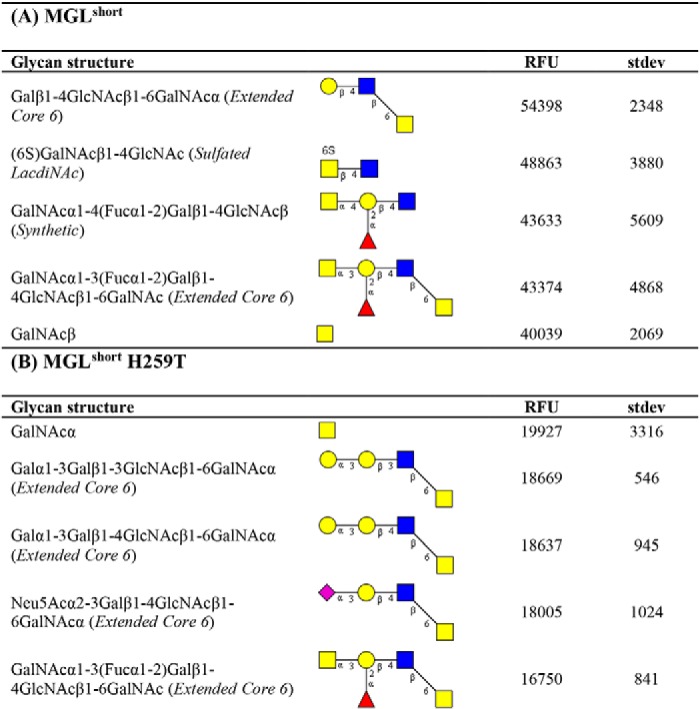
To further extract carbohydrate determinants associated with MGL binding, we analyzed our array results using the publicly available Glycopattern web-based resource (https://glycopattern.emory.edu)7 (18). We first employed the GlycanMotifMiner to discover glycan binding motifs for MGLshort and MGLshort H259T. A binding motif was defined as a structural glycan element that contained the motif and was classified as an MGL-binding glycan. For both MGL variants, we identified the extended core 6 O-glycan Galβ1–3GlcNAcβ1–6GalNAc as the main glycan-binding motif (Table 1). In addition, MGLshort recognized the LDN motif and the globoside glycolipid GalNAcβ1–3Gal (Gb4Cer). The MGLshort H259T only seemed to favor core 6–derived structures (Table 1). Although not identified as a glycan-binding motif, some additional glycans emerged as high-affinity ligands for MGL on the glycan microarray. These included the single α- and β-GalNAc and 6-sulfated LDN (Table 2).
Table 1.
Glycan binding motifs of MGLshort and MGLshort H259T
The CFG glycan array data of MGLshort (A) and MGLshort H259T (B) (200 and 2 μg/ml) were analyzed using the data mining GlycoPattern software (https://glycopattern.emory.edu)7 (18), and the identified glycan-binding motifs are listed below. Associated with each motif are the number of “binders” and “non-binders,” which were determined using a statistical analysis (z-score) of the glycan array data based on the relative fluorescence units. A binder was defined as an MGL-binding structure that contains the motif. A non-binder contains the glycan motif but failed to interact with MGL. Yellow squares, GalNAc; blue squares, GlcNAc; yellow circles, galactose.
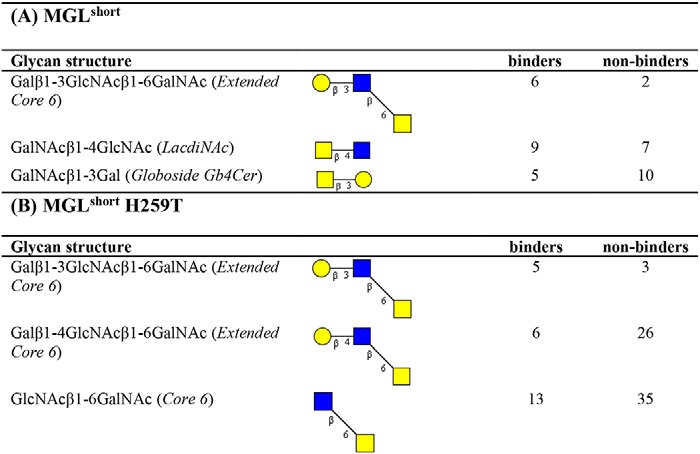
Table 2.
High-affinity ligands for MGLshort and MGLshort H259T
Listed are the top five binding glycans (at 2 μg/ml) on the CFG glycan array for MGLshort (A) and MGLshort H259T (B). RFU, relative fluorescence units. Yellow squares, GalNAc; blue squares, GlcNAc; yellow circles, galactose; red triangles, fucose; purple diamonds, sialic acid.
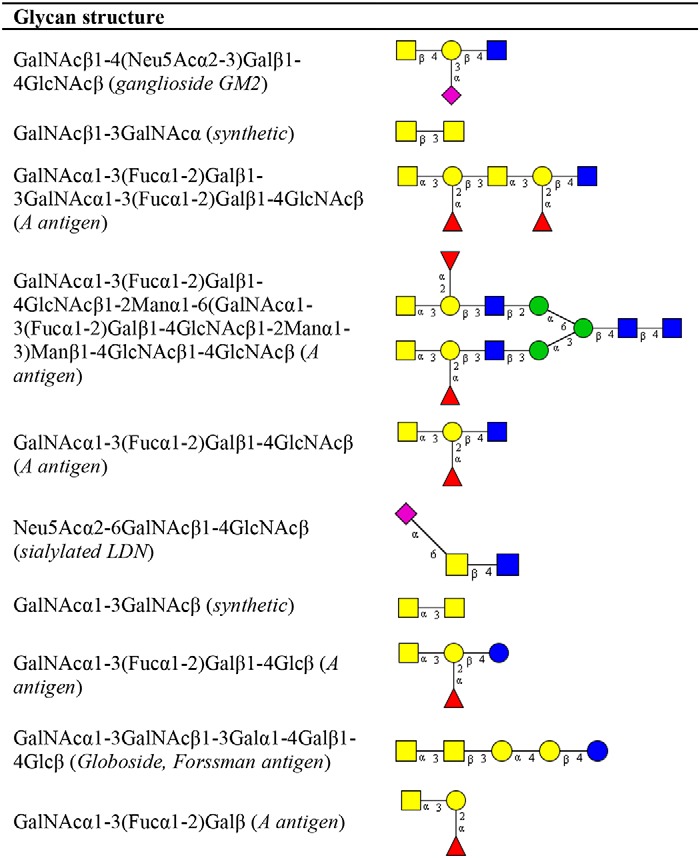
To evaluate the differential glycan binding of MGLshort and MGLshort H259T, we used the side-by-side comparison tool in the Glycopattern software. Here, we identified 10 glycan structures that displayed high binding to MGLshort, but failed to interact with the MGLshort H259T (Table 3). Five of these contain the GalNAcα1–3(Fucα1–2)Galβ blood group A antigen. Furthermore, we observed diminished binding to the synthetic GalNAcβ1–3GalNAcα epitope, the ganglioside GM2, sialylated LDN, a synthetic ganglioside containing polysialic acid, and the Forssman antigen (GalNAcα1–3GalNAcβ) (Table 3).
Table 3.
Differential glycan binding of MGLshort H259T
The CFG glycan array data of the MGLshort and MGLshort H259T (200 and 2 μg/ml) were analyzed using the data mining GlycoPattern software (https://glycopattern.emory.edu)7 (18). Listed are glycans that failed to bind MGLshort H259T and showed binding of >10,000 relative fluorescence units for MGLshort. Yellow squares, GalNAc; blue squares, GlcNAc; yellow circles, galactose; red triangles, fucose; purple diamonds, sialic acid; blue circles, glucose; green circles, mannose.
Tn-peptide specificity of MGL variants
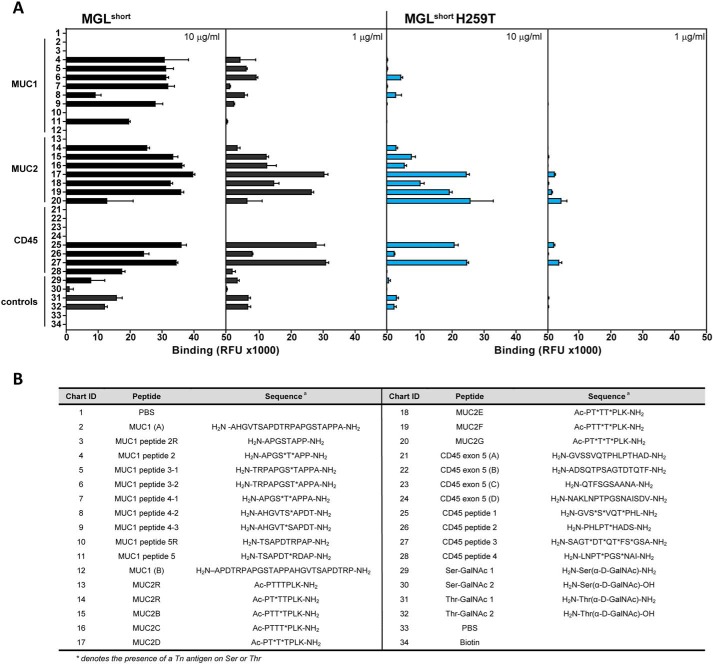
The lectin and antibody-mediated recognition of GalNAc moieties in Tn-containing mucin structures seems to be fine-tuned by the peptide backbone (19). Moreover, the underlying Ser of Thr residues in Tn antigen play a key role in the recognition process (20, 21). In the case of MGL, additional contacts with the peptide backbone upon binding of MUC1 Tn-glycopeptides have been described previously (17, 22). To address whether the His259 plays a role in the fine specificity of MGL toward Tn-containing glycopeptides, we employed a mucin glycopeptide array that included the known MGL ligands MUC1, MUC2, and CD45, which we interrogated using the MGLshort- and MGLshort H259T-Fc proteins (Fig. 2) (19). We chose to probe these MGL ligands based on their prior identification as major MGL ligands (6, 23, 24).
Figure 2.
Binding profile of MGL variants to the Tn-glycopeptide array. A, glycopeptides and corresponding parental peptides were printed at 100 μm and probed using the MGLshort-Fc (left panels) and MGLshort H259T-Fc (right panels) at the indicated concentrations. Relevant protein carriers are indicated in the figure. Data represent mean ± S.D. (error bars). RFU, relative fluorescence units. B, list of peptide backbones and Tn attachment sites of glycopeptides present on the Tn-glycopeptide array. *, presence of a Tn antigen on Ser or Thr.
At low concentrations, binding of MGLshort was avid for CD45 and MUC2 glycopeptides, whereas the well-known MGL ligand MUC1 (17, 22, 23) displayed weaker binding to MGLshort (Fig. 2A). Curiously, MGLshort preferred the diglycosylated T*T* or T*XT* MUC2 peptides (where the asterisk denotes the presence of a Tn antigen on Ser or Thr) compared with the mono- and triglycosylated peptides, whereas the diglycopeptide S*T* MUC1 displayed much weaker binding to MGLshort (Fig. 2A). Additionally, MGLshort may favor a Thr-based Tn antigen compared with the Ser-linked Tn antigen on this array (compare peptides 29/30 with 31/32).
Interestingly, in the glycopeptide array, MGLshort H259T lost high-affinity binding toward MUC1 structures, even at high probing concentrations, while displaying a residual affinity toward MUC2- and CD45-derived Tn-glycopeptides (Fig. 2A), indicating that replacement of His259 by a threonine residue has an impact on the Tn-glycopeptide recognition by MGL. Indeed, the loss in affinity of the MGLshort H259T mutant toward Tn-glycopeptides could imply that the His259 residue interacts with the surrounding peptide backbone of Tn-glycopeptides.
MGLshort H259T has a diminished capacity to recognize cellular ligands
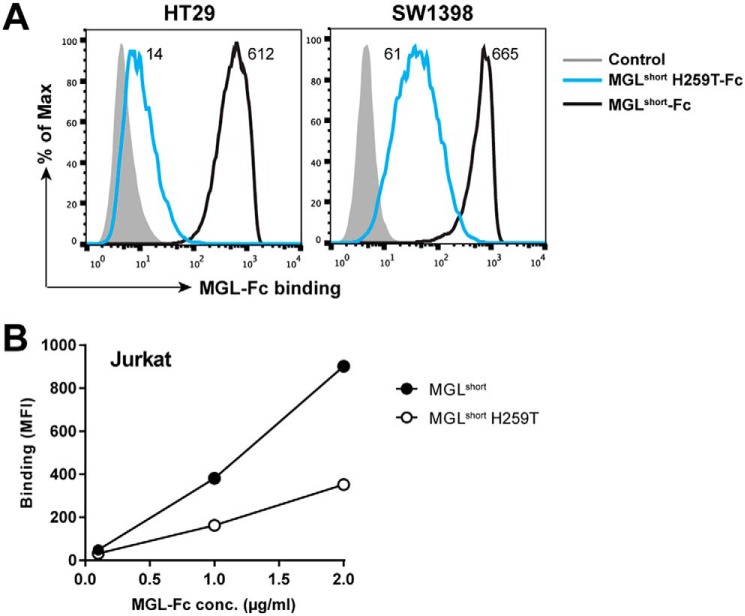
To confirm that the MGLshort H259T mutant also has a diminished capacity to interact with cellular MGL ligands, we performed flow cytometry–based MGL-Fc binding assays to colorectal carcinoma and Jurkat cell lines. The colorectal carcinoma cell lines HT29 and SW1398 were previously shown to contain large numbers of MGL-binding ligands (25). Whereas MGLshort displayed high binding to the colorectal carcinoma cell lines, the MGLshort H259T failed to recognize the HT29 cells and displayed a 10-fold decreased binding to the SW1398 cells (Fig. 3A). The Jurkat cell line has a deficiency in the COSMC chaperone leading to an absence of T-synthase activity and thus a complete lack of O-glycan elongation beyond the Tn stage (26). Again, we observed a reduced binding of MGLshort H259T to Jurkat cells (Fig. 3B), verifying that also on cells, MGLshort H259T has a reduced ability to engage Tn antigen. Together, our data indicate that although the H259T MGL mutant can still recognize Tn antigen, it does so with a strongly reduced affinity.
Figure 3.
MGLshort H259T displays poor recognition of GalNAc-containing glycan epitopes on tumor cells. Colorectal cancer cell lines HT29 and SW1398 (A) and the COSMC-deficient Jurkat cell line (B) were incubated with 5 μg/ml or the indicated concentrations of the MGL-Fc variants for 30 min at 4 °C. Binding was detected using an FITC-labeled goat anti-human Fc secondary antibody and analyzed by flow cytometry. One representative experiment of three is shown.
Molecular dynamics simulations of MGL and the H259T mutant
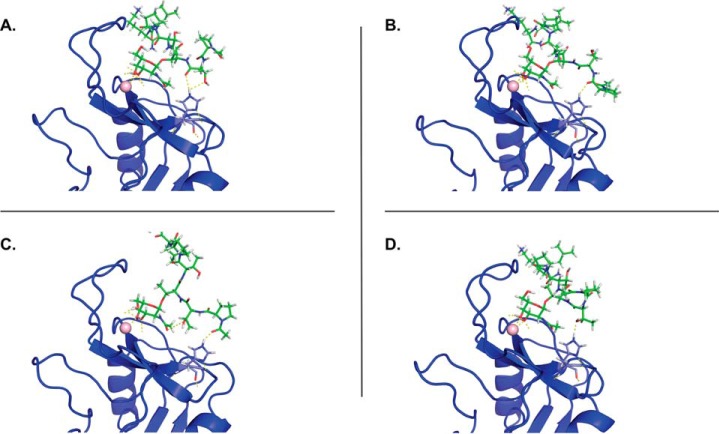
The involvement of His259 in the MGL-mediated recognition of Tn-glycopeptides seems to indicate that this residue is part of what we dubbed the secondary binding site in the MGL CRD. To check whether this secondary binding site establishes additional interactions with the surrounding peptide backbone of Tn-glycopeptides, we conducted molecular dynamics (MD) simulations on both MGL/Tn-glycopeptide and MGL H259T/Tn-glycopeptide complexes. The Tn-glycopeptides MUC2B Ac-PTT*TPLK-NH2 (peptide 15 in Fig. 2B) and MUC1 H2N-TRPAPGST*APPA-NH2 (peptide 6 in Fig. 2B) were selected based on the array results. A 3D model of the MGL CRD construct and the corresponding H259T mutant was built employing the homology model generated for MGL (residues Cys181–Leu308), as previously reported by us (17). Intensive 1-μs MD simulation was initially accomplished on both the WT MGLshort (MGL-His259) and the H259T mutant (MGL-Thr259) in explicit water and Ca2+ ions to evaluate the stability of both models. A structural ensemble of both proteins is depicted in Fig. S2. The calculated root mean square deviation shows that these molecules are equally stable and that the replacement of His259 by Thr does not induce major conformational perturbations (Fig. S2). Complexes of MGL-His259 and MGL-Thr259 with MUC2B and MUC1 glycopeptides were further generated as described by Marcelo et al. (17). Our previous MD and CORCEMA-ST analysis allowed us to conclude that the GalNAc monosaccharide interacts with MGL, employing two distinct binding modes (A and B), with alternative relative orientations of the galactosyl ring toward MGL (17). From a recognition point of view, histidine residues are commonly involved in H bonds and water-mediated interactions (27), and in GalNAc-binding mode A, the NHAc fragment of GalNAc is close to the His259 to potentially establish a stable H bond. Thus, MD simulations of the generated complexes in binding mode A were further submitted to evaluate their stability and the existence of intermolecular interactions involving either His259 (WT MGL) or Thr259 (mutant) and the surrounding peptide structure in GalNAc-containing peptides. In the case of MUC2B, transient H bonds were detected with different peptide residues (Fig. 4, A–D). Despite the fact that these H bonds are not highly populated, the sum of these H bonds in the surrounding peptide sequence can help to stabilize the complex and thereby significantly increase the binding affinity of MGL toward Tn-glycopeptides. No H bonds were detected between Thr259 and peptide backbone in the MD simulation analysis of mutant MGL-Thr259/MUC2B complex. The replacement of His259 by Thr thus may abolish these additional interactions. Furthermore, as expected, a stable H bond was detected between the CO of the NHAc group of GalNAc and the polar hydrogen Hϵ of His259.
Figure 4.
Selected frames of the MGL-His259/MUC2B complex extracted from 200-ns MD simulations. A–D, transient H bonds are established between the His259 and the peptide backbone (shown as yellow dashed lines). No H bonds were detected in the MGL-Thr259/MUC2B complex.
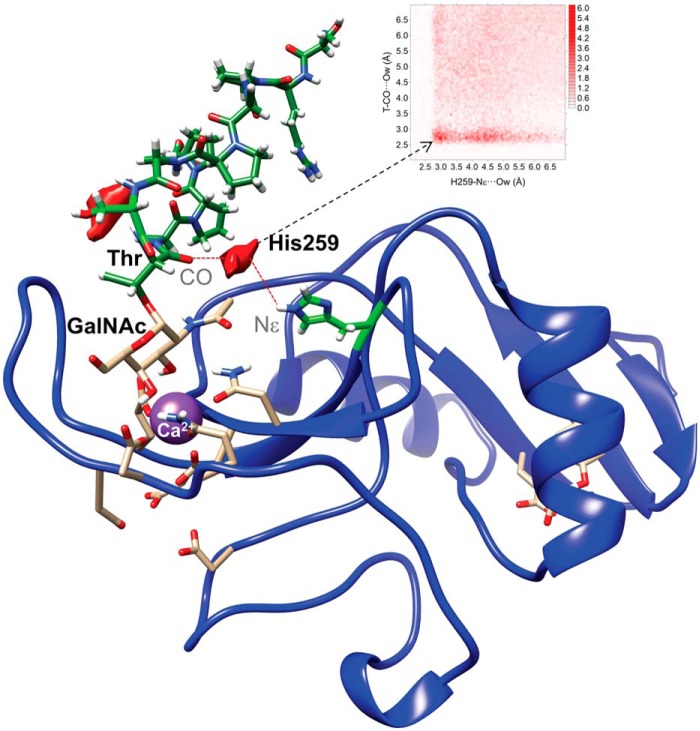
Interestingly, stable H bonds were not present between His259 and the peptide backbone of the MUC1-derived Tn-glycopeptide; however, a water pocket could be observed between the carbonyl CO of Thr linked to GalNAc and the polar hydrogen of the aromatic ring of His259 (Fig. 5). In contrast, no water oxygen density was detected involving the Thr259 amino acid for the complex generated using the MGLshort H259T mutant model. Overall, the MD results suggest that His259 plays a critical role in the MGL recognition of Tn-glycopeptides by establishing intermolecular interactions with the peptide backbone.
Figure 5.
Water-mediated binding between Tn-glycopeptide MUC1 (H2N-TRPAPGST*APPA-NH2) and His259 of MGLshort. Shown is the 2D-rdf function derived from 200-ns MD simulations between carbonyl (CO) of the glycosylated Thr and the polar nitrogen Nϵ of the aromatic ring of His259 (top), together with the water density (shown in red) calculated for the MGL-His259/MUC1–derived Tn-glycopeptide complex. No water density was detected in the case of MGL-Thr259/MUC1-derived Tn-glycopeptide complex.
Discussion
In this paper, we further defined the fine specificity of the human C-type lectin MGL through microarray analysis and MD simulations. Although MGL binding to the CFG glycan array has been analyzed before (8, 28) (see also the CFG glycan array data website: http://www.functionalglycomics.org),7 the carbohydrate recognition profiles of the different human MGL splice variants and the H259T mutant have never been directly compared. In our glycan array analyses, all identified MGL ligands contained a nonreducing terminal GalNAc with the equatorial/axial 3-OH and 4-OH groups exposed, allowing the interaction with Ca2+ at the primary binding site. None of the 3- or 4-extended GalNAc structures were recognized by MGL in the glycan microarray, confirming its unique preference for terminal GalNAc residues (8–10, 12, 29). It is noteworthy that extension at C-6 was allowed. Elongation or sulfation of the 3-OH or 4-OH abrogated MGL binding, thus explaining why some structures that do contain an identified MGL-binding motif were still categorized as nonbinders by the Glycopattern software (Table 1).
Strikingly, the identified MGL-binding elements are highly restricted in their expression pattern, suggesting that MGL may have some unique and distinctive functions within the human body. The LDN motif is mainly found on intestinal helminths (8) and in humans on glycodelin, a human glycoprotein with potent immunosuppressive and contraceptive activities (30). Sulfated LDN is a marker for pituitary glycoprotein hormones, such as lutropin (31). The enzyme that synthesizes LDN, β1,4-N-acetylgalactosaminyltransferase III (B4GALNT3) is overexpressed in colon cancer (32). Noticeably, both high MGL binding and overexpression of B4GALNT3 are associated with poor survival of colon cancer patients (25, 32). The preferred core 6 ligand has only been detected on human intestinal mucins and in the human ovary (33) (http://www.functionalglycomics.org),7 whereas the Gb4Cer glycolipid is most abundant on human embryonic stem cells and up-regulated on colon cancer (34). Together, these studies indicate that MGL may recognize cancer cells through their glycolipid framework and/or LDN expression and may be involved in hormone action or turnover. Both potential roles have so far not been studied in relation to MGL biology, although the mouse MGL1 was recently shown to clear WT and hyposialylated von Willebrand factor (35), indicating that nonimmune-related functions might exist for human MGL as well.
Intriguingly, we could detect some subtle nuances in glycan specificity between the MGLshort and MGLlong splice variants, even though their CRDs are completely identical. The additional 27 amino acids in MGLlong stalk region could potentially affect multimerization of the receptor. The neck region of MGLshort has been shown to form trimers, generating a cluster of binding sites for glycans abundant on pathogens and tumor cells (28). So far, there are no indications that multimerization might be different for MGLlong. The MGL CRDs have some flexibility to engage differentially spaced glycans on the counteracting interface (28). It is therefore tempting to speculate that the 27-amino acid region alters the orientation and spacing of the individual MGL CRDs, thereby modulating the range of ligands that are able to interact. This phenomenon has already been demonstrated for DC-SIGNR, where the variable number of neck repeats in DC-SIGNR affects the orientation of the individual CRDs and subsequently the relative binding affinities for surface glycoproteins (36). Structural studies of MGLshort and MGLlong trimers are required to formally prove the effect of the extended neck region in MGLlong, as well as a more quantitative analysis of binding affinities of each interacting glycan with surface plasmon resonance.
Engagement of Tn antigens by MGL on human dendritic cells is thought to lead to immune tolerance via the MGL-induced secretion of the anti-inflammatory cytokine IL-10 and the ability to instruct the differentiation of suppressive CD4+ T cells (4, 5). Moreover, through direct engagement of CD45 on activated T cells, MGL is able to inhibit T cell proliferation and cytokine release, while at the same time promoting T cell apoptosis (6). MGL also specifically recognizes the cancer-associated MUC1 and MUC2 (12, 23, 24), suggesting that MGL might promote immune evasion by Tn antigen–positive tumors as well (37). Therefore, we constructed a Tn-glycopeptide array harboring these immune-relevant MGL ligands. On our Tn-glycopeptide array, WT MGLshort seemed to prefer MUC2- and CD45-derived glycopeptides over the MUC1 glycopeptides. However, the MUC1 glycopeptides in this particular array represent only a subset of possible glycosylation patterns of the MUC1 repeats; thus, we cannot exclude the possibility that other MUC1 glycopeptides may be dominant for MGL binding. MGLshort also appeared to favor diglycosylated peptides, as well as GalNAc-Thr over GalNAc-Ser. In general, lectin affinity increases with the increased valency of glycan epitopes in a molecule (38). From the four CD45 glycopeptides tested, MGLshort preferred the tri- and tetraglycosylated peptides over the mono- and diglycosylated peptides. The preference for Tn clustering by MGL might be caused by the use of dimeric MGL-Fc constructs, which contain two MGL molecules coupled to one Fc-tail. However, further experiments are required to determine the minimal distance required in a multi-Tn-peptide to accommodate two MGL proteins. Differential recognition of GalNAc-Ser and GalNAc-Thr has been observed before and might be explained by the relatively fixed position of the GalNAc-Thr compared with the more flexible GalNAc-Ser (19, 39–43).
We did, however, find major differences in the glycan-binding profiles of the MGLshort and its H259T mutant, whereby the MGLshort H259T was still able to recognize a single GalNAc monosaccharide (Fig. 1C) and core 6 structures (Table 2). Nevertheless, the H259T mutant displayed a diminished capacity to bind elongated structures, such as the LDN epitope and sialyl-Tn (Fig. 1), as well as the blood group A determinant, the ganglioside GM2, sialylated LDN, and the Forsmann antigen (Table 3). Blood group antigens are highly expressed on erythrocytes and in a variety of other tissues, including the vascular endothelium, mucus secretions, and epithelial surfaces. Intriguingly, in a rat model for colon cancer, expression of blood group A antigen increased apoptosis resistance and facilitated immune escape (44), a feature that may be linked to the immunosuppressive properties of MGL (3, 5). Forssman antigen has been demonstrated in stomach and colon cancer (45) and sialylated LDN in prostate cancer (46), again pointing to an influential role for MGL in tumor biology (12, 23, 25).
Also on the Tn-glycopeptide array, a reduced binding capacity of the MGLshort H259T mutant could be observed. The local sequence context around the GalNAc-modified Ser or Thr residue thus emerges as an important factor for MGL binding specificity, which is further corroborated by the loss of affinity of MGLshort H259T to particular glycan structures as well as Tn-containing glycopeptides and Tn-expressing cells. This loss of affinity directly implies that MGL recognizes not only the GalNAc monosaccharide, but also the underlying glycan or protein backbone, through a secondary binding site, of which the His259 amino acid is a key element. Through MD simulations, we could confirm the MGL-mediated recognition of the peptide backbone in MUC1 (17, 22) as well as in MUC2 through the His259 residue. Strikingly, the type of interactions, either through H bonds or water bridges, depended on the peptide sequence of the glycopeptide. We assume that also in the recognition of GalNAc-Tyr (11), MGL is able to engage the underlying tyrosine residue. Actually, the MGL binding mode shows clear parallels to the first structurally characterized Tn-glycopeptide–specific 237mAb antibody (21). Similar to MGL, the primary binding site of 237mAb is anchored by the Tn antigen, whereas the peptide backbone of podoplanin provides the additional specificity, thus conveying a dual recognition mode toward both Tn antigen and the podoplanin peptide backbone (21). The additional interactions involving the His259 residue, conveying fine specificity toward Tn-containing proteins or peptides, clearly illustrate the importance of exploring molecular recognition events for the optimal design of MGL-targeting structures for anti-tumor vaccines in light of the proposed role of MGL in impacting immunological responses through its highly exclusive recognition of tumor-associated glycan structures (47).
Experimental procedures
Generation and production of MGL-Fc constructs
The MGL-Fcshort variant (corresponding to the 6C splice variant (7)) was constructed as described (8). cDNA encoding the extracellular domains of the MGLlong isomer (splice variant 6B (7)) was bought from Baseclear and cloned into the Sig-pIgG1-Fc vector. The MGLshort H259T mutant was generated by site-directed mutagenesis using the QuikChange II site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. All MGL-Fc proteins were produced in CHO-K1 cells and purified from supernatants using HiTrap Protein A HP columns (GE Healthcare). MGL-Fc concentrations were determined by ELISA using human IgG1 as a standard, and purity was verified by Western blot analysis (Fig. S1A).
Antibody- and lectin-binding ELISAs
Anti-MGL antibodies 18E4 and 1G6.6 were generated as described before (3). Anti-MGL clone 125A10 was obtained from Dendritics. Isotype-matched control antibodies were purchased from eBioscience. To test the reactivity of these anti-MGL antibodies toward the different MGL-Fc proteins, NUNC MaxiSorp plates were coated overnight at room temperature with 5 μg/ml of the purified MGL-Fc proteins in 0.2 m NaHCO3 (pH 9.2). After washing with TSM (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 1 mm CaCl2), plates were blocked for 30 min at 37 °C with TSM containing 1% BSA. Anti-MGL antibodies and isotype controls were added to the wells at 5 μg/ml in TSM plus 0.5% BSA and incubated for 1 h at room temperature. Plates were washed with TSM plus 0.05% Tween 20, and bound antibodies were detected using peroxidase-labeled goat anti-mouse IgG Fc-specific secondary antibodies (0.3 μg/ml in TSM plus 0.05% Tween 20). TMB (100 μg/ml TMB and 0.003% H2O2 in 0.1 m NaAc, pH 4) was used as a substrate to visualize the presence of bound peroxidase, and optical densities were measured at 450 nm.
To investigate MGL-Fc binding properties, NUNC MaxiSorp plates were coated overnight at room temperature with the indicated concentrations of polyacrylamide-coupled glycoconjugates (Lectinity) in 0.2 m NaHCO3 (pH 9.2). After washing with TSM, plates were blocked with 1% BSA plus TSM. MGL-Fc proteins (0.5 μg/ml) were added and incubated for 2 h at room temperature. Plates were washed with TSM plus 0.05% Tween 20, and bound MGL-Fc was detected using a peroxidase-labeled goat anti-human IgG-Fc (Jackson Immunoresearch Laboratories; 0.3 μg/ml in TSM plus 0.05% Tween 20). The reaction was developed using TMB (100 μg/ml TMB and 0.003% H2O2 in 0.1 m NaAc, pH 4), and optical densities were measured at 450 nm.
Glycan microarray analysis
The mammalian glycan array (version 5.1, containing 610 glycan structures) was developed by the Consortium of Functional Glycomics (www.functionalglycomics.org) from a library of natural and synthetic mammalian glycans with amino-linkers and printed in replicates of six onto N-hydroxysuccinimide–activated glass slides, forming covalent amide linkages. 70 μl of the MGL-Fc proteins (2 and 200 μg/ml in TSM containing 1% BSA and 0.05% Tween 20) was applied to the array and incubated at room temperature in a humidified chamber for 1 h. After extensive washing in TSM, bound MGL-Fc was detected using an Alexa 488–labeled anti-human IgG secondary antibody (1 μg/ml in TSM containing 1% BSA and 0.05% Tween 20). Fluorescence intensities were detected using a PerkinElmer Life Sciences ScanArray 5000 scanner, followed by image analyses using IMAGENE image analysis software (for representative images, see Fig. S3). The glycan array data (2 and 200 μg/ml) was analyzed using the Data mining feature in the GlycoPattern (https://glycopattern.emory.edu)7 software (18) to identify glycan binding motifs and glycans that differentially bound the MGL variants. Glycan structures were drawn using the Glycoworkbench software (48). All glycan microarray analyses were carried out in accordance with the MIRAGE (minimum information required for a glycomics experiment) guidelines for reporting glycan microarray-based data (49).
Tn-glycopeptide array
Glycopeptides were synthesized by solid-phase peptide synthesis as carboxamides at the C terminus following published procedures (50). Those incorporating lysine were acetylated at the N terminus with the lysine side chain providing the amino group for the reaction with the N-hydroxysuccinimide–functionalized slide. The others had a free N terminus through which they were immobilized on the slide (19, 51). The glycopeptides were adjusted to the same concentration (100 μm) in printing buffer (300 mm sodium phosphate, pH 8.5) and spotted in replicates of 6. The Tn-glycopeptide was probed using MGL-Fc at a concentration of 10 and 1 μg/ml. Bound MGL-Fc was detected using an Alexa 488–labeled anti-human IgG secondary antibody at 5 μg/ml. Other assay conditions, buffers, and methods for imaging and processing of the data were identical to the screening of the glycan microarray and as described previously (Fig. S3) (19).
MGL-Fc staining of tumor cells
The human colon cancer cell lines SW1398 and HT29 were maintained in Dulbecco's modified Eagle's medium (Thermo Fischer Scientific) supplemented with 10% fetal calf serum and antibiotics. The Jurkat cell line was cultured in RPMI (Thermo Fisher Scientific) supplemented with 10% fetal calf serum and antibiotics. Colon cancer and Jurkat cell lines were incubated for 30 min at 4 °C with 5 μg/ml or the indicated concentrations of the different MGL-Fc variants. After washing with Hanks' buffered saline solution containing 0.5% BSA, the bound MGL-Fc was counterstained for 30 min at 4 °C using an FITC-labeled goat anti-human IgG Fc (Jackson Immunoresearch). Cells were analyzed by flow cytometry on a CyAn flow cytometer (Beckman Coulter). Dead cells were excluded by 7-aminoactinomycin D discrimination (Molecular Probes).
MD simulations
MD simulations were performed using Amber16 implemented with the force field ff14SB (52) and GLYCAM 06j-1 (53) to properly reproduce the conformational behavior of the glycopeptides. The 3D models of the MGLshort and the corresponding mutant H259T were generated according to Marcelo et al. (17). 1-μs MD simulations were performed to compare the stability of both models for MGLshort and the MGLshort H259T. The MUC2 glycopeptide Ac-PTT*TPLK-NH2 (peptide 15; Fig. 2B) and the MUC1 glycopeptide H2N-TRPAPGST*APPA-NH2 (peptide 6; Fig. 2B) were selected to generate the corresponding MGLshort/Tn-glycopeptide and MGLshort H259T/Tn-glycopeptide complexes.
The 3D models of MGLshort and corresponding H259T mutant, as well as the MGL/Tn-glycopeptide complexes, were immersed in a box with a 10-Å buffer of TIP3P (54) water molecules plus Ca2+ and neutralized by adding explicit counterions (Na+). A two-stage geometry optimization approach was used. The first stage minimizes only the positions of solvent molecules and ions, and the second stage is an unrestrained minimization of all of the atoms in the simulation cell. The systems were then gently heated by incrementing the temperature from 0 to 300 K under a constant pressure of 1 atm and periodic boundary conditions. Harmonic restraints of 30 kcal/mol were applied to the solute, and the Andersen temperature coupling scheme (54) was used to control and equalize the temperature. The time step was kept at 1 fs during the heating stages. Water molecules are treated with the SHAKE algorithm, such that the angle between the hydrogen atoms is kept fixed. Long-range electrostatic effects are modeled using the particle mesh–Ewald method (55). An 8-Å cut-off was applied to Lennard-Jones interactions. Each system was equilibrated for 2 ns with a 2-fs time step at a constant volume and temperature of 300 K. Production trajectories, under the same simulation conditions, were then run for an additional 1 μs in the case of MGLshort and MGLshort H259T and 200 ns in the case of Tn-glycopeptide–containing complexes.
Statistics
Significant differences were evaluated using the GraphPad Prism software. A p value < 0.05 was considered to be significant.
Author contributions
F. M., F. C., D. L., and S. J. v. V. conceptualization; F. M., F. C., D. L., and S. J. v. V. formal analysis; F. M., N. S., F. C., J. C. v. d. H., I. M. V., D. L., D. F. S., and S. J. v. V. investigation; F. M., N. S., F. C., J. C. v. d. H., I. M. V., D. L., G.-J. P. B., D. F. S., and S. J. v. V. methodology; F. M., F. C., D. L., and S. J. v. V. writing-original draft; N. S., J. C. v. d. H., I. M. V., G.-J. P. B., and D. F. S. writing-review and editing; D. L. and S. J. v. V. funding acquisition; G.-J. P. B. supervision.
Supplementary Material
This work was supported by a bridging grant under National Institutes of Health Grant U54GM62116 (to the Consortium for Functional Glycomics) and P41GM103390 (to the Research Resource for Integrated Glycotechnology). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S3.
The MGL array data for this paper are available from the Glycan array data Consortium for Functional Glycomics.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CRD
- carbohydrate recognition domain
- MGL
- macrophage galactose-type lectin
- LDN
- LacdiNAc
- LacNAc
- N-acetyl-d-lactosamine
- PAA
- polyacrylamide
- MD
- molecular dynamics
- 3D
- three-dimensional
- B4GALNT3
- β1,4-N-acetylgalactosaminyltransferase III
- GM2
- GalNAcβ1–4(Neu5Acα2–3)Galβ1–4Glc
- GM3
- NeuAcα 2,3Galβ1,4Glc-ceramide
- GD2
- GalNAcβ1–4(Neu5Acα2–8Neu5Acα2–3)Galβ1–4Glc
- Gb4Cer
- GalNAcβ1–3Gal.
References
- 1. Drickamer K., and Taylor M. E. (2015) Recent insights into structures and functions of C-type lectins in the immune system. Curr. Opin. Struct. Biol. 34, 26–34 10.1016/j.sbi.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zelensky A. N., and Gready J. E. (2005) The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217 10.1111/j.1742-4658.2005.05031.x [DOI] [PubMed] [Google Scholar]
- 3. van Vliet S. J., van Liempt E., Geijtenbeek T. B., and van Kooyk Y. (2006) Differential regulation of C-type lectin expression on tolerogenic dendritic cell subsets. Immunobiology 211, 577–585 10.1016/j.imbio.2006.05.022 [DOI] [PubMed] [Google Scholar]
- 4. van Vliet S. J., Vuist I. M., Lenos K., Tefsen B., Kalay H., García-Vallejo J. J., and van Kooyk Y. (2013) Human T cell activation results in extracellular signal-regulated kinase (ERK)-calcineurin-dependent exposure of Tn antigen on the cell surface and binding of the macrophage galactose-type lectin (MGL). J. Biol. Chem. 288, 27519–27532 10.1074/jbc.M113.471045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li D., Romain G., Flamar A. L., Duluc D., Dullaers M., Li X. H., Zurawski S., Bosquet N., Palucka A. K., Le Grand R., O'Garra A., Zurawski G., Banchereau J., and Oh S. (2012) Targeting self- and foreign antigens to dendritic cells via DC-ASGPR generates IL-10-producing suppressive CD4+ T cells. J. Exp. Med. 209, 109–121 10.1084/jem.20110399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Vliet S. J., Gringhuis S. I., Geijtenbeek T. B., and van Kooyk Y. (2006) Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 7, 1200–1208 10.1038/ni1390 [DOI] [PubMed] [Google Scholar]
- 7. Higashi N., Fujioka K., Denda-Nagai K., Hashimoto S., Nagai S., Sato T., Fujita Y., Morikawa A., Tsuiji M., Miyata-Takeuchi M., Sano Y., Suzuki N., Yamamoto K., Matsushima K., and Irimura T. (2002) The macrophage C-type lectin specific for galactose/N-acetylgalactosamine is an endocytic receptor expressed on monocyte-derived immature dendritic cells. J. Biol. Chem. 277, 20686–20693 10.1074/jbc.M202104200 [DOI] [PubMed] [Google Scholar]
- 8. van Vliet S. J., van Liempt E., Saeland E., Aarnoudse C. A., Appelmelk B., Irimura T., Geijtenbeek T. B., Blixt O., Alvarez R., van Die I., and van Kooyk Y. (2005) Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int. Immunol. 17, 661–669 10.1093/intimm/dxh246 [DOI] [PubMed] [Google Scholar]
- 9. Suzuki N., Yamamoto K., Toyoshima S., Osawa T., and Irimura T. (1996) Molecular cloning and expression of cDNA encoding human macrophage C-type lectin: its unique carbohydrate binding specificity for Tn antigen. J. Immunol. 156, 128–135 [PubMed] [Google Scholar]
- 10. Mortezai N., Behnken H. N., Kurze A. K., Ludewig P., Buck F., Meyer B., and Wagener C. (2013) Tumor-associated Neu5Ac-Tn and Neu5Gc-Tn antigens bind to C-type lectin CLEC10A (CD301, MGL). Glycobiology 23, 844–852 10.1093/glycob/cwt021 [DOI] [PubMed] [Google Scholar]
- 11. Gibadullin R., Farnsworth D. W., Barchi J. J. Jr., and Gildersleeve J. C. (2017) GalNAc-tyrosine is a ligand of plant lectins, antibodies, and human and murine macrophage galactose-type lectins. ACS Chem. Biol. 12, 2172–2182 10.1021/acschembio.7b00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beatson R., Maurstad G., Picco G., Arulappu A., Coleman J., Wandell H. H., Clausen H., Mandel U., Taylor-Papadimitriou J., Sletmoen M., and Burchell J. M. (2015) The breast cancer-associated glycoforms of MUC1, MUC1-Tn and sialyl-Tn, are expressed in COSMC wild-type cells and bind the C-type lectin MGL. PLoS One 10, e0125994 10.1371/journal.pone.0125994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kolatkar A. R., and Weis W. I. (1996) Structural basis of galactose recognition by C-type animal lectins. J. Biol. Chem. 271, 6679–6685 10.1074/jbc.271.12.6679 [DOI] [PubMed] [Google Scholar]
- 14. Tsuiji M., Fujimori M., Ohashi Y., Higashi N., Onami T. M., Hedrick S. M., and Irimura T. (2002) Molecular cloning and characterization of a novel mouse macrophage C-type lectin, mMGL2, which has a distinct carbohydrate specificity from mMGL1. J. Biol. Chem. 277, 28892–28901 10.1074/jbc.M203774200 [DOI] [PubMed] [Google Scholar]
- 15. Singh S. K., Streng-Ouwehand I., Litjens M., Weelij D. R., García-Vallejo J. J., van Vliet S. J., Saeland E., and van Kooyk Y. (2009) Characterization of murine MGL1 and MGL2 C-type lectins: distinct glycan specificities and tumor binding properties. Mol. Immunol. 46, 1240–1249 10.1016/j.molimm.2008.11.021 [DOI] [PubMed] [Google Scholar]
- 16. Sakakura M., Oo-Puthinan S., Moriyama C., Kimura T., Moriya J., Irimura T., and Shimada I. (2008) Carbohydrate binding mechanism of the macrophage galactose-type C-type lectin 1 revealed by saturation transfer experiments. J. Biol. Chem. 283, 33665–33673 10.1074/jbc.M804067200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marcelo F., Garcia-Martin F., Matsushita T., Sardinha J., Coelho H., Oude-Vrielink A., Koller C., André S., Cabrita E. J., Gabius H. J., Nishimura S., Jiménez-Barbero J., and Cañada F. J. (2014) Delineating binding modes of Gal/GalNAc and structural elements of the molecular recognition of tumor-associated mucin glycopeptides by the human macrophage galactose-type lectin. Chemistry 20, 16147–16155 10.1002/chem.201404566 [DOI] [PubMed] [Google Scholar]
- 18. Agravat S. B., Saltz J. H., Cummings R. D., and Smith D. F. (2014) GlycoPattern: a web platform for glycan array mining. Bioinformatics 30, 3417–3418 10.1093/bioinformatics/btu559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borgert A., Heimburg-Molinaro J., Song X., Lasanajak Y., Ju T., Liu M., Thompson P., Ragupathi G., Barany G., Smith D. F., Cummings R. D., and Live D. (2012) Deciphering structural elements of mucin glycoprotein recognition. ACS Chem. Biol. 7, 1031–1039 10.1021/cb300076s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Madariaga D., Martínez-Sáez N., Somovilla V. J., Coelho H., Valero-González J., Castro-López J., Asensio J. L., Jiménez-Barbero J., Busto J. H., Avenoza A., Marcelo F., Hurtado-Guerrero R., Corzana F., and Peregrina J. M. (2015) Detection of tumor-associated glycopeptides by lectins: the peptide context modulates carbohydrate recognition. ACS Chem. Biol. 10, 747–756 10.1021/cb500855x [DOI] [PubMed] [Google Scholar]
- 21. Brooks C. L., Schietinger A., Borisova S. N., Kufer P., Okon M., Hirama T., Mackenzie C. R., Wang L. X., Schreiber H., and Evans S. V. (2010) Antibody recognition of a unique tumor-specific glycopeptide antigen. Proc. Natl. Acad. Sci. U.S.A. 107, 10056–10061 10.1073/pnas.0915176107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Artigas G., Monteiro J. T., Hinou H., Nishimura S. I., Lepenies B., and Garcia-Martin F. (2017) Glycopeptides as targets for dendritic cells: exploring MUC1 glycopeptides binding profile toward macrophage galactose-type lectin (MGL) orthologs. J. Med. Chem. 60, 9012–9021 10.1021/acs.jmedchem.7b01242 [DOI] [PubMed] [Google Scholar]
- 23. Saeland E., van Vliet S. J., Bäckström M., van den Berg V. C., Geijtenbeek T. B., Meijer G. A., and van Kooyk Y. (2007) The C-type lectin MGL expressed by dendritic cells detects glycan changes on MUC1 in colon carcinoma. Cancer Immunol. Immunother. 56, 1225–1236 10.1007/s00262-006-0274-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Napoletano C., Rughetti A., Agervig Tarp M. P., Coleman J., Bennett E. P., Picco G., Sale P., Denda-Nagai K., Irimura T., Mandel U., Clausen H., Frati L., Taylor-Papadimitriou J., Burchell J., and Nuti M. (2007) Tumor-associated Tn-MUC1 glycoform is internalized through the macrophage galactose-type C-type lectin and delivered to the HLA class I and II compartments in dendritic cells. Cancer Res. 67, 8358–8367 10.1158/0008-5472.CAN-07-1035 [DOI] [PubMed] [Google Scholar]
- 25. Lenos K., Goos J. A., Vuist I. M., den Uil S. H., Delis-van Diemen P. M., Belt E. J., Stockmann H. B., Bril H., de Wit M., Carvalho B., Giblett S., Pritchard C. A., Meijer G. A., van Kooyk Y., Fijneman R. J., and van Vliet S. J. (2015) MGL ligand expression is correlated to BRAF mutation and associated with poor survival of stage III colon cancer patients. Oncotarget 6, 26278–26290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ju T., and Cummings R. D. (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 10.1073/pnas.262438199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao J. H., Chien C. T., Wu H. Y., Huang K. F., Wang I., Ho M. R., Tu I. F., Lee I. M., Li W., Shih Y. L., Wu C. Y., Lukyanov P. A., Hsu S. T., and Wu S. H. (2016) A multivalent marine lectin from Crenomytilus grayanus possesses anti-cancer activity through recognizing globotriose Gb3. J. Am. Chem. Soc. 138, 4787–4795 10.1021/jacs.6b00111 [DOI] [PubMed] [Google Scholar]
- 28. Jégouzo S. A., Quintero-Martínez A., Ouyang X., dos Santos Á., Taylor M. E., and Drickamer K. (2013) Organization of the extracellular portion of the macrophage galactose receptor: a trimeric cluster of simple binding sites for N-acetylgalactosamine. Glycobiology 23, 853–864 10.1093/glycob/cwt022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iida S., Yamamoto K., and Irimura T. (1999) Interaction of human macrophage C-type lectin with O-linked N-acetylgalactosamine residues on mucin glycopeptides. J. Biol. Chem. 274, 10697–10705 10.1074/jbc.274.16.10697 [DOI] [PubMed] [Google Scholar]
- 30. Dell A., Morris H. R., Easton R. L., Panico M., Patankar M., Oehniger S., Koistinen R., Koistinen H., Seppala M., and Clark G. F. (1995) Structural analysis of the oligosaccharides derived from glycodelin, a human glycoprotein with potent immunosuppressive and contraceptive activities. J. Biol. Chem. 270, 24116–24126 10.1074/jbc.270.41.24116 [DOI] [PubMed] [Google Scholar]
- 31. Fiete D., Srivastava V., Hindsgaul O., and Baenziger J. U. (1991) A hepatic reticuloendothelial cell receptor specific for SO4–4GalNAc β1,4GlcNAcβ1,2Manα that mediates rapid clearance of lutropin. Cell 67, 1103–1110 10.1016/0092-8674(91)90287-9 [DOI] [PubMed] [Google Scholar]
- 32. Che M. I., Huang J., Hung J. S., Lin Y. C., Huang M. J., Lai H. S., Hsu W. M., Liang J. T., and Huang M. C. (2014) β1,4-N-acetylgalactosaminyltransferase III modulates cancer stemness through EGFR signaling pathway in colon cancer cells. Oncotarget 5, 3673–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brockhausen I., Schachter H., and Stanley P. (2009) O-GalNAc glycans. In Essentials of Glycobiology, 2nd Ed. (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E., eds) Chapter 9, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 34. Joo E. J., Weyers A., Li G., Gasimli L., Li L., Choi W. J., Lee K. B., and Linhardt R. J. (2014) Carbohydrate-containing molecules as potential biomarkers in colon cancer. OMICS 18, 231–241 10.1089/omi.2013.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward S. E., O'Sullivan J. M., Drakeford C., Aguila S., Jondle C. N., Sharma J., Fallon P. G., Brophy T. M., Preston R. J. S., Smyth P., Sheils O., Chion A., and O'Donnell J. S. (2018) A novel role for the macrophage galactose-type lectin receptor in mediating von Willebrand factor clearance. Blood 131, 911–916 10.1182/blood-2017-06-787853 [DOI] [PubMed] [Google Scholar]
- 36. Feinberg H., Tso C. K., Taylor M. E., Drickamer K., and Weis W. I. (2009) Segmented helical structure of the neck region of the glycan-binding receptor DC-SIGNR. J. Mol. Biol. 394, 613–620 10.1016/j.jmb.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodríguez E., Schetters S. T. T., and van Kooyk Y. (2018) The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211 10.1038/nri.2018.3 [DOI] [PubMed] [Google Scholar]
- 38. Dam T. K., and Brewer C. F. (2010) Lectins as pattern recognition molecules: the effects of epitope density in innate immunity. Glycobiology 20, 270–279 10.1093/glycob/cwp186 [DOI] [PubMed] [Google Scholar]
- 39. Madariaga D., Martínez-Sáez N., Somovilla V. J., García-García L., Berbis M. Á., Valero-Gónzalez J., Martín-Santamaría S., Hurtado-Guerrero R., Asensio J. L., Jiménez-Barbero J., Avenoza A., Busto J. H., Corzana F., and Peregrina J. M. (2014) Serine versus threonine glycosylation with α-O-GalNAc: unexpected selectivity in their molecular recognition with lectins. Chemistry 20, 12616–12627 10.1002/chem.201403700 [DOI] [PubMed] [Google Scholar]
- 40. Corzana F., Busto J. H., Jiménez-Osés G., Asensio J. L., Jiménez-Barbero J., Peregrina J. M., and Avenoza A. (2006) New insights into α-GalNAc-Ser motif: influence of hydrogen bonding versus solvent interactions on the preferred conformation. J. Am. Chem. Soc. 128, 14640–14648 10.1021/ja064539u [DOI] [PubMed] [Google Scholar]
- 41. Corzana F., Busto J. H., Jiménez-Osés G., García de Luis M., Asensio J. L., Jiménez-Barbero J., Peregrina J. M., and Avenoza A. (2007) Serine versus threonine glycosylation: the methyl group causes a drastic alteration on the carbohydrate orientation and on the surrounding water shell. J. Am. Chem. Soc. 129, 9458–9467 10.1021/ja072181b [DOI] [PubMed] [Google Scholar]
- 42. Martínez-Sáez N., Peregrina J. M., and Corzana F. (2017) Principles of mucin structure: implications for the rational design of cancer vaccines derived from MUC1-glycopeptides. Chem. Soc. Rev. 46, 7154–7175 10.1039/C6CS00858E [DOI] [PubMed] [Google Scholar]
- 43. Bermejo I. A., Usabiaga I., Compañón I., Castro-López J., Insausti A., Fernández J. A., Avenoza A., Busto J. H., Jiménez-Barbero J., Asensio J. L., Peregrina J. M., Jiménez-Osés G., Hurtado-Guerrero R., Cocinero E. J., and Corzana F. (2018) Water sculpts the distinctive shapes and dynamics of the Tn antigens: implications for their molecular recognition. J. Am. Chem. Soc. 140, 9952–9960 10.1021/jacs.8b04801 [DOI] [PubMed] [Google Scholar]
- 44. Marionneau S., Le Moullac-Vaidye B., and Le Pendu J. (2002) Expression of histo-blood group A antigen increases resistance to apoptosis and facilitates escape from immune control of rat colon carcinoma cells. Glycobiology 12, 851–856 10.1093/glycob/cwf103 [DOI] [PubMed] [Google Scholar]
- 45. Hakomori S., Wang S. M., and Young W. W. Jr. (1977) Isoantigenic expression of Forssman glycolipid in human gastric and colonic mucosa: its possible identity with “A-like antigen” in human cancer. Proc. Natl. Acad. Sci. U.S.A. 74, 3023–3027 10.1073/pnas.74.7.3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hirano K., Matsuda A., Shirai T., and Furukawa K. (2014) Expression of LacdiNAc groups on N-glycans among human tumors is complex. Biomed. Res. Int. 2014, 981627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Freire T., Zhang X., Dériaud E., Ganneau C., Vichier-Guerre S., Azria E., Launay O., Lo-Man R., Bay S., and Leclerc C. (2010) Glycosidic Tn-based vaccines targeting dermal dendritic cells favor germinal center B-cell development and potent antibody response in the absence of adjuvant. Blood 116, 3526–3536 10.1182/blood-2010-04-279133 [DOI] [PubMed] [Google Scholar]
- 48. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 10.1021/pr7008252 [DOI] [PubMed] [Google Scholar]
- 49. York W. S., Agravat S., Aoki-Kinoshita K. F., McBride R., Campbell M. P., Costello C. E., Dell A., Feizi T., Haslam S. M., Karlsson N., Khoo K. H., Kolarich D., Liu Y., Novotny M., Packer N. H., et al. (2014) MIRAGE: the minimum information required for a glycomics experiment. Glycobiology 24, 402–406 10.1093/glycob/cwu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Movahedin M., Brooks T. M., Supekar N. T., Gokanapudi N., Boons G. J., and Brooks C. L. (2017) Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen. Glycobiology 27, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu M., Barany G., and Live D. (2005) Parallel solid-phase synthesis of mucin-like glycopeptides. Carbohydr. Res. 340, 2111–2122 10.1016/j.carres.2005.05.023 [DOI] [PubMed] [Google Scholar]
- 52. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., and Woods R. J. (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 10.1002/jcc.20820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andrea T. A., Swope W. C., and Andersen H. C. (1983) The role of long ranged forces in determining the structure and properties of liquid water. J. Chem. Phys. 79, 4576–4584 10.1063/1.446373 [DOI] [Google Scholar]
- 55. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 10.1063/1.464397 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.