Abstract
The cyanoacrylate compound phenamacril (also known as JS399–19) is a recently identified fungicide that exerts its antifungal effect on susceptible Fusarium species by inhibiting the ATPase activity of their myosin class I motor domains. Although much is known about the antifungal spectrum of phenamacril, the exact mechanism behind the phenamacril-mediated inhibition remains to be resolved. Here, we describe the characterization of the effect of phenamacril on purified myosin motor constructs from the model plant pathogen and phenamacril-susceptible species Fusarium graminearum, phenamacril-resistant Fusarium species, and the mycetozoan model organism Dictyostelium discoideum. Our results show that phenamacril potently (IC50 ∼360 nm), reversibly, and noncompetitively inhibits ATP turnover, actin binding during ATP turnover, and motor activity of F. graminearum myosin-1. Phenamacril also inhibits the ATPase activity of Fusarium avenaceum myosin-1 but has little or no inhibitory effect on the motor activity of Fusarium solani myosin-1, human myosin-1c, and D. discoideum myosin isoforms 1B, 1E, and 2. Our findings indicate that phenamacril is a species-specific, noncompetitive inhibitor of class I myosin in susceptible Fusarium sp.
Keywords: fungi, myosin, ATPase, inhibitor, allosteric regulation, inhibition mechanism, cytoskeleton, fungicide, motor protein, phenamacril
Introduction
The cyanoacrylate phenamacril (Fig. 1) (1) (formerly known as JS399–19) has been presented as a novel and environmentally benign fungicide. A growth-inhibitory effect of phenamacril against certain economically devastating phytopathogenic fungi within the genus Fusarium is well-established (2–6). By inhibiting the ATPase activity of class I myosin in susceptible Fusarium spp., phenamacril disrupts the activity of an essential actin-associated motor protein (3, 4). Myosins are ubiquitous eukaryotic motor proteins, which can be divided into ∼35 classes (7). Although several classes and isoforms may be present in a given organism, Fusarium only encodes single myosin heavy chains (MHC) from class I (4), class II (8), class V (9), and class XVII (10). All myosin isoforms share a functionally and structurally conserved N-terminal motor domain, a neck region which binds EF-hand proteins such as myosin light chains or calmodulin (11, 12) and class-specific C-terminal dimerization and/or cargo-binding domains. The Mg2+-dependent ATPase activity of the motor domain utilizes the energy stored in ATP to produce unidirectional movement along polar actin filaments. Thereby, myosin isoforms facilitate directional cargo-transport processes, local constriction, and other specialized energy-requiring tasks within the cell (8, 13–17).
Figure 1.
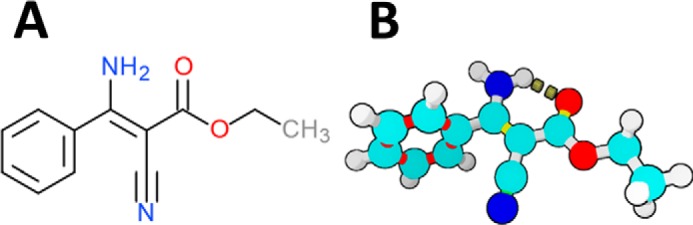
Structure of phenamacril. A, structural formula of phenamacril ((Z)-ethyl 2-cyano-3-amino-3-phenylacrylate). B, energy-minimized ball-and-stick model. Empirical evidence suggests that an intramolecular hydrogen-bond between the amine proton and the oxo-group stabilizes the Z-configuration.
Since the establishment of baseline sensitivity of Fusarium graminearum to phenamacril in 2008 (18), both laboratory (3, 4, 18–20) and field-resistant strains (5) have been characterized in China, where the compound is widely used to control Fusarium-induced infections of cereals (5, 6). Resistance development was observed to correlate with amino acid mutations, which are primarily located within the actin-binding cleft of the myosin motor domain (4, 5, 19). This region is known to play a pivotal role in mediating allosteric communication between the nucleotide- and actin-binding sites (21–23). It is a well-characterized and conserved allosteric site targeted by other class-specific myosin inhibitors such as blebbistatin (24) and the halogenated pseudilins (21, 23, 25). Although much is known about the effect of blebbistatin and the halogenated pseudilins on actomyosin kinetics (21, 23, 24, 26, 27), the biochemical parameters associated with the phenamacril-mediated inhibition of Fusarium class I myosin have not been characterized.
Here, we describe the elucidation of the mechanism underlying phenamacril-mediated inhibition of Fusarium spp. class I myosin and provide insights into its effect on actomyosin kinetics. To this end, we undertook the production of four active myosin motor domain constructs from both susceptible and phenamacril-resistant species of Fusarium. We copurified the constructs with F. graminearum calmodulin (FgCaM)4 bound to the lever arm region (28). The soluble and active protein preparations were used for functional analyses. We used an in vitro motility assay (29) to assess the effect of phenamacril on the capacity of the myosin head construct to translocate fluorescently labeled F-actin filaments before and after inhibitor washout. This allowed us to demonstrate that phenamacril acts as a reversible effector of motor function. Finally, we used an NADH-coupled ATPase assay and stopped-flow measurements to establish a nanomolar IC50 value for the phenamacril-mediated inhibition of F. graminearum class I myosin (FgMyo1) (30) and to demonstrate that phenamacril is a specific and noncompetitive inhibitor of myosin ATPase activity.
Results
Phenamacril reversibly inhibits the motor function of the FgMyo1-FgCaM complex
Using the baculovirus expression system, we produced and purified myosin constructs from F. graminearum, F. avenaceum, and F. solani in Sf9 insect cells. Myosin heavy chain constructs encompassing two IQ-motifs were coproduced with FgCaM. Partial loss of FgCaM during the final size-exclusion chromatography step (Fig. S1) did not affect the solubility and stability of the purified protein. However, motor activity was greatly reduced. The protein could be stored at −80 °C in buffer 30% trehalose in this form. Motor activity was restored when recombinant FgCaM produced in Escherichia coli was added to FaMyo1IQ2, FgMyo1IQ2, or FsMyo1IQ2 after thawing. Typically, substoichiometric additions of FgCaM were sufficient for maximal activation.
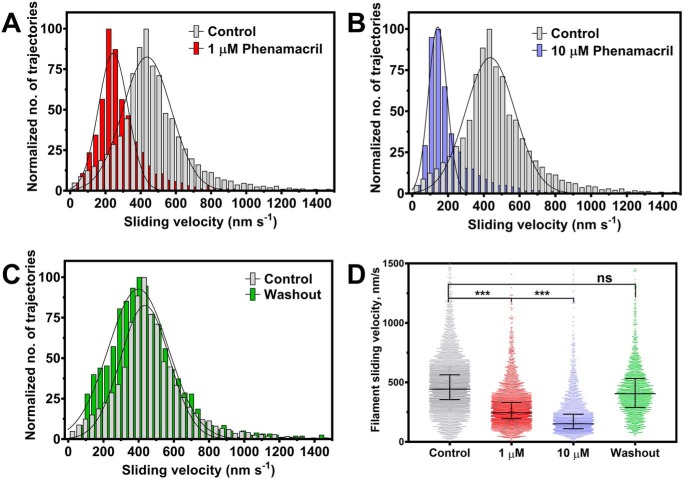
To assess if phenamacril-mediated inhibition of F. graminearum class I myosin is reversible, we conducted in vitro motility assays, where F-actin filaments move in an ATP-dependent manner on nitrocellulose-coated glass slides decorated with FgMyo1IQ2. More than 600 rhodamine-phalloidin–labeled F-actin filaments were tracked, both before and after the infusion of phenamacril, as well as after inhibitor washout. The resulting trajectory-associated velocities could be fitted to Gaussian distributions (Fig. 2). Specifically, we found that phenamacril inhibits the movement of F-actin filaments. In the absence of the inhibitor, actin filaments moved with an average velocity of 436 ± 165 nm·s−1. In the presence of 1 μm and 10 μm phenamacril, we observed average velocities of 234 ± 100 nm·s−1 and 133 ± 64 nm·s−1, respectively. Washout of the inhibitor restored the average sliding velocity to 389 ± 201 nm·s−1.
Figure 2.
Functional inhibition of FgMyo1IQ2 by phenamacril. A–C, trajectory-normalized distribution of the pooled sliding velocities of rhodamine-phalloidin–labeled F-actin filaments on a lawn of FgMyo1IQ2 before and after the infusion of phenamacril. The average sliding velocity decreases with the sequential infusion of 1–10 μm phenamacril (A and B) and is restored upon inhibitor washout (C). D, scatter plot of filament sliding velocities with median and interquartile range from three independent experiments. *** and ns denote that the differences between experiments were significant (p < 0.0005) or not significant, respectively.
Phenamacril is a noncompetitive inhibitor of FgMyo1
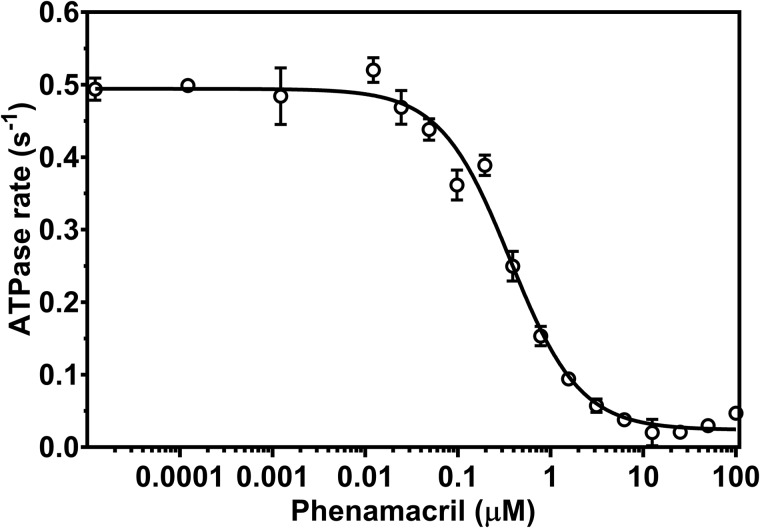
To further characterize the inhibitory potential of phenamacril, we established the half-maximal inhibitory concentration (IC50 value) by using a steady-state NADH-coupled ATPase assay in the presence of 20 μm F-actin and increasing concentrations of phenamacril in the range from 0.1 nm to 100 μm. To simplify the assay, we used motor domain construct FgMyo1, which lacks both IQ-motifs. FgMyo1 displays the same ATPase activity as FgCaM-saturated construct FgMyo1IQ2. Consistent with the data from the in vitro motility assay, phenamacril inhibited the ATPase activity in a dose-dependent manner. By nonlinear regression, we determined the relative IC50 value of the phenamacril-mediated inhibition of FgMyo1 to 365 ± 39 nm with 0–10% residual ATPase activity at ∼10 μm phenamacril (Fig. 3).
Figure 3.
Phenamacril is a potent inhibitor of FgMyo1 ATPase activity. The steady-state actin-activated ATPase rate of FgMyo1 was measured in the presence of 20 μm F-actin and 0.1 to 100 μm phenamacril. A four-parameter logistic curve with variable slope (Hill equation) was used to establish an IC50 value of 365 ± 39 nm (n = 6 reactions, three replicate series).
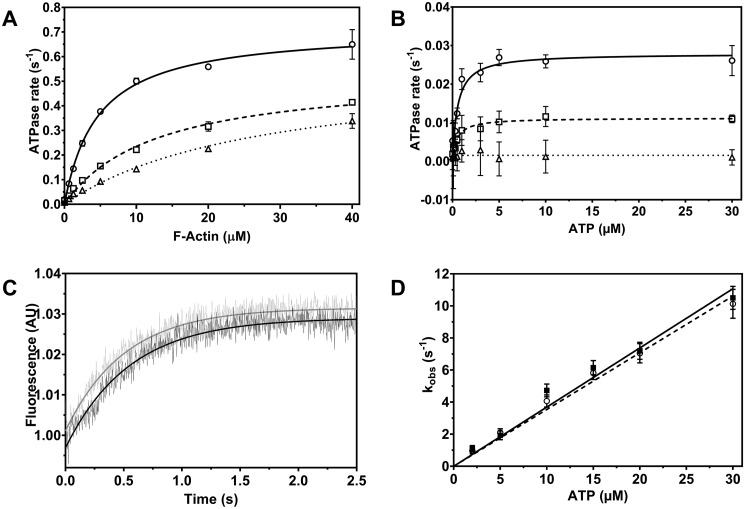
To elucidate the mechanism of inhibition, we measured the inhibited and noninhibited steady-state ATPase activity of FgMyo1 as a function of the F-actin and ATP concentration (Fig. 4, A and B). Fitting the data with hyperbolic functions demonstrated that the effect of phenamacril on the actin-activated ATPase activity was a lowering of kcat,actin (maximum ATP turnover in the presence of saturating concentrations of F-actin) and an increase in the value for Kapp,actin (apparent F-actin affinity in the presence of ATP). Specifically, in the presence of 300 and 600 nm phenamacril, Kapp,actin increased from 4.8 ± 0.5 μm to 11.7 ± 0.9 μm and 24.9 ± 3.5 μm, respectively. kcat,actin decreased from 0.72 ± 0.04 s−1 in the noninhibited state to 0.51 ± 0.03 s−1 and 0.49 ± 0.05 s−1 in the presence of 300 and 600 nm phenamacril, respectively. ATP titration in the presence of phenamacril further demonstrated that although phenamacril reduces kcat,basal (maximum ATP turnover in the absence of F-actin), it does not significantly affect the apparent affinity of FgMyo1 for ATP (Kapp,basal) (Fig. 4B and Fig. S2). This result confirms that phenamacril is a noncompetitive inhibitor of FgMyo1.
Figure 4.
Phenamacril-induced changes in myosin function. A, actin-activated ATPase activity of FgMyo1 was measured over the range from 0 to 40 μm F-actin in the absence and presence of phenamacril. Open circle, ethanol control (0 μm phenamacril) (kcat,actin = 0.72 ± 0.04 s−1, Kapp,actin = 4.80 ± 0.5 μm); open square, 300 nm (kcat,actin = 0.51 ± 0.03 s−1, Kapp,actin = 11.7 ± 0.09 μm); open triangle, 600 nm phenamacril (kcat,actin = 0.49 ± 0.05 s−1, Kapp,actin = 24.9 ± 3.5 μm). B, basal ATPase activity of FgMyo1 was measured in assay buffer containing Ca2+ and ATP in the range from 0 to 30 μm. Open circle, ethanol control (0 μm phenamacril): kcat,basal = 0.028 ± 0.0009 s−1, Kapp,basal = 0.60 ± 0.09 μm); open square, 600 nm phenamacril: kcat,basal = 0.011 ± 0.0004 s−1, Kapp,basal = 0.53 ± 0.09 μm); open triangle, 10 μm phenamacril: no ATP turnover. C, inhibition of ATP-induced dissociation of the acto-FgMyo1-MD complex are monitored by the associated change in pyrene-actin fluorescence. The transients shown were obtained after mixing a complex of 0.4 μm FgMyo1-MD and 0.2 μm pyrene-actin with 10 μm ATP in the presence of 2% ethanol (control, black transient) or 10 μm phenamacril (gray transient). The rate constants (kobs) for the dissociation of the actomyosin complex are obtained from exponential fits to the data and correspond to 1.93 s−1 (control) and 2.11 s−1 (10 μm phenamacril). Traces were normalized to their starting values. D, the kobs values of the ATP-induced dissociation plotted against a series of ATP concentrations in the absence (control, filled square) and presence (open circle) of 10 μm phenamacril (in both syringes) are shown as a linear fit to the data. The apparent second-order rate constant (K1k+2) for ATP binding to acto-FgMyo1 is 0.37 ± 0.02 μm−1s−1 in the absence and 0.35 ± 0.01 μm−1s−1 in the presence of phenamacril. Data in panels A and B were fitted by the hyperbolic Michaelis-Menten function. Error bars denote the S.D. around the mean (A and B: n = 4 reactions, two replicate series; D: n = 4–6 measurements, two independent experiments).
We used stopped-flow experiments to probe the impact of phenamacril on the population of the weak actin-binding myosin states. In the absence of ATP, the association of pyrene-labeled fluorescent F-actin with the myosin motor (rigor state) results in quenching of the fluorescence signal. We measured the rate of the ATP-induced increase in fluorescence that results from actomyosin dissociation (Fig. 4C). Exponential fits to the fluorescence transients yielded kobs values that were plotted against the respective ATP concentration (Fig. 4D). Linear fitting of the data gave the second-order rate constant (K1k+2), which in the absence and presence of 10 μm phenamacril was 0.37 ± 0.02 μm−1·s−1 and 0.35 ± 0.01 μm−1·s−1, respectively. This shows that phenamacril, like blebbistatin and the halogenated pseudilins, does not inhibit or interfere with ATP-induced dissociation of the actomyosin complex. In line with the steady-state ATPase experiments, it further shows that phenamacril does not compete with ATP for the nucleotide-binding site.
Specificity of Fusarium myosin-1 inhibition by phenamacril
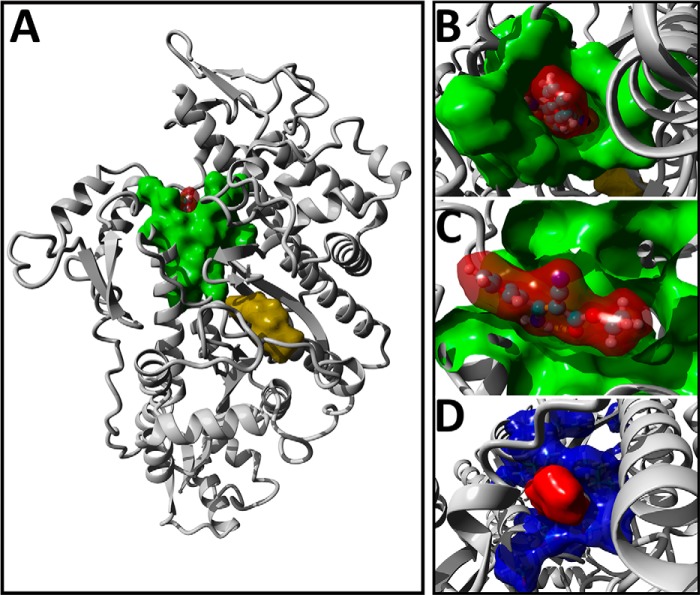
To characterize the specificity of phenamacril, we initially modeled the pre–power stroke state of FgMyo1 and docked phenamacril into 20 conformers covering a 500-ps molecular dynamics trajectory. In line with the noncompetitive mode of inhibition, the best-ranking docking poses clustered at the apex of the actin-binding cleft (Fig. 5, A–C and Fig. S3), in the immediate vicinity of residues that have been associated with resistance development (Fig. 5D). Residues define the allosteric binding pocket and are predicted to pack tightly around phenamacril. In particular, the aliphatic ethyl-ester chain of phenamacril is predicted to make close contact with residues at the rear of the binding pocket. In contrast, the aromatic moiety of the inhibitor occupies a more solvent-exposed position. Most of the interactions are hydrophobic in nature. Hydrogen-bond interactions are contributed by main-chain residues Ser-217, Lys-537, and Cys-423 and the side chain of Cys-423 (Fig. S3).
Figure 5.
Blind-docking of phenamacril is consistent with a noncompetitive mode of inhibition. A, the position of the top-ranking docking pose of phenamacril (red surface-rendering) in FgMyo1 is shown, highlighting contact residues (green). For reference, the position of ADP-vanadate (yellow) was superimposed into the model. B and C, top view (B) and cut-plane side view (C) of phenamacril and the molecular surface of amino acid residues involved in the FgMyo1-phenamacril interaction. D, surface-rendering of the phenamacril docking-pose (red) and position of residues (blue), which are known to mutate and confer resistance to phenamacril.
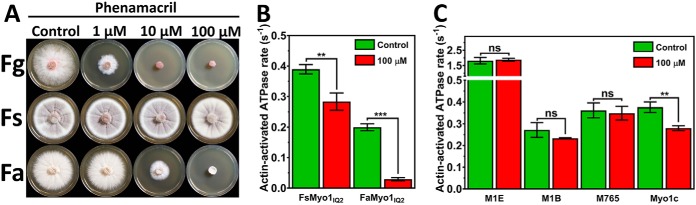
When compared to F. graminearum, F. avenaceum and F. solani show reduced susceptibility and resistance (respectively) to Phenamacril on Phenamacril-amended agar plates (Fig. 6A). To further assess whether natural resistance in F. solani can be attributed to myosin sequence divergence (Fig. S4) or whether other mechanisms of resistance are involved, we examined the effect phenamacril on actin-activated turnover of ATP by F. solani myosin head construct FsMyo1IQ2 (Fig. 6B). Consistent with the amended agar assay, our results show that the growth-inhibitory effect on F. avenaceum correlates with the inhibition of the enzymatic activity of FaMyo1IQ2 in vitro. Conversely, although phenamacril also significantly inhibits FsMyo1IQ2 (p < 0.005), it still retained 78% ATPase activity at the extreme of 100 μm phenamacril. FsMyo1IQ2 has two S217T and M375K substitutions in the immediate vicinity of the proposed phenamacril-binding pocket of F. graminearum. Introduction of either or both substitutions into the docked homology model resulted in a spatial overlap of phenamacril with the exchanged side chains (Fig. S4), suggesting this to be the resistance-conferring mechanism in F. solani.
Figure 6.
Specificity of phenamacril-mediated inhibition. A, phenamacril-amended agar plates inoculated with 5 mm plugs of F. graminearum (Fg), F. solani (Fs), and F. avenaceum (Fa). B, steady-state actin-activated ATPase activity of FsMyo1IQ2 and FaMyo1IQ2 in the presence of either 0 μm (ethanol control) or 100 μm phenamacril and 20 μm F-actin (n = 6 measurements, three replicate series). C, steady-state actin-activated ATPase activity of D. discoideum myosin-1B (M1B, 10 μm F-actin), myosin-1E (M1E, 10 μm F-actin), myosin 2 (M765, 10 μm F-actin), and human myosin-1c (Myo1c, 20 μm F-actin), in the presence of 0 μm (ethanol control) or 100 μm phenamacril (n = 3–6 measurements, two replicate series). Error bars denote the S.D. around the mean. The difference between sample means were either statistically significant (** for p < 0.005), highly significant (*** for p ≤ 0.0005), or not significant (ns).
To further assess the proposed species-specificity of phenamacril for Fusarium, we decided to include a selection of important and well-characterized myosin constructs in our study. These included human myosin-1c (Myo1c) and D. discoideum myosin-1E, myosin-1B (M1B), and myosin-2 (M765) (an amino acid alignment is shown in Fig. S5). Human Myo1c is involved in insulin uptake (12); myosin-1E is involved in phagocytosis (31); myosin-1B in cell translocation and intracellular particle motility (15); and myosin-2 is essential for cell movement, efficient chemotaxis, capping of cell surface receptors, and cytokinesis (13, 14). We measured the effect of 100 μm phenamacril on their actin-activated motor domain steady-state ATPase activities but found no statistically significant difference between the controls and the phenamacril-amended samples for the D. discoideum myosins (Fig. 6C). Human Myo1c activity is inhibited to a similar extent as FsMyo1IQ2. Overall our results indicate that phenamacril is a potent noncompetitive inhibitor of F. graminearum class I myosin but that further studies are warranted in regard to its environmental impact.
Discussion
We purified active Fusarium myosin motor domain constructs and characterized the mechanism and parameters underlying the phenamacril-induced inhibition of myosin ATPase and motor activity. Phenamacril is currently being applied as a fungicide in China for agricultural management of Fusarium-induced plant diseases (5, 6). Although the seemingly high specificity associated with phenamacril indicates a noncompetitive mode of inhibition, the mechanism has remained uncharacterized for more than a decade. From an agricultural perspective, insight into this mechanism is of relevance for evaluating the potential risks associated with the application of a fungicide that targets a ubiquitous and structurally conserved domain. ATP-binding motifs including the P-loop, switch-1, and switch-2 are not only highly conserved within the myosin family but among all P-loop nucleotide triphosphatases, such as microtubule-based motor proteins, protein kinases, and G-proteins (7, 32). In contrast, allosteric effector-binding sites are less well-conserved (33–35). In the case of myosin motor domains, at least four nonoverlapping binding sites for druglike modulators of myosin function have been identified (23, 26, 36–38).
To resolve the involvement of either a competitive or a noncompetitive mode of inhibition, we produced soluble and active Fusarium myosin-1 head constructs in complex with FgCaM and demonstrated that the functional inhibition of FgMyoIQ2 is a reversible process. Moreover, our results show that FgCaM functions as a myosin light chain and binds to IQ-motifs from F. graminearum, F. avenaceum, and F. solani myosin-1. Moreover, we were able to purify an active truncated version of the motor domain (FgMyo1), which allowed for analysis in the absence of the myosin light chain. This allowed us to demonstrate that phenamacril has an inhibitory potency (IC50 ∼360 nm) comparable to other known reversible and noncompetitive myosin inhibitors such as blebbistatin (0.5–5 μm) (24), pentachloropseudilin (1–5 μm) (21), and pentabromopseudilin (1.2 μm) (23). More importantly, we could directly show that phenamacril does not compete with ATP for binding to the active site of FgMyo1 nor does it interfere with the ATP-induced dissociation of the actomyosin complex.
It has previously been shown that small molecule inhibitors can reduce myosin-mediated force generation by means of different mechanisms. For example, the more than 50-fold reduction of actin filament sliding velocity observed for D. discoideum myosin-5b in the presence of 10 μm of the inhibitor pentabromopseudilin has been shown to be the result of a combined effect of the drug on ATP binding, ATP hydrolysis, and ADP dissociation (23). Similarly, the blebbistatin-induced inhibition of the actin filament sliding velocity for skeletal muscle myosin-2 (40) has been attributed to a reduction of the phosphate release step (24). For both inhibitors, no significant effect on the rate of the ATP-induced dissociation was observed. In line with these studies, phenamacril seems to exert its effect not via reducing the rate of ATP-induced dissociation but rather affects one or more of the other steps in the actomyosin ATPase cycle.
Collectively, our study classifies phenamacril as a reversible-binding and noncompetitive myosin inhibitor. This is in good agreement with blind-docking studies, which predict phenamacril to bind in close proximity to the allosteric binding pockets associated with the halogenated pseudilins (21, 23, 25) and blebbistatin (26). This region includes several of the residues that are implicated in phenamacril resistance (3–5, 19, 20). This finding shows a path toward the rational design of new and improved cyanoacrylate fungicides based on the phenamacril scaffold.
Although blebbistatin and some halogenated pseudilins are known to be highly class specific, noncompetitive inhibitors of myosin ATPase and motor activity, they are not necessarily species specific (21, 23–26). The lack of any profound inhibitory activity against human Myo1c and the Dictyostelium myosins belonging to classes I and II seems to confirm the high degree of specificity reported for phenamacril (3, 4, 6, 18). Nonetheless, our results do not exclude toxicological implications for the application of phenamacril as a fungicide. Caution remains warranted in regard to its wider user (5, 6).
The correspondence between the amended agar assay and the in vitro ATPase assays indicates that the effect of phenamacril on Fusarium is tied to structural aspects of their myosins. In F. solani, we speculate that a S217T substitution is implicated in its resistance to phenamacril. Although a conservative substitution, this amino acid position has previously been linked to phenamacril resistance in Fusarium (3–5, 19). The myosins from D. discoideum and human Myo1c do not contain substitutions at the aforementioned position but are otherwise highly divergent, have numerous substitutions in the immediate vicinity of the proposed binding pocket, and have three to five substitutions throughout the residues known to be implicated in phenamacril resistance (cf. Fig. S5). Therefore, it seems likely that the lack of inhibitory activity against these myosin constructs is the combined effect of overall sequence divergence and specific amino acid substitutions.
In conclusion, phenamacril is a potent and reversible noncompetitive inhibitor of class I myosin in the model plant pathogen F. graminearum. Identification of its mode of action and allosteric binding pocket will serve not only as a reference foundation for future studies aimed at characterizing myosins in Fusarium but also facilitate the design of novel inhibitors or fungicides based on the phenamacril scaffold.
Experimental procedures
Phenamacril
Phenamacril ((Z)-ethyl 2-cyano-3-amino-3-phenylacrylate) was obtained in accordance with the experimental procedures described previously (41).
Species and strains
F. graminearum PH-1 (NRRL 31084), F. solani f. sp. pisi 77–14-4 (FGSC 9596), and F. avenaceum 05001 were acquired from the Agricultural Research Service Culture Collection, the Fungal Genetics Stock Center (42), and IBT culture collection at the Danish Technical University (Kgs. Lyngby, Denmark) (43), respectively. D. discoideum AX2 was supplied by the Institute for Biophysical Chemistry (Hannover Medical School, Germany) (44).
RNA purification cDNA library preparation
Fifty ml YPG medium, pH 6.5 ± 0.1 (20 g·liter−1 soya peptone, 10 g·liter−1 yeast extract, and 50 ml·liter−1 50% (w/v) D(+)-glucose) was inoculated with five 5 mm mycelia plugs from an actively growing culture and incubated for 5 days at 25 °C and 130 rpm. Mycelia was harvested with Miracloth (Merck), washed twice with double-distilled water and freeze-dried overnight. RNA was subsequently purified from ∼25 mg freeze-dried mycelium using the RNeasy Plant kit (Qiagen, Hilden, Germany), including the optional on-column DNase treatment. The cDNA library was synthesized with SuperScriptTM III Reverse Transcriptase (Thermo Fisher Scientific) and oligo T primers according to manufacturer's protocol.
Bioinformatics
DNA oligo primers (Eurofins Genomics, Ebersberg, Germany) (Table S1) for amplification of the protein-encoding region of calmodulin from F. graminearum PH-1, myosin class I motor domain constructs from F. graminearum PH-1, F. solani f. sp. pisi 77–14-4, and F. avenaceum 05001 were designed based on locus tags FGSG_01891, FGSG_01409, NECHADRAFT_103022, and FAVG1_11042, respectively. DNA oligo primers for amplification of motor domain encoding regions incorporated an in-frame N-terminal FLAG tag (MDYKDDDDK) and a C-terminal hexa-histidine tag (His6), the latter preceded by a flexible Ser-Gly-Ser linker (SGS).
In silico modeling
The crystal structure of D. discoideum myosin-1E (PDB ID 1LKX) (45) was used as template for homology modeling of the F. graminearum and F. solani class I myosin motor domains in the pre–power stroke state. The C chain of 1LKX with ADP-vanadate was energy-minimized and refined in YASARA Structure and WHAT IF version 17.8.15 (Yasara2 force field, 25 °C, TIP3P water model, 1000-ps simulation, 40 snapshots) (46, 47). The lowest energy conformer was subsequently used as a template in SWISS-MODEL (48). Following repositioning of the ADP-vanadate, the model was refined using the above parameters.
The last 20 refined structures (covering a 500-ps trajectory) were superpositioned and prepared for global ensemble docking with energy-minimized phenamacril. Following docking in YASARA with AutoDock VINA (100 docking runs per conformer) (49), the best-ranking docking pose was energy-minimized and redocked to visualize and identify interaction partners.
Vector construction
All coding sequences were amplified from the cDNA libraries using Phusion Green High-Fidelity DNA polymerase (Thermo Fischer Scientific). The coding region of FgCaM was cloned into the pFastBacTM Dual (pFBD) Expression Vector (Thermo Fischer Scientific) under control of the p10 promotor. The coding regions of FgMyo1 (amino acid residues 1–729), FgMyo1IQ2 (amino acid residues 1–776), FsMyo1IQ2 (amino acid residues 1–772), and FaMyo1IQ2 (amino acid residues 1–776) were subsequently cloned into the pFBD::FgCaM vector under the control of the polyhedrin promotor.
The FgCaM coding sequence for heterologous protein expression in E. coli BL21(DE3) was cloned into the malE-vector with ligase-independent cloning using LIC-qualified T4 DNA polymerase (Merck) to generate overhangs. The construct was fused in-frame with an N-terminally located maltose-binding protein CDS and an interspaced tobacco etch virus (TEV) protease site CDS. All coding regions were verified by Sanger DNA sequencing (Eurofins Genomics, Germany).
Protein expression and purification
FgMyo1, FgMyo1IQ2, FsMyo1IQ2, and FaMyo1IQ2 (collectively referred to as FMD) were overexpressed in the Bac-to-Bac® baculovirus expression system (Thermo Fisher Scientific) by cotransfecting Sf9 insect cells with FMD/FgCaM and HSP90/UNC45 (50) viral titer. Four liters of culture in Sf-900 III SFM (Thermo Fischer Scientific) was transfected and incubated at 27 °C and 130 rpm for ∼48 h. Cell pellets were subsequently washed once with phosphate buffer saline and ultrasonically lysed (2 × 30 s, 40% power, 50% duty cycle) in 160 ml ice-cold lysis buffer (50 mm HEPES, pH 7.3, 400 mm NaCl, 4 mm MgCl2, 0.1 mm EGTA, 5% glycerol, 2 mm ATP, 2.5 mm β-mercaptoethanol, 5 mm benzamidine (Sigma-Aldrich), three cOmplete Protease Inhibitor Mixture tablets (Roche), 0.1 mm PMSF, 100 μg/ml p-tosyl-l-arginine methyl ester (TAME), 80 μg/ml TPCK, 2 μg/ml pepstatin, and 5 μg/ml leupeptin). Following 30 min of gentle stirring at 4 °C, the supernatant was incubated for 2 h at 4 °C with 5 ml anti-FLAG® M2 resin (Sigma-Aldrich) on a rotating wheel. Following resuspension in an ATP wash buffer (50 mm HEPES, pH 7.3, 400 mm NaCl, 4 mm MgCl2, 0.1 mm EGTA, 0.5 mm ATP, 3 mm benzamidine, 0.2% Triton X-100), resins were transferred to a gravity flow column and washed with ∼50 column volumes of wash buffer I (50 mm HEPES, pH 7.3, 300 mm NaCl, 4 mm MgCl2, 0.1 mm EGTA), wash buffer II (50 mm HEPES, pH 7.3, 600 mm NaCl, 4 mm MgCl2, 0.1 mm EGTA), and finally eluted in a FLAG-elution buffer (50 mm HEPES, pH 7.3, 400 mm NaCl, 4 mm MgCl2, 0.1 mm EGTA, 3 mm benzamidine, 100 μg/ml FLAG peptide). Following overnight dialysis (50 mm HEPES, pH 7.3, 400 mm NaCl, 4 mm MgCl2, 0.5 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 3 mm benzamidine, 1% trehalose), the retentate was concentrated using an Amicon 50 kDa molecular weight cut-off filter (Merck). Motor domains with IQ-motifs were further purified on a pre-equilibrated Superdex 200 10/30 GL size-exclusion column and finally stored at −80 °C in 30% trehalose (Fig. S1A). Based on at least two independent purifications, the average final yields of FgMD (n = 3), FgMyo1IQ2 (n = 3), FaMyo1IQ2 (n = 2), and FsMyo1IQ2 (n = 2) were 8.0 mg, 6.0 mg, 8.0 mg, and 7.6 mg, respectively.
The expression and purification of D. discoideum myosin-1E, myosin-1B, and myosin-2 (M765) motor domain constructs with and without lever arm regions have been described elsewhere (31, 51, 52). Human Myo1c was obtained according to Münnich et al. (12). G-actin from Gallus (chicken pectoralis major muscle) was purified according to Lehrer and Kerwar (53) with minor modifications.
FgCaM was overexpressed in E. coli BL21(DE3) (Thermo Scientific) in 1 liter Lennox Luria Bertani broth, pH 7.0 (Alfa Aesar, Thermo Scientific). Expression was induced by 1 mm IPTG at A600 ∼ 0.5 and cultures were subsequently incubated overnight at 25 °C and 150 rpm. Cells were ultrasonically lysed (10 bursts, 10-s duration, 60 pause intervals, 40% power) on ice in the presence of 0.1 mm PMSF and the P8340 Protease Inhibitor Mixture (Sigma-Aldrich). Lysate was centrifuged at 30,000 relative centrifugal force for 45 min and the supernatant was then filtered through a 0.45 μm filter before being loaded onto a pre-equilibrated (20 mm Tri-HCl, pH 7.2, 100 mm NaCl, and 1 mm DTT) amylose resin–packed column on an Äkta Prime system (GE Healthcare). Elution was started using an amylose buffer (20 mm Tris-HCl, pH 7.2, 100 mm NaCl, 10 mm maltose, 1 mm DTT). MBP-FgCaM containing fractions were cleaved with 1:100 (w/w) TEV-protease produced in-house, and dialyzed overnight at 4 °C. The retentate was subsequently loaded onto a pre-equilibrated (20 mm Tri-HCl, pH 7.2, 50 mm NaCl, and 1 mm DTT) Q-Sepharose anion-exchange column and eluted with a gradient IEX buffer (20 mm Tris-HCl, pH 7.2, 50 mm NaCl, 500 mm NaCl, 1 mm DTT). Calmodulin-containing fractions were finally separated on a Superdex 16/60 pg 200 size-exclusion column pre-equilibrated in 20 mm HEPES, pH 7.2, 100 mm NaCl, and 1 mm DTT (Fig. S1B).
ATPase assays
Steady-state NADH-coupled ATPase assays in the presence of variable concentrations of F-actin were performed at 25 °C in standard ATPase-buffer (25 mm HEPES, pH 7.3, 5 mm MgCl2, 25 mm KCl, 0.5 mm DTT) containing 0.4 mm NADH (Roche), 0.02 mg·ml−1 lactate dehydrogenase (Roche), 0.5 mm phosphoenolpyruvate (Roche), 0.05 mg·ml−1 pyruvate kinase (Roche), 1 mm ATP (Sigma-Aldrich), and 0–100 μm phenamacril (30). Basal steady-state ATPase rate of the truncated version of FgMyo1 in the presence of Ca2+ was measured in an assay solution containing 5 mm CaCl2, 0.6 m KCl, 1 mm ATP, and 25 mm HEPES, pH 7.4. Reactions were started by the addition of 0.1 μm FgMyo1. Basal ATP turnover of FgMyo1 in the presence of Ca2+ is ∼4-fold faster compared with basal ATP turnover in standard ATPase-buffer. Measurements were performed in a Tecan Infinite M1000 Pro (Tecan Trading AG, Switzerland) or a MultiskanFC Microplate Photometer (Thermo Fisher Scientific) at 340 nm in Costar Corning half-area plates (Sigma-Aldrich) at 10- or 20-s intervals (180–360 intervals).
Stopped-flow kinetics
Transient kinetic experiments were performed at 20 °C with a Hi-tech Scientific SF-61 SX single mixing stopped-flow system (TgK Scientific Limited, Bradford on Avon, UK) in assay buffer (20 mm MOPS, pH 7.0, 100 mm KCl, 5 mm MgCl2, 1 mm DTT). Pyrene fluorescence was excited at 365 nm and detected after passing through a KV 389 nm cut-off filter. Fluorescence transients were fitted with single exponential equations and the resulting kinetic data were analyzed according to models described previously (17, 54). The mechanism of ATP-induced pyrene fluorescence enhancement was modeled according to Equation 1, which describes a two-step mechanism for ATP binding to actomyosin:
| (Eq. 1) |
At lower ATP concentrations (<50 μm), the observed rate constants increased linearly, defining the second-order rate binding constant K1k+2.
In vitro motility assays
The in vitro motility experiments were performed according to Kron and Spudich (29) with modifications described by Taft and coworkers (17). Approximately 7.7 μm FgMyo1IQ2 in assay buffer (25 mm imidazole, pH 7.4, 25 mm KCl, 4 mm MgCl2, 1 mm EGTA) containing 5 μm FgCaM, 12 μm sheared F-actin, 10 mm DTT, and 2 mm ATP was centrifuged for 20 min at 100,000 × g and 4 °C. The supernatant contains the active myosin, which was bound via an anti-His antibody (Penta-His, Qiagen, Hilden, Germany) to the flow-cell surface. 20 nm rhodamine-phalloidin–labeled F-actin was subsequently added to the flow cell (∼10 μl capacity). All buffer solutions used after this step contained 7.5 μm BSA and 5 μm FgCaM. To probe the effect of phenamacril on motile activity, the flow cells were consecutively infused with 2 mm Mg2+-ATP and 0, 1, 10, and 0 μm (washout) phenamacril. Movement of the rhodamine-phalloidin–labeled F-actin was monitored at 25 °C at 300-ms intervals for 30 s with an Olympus IX70 epifluorescence microscope equipped with a 60× Plan Apo 1.49 NA oil immersion objective lens (ApoN, Olympus) and an OrcaFlash 4.0 CMOS camera (Hamamatsu Photonics, Iwata City, Japan). For each flow-cell, filament sliding velocity was tracked in three independent areas with DiaTrack 3.05 (39) (Semasopht, Switzerland). GraphPad Prism 7.03 was used for final binning and data analysis.
Amended agar assay
Mycelia plugs (5 mm) taken from actively growing colonies were placed in the center of 5.5-cm vented Petri dishes containing YPG agar media, pH 6.5 ± 0.1. Plates were amended with either 1–100 μm phenamacril or 0.5% ethanol (control). Growth of three replicates was monitored at 25 °C until the control reached the edge of the plate (42).
Statistical analysis
Unless otherwise stated, error bars denote mean ± S.D. In vitro motility data are reported as mean ± ½FWHM (full width at half maximum). Student's two-tailed t test was used to compare sample means, with statistical significance assigned as ns (not significant) for p > 0.05, * for p ≤ 0.05, ** for p ≤ 0.005, and *** for p ≤ 0.0005.
Author contributions
R. D. W. and M. H. T. data curation; R. D. W., M. H. T., S. G., C. T., Z. B., and D. J. M. formal analysis; R. D. W., M. H. T., D. J. M., and T. E. S. investigation; R. D. W. writing-original draft; M. H. T., H. G., and D. J. M. funding acquisition; M. H. T., H. G., D. J. M., and T. E. S. methodology; M. H. T., H. G., D. J. M., and T. E. S. writing-review and editing; D. J. M. and T. E. S. conceptualization; D. J. M. and T. E. S. supervision; D. J. M. and T. E. S. project administration.
Supplementary Material
Acknowledgments
We thank Michael Toft Overgaard for providing us with the malE-vector, Christian Holt for assistance during the purification of Fusarium calmodulin. Joanna Berger for expert secretarial assistance. Special thanks to Reinhard Wimmer, Thorbjørn T. Nielsen, and Søren S. Donau for input on the chemical aspects of phenamacril.
This work was supported by The Danish Ministry of Higher Education and Science Grant 4005-00204B (to T. E. S.), Deutsche Forschungsgemeinschaft Grant MA1081/22-01 (to D. J. M), and Volkswagen Foundation Grant VWZN3012 (to M. H. T. and D. J. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
- FgCaM
- Fusarium graminearum calmodulin
- FgMyo1
- F. graminearum class I myosin motor domain construct
- FaMyo1IQ2
- F. avenaceum myosin head construct including 2 IQ-motifs
- FgMyo1IQ2
- F. graminearum myosin head construct including 2 IQ-motifs
- FsMyo1IQ2
- F. solani myosin head construct including 2 IQ-motifs
- A·M
- actomyosin complex
- TPCK
- l-1-tosylamido-2-phenylethyl chloromethyl ketone
- TEV
- tobacco etch virus.
References
- 1. Donau S. S., Bechmann M., Müller N., Nielsen T. T., and Wimmer R. (2017) (Z), not (E)—an end to a century of confusion about the double-bond stereoisomers of 3-amino-2-cyanoacrylates. Eur. J. Org. Chem. 2017, 6408–6412 10.1002/ejoc.201701235 [DOI] [Google Scholar]
- 2. Chen Y., Wang W. X., Zhang A. F., Gu C. Y., Zhou M. G., and Gao T. C. (2011) Activity of the fungicide JS399–19 against Fusarium head blight of wheat and the risk of resistance. Agric. Sci. China. 10, 1906–1913 10.1016/S1671-2927(11)60191-0 [DOI] [Google Scholar]
- 3. Zhang C., Chen Y., Yin Y., Ji H.-H., Shim W.-B., Hou Y., Zhou M., Li X., and Ma Z. (2015) A small molecule species specifically inhibits Fusarium myosin I. Environ. Microbiol. 17, 2735–2746 10.1111/1462-2920.12711 [DOI] [PubMed] [Google Scholar]
- 4. Zheng Z., Hou Y., Cai Y., Zhang Y., Li Y., and Zhou M. (2015) Whole-genome sequencing reveals that mutations in myosin-5 confer resistance to the fungicide phenamacril in Fusarium graminearum. Sci. Rep. 5, 8248 10.1038/srep08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hou Y.-P., Qu X.-P., Mao X.-W., Kuang J., Duan Y.-B., Song X. S., Wang J.-X., Chen C.-J., and Zhou M.-G. (2018) Resistance mechanism of Fusarium fujikuroi to phenamacril in the field. Pest Manag. Sci. 74, 607–616 10.1002/ps.4742 [DOI] [PubMed] [Google Scholar]
- 6. Li H., Diao Y., Wang J., Chen C., Ni J., and Zhou M. (2008) JS399–19, a new fungicide against wheat scab. Crop Prot. 27, 90–95 10.1016/j.cropro.2007.04.010 [DOI] [Google Scholar]
- 7. Odronitz F., and Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R106 10.1186/gb-2007-8-9-r196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song B., Li H. P., Zhang J. B., Wang J. H., Gong A. D., Song X. S., Chen T., and Liao Y. C. (2013) Type II myosin gene in Fusarium graminearum is required for septation, development, mycotoxin biosynthesis and pathogenicity. Fungal Genet. Biol. 54, 60–70 10.1016/j.fgb.2013.02.010 [DOI] [PubMed] [Google Scholar]
- 9. Zheng Z., Liu X., Li B., Cai Y., Zhu Y., and Zhou M. (2016) Myosins FaMyo2B and Famyo2 affect asexual and sexual development, reduces pathogenicity, and FaMyo2B acts jointly with the myosin passenger protein FaSmy1 to affect resistance to phenamacril in Fusarium asiaticum. PLoS One 11, e0154058 10.1371/journal.pone.0154058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Madrid M. P., Di Pietro A., and Roncero M. I. G. (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47, 257–266 [DOI] [PubMed] [Google Scholar]
- 11. Wolenski J. S. (1995) Regulation of calmodulin-binding myosins. Trends Cell Biol. 5, 310–316 10.1016/S0962-8924(00)89053-4 [DOI] [PubMed] [Google Scholar]
- 12. Münnich S., Taft M. H., and Manstein D. J. (2014) Crystal structure of human myosin-1c—the motor in GLUT4 exocytosis: Implications for Ca2+ regulation and 14–3-3 binding. J. Mol. Biol. 426, 2070–2081 10.1016/j.jmb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 13. Manstein D. J., Titus M. A., De Lozanne A., and Spudich J. A. (1989) Gene replacement in Dictyostelium: Generation of myosin null mutants. EMBO J. 8, 923–932 10.1002/j.1460-2075.1989.tb03453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Lozanne A., and Spudich J. A. (1987) Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086–1091 10.1126/science.3576222 [DOI] [PubMed] [Google Scholar]
- 15. Wessels D., Murray J., Jung G., Hammer J. A. 3rd, and Soll D. R. (1991) Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil. Cytoskeleton 20, 301–315 10.1002/cm.970200406 [DOI] [PubMed] [Google Scholar]
- 16. Makowska K. A., Hughes R. E., White K. J., Wells C. M., and Peckham M. (2015) Specific myosins control actin organization, cell morphology, and migration in prostate cancer cells. Cell Rep. 13, 2118–2125 10.1016/j.celrep.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taft M. H., Hartmann F. K., Rump A., Keller H., Chizhov I., Manstein D. J., and Tsiavaliaris G. (2008) Dictyostelium myosin-5b is a conditional processive motor. J. Biol. Chem. 283, 26902–26910 10.1074/jbc.M802957200 [DOI] [PubMed] [Google Scholar]
- 18. Chen Y., Li H., Chen C., and Zhou M. (2008) Sensitivity of Fusarium graminearum to fungicide JS399–19: In vitro determination of baseline sensitivity and the risk of developing fungicide resistance. Phytoparasitica 36, 326–337 10.1007/BF02980812 [DOI] [Google Scholar]
- 19. Li B., Zheng Z., Liu X., Cai Y., Mao X., and Zhou M. (2016) Genotypes and characteristics of phenamacril-resistant mutants in Fusarium asiaticum. Plant Dis. 100, 1754–1761 10.1094/PDIS-02-16-0169-RE [DOI] [PubMed] [Google Scholar]
- 20. Zheng Z., Zhang Y., Wu X., Yang H., Ma L., and Zhou M. (2018) FoMyo5 motor domain substitutions (Val151 to Ala and Ser418 to Thr) cause natural resistance to fungicide phenamacril in Fusarium oxysporum. Pestic. Biochem. Physiol. 147, 119–126 10.1016/j.pestbp.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 21. Chinthalapudi K., Taft M. H., Martin R., Heissler S. M., Preller M., Hartmann F. K., Brandstaetter H., Kendrick-Jones J., Tsiavaliaris G., Gutzeit H. O., Fedorov R., Buss F., Knölker H. J., Coluccio L. M., and Manstein D. J. (2011) Mechanism and specificity of pentachloropseudilin-mediated inhibition of myosin motor activity. J. Biol. Chem. 286, 29700–29708 10.1074/jbc.M111.239210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Behrens V. A., Münnich S., Adler-Gunzelmann G., Thiel C., Henn A., Latham S. L., and Taft M. H. (2017) The conserved lysine-265 allosterically modulates nucleotide- and actin-binding site coupling in myosin-2. Sci. Rep. 7, 7650 10.1038/s41598-017-07933-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fedorov R., Böhl M., Tsiavaliaris G., Hartmann F. K., Taft M. H., Baruch P., Brenner B., Martin R., Knölker H. J., Gutzeit H. O., and Manstein D. J. (2009) The mechanism of pentabromopseudilin inhibition of myosin motor activity. Nat. Struct. Mol. Biol. 16, 80–88 10.1038/nsmb.1542 [DOI] [PubMed] [Google Scholar]
- 24. Kovács M., Tóth J., Hetényi C., Málnási-Csizmadia A., and Seller J. R. (2004) Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279, 35557–35563 10.1074/jbc.M405319200 [DOI] [PubMed] [Google Scholar]
- 25. Preller M., Chinthalapudi K., Martin R., Knölker H. J., and Manstein D. J. (2011) Inhibition of myosin ATPase activity by halogenated pseudilins: A structure-activity study. J. Med. Chem. 54, 3675–3685 10.1021/jm200259f [DOI] [PubMed] [Google Scholar]
- 26. Allingham J. S., Smith R., and Rayment I. (2005) The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 12, 378–379 10.1038/nsmb908 [DOI] [PubMed] [Google Scholar]
- 27. Straight A. F., Cheung A., Limouze J., Chen I., Westwood N. J., Sellers J. R., and Mitchison T. J. (2003) Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 299, 1743–1747 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- 28. Anson M., Geeves M. A., Kurzawa S. E., and Manstein D. J. (1996) Myosin motors with artificial lever arms. EMBO J. 15, 6069–6074 10.1002/j.1460-2075.1996.tb00995.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kron S. J., and Spudich J. A. (1986) Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. U.S.A. 83, 6272–6276 10.1073/pnas.83.17.6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Furch M., Geeves M. A., and Manstein D. J. (1998) Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry 37, 6317–6326 10.1021/bi972851y [DOI] [PubMed] [Google Scholar]
- 31. Dürrwang U., Fujita-Becker S., Erent M., Kull F. J., Tsiavaliaris G., Geeves M. A., and Manstein D. J. (2006) Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J. Cell Sci. 119, 550–558 10.1242/jcs.02774 [DOI] [PubMed] [Google Scholar]
- 32. Saraste M., Sibbald P. R., and Wittinghofer A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 10.1016/0968-0004(90)90281-F [DOI] [PubMed] [Google Scholar]
- 33. Kuriyan J., and Eisenberg D. (2007) The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 10.1038/nature06524 [DOI] [PubMed] [Google Scholar]
- 34. Salvesen G. S., and Riedl S. J. (2007) Caspase inhibition, specifically. Structure 15, 513–514 10.1016/j.str.2007.04.006 [DOI] [PubMed] [Google Scholar]
- 35. Schweizer A., Roschitzki-Voser H., Amstutz P., Briand C., Gulotti-Georgieva M., Prenosil E., Binz H. K., Capitani G., Baici A., Plückthun A., and Grütter M. G. (2007) Inhibition of caspase-2 by a designed ankyrin repeat protein: Specificity, structure, and inhibition mechanism. Structure 15, 625–636 10.1016/j.str.2007.03.014 [DOI] [PubMed] [Google Scholar]
- 36. Planelles-Herrero V. J., Hartman J. J., Robert-Paganin J., Malik F. I., and Houdusse A. (2017) Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 8, 190 10.1038/s41467-017-00176-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sirigu S., Hartman J. J., Planelles-Herrero V. J., Ropars V., Clancy S., Wang X., Chuang G., Qian X., Lu P.-P., Barrett E., Rudolph K., Royer C., Morgan B. P., Stura E. A., Malik F. I., and Houdusse A. M. (2016) Highly selective inhibition of myosin motors provides the basis of potential therapeutic application. Proc. Natl. Acad. Sci. 113, E7448–E7455 10.1073/pnas.1609342113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Preller M., and Manstein D. J. (2013) Myosin structure, allostery, and mechano-chemistry. Structure 21, 1911–1922 10.1016/j.str.2013.09.015 [DOI] [PubMed] [Google Scholar]
- 39. Vallotton P., and Olivier S. (2013) Tri-track: Free software for large-scale particle tracking. Microsc. Microanal. 19, 451–460 10.1017/S1431927612014328 [DOI] [PubMed] [Google Scholar]
- 40. Limouze J., Straight A. F., Mitchison T., and Sellers J. R. (2004) Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 25, 337–341 10.1007/s10974-004-6060-7 [DOI] [PubMed] [Google Scholar]
- 41. Wollenberg R. D., Donau S. S., Nielsen T. T., Sørensen J. L., Giese H., Wimmer R., and Søndergaard T. E. (2016) Real-time imaging of the growth-inhibitory effect of JS399–19 on Fusarium. Pestic. Biochem. Physiol. 134, 24–30 10.1016/j.pestbp.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 42. McCluskey K., Wiest A., and Plamann M. (2010) The fungal genetics stock center: A repository for 50 years of fungal genetics research. J. Biosci. 35, 119–126 10.1007/s12038-010-0014-6 [DOI] [PubMed] [Google Scholar]
- 43. Lysøe E., Harris L. J., Walkowiak S., Subramaniam R., Divon H. H., Riiser E. S., Llorens C., Gabaldón T., Kistler H. C., Jonkers W., Kolseth A.-K., Nielsen K. F., Thrane U., and Frandsen R. J. N. (2014) The genome of the generalist plant pathogen Fusarium avenaceum is enriched with genes involved in redox, signaling and secondary metabolism. PLoS One 9, e112703 10.1371/journal.pone.0112703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manstein D. J., Ruppel K. M., and Spudich J. A. (1989) Expression and characterization of a functional myosin head fragment in Dictyostelium discoideum. Science 246, 656–658 10.1126/science.2530629 [DOI] [PubMed] [Google Scholar]
- 45. Kollmar M., Dürrwang U., Kliche W., Manstein D. J., and Kull F. J. (2002) Crystal structure of the motor domain of a class-I myosin. EMBO J. 21, 2517–2525 10.1093/emboj/21.11.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krieger E., and Vriend G. (2014) YASARA View—molecular graphics for all devices - from smartphones to workstations. Bioinformatics. 30, 2981–2982 10.1093/bioinformatics/btu426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., and Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinformat. 77, 114–122 10.1002/prot.22570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., and Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13 10.1038/nprot.2008.197 [DOI] [PubMed] [Google Scholar]
- 49. Trott O., and Olson A. J. (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bird J. E., Takagi Y., Billington N., Strub M.-P., Sellers J. R., and Friedman T. B. (2014) Chaperone-enhanced purification of unconventional myosin-15, a molecular motor specialized for stereocilia protein trafficking. Proc. Natl. Acad. Sci. 111, 12390–12395 10.1073/pnas.1409459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsiavaliaris G., Fujita-Becker S., Dürrwang U., Diensthuber R. P., Geeves M. A., and Manstein D. J. (2008) Mechanism, regulation, and functional properties of Dictyostelium myosin-1B. J. Biol. Chem. 283, 4520–4527 10.1074/jbc.M708113200 [DOI] [PubMed] [Google Scholar]
- 52. Kurzawa S. E., Manstein D. J., and Geeves M. A. (1997) Dictyostelium discoideum myosin II: Characterization of functional myosin motor fragments. Biochemistry 36, 317–323 10.1021/bi962166b [DOI] [PubMed] [Google Scholar]
- 53. Lehrer S. S., and Kerwar G. (1972) Intrinsic fluorescence of actin. Biochemistry 11, 1211–1217 10.1021/bi00757a015 [DOI] [PubMed] [Google Scholar]
- 54. Bagshaw C. R. (1975) The kinetic mechanism of the manganous ion-dependent adenosine triphosphatase of myosin subfragment 1. FEBS Lett. 58, 197–201 10.1016/0014-5793(75)80258-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.