Abstract
Recent studies provide compelling evidence to suggest that the tight junction protein claudin 1, aberrantly expressed in several cancer types, plays an important role in cancer progression. Dysregulation of claudin 1 has been shown to induce epithelial mesenchymal transition (EMT). Furthermore, activation of the ERK signaling pathway by protein kinase C (PKC) was shown to be necessary for EMT induction. Whether PKC is involved in regulating breast cancer progression has not been addressed. The PKC activator 12-O-tetradecanoylphorbol 13-acetate (TPA) was used to investigate the effect of PKC activity on claudin 1 transcription and protein levels, subcellular distribution, and alterations in EMT markers in human breast cancer (HBC) cell lines. As well, tissue microarray analysis (TMA) of a large cohort of invasive HBC biopsies was conducted to investigate correlations between claudin 1 and PKC isomers. TPA upregulated claudin 1 levels in all HBC cell lines analyzed. In particular, a high induction of claudin 1 protein was observed in the MCF7 cell line. TPA treatment also led to an accumulation of claudin 1 in the cytoplasm. Additionally, we demonstrated that the upregulation of claudin 1 was through the ERK signaling pathway. In patient biopsies, we identified a significant positive correlation between claudin 1, PKCα, and PKCε in ER+ tumors. A similar correlation between claudin 1 and PKCε was identified in ER− tumors, and high PKCε was associated with shorter disease-free survival. Collectively, these studies demonstrate that claudin 1 and the ERK signaling pathway are important players in HBC progression.
Introduction
The claudins are a family of integral membrane proteins central to the formation of the tight junctions (TJs) of epithelial cells [1], [2], [3], [4]. These TJ proteins are directly involved in the paracellular sealing between adjacent cells [1], [2], [3], [4] where they provide a “fence” and a “barrier” function, facilitating the active transport of small ions and nutrients between these cells [5]. As well, TJ proteins are also considered key players in maintaining apical and basolateral polarity across the plasma domains [6], [7], [8], [9], [10], [11], for review: [12], [13], [14]. Claudin 1, the first of 24 members of this family of proteins to be identified [1], [2], forms the backbone of the TJ in epithelial cells [15] and plays a vital role in regulating epithelial barrier function. Claudin 1–deficient mice die within 1 day of birth [15].
Currently, there exists a wealth of accumulating evidence which shows that some members of the claudin family, in particular claudin 1, exhibit abnormal gene expression and are associated with the cellular dysregulation and progression in human cancers [13], [14], [16], [17], [18], [19], [20], [21], [22]. During cancer progression, the upregulation of claudin 1 has been shown to lead to the promotion of epithelial mesenchymal transition, EMT [23], [24], [25], cellular invasion and migration [21], [24], [25], [26], [27], [28], [29], [30], as well as an accumulation or mislocalization of the claudin 1 protein in the cytoplasm [21], [24], [25], [28], [29], [31], [32], [33]. The more recent observation that some aggressive breast cancers are associated with low levels of claudin protein family members, 3, 4, 5, and 7 has now led to the consensus to define a new molecular subtype of breast cancers, the “claudin low” subtype [34], [35]. These claudin low breast tumors were generally derived from patients diagnosed with poor prognoses [36]. Conversely, high levels of claudin 1 have also been identified in, and associated with, the aggressive breast cancer phenotype. Original studies from our laboratory [31], [37], [38] and later others [39] identified an association between high claudin 1 expression/levels and breast cancer invasiveness. In a large cohort of human breast cancers of mixed pathologies, we found a significant correlation between high claudin 1 levels and the basal-like subtype, an aggressive form of breast cancer [31], [37]. High levels of claudin 1 have also been identified in the BRCA1 breast cancers, a tumor type that is linked to poor prognosis [40]. Additionally, tumors of the luminal subtype have been reported to exhibit high claudin 1 levels [39]. Whether these tumors are yet another new subtype of breast cancer warrants further investigations. Thus, the role of claudin 1 in breast cancer appears to be quite complex, and the range of levels reported among the different subtypes suggest that other mitigating factors, including the interaction with mediators in signaling pathways, such as the protein kinases, that play a role in cancer, may also impact the role of claudin 1 during breast cancer progression.
The multi-isomer protein kinase C (PKC) family of serine-threonine kinases, 12 identified to date [41], [42], plays regulatory roles in normal tissue as well as cancer. The most studied conventional isomers are PKCα, PKCδ, PKCε, and PKCγ, which, in healthy tissues, have been shown to be important in regulating epithelial barrier function and mammary gland development [43], [44], [45]; for review, 46]. Among the PKC isomers, much variation exists in terms of expression profile and mechanism of action [47], [48]. As well, many PKC isomers have also been shown to be involved in cancer progression and metastasis [49], [50], [51]; for review, 52] and play roles in both tumor suppression and/or promotion [53], [54], [55]. Notably, aberrant kinase activity to date has been linked to nearly 25% of all cancers [52].
A molecular connection between PKC and claudin 1 has now also been identified in several cancers, as an increasing number of studies show that PKC activity is important in regulating claudin 1 expression [21], [43], [56], [57], [58]. In these cancers, PKC activation led to increases in claudin 1 expression, alterations in EMT markers, increased cell motility, and enhanced aggressiveness [18], [21], [23], [26], [59], all of which have been proposed as possible mechanisms underlying claudin 1–dependent changes. PKC activation has also been associated with a mislocalization of claudin 1 from the cell membrane to the cytoplasm during cancer progression, and such mislocalization has been linked to enhanced metastatic potential [18], [21], [59]. However, a link between PKC and claudin 1 has not been demonstrated in breast cancer.
In the present study we investigated the effect of PKC activity on claudin 1 regulation in breast cancer. Using the PKC activator 12-O-tetradecanoylphorbol 13-acetate (TPA) and a panel of human breast cancer cell lines, we examined TPA effects on claudin 1 transcription and protein levels, subcellular distribution and cell proliferation, as well as alterations in EMT markers. We showed that PKC activation highly upregulated claudin 1 expression in MCF7 human breast cancer cells in vitro and that activation of PKCε was involved. We also determined that the TPA-stimulated claudin 1 upregulation was through the ERK pathway. Furthermore, using a large cohort of human breast cancer biopsies, we have identified a positive correlation between claudin 1 and PKCε. Collectively, these studies point to an important relationship between claudin 1, PKCε, and the ERK pathway during breast cancer progression and provide evidence to suggest that PKC activation is involved in the increased claudin 1 levels associated with some breast cancer subtypes.
Materials and Methods
Cell Culture
The human breast cancer (HBC) cell lines MCF7, BT-20, MDA-MB231, ZR75, and T47D and the non-tumorigenic human breast epithelial cell line MCF10A were obtained from the American Type Culture Collection (ATCC) and authenticated by Genetica DNA Laboratories (Burlington, NC). The Madin-Darby Canine Kidney (MDCKII) epithelial cell line was also purchased from ATCC. MCF7, MDA-MB231, T47D, ZR75, and MDCKII cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; 4.5 g/l glucose, Hyclone Laboratories Inc., Logan, UT) with 10% fetal bovine serum supplemented with 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM glutamine (all purchased from Hyclone Laboratories Inc.), and 10 μg/ml bovine insulin (Sigma-Aldrich Canada Co., Oakville, ON, Canada). BT-20 cells were cultured in Eagle’s Minimum Essential Medium (Lonza Inc., Williamsport, PA) with 10% fetal bovine serum and supplemented with 50 U/ml penicillin, 50 mg/ml streptomycin, and 1 mM pyruvate (all purchased from Hyclone Laboratories Inc.). MCF10A cells were grown in DMEM (4.5 g/L glucose) supplemented with 50 U/mL penicillin, 50 mg/mL streptomycin, 2 mM glutamine, and 5% horse serum (Hyclone Laboratories Inc.); 10 μg/ml bovine insulin, 1μM hydrocortisone, and 0.1 μg/ml cholera toxin (Sigma-Aldrich Canada Co.); and 20 ng/ml human epidermal growth factor (EGF; EMD Millipore Corp., Temecula, CA). All cells were grown at 37°C in an atmosphere of 95% air and 5% CO2.
Western Blot Analysis
Cells were lysed in isolation buffer [50 mM Tris-Cl pH 6.8; 5% sodium dodecyl sulfate (SDS); 20 mM EDTA; 5 mM β-glycerophosphate] containing complete mini protease inhibitor cocktail (Roche Diagnostics, Laval, QC, Canada). Protein concentration was determined using the mini BCA assay (Pierce Biotechnology, Rockford, IL). Samples were mixed 3:1 with 4× SDS buffer [(500 mM Tris, pH 6.8) 40% glycerol, 8% SDS, 0.04% (w/v) bromophenol blue and 0.4 M dithiothreitol], boiled for 5 minutes at 100°C, and electrophoresed in SDS polyacrylamide gels.
Western blotting was performed as previously described [33] using the following primary antibodies: rabbit claudin 1 (1:200; Life Technologies Inc., Burlington, ON, Canada; cat. #51-9000); mouse β-actin, (1:5000, Abcam Inc., Toronto, ON, Canada; cat. #ab8226); mouse E-cadherin (1:500, Life Technologies Inc.; cat. #33-4000); rabbit β-catenin (1:1000; cat. #8480), rabbit P44/42 MAPK (ERK1,2; 1:1500; cat. #4695), rabbit phospho-p44/42 MAPK (THR202/Tyr204; 1:1000; cat. #4370), rabbit SAPK/JNK (1:1000; cat. #9258), and rabbit phospho-SAPK/JNK (Thr183/Tyr185; 1:1000; cat. #4668), all purchased from Cell Signaling Technology (Danvers, MA).
Signals were developed with Pico chemiluminescence substrate (Pierce Biotechnology) and visualized on X-ray films. Densitometry of scanned images was carried out using the Image Studio Lite software v.5.2 (LI-COR Biosciences, Lincoln, NE).
Subcellular Fractionation
Cells were grown to 90% confluency, and subcellular fractions were isolated using the ProteoExtract Subcellular Proteome Extraction Kit (S-PEK, Calbiochem) according to the manufacturer’s instructions. Protein fractions were subjected to acetone precipitation, and pellets were reconstituted in sample isolation buffer (50 mM Tris-Cl pH 6.8; 5% SDS; 20 mM EDTA; 5 mM β-glycerophosphate, containing complete mini protease inhibitor cocktail). The mini BCA assay (Pierce Biotechnology) was used to determine the protein concentration of each fraction, prior to equal loading in 15% SDS-polyacrylamide electrophoresis gel and Western blotting with the claudin 1 (Life Technologies Inc.), E-cadherin (Life Technologies Inc.), and the steroid receptor activator protein (SRAP, 743A; Bethyl Laboratories, Montgomery, TX).
Stable Transfection of MCF7 Cells with the Claudin 1-GFP Construct
The full-length coding sequence of the human claudin 1 gene was cloned in frame with an N-terminally fused GFP-sequence of the pEGFP-C1 vector (a gift from Dr. Claas Ruffer, Institute of Medical Biochemistry, University of Munster, Germany). MCF7 cells were stably transfected using Lipofectamine 2000 (Life Technologies Inc). Single clones were selected using Geneticin (Life Technologies Inc.), and exogenous claudin 1 was confirmed by Western blotting and immunofluorescence (IF) analysis.
IF Microscopy
For IF staining, cells were cultured on glass coverslips and fixed with ice-cold 100% methanol. Cells were permeabilized with 0.2% Tween-20 in PBS, blocked with 1% BSA in PBS, and incubated with the E-cadherin mouse polyclonal primary antibody or the mouse zona occludin 1 (ZO-1) antibody (Life Technologies Inc.; 1:50 dilution for both antibodies). The cells were washed with PBS and incubated with the claudin 1 rabbit primary antibody (Life Technologies Inc., dilution 1:50) overnight at 4°C in a humid chamber. They were then incubated with secondary anti-rabbit antibody conjugated with AlexaFluor 488 and anti-mouse antibody conjugate with AlexaFluor 555 (dilution 1:1000 each, Life Technologies Inc.) for 1 hour at room temperature. Cells were washed three times with PBS and incubated with 4′,6-diamidino-2-phenylindole-dihydrochloride and mounted in FluorSave (Calbiochem, Billerica, MA). Fluorescent images were captured with a Zeiss AxioObserver Z1 microscope using AxioVision Rel.4.8 software (Carl Zeiss, Jena, Germany).
qPCR
RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories Inc.) according to the manufacturer’s instructions. For assays on the ABI 7500 qPCR system, 2 μl of the RT reaction (50 ng total RNA) was added to a 96-well plate (MicroAmp Optical 96-well reaction plate, Applied Bisosystems) together with 18 μl of a reaction mix consisting of 10 μl SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories Inc.) and 0.5 μl (250 nM final concentration) of claudin 1 forward (5’cgggttgcttgcaatgtgc3’) and reverse (5’ccggcgacaacatcgtgac3’) primers and 7 μl nuclease-free water. Thermal cycling profile consisted of polymerase activation and DNA denaturation; 98°C for 30 seconds followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Cycle threshold (Ct) and relative quantitation values were calculated automatically by the ABI 7500 instrument software. Normalization was carried out using the Cts derived from qPCR with cyclophilin primers (forward; 5’ccagttcatgtgccagggcggtga3’; reverse; 5’aagaactgagagccattggtgtttggg 3’) and was calculated via the ∆∆Ct method [60].
TPA Treatment
PKC Inhibitor Studies
Pretreatment with the pan PKC inhibitor (GF109203x, 3 μM, Sigma-Aldrich Canada Co.), the ERK inhibitor (U0126, 10 μM, EMD Millipore, Etobicoke, Canada), the JNK inhibitor II (20 μM, EMD Millipore), the PKC α/β/μ inhibitor (Go6976, 1 μM, EMD Millipore), or the PKC δ inhibitor (Rottlerin, 5 μM, EMD Millipore) was initiated 1 hour prior to TPA treatment. Cells were then treated with 100 nM TPA (Sigma-Aldrich Canada Co.) or vehicle control (dimethyl sulfoxide, DMSO).
Cell Counting Assays
Cells were plated in 24-well cell culture dishes in DMEM. After 24 hours, the medium was changed to DMEM supplemented with 100 nM TPA (Sigma-Aldrich Canada Co.) or vehicle control (DMSO). The medium was then replaced every 2 days for 6 days. On day 6, cells were detached from three separate wells using 0.05% trypsin (Hyclone Laboratories Inc.) and counted using the TC-1 counter (Bio-Rad Laboratories Inc.).
Cellular Localization Studies
Cells stably transfected with the claudin 1-GFP construct were treated with 100 nM TPA (Sigma-Aldrich Canada Co.) or vehicle control (DMSO) for 18 hours. GFP signal was visualized using a Zeiss Axio Observer D1 inverted microscope and an Axio Cam MR3 camera. Images were acquired with a LD Plan-Neofluar 40×/0.6 objective and 62HE filter with ex 493 nm and em 517 nm with an exposure time of 548 milliseconds for all samples.
Tissue Microarray Analysis
TMAs of breast tumor biopsies were obtained from the Manitoba Breast Tumour Bank (MBTB, University of Manitoba), which operates with the approval from the Faculty of Health Sciences, University of Manitoba, Research Ethics Board at the Bannatyne Campus. Collection, handling, and histopathological assessment of tumor tissues have been previously described [31], [61]. The breast cancer TMAs were comprised of a cohort of 768 breast tumor biopsies/specimens (consisting of 447 ER− and 321 ER+ tumors). The estrogen receptor (ER) and progesterone receptor (PR) status was determined by the ligand binding assay (ER+ ≥3 fmol/mg protein, PR+ ≥10 fmol/mg protein). The clinicopathological characteristics of the patient cohorts were provided by the MBTB and used for statistical analyses.
Immunohistochemistry
TMAs
Immunohistochemistry (IHC) was performed on TMAs using the methods as described previously [31], [61]. Briefly, serial sections (5 μm) of the TMAs were stained with rabbit polyclonal antibodies to claudin 1 at a dilution of 1:150 (Life Technologies Inc., Burlington, ON, Canada), rabbit polyclonal PKCα C-20 (Santa Cruz Biotechnology Inc.) at a dilution of 1:750, or the rabbit polyclonal PKCε C-20 (Santa Cruz Biotechnology Inc.) at a dilution of 1:750. The paraffin-embedded tissue sections were processed using the automated Leica BOND Rx system. Tissues were processed and incubated for 60 minutes with the primary antibody and 30 minutes with the secondary antibody following standard protocol. Validation of the claudin 1 antibody has been described previously [31]. The validation of the PKCα antibodies included using PKCα knockout mouse material, blocks provided by Michael Leitges, Biotechnology Centre of Oslo, University of Oslo [62].
Antibodies to CK5/6 (D5/16B4, Life Technologies Inc.), EGFR (3C6, Ventana Systems), and HER2 (Cb11, NovaCastra, Concord, ON, Canada) were used as already detailed [37]. Only those tumors from which we were able to retrieve interpretable data (intact, unfolded tumor sections) were considered for analysis. As well, of our 768 patient sample cohort, some tumor cores were unavailable for analysis due to exhaustion of the TMA. All IHC data were compiled into the database maintained by the MBTB and made available for correlation analyses and other statistical comparisons.
Quantification and Cutoff Selection
Positive staining of antibody was assessed by light microscopy. A semiquantitative assessment strategy of the stained tissues was employed as previously described [31], [37]. Both staining intensity (scale 0-3) and the percentage of positive cells (0-100%) were multiplied to generate an H score ranging from 0 to 300. TMA staining was evaluated independently by two investigators (A.B. and C.P.). Where discordance (i.e., different scores given by different investigators) was found, cases were reevaluated and discussed and a consensus reached. Only tumor biopsies whose ER/PR status was determined by both ligand-binding assay (ER− <3 fmol/mg protein, PR− <10 fmol/mg protein) and by IHC (ER−/PR− <10% positive cells) were considered as negative in this study. Primary categorical analysis was as follows: positivity for CK5/6 and EGFR was set as ≥10% of cells staining, and for HER2, tumor cores that showed membrane-staining intensity of 2 or 3 were considered positive. The ER+ cohort was comprised of 447 breast tumors. Further, using a strict criteria for the basal-like subtype (ER− PR−, HER2− and EGFR and/or CK5/6 +), 128 tumors were identified by IHC as having the BLBC phenotype. Additionally, the 193 ER− tumors were designated as “ER−, nonbasal”.
Statistical Analysis
Analysis was carried out as previously described [25], [26] using SAS 9.2 (SAS, Cary, NC) statistical software. The Kruskal-Wallis test was used to compare the median H-scores for claudin 1. Associations between claudin 1 and other clinical-pathological variables were tested using contingency methods (continuity adjusted chi-square was used for node, age, and size data; exact linear association was used for grade). Pearson correlation coefficients of claudin 1 protein with PKCα, PKCε, and clinical-pathological characteristics of the breast tumor cohorts were calculated. Univariate survival analyses were performed using Cox regression to generate Kaplan-Meier curves. Overall survival (OS) was defined as the time from initial surgery to the date of death attributable to breast cancer only. Recurrence time was defined as the time from initial surgery to the date of clinically documented local or distant disease recurrence. The Student’s t test was used to interrogate the effect of TPA treatment on claudin1, E-cadherin, and β-catenin protein levels in cell lines (GraphPad Prism 7, GraphPad Software Inc.). Analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test was used to assess difference in claudin 1 levels in the presence of inhibitors.
Results
Localization of Claudin 1 in Both the Membrane and Cytoplasm of Human Breast Cancer Cell Lines and Tumor Biopsies
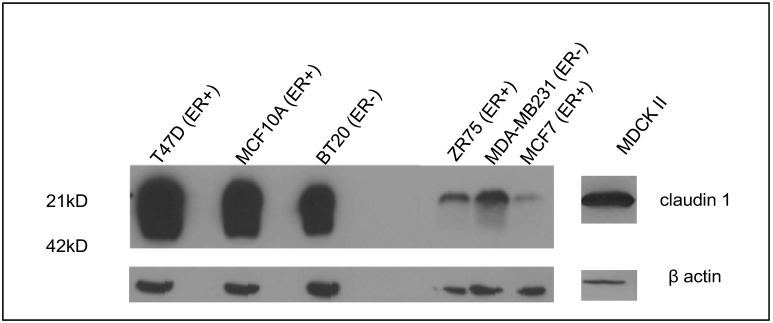
To understand the regulation of claudin 1 in HBC, baseline endogenous claudin 1 levels were examined in a panel of breast cancer cell lines. Western blot analysis showed varying levels of claudin 1 in the different cell lines. Whereas the MCF7, ZR75, and MDA-MB231 exhibited low levels of endogenous claudin 1, very high levels were observed in T47D and the BT20 HBC cell lines (Figure 1). As a positive control for membrane claudin 1, we used the MDCKII epithelial cell line, which exhibits high levels of claudin 1 in the membrane. We found that this cell line, as well as the nontumorigenic human breast epithelial cell line MCF10A, both displayed high endogenous levels of membrane bound claudin 1.
Figure 1.
Endogenous claudin 1 protein levels in human breast cancer cell lines. Western blot analyses showing claudin 1 protein in a panel of human breast cancer cell lines. High levels of claudin 1 protein are shown in the T47D, MCF10A, and BT20 cell lines, whereas ZR75, MDA-MB231, and MCF7 cells displayed low levels of endogenous claudin 1. The nontumorigenic MDCKII cell line which exhibits high endogenous levels of claudin 1 was used as a positive control for claudin 1. β-Actin was used as a control for protein loading. ER+, estrogen receptor positive; ER−, estrogen receptor negative.
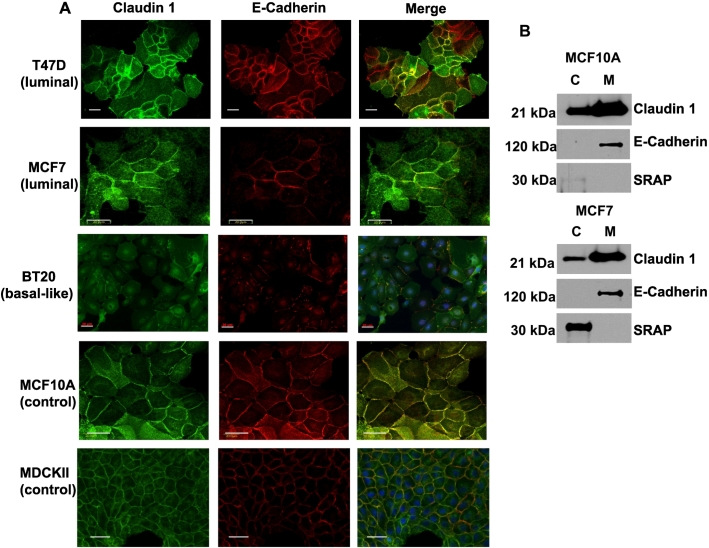
IF staining and subcellular fractionation strategies were further carried out to examine claudin 1 cellular distribution in the cell lines. A tight junction protein, claudin 1 is typically localized in the cell membrane. However, IF data revealed varying levels of both membranous and cytoplasmic claudin 1 staining in all the breast cancer cell lines analyzed (Figure 2). Interestingly, cytoplasmic staining was clearly visible in T47D cells, although these cells are known to have high endogenous claudin 1, very much comparable to the levels observed in the nontumorigenic cell line MCF10A. Moreover, MCF7, which has little or no tight junctions [63], and the BT20 cell line, both clearly displayed membrane staining for claudin 1. The MDCKII cells, known to exhibit well-defined tight junctions, revealed a clear chicken wire punctate pattern of membrane staining for claudin 1 but no cytoplasmic staining (Figure 2). Subcellular fractionation studies were informative, primarily in confirming the presence or absence of claudin 1 within the two compartments, but however were not reliable for precise quantification of the claudin 1 protein. The SRAP, a cytoplasmic protein [64], and E-cadherin, a membrane bound adhesion protein, were used as controls to confirm cytoplasmic and membrane fractions, respectively (Figure 2).
Figure 2.
Subcellular localization of claudin 1 in human breast cancer cell lines. (A) IF microscopy was used to detect claudin 1 in breast cancer cell lines. The adheren junction protein E-cadherin was used as a positive control for membrane staining, and the MDCKII cells were used as a positive control for claudin 1 membrane localization. (B) Western blot analysis showing relative levels of claudin 1 in different subcellular fractions from two representative cell lines. SRA (steroid receptor RNA activator) and E-cadherin were used to confirm cytoplasmic (C) and membrane (M) fractions, respectively. Scale bars = 20μm.
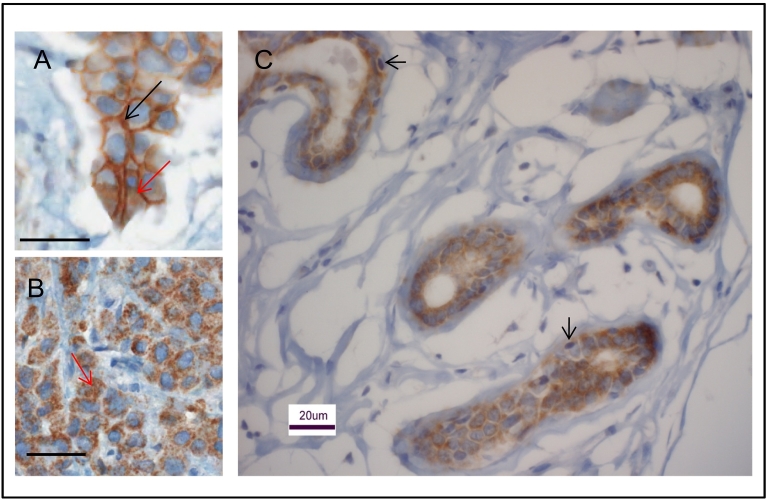
IHC analysis of human invasive breast cancer biopsies revealed a diffuse pattern of staining for claudin 1 in the tumor cells (Figure 3). Moreover, in this study as well as in an earlier study [37], we found that some breast tumors displayed both membranous and cytoplasmic staining for claudin 1 (Figure 3A), while others showed only cytoplasmic staining (Figure 3B). In the normal human mammary gland tissue, notably, claudin 1 staining was observed only in the ductal epithelial cells but not in the basal-like myoepithelial cells (Figure 3C).
Figure 3.
Immunohistochemical analysis of the claudin 1 protein in normal breast tissue and breast cancer. Representative claudin 1 staining in human invasive breast cancer, and healthy normal mammary ductal epithelial cells are shown. Tumor depicted in panel A shows both membrane (black arrows) and cytoplasmic staining (red arrows), while the tumor in panel B shows only cytoplasmic staining (red arrows). (C) Normal ductal epithelial cells in a healthy mammary gland tissue show high levels of claudin 1, while the basal-like myoepithelial cells (depicted by the arrows) are primarily negative. Scale bars = 20μm.
Upregulation of Claudin 1 at Both the Transcriptional and Protein Levels and Retention in the Cytoplasm
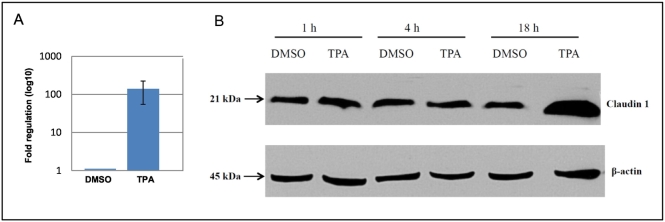
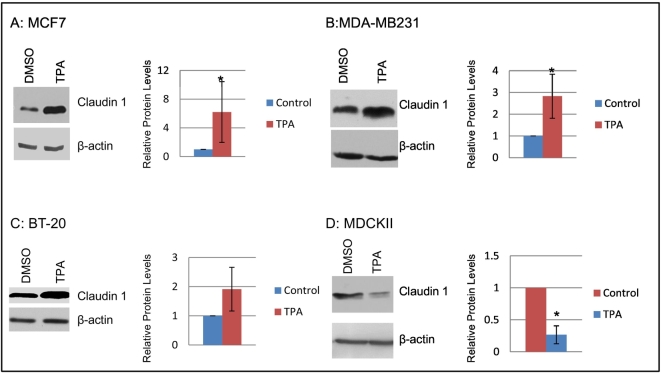
To interrogate PKC upregulation of claudin 1 in breast cancer, the HBC cell line MCF7, which exhibits low levels of endogenous claudin 1 expression, was treated with TPA. TPA induction at 18 hours resulted in a dramatic upregulation of claudin 1 (Figure 4) at both the transcriptional (>100-fold, Figure 4A) and protein level (Figure 4B). TPA upregulation of claudin 1 was also observed in the other HBC cell lines examined in our panel, irrespective of their endogenous claudin 1 levels (Figure 5, A-C). Conversely, in the nontumorigenic control kidney epithelial MDCKII cells, which have high endogenous levels of claudin 1, a reduction in claudin 1 protein upon TPA treatment (Figure 5D) was observed.
Figure 4.
Activation of PKC by TPA in MCF7 cells led to the upregulation of claudin 1 expression and increased protein level in a time-dependent manner. Real-time PCR and Western blot analyses were carried out on MCF7 cells treated with TPA (100 nM TPA for up to 18 hours, as described in Materials and Methods). PKC activation led to (A) a large fold increase in claudin 1 transcript and (B) protein levels (n = 3 experimental replicates).
Figure 5.
PKC activation leads to an increase in claudin 1 expression in several breast cancer cell lines but to a decrease in the nontumorigenic MDCKII cell line. Cells were treated with 100 nM TPA or vehicle control (DMSO) for 18 hours. Western blots of a representative experiment are shown for each cell line (left panels). The relative protein levels were determined by densitometry (right panels, mean ± SD for experimental replicates). (A) MCF7 cells; n=6. (B) MDA-MB231 cells; n=5. (C) BT-20 cells; n=4. (D) MDCKII cells, n=4. *P<.05; Student’s t test.
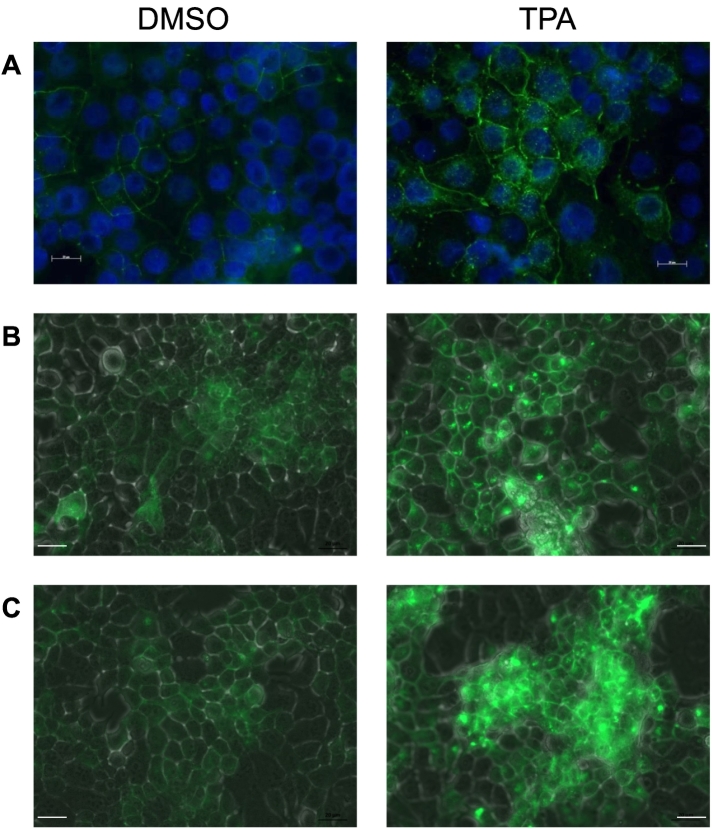
TPA treatment also led to an accumulation/retention of claudin 1 in the cytoplasm of the MCF7 cells (Figure 6A). This mislocalization of claudin 1 was also observed when claudin 1 was overexpressed in MCF7 cells (Figure 6, B and C).
Figure 6.
PKC increases accumulation of claudin 1 protein in the cytoplasm of MCF7 cells. (A) IF with the claudin 1 antibody was used to show TPA treatment increased accumulation of endogenous claudin 1 in the cytoplasm of MCF7 cells. (B, C) Exogenous GFP-tagged claudin 1, visualized using a Zeiss Axio Observer D1 inverted microscope, was also increased in the cytoplasm following TPA treatment. Scale bars = 20 μm.
Inhibition of MCF7 HBC Cell Proliferation Following Prolonged TPA Treatment
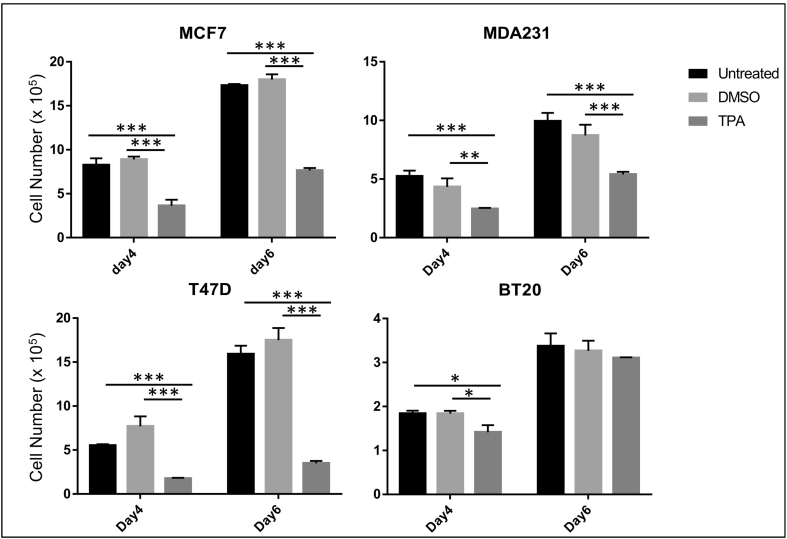
Recently, it has been shown that short-term TPA treatment (24 hours) led to an antiproliferative effect in MCF7 cells [65], and this was attributed to a prosurvival response to TPA. In the present study, we interrogated whether long-term TPA treatment would have a similar effect on MCF7 cell proliferation. Cells were treated with 100 nM TPA over a period of 6 days (as described in Materials and Methods, 2.7.2). In line with studies by Fortino et al. [65], we found that long-term treatment with TPA as well significantly inhibited MCF7 cell numbers (Figure 7). Interestingly, a similar response to long-term TPA treatment was also observed in the other breast cancer cell lines (T47D, BT20, and MDA-231) as well (Figure 7). Thus, it appears that either short-term or long-term TPA treatment leads to an antiproliferative effect on HBC cells.
Figure 7.
Long-term TPA treatment inhibited expansion of breast cancer cell numbers. Cells were treated with 100 nM TPA over a period of 6 days (as described in Materials and Methods). TPA significantly inhibited the HBC MCF7, T47D, BT20, and MDA-231 cell numbers. n=3. Values are mean ± SD for experimental replicates, statistical analyses, two-way ANOVA and Tukey’s multiple comparison tests, *P<.05; **P<.01; ***P<.001.
Downregulation of E-cadherin and β-Catenin in MCF7 Cells Following PKC Activation
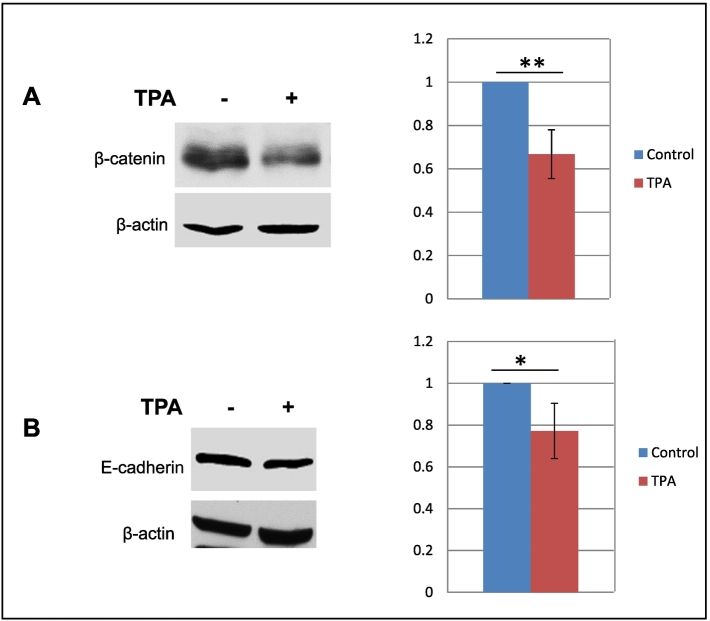
The association between PKC activity and cancer progression leading to metastasis is well established [55], [66], [67]. Enhanced aggressiveness has been associated with a loss of E-cadherin and alteration in β-catenin expression [23], [26], [68]. In an earlier study, we showed that a number of EMT-related genes including E-cadherin were significantly downregulated when MCF7 cells overexpressing claudin 1 were compared to control cells [33]. However, primers for β-catenin were not included in the commercial PCR array platform used for those studies. Thus, in this study, Western blot analysis to determine the effect of TPA treatment on the regulation of the β-catenin protein was conducted. A modest, however significant, downregulation of β-catenin protein upon TPA treatment was observed (Figure 8). In this same study, E-cadherin was also modestly but significantly downregulated in response to TPA treatment. This downregulation of E-cadherin in response to increased claudin 1 levels was consistent with our previous studies [33].
Figure 8.
Activation of PKC leads to a decrease in E-cadherin and β-catenin levels in MCF7 cells. Cells were treated with 100 nM TPA or vehicle control (DMSO) for 18 hours. Western blots of a representative experiment are shown for each cell line (left panels). The relative protein levels were determined by densitometry (right panels, mean ± SD for experimental replicates). (A) E-cadherin; n=4. (B) β-catenin; n=4. *P<.05; **P<.01; Student’s t test.
PKC and ERK Inhibitors Block Claudin 1 Upregulation
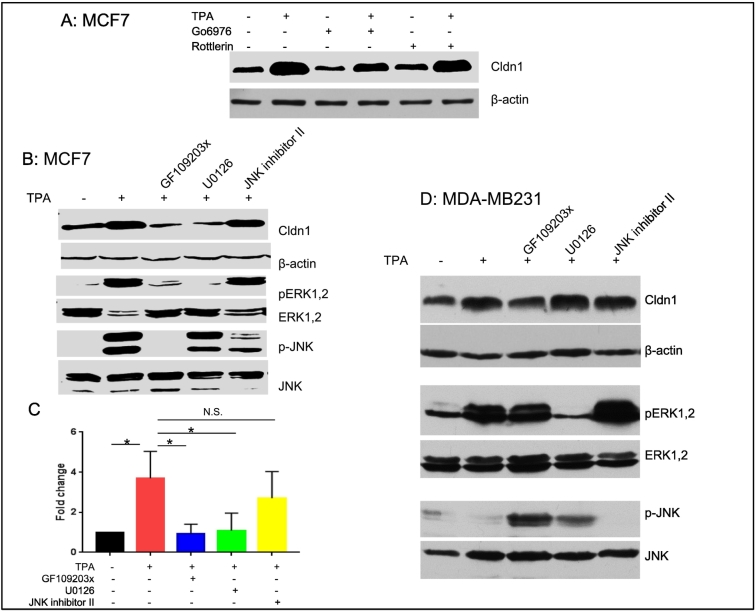
Using commercially available PKC inhibitors [PKC α/β/μ inhibitor (Go6976, 1 μM), the PKC δ inhibitor (Rottlerin, 5 μM), and the pan PKC inhibitor (GF109203x, 3 μM)], we examined their effectiveness in blocking claudin 1 upregulation by TPA. Go6976, which is known to inhibit PKCα and PKCβ, only partially inhibited claudin 1 upregulation, whereas rottlerin, the PKCδ inhibitor, appeared to have no effect. However, the PKC inhibitor GF109203, which is known to inhibit PKCα, β, δ, and ε, significantly inhibited claudin 1 induction by TPA in the MCF7 cells (P<.05) as shown in Figure 9, A and B, suggesting that PKCε may be involved in the upregulation of claudin 1.
Figure 9.
Upregulation of claudin 1 by TPA was significantly inhibited by the PKC inhibitor GF109203 and mediated by the ERK pathway in MCF7 cells but not in MDA-MB231 cells. (A) MCF7 cells were treated with 100 nM TPA or vehicle control (DMSO) for 18 hours. Pretreatment with the PKC α/β/μ inhibitor (Go6976, 1 μM) or the PKC δ inhibitor (Rottlerin, 5 μM) was initiated 1 hour prior to TPA treatment as indicated and continued for 19 hours. A Western blot of a representative experiment is shown, with β-actin as a loading control. In three experimental replicates, there was a consistent trend (not statistically significant, ANOVA, P=.148) towards reduced claudin 1 upregulation by TPA treatment in the presence of the inhibitors. (B) Cells were treated with 100 nM TPA or vehicle control (DMSO) for 18 hours. Pretreatment with the PKC inhibitor (GF109203x, 3 μM), the ERK inhibitor (U0126, 10 μM), or the JNK inhibitor II (20 μM) was initiated 1 hour prior to TPA treatment as indicated and continued for 19 hours. Western blots of a representative experiment are shown. Upregulation of claudin 1 in MCF7 cells was blocked by both the PKC and ERK inhibitors, but not the JNK inhibitor. (C) The relative protein levels of claudin 1 (panel B) were determined by densitometry (mean ± SE for n=3 experimental replicates). ANOVA P<.05; *P<.05 Dunnett’s multiple comparison test. (D) The upregulation of claudin 1 by TPA in the MDA-MB231 cells was not blocked by any of the inhibitors used for pre-treatment.
We further demonstrated that upregulation of claudin 1 in MCF7 cells was blocked by an ERK inhibitor but not a JNK inhibitor (Figure 9, B and C). Interestingly, in the MDA-MB231 cell line, although the ERK and JNK pathways were also activated by PKC, the upregulation of claudin 1 was not blocked by any of these inhibitors (Figure 9D).
Correlation of PKCε with Claudin 1 in Human Invasive Breast Cancer Biopsies
Based on the observations suggesting that PKCε may play a central role in the PKC/claudin 1 pathway, TMA analysis was carried out on a large cohort of human breast tumor patient biopsies to examine whether there was an association between PKC isomers and claudin 1 (Table 2, Figure 10). The cohort was comprised of ER− (basal and nonbasal) and ER+ human breast tumors (a total of 768 biopsies; see Materials and Methods.). We observed a significant correlation between PKCε and claudin 1 in both the ER+ and ER− tumors (basal-like and non–basal-like) subgroups in this cohort (Table 2). Interestingly, we also found a significant positive correlation between claudin 1 and PKCα in ER+ tumors (Table 2).
Table 2.
Correlation Analysis of Claudin 1 Protein with PKCα, PKCε, and Clinical-Pathological Characteristics of the Breast Tumor Cohorts.
| Age at Diagnosis | Ki67 | ER | PR | Total PKCα H-SCORE | Total PKCε H-SCORE | ||
|---|---|---|---|---|---|---|---|
| Group | |||||||
| Total | 0.01092 | 0.01045 | −0.23663 | −0.1209 | 0.33851 | 0.32725 | |
| P value | .7823 | .8112 | <.0001 | .0048 | <.0001 | <.0001 | |
| n | 643 | 525 | 542 | 542 | 464 | 632 | |
| ER+ | 0.08539 | 0.19297 | −0.12005 | −0.05074 | 0.33714 | 0.35677 | |
| P value | .1005 | .0003 | .0209 | .3304 | <.0001 | <.0001 | |
| n | 371 | 355 | 370 | 370 | 367 | 364 | |
| ER− | 0.09179 | −0.1526 | ND | 0.36786 | |||
| Nonbasal | P value | .2424 | .0907 | - | - | <.0001 | |
| n | 164 | 124 | 162 | ||||
| ER− | 0.21541 | −0.22688 | 0.45369 | 0.34034 | |||
| Basal | P value | .0252 | .1294 | - | - | .0002 | .0004 |
| n | 108 | 46 | 61 | 106 | |||
Values are Pearson correlation coefficients. ND, not determined.
Statistical significance is highlighted in bold type.
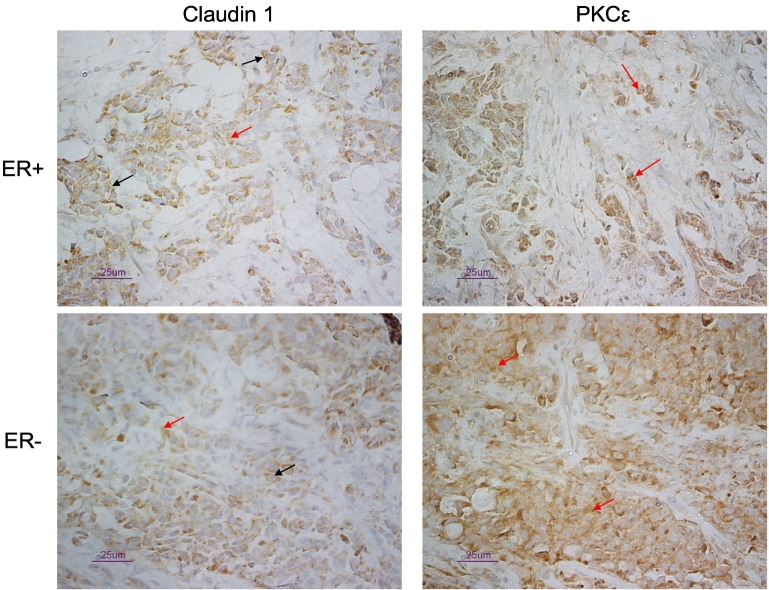
Figure 10.
Representative immunostaining of ER+ and ER− breast tumors with the claudin 1, and PKCε antibodies. Staining was specific for tumor tissue, and while both membrane (black arrows) and cytoplasmic staining (red arrows) were observed with the claudin 1 antibody, staining with the PKCε antibody was confined to the cytoplasm (red arrows). Scale bar = 25 μm.
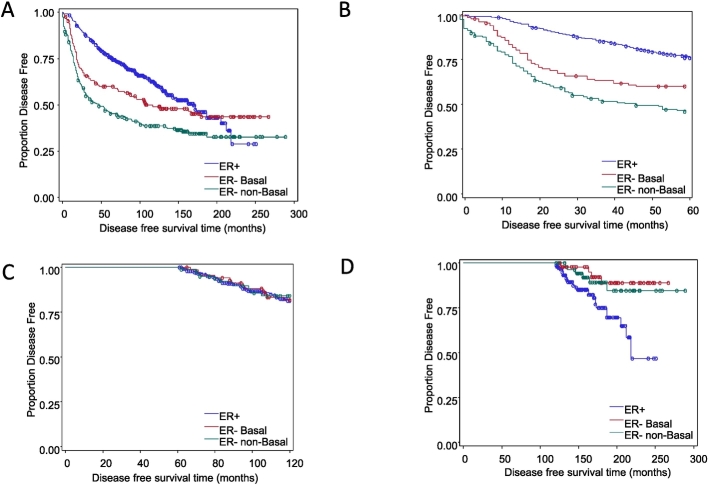
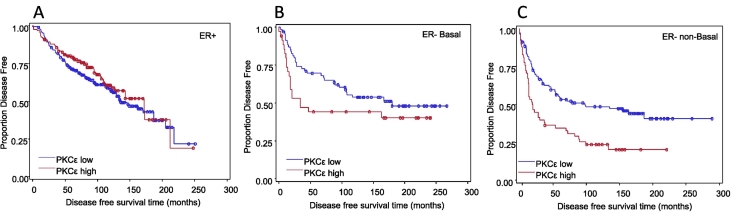
Also noted in this study was that when patient survival was examined over a 20-year period in this cohort (Table 3; Figure 11A), we observed that, within the first 5 years following diagnosis (Table 1; Figure 11B), patients with ER+ tumors had significantly longer disease-free survival compared with ER− basal (P<.01) and ER− nonbasal (P<.01) groups. Interestingly, we found that 5 to 10 years postdiagnosis, there were no significant differences in disease-free survival among the groups (Figure 11C). Moreover, 10 years postdiagnosis, patients in the ER+ group had a significantly worse disease-free survival prognosis than the other two ER− groups (P<.001; Figure 11D). Although claudin 1 levels were not a prognostic indicator of disease recurrence or survival (Table 3), intriguingly, high PKCε was associated with shorter disease-free survival in the ER− breast tumor cohort (Table 3; Figure 12).
Table 3.
Patient Survival/Recurrence Related to Claudin 1 and PKCε Levels
| 5-Year Survival/Recurrence |
20-Year Survival/Recurrence |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | ER+ | ER− Nonbasal | ER− Basal | Total | ER+ | ER− Nonbasal | ER− Basal | ||
| Claudin 1 (H-score >40 vs. H-score ≤40) | n= | 643 | 371 | 164 | 108 | 643 | 371 | 164 | 108 |
| Survival | 1.46⁎ | NS | NS | NS | NS | NS | NS | NS | |
| Recurrence | NS | NS | NS | NS | NS | NS | NS | NS | |
| Survival or recurrence | NS | NS | NS | NS | NS | NS | NS | NS | |
| PKCε intensity (2, 3, vs. 1) | n= | 685 | 399 | 172 | 114 | 685 | 399 | 172 | 114 |
| Survival | 1.38⁎ | NS | 1.79⁎ | 2.35⁎⁎ | NS | NS | 1.79⁎⁎ | 2.02⁎ | |
| Recurrence | NS | NS | 1.80⁎ | 2.12⁎ | NS | NS | 1.96⁎ | NS | |
| Survival or recurrence | NS | NS | 1.76⁎⁎ | 2.33⁎⁎ | NS | NS | 1.91⁎⁎ | NS | |
Claudin 1 was not a prognostic indicator of disease recurrence/survival; however, high PKCε was associated with shorter survival in patients with ER− tumors. Values are hazard ratios. NS, not statistically significant.
P<.05.
P<.01.
Figure 11.
Kaplan-Meier graphs for disease free survival in the breast tumor cohort. Univariate survival analyses were performed using Cox regression. Symbols on the graph lines represent censored data (recurrence or death from the disease). ER+ group n=447; events=173. ER− basal group n=128; events=66. ER− nonbasal group n=193; events=119. (A) Disease-free survival from time of diagnosis to 20+ years. ER− basal versus ER+, HR=1.39, P<.05; ER− nonbasal versus ER+ HR=2.04, P<.001. (B) Disease-free survival in the first 5 years after diagnosis. ER− basal versus ER+, HR=2.02, P<.001; ER− nonbasal versus ER+ HR= 3.12, P<.001. (C). Disease-free survival between 5 and 10 years after diagnosis. No significant differences among the groups. (D) Disease-free survival between 10 and 20+ years following diagnosis. ER− basal versus ER+, HR=0.24, P<.001; ER− nonbasal versus ER+ HR=0.36, P<.001.
Table 1.
Comparison of the Clinical-Pathological Characteristics of the Breast Tumor Cohorts.
| Total (n=768) | ER+ (n=447) | ER−, Nonbasal (n=193) | ER−, Basal (n=128) | P Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Patient age | |||||||||
| Median | 65 | 68 | 56 | 57.5 | |||||
| Tumor grade | # cases | % | # cases | % | # cases | % | # cases | % | |
| 0-5 | 34 | 6 | 21 | 6 | 9 | 7 | 4 | 8 | <.0001 |
| 6-7 | 362 | 66 | 298 | 82 | 53 | 39 | 11 | 23 | |
| 8-10 | 151 | 28 | 44 | 12 | 74 | 54 | 33 | 69 | |
| Nodal status | |||||||||
| +ve | 356 | 46.8% | 204 | 45.6% | 92 | 48.8% | 60 | 48.4% | .7275 |
| −ve | 404 | 53.2% | 243 | 54.4% | 98 | 51.2% | 63 | 51.6% | |
| Tumor size | |||||||||
| >2.5 cm | 301 | 48% | 195 | 45% | 26 | 50% | 80 | 56% | .0589 |
| <=2.5 cm | 331 | 52% | 242 | 55% | 26 | 50% | 63 | 44% | |
| Claudin 1 (H-score) | # cases | median | # cases | median | # cases | median | # cases | median | <.0001 |
| 643 | 30 | 371 | 15 | 164 | 70 | 108 | 62.50 | ||
| PKCα (intensity) | # cases | % | # cases | % | # cases | % | # cases | % | |
| 1 | 387 | 73.6% | 307 | 73.6% | 31 | 77.5% | 49 | 71.0% | .7595 |
| 2, 3 | 139 | 26.4% | 110 | 26.4% | 9 | 22.5% | 20 | 29.0% | |
| PKCε (intensity) | # cases | % | # cases | % | # cases | % | # cases | % | |
| 1 | 449 | 69.0% | 259 | 64.9% | 112 | 71.8% | 78 | 81.3% | <.01 |
| 2, 3 | 202 | 31.0% | 140 | 35.1% | 44 | 28.2% | 18 | 18.8% | |
| Recurrence (5 years) | 32.0% | 22.8% | 50.3% | 37.5% | <.001 | ||||
| Survival (5 years) | 76.5% | 89.0% | 54.5% | 65.4% | |||||
Figure 12.
High PKCε expression was associated with shorter disease-free survival in the ER− breast tumor cohort. Kaplan-Meier graphs are shown. For each case, an immunohistochemical staining intensity of 1 was defined as low, and high was defined as a staining intensity of 2 or 3. Univariate survival analyses were performed using Cox regression. Hazard ratios are listed in Table 3. Symbols on the graph lines represent censored data (recurrence or death from the disease). (A) ER+; n=399; low PKCε events=110; high PKCε events=48 (NS). (B) ER− basal; n=114; low PKCε events=37; high PKCε events=21 (NS). (C) ER− nonbasal; n=172; low PKCε events=61; high PKCε events=45, P<.01.
Discussion
Protein kinases have long been shown to play significant roles in the progression of many cancers [51]. Furthermore, alteration in the activity of particular members of this family of isomers has been directly linked to phenotypic and behavioral changes associated with enhanced metastatic aggressiveness [51], [69], [70], [71]. Such changes include alterations in EMT markers, increased cell motility and invasiveness, and a mislocalization of tight junction proteins from the cell membrane to the cytoplasm [72]. The loss of tight junction proteins from the membrane facilitates the loss of cellular integrity, an integral player of the metastatic process [8], [12], [73].
Claudin 1 is an important tight junction protein as it forms the backbone of the tight junction in epithelial cells [15] and plays a major role in the control of nutrients and small ions between these cells. It is now well documented that claudin 1 is altered in several cancers [9], [18], [21] and has been shown to be upregulated by PKC activity [21], [23], [26], [32], [59]. However, claudin 1 is one of only two known claudin TJ proteins, the other claudin 18a, [74] shown to be induced by PKC during cancer progression. The regulation of claudin 1 by PKC has not been previously addressed in breast cancer.
In this study, we investigated the regulation of claudin 1 by PKC in breast cancer. We first established the baseline levels of claudin 1 in different human breast cancer cell lines. The MCF7 cell line was identified as an appropriate model to address PKC upregulation of claudin 1 in breast cancer as it is known to exhibit low endogenous levels. Additionally, it has previously been demonstrated that PKC can enhance the metastatic potential of the MCF7 cell line [65].
Following TPA treatment, claudin 1 expression was shown to be significantly upregulated in MCF7 breast cancer cells, whereas in the nontumorigenic MDCKII cells, claudin 1 protein levels were downregulated upon PKC activation by TPA. In MDCKII cells, claudin 1 is localized in the cell membrane. Thus, it is therefore plausible that, in MDCKII cells, where claudin 1 primarily has a barrier function [75], its interaction with the PKC signaling pathway may be very different from its interaction with the very same pathway in the MCF7 breast cancer cells where it is localized in the cytoplasm as well.
We further showed that MCF7 cells treated with TPA or which overexpressed claudin 1 led to increased retention of the protein in the cytoplasm. Increased accumulation of claudin 1 in the cytoplasm during cancer progression has been reported in several other cancers, and cytoplasmic mislocalization has frequently been associated with enhanced metastatic potential [18], [21], [59]. It has been suggested that claudin 1 retained in the cytoplasm may serve another function at the expense of a barrier function. One such function may be related to the activation of autophagy, which in turn promotes tumor survival and proliferation as suggested by a recent report [76]. It is also plausible that TPA may indirectly affect claudin 1 through regulation of another interacting partner in the cytoplasm. When not in the membrane, claudin 1 has been shown to interact with ephrin B1 [77], ESCRT [78], EpCAM [79], and CD9, the latter which has been shown to stabilize cytoplasmic claudin 1 [63]. However, PKC upregulation of claudin 1 at the level of transcription in MCF7 cells demonstrates that PKC activation in these breast cancer cells also occurs not only at the protein level but at the level of gene expression as well.
In addition, our studies also demonstrated that, at the protein level, there was a small, nonetheless significant, decrease in both E-cadherin and β-catenin protein upon PKC activation in MCF7 cells. This inverse relationship between these EMT regulators was consistent with similar results described in the other cancers [18], [21], [59].
Long-term PKC activation of MCF7 cells also resulted in significant growth inhibition in all breast cancer cell lines examined (Figure 8). Inhibition of MCF7 cell growth has been previously described by Fortino et al. [65] following a shorter period of TPA treatment, which these authors further demonstrated was attributed to prosurvival and antiproliferative properties of TPA in these cells.
To further define the PKC isomers and signaling pathways involved in claudin 1 upregulation in MCF7 cells, we used inhibitors of PKC to block PKC activation by TPA. The pan-inhibitor GF109203, which inhibits PKCα, β, δ, and ε, was most effective in blocking claudin 1 upregulation by TPA, whereas rottlerin, which inhibits PKCδ, appeared to have had no effect. Go6976, which inhibits PKCα and PKCβ, was only partially effective. Importantly, these results pointed to PKCε and, to a lesser extent, PKCα as possible regulators of claudin 1 in MCF7 cells.
Increased PKCα expression has been reported in ER− breast tumor samples [80], and PKCα has also been identified as a marker of poor prognosis independent of other factors [70]. Patients with PKCα-negative tumors were also shown to have better responses to endocrine therapy [80], [81]. Furthermore, in vitro, PKCα has been shown to enhance migratory potential and promote more aggressive behavior in MCF7 [82] and MDA-MB231 cells [51], [70].
Similarly, an oncogenic role has frequently been assigned to PKCε in breast cancer, and it is considered a marker of aggressiveness [72]. PKCε has been shown to be essential for enhanced cell migration and invasion and promote breast cancer survival [71]. High expression levels of PKCε have also been shown to correlate with high tumor grade, HER2 expression, ER negativity, and poor survival in patients with breast cancer [83]. In vitro, PKCε has been shown to promote cell survival by inhibiting apoptosis of MCF7 cells and to promote EMT [72]. It has been demonstrated that when PKCε was overexpressed in the nonmalignant immortalized breast epithelial cell line MCF10A, these cells lost their phenotypic epithelial characteristics and became fibroblastic and spindly, increased cell migration, and were protected from anoikis [72].
We demonstrated that the upregulation of claudin 1 by TPA was effectively blocked by PKC and ERK inhibitors, but not by JNK inhibitors, providing strong evidence that claudin 1 induction in MCF7 cells was through the activation of the ERK pathway. Interestingly, the inhibition of the ERK signaling pathway did not block the upregulation of claudin 1 in the MDA-MB231 cell line, leading us to suggest that these two cell lines may have different PKC signaling pathways. Indeed, Platet et al. [84] previously demonstrated that TPA increased the invasiveness of MCF7 cells but decreased the invasiveness of MDA-MB231 cells and attributed this to the ER status and abnormal TPA regulation of PKCα activity in the MDA-MB231 cells. Another plausible explanation is that the differences observed in the responses of these cells to the PKC inhibitors may reflect the extent of leakiness at their tight junctions. Sjo et al. [85] demonstrated that the leakiness or tightness of the membrane barrier of cells affects the response of claudin family members to PKC activation.
We also investigated whether there was a link between PKCε, PKCα, and claudin 1 in a large cohort of breast cancer biopsies (768 samples) of mixed pathologies. We further divided these samples into ER+ and ER− subgroups. The ER− tumors were then yet again divided into basal and nonbasal since, in an earlier study, we identified a relationship between high claudin 1 expression and the basal-like subtype of breast cancers. In the present study, we found a correlation between claudin 1, PKCα, and PKCε in patient biopsies. Both PKCε and PKCα positively correlated with claudin 1 in the ER+ tumors. We also identified a further significant correlation between claudin 1 and PKCε in ER− tumors and basal and non–basal-like subtype of tumors. (Table 2). Additionally, our analysis showed that overall PKCε levels and claudin 1 were higher in the ER− tumors compared to ER+ tumors (Table 3).
Interestingly, we identified a negative association between patient’s survival and PKCε. High PKCε levels significantly correlated with decreased survival and/or recurrence rate in both the basal and nonbasal ER− tumors. However, claudin 1 alone, whether high or low, had no effects on patient survival.
Conclusions
Our studies demonstrated that PKC activation strongly upregulates claudin 1 in MCF7 HBC cells and that overexpression of claudin 1 in MCF7 cells led to increased mislocalization of the protein from the membrane to the cytoplasm. We also show that claudin 1 upregulation is linked to the downregulation of EMT markers, the process that leads to metastasis. Furthermore, we identified a correlation between patient survival and PKCε. There was a decrease in the survival rates of patients if the tumors were ER− and PKCε levels were high. This study therefore provides further insights into a role for claudin 1 in enhancing the metastatic potential of some breast cancers.
In conclusion, our studies show supporting evidence to suggest claudin 1 as a target for therapeutic management of ER− breast cancers and should be considered in conjunction with currently used PKC inhibitors.
Acknowledgments
Acknowledgements
The authors would like to thank the personnel at the MBTB for technical support, particularly Andrea Fritensky and Michelle Parisien. Biostatistical consultation was kindly provided by Dr. Z. Nugent, Senior Health Outcomes Analyst, Manitoba Institute of Cell Biology, University of Manitoba. The pEGFP-C1-claudin 1 vector was a gift from Dr. Claas Ruffer, Institute of Medical Biochemistry, University of Munster, Germany.
Authors’ Contributions
++YM, designed, and supervised the study. YM, AO and AAB co-wrote the manuscript. AAB, and CP carried out H scoring evaluation of the TMAs. XM generated stable clonal cell lines. XM, NW, and AAB carried out assays in the human breast cancer cell lines. NW was also responsible for the subcellular fractionation assays. NW and SH carried out the IF studies. EL, TK, MP, and LCM contributed intellectually to many aspects of the study, and EL reviewed and edited the manuscript. All authors read and approved the final manuscript.
Footnotes
Funding: This study was supported by a grant from the Canadian Breast Cancer Foundation and CancerCare Manitoba Foundation. The funding institutions had no role in the design of the study; collection, analysis, and interpretation of data; and writing the manuscript.
Contributor Information
Anne A. Blanchard, Email: Anne.Blanchard@umanitoba.ca.
Xiuli Ma, Email: Xiuli.ma@umanitoba.ca.
Nan Wang, Email: nwang530@gmail.com.
Sabine Hombach-Klonisch, Email: Sabine.Hombach@umanitoba.ca.
Carla Penner, Email: crpenner@dsmanitoba.ca.
Arzu Ozturk, Email: Arzu.Ozturk@umanitoba.ca.
Thomas Klonisch, Email: Thomas.Klonisch@umanitoba.ca.
Marshall Pitz, Email: marshall.pitz@cancercare.mb.ca.
Leigh Murphy, Email: Leigh.Murphy@umanitoba.ca.
Etienne Leygue, Email: Etienne.Leygue@umanitoba.ca.
Yvonne Myal, Email: Yvonne.myal@umanitoba.ca.
References
- 1.Gonzales-Mariscal L. CRC Press; Boca Raton: 2001. Tight Junctions. Vol. [Google Scholar]
- 2.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 3.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 4.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 5.Diamond JM. Twenty-first Bowditch lecture. The epithelial junction: bridge, gate, and fence. Physiologist. 1977;20:10–18. [PubMed] [Google Scholar]
- 6.Eaton S, Simons K. Apical, basal, and lateral cues for epithelial polarization. Cell. 1995;82:5–8. doi: 10.1016/0092-8674(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 7.Gumbiner B. Structure, biochemistry, and assembly of epithelial tight junctions. Am J Phys. 1987;253:C749–C758. doi: 10.1152/ajpcell.1987.253.6.C749. [DOI] [PubMed] [Google Scholar]
- 8.Itoh M, Bissell MJ. The organization of tight junctions in epithelia: implications for mammary gland biology and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:449–462. doi: 10.1023/B:JOMG.0000017431.45314.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 10.Simons K, Fuller SD. Cell surface polarity in epithelia. Annu Rev Cell Biol. 1985;1:243–288. doi: 10.1146/annurev.cb.01.110185.001331. [DOI] [PubMed] [Google Scholar]
- 11.Tsukita S, Furuse M. Claudin-based barrier in simple and stratified cellular sheets. Curr Opin Cell Biol. 2002;14:531–536. doi: 10.1016/s0955-0674(02)00362-9. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Mariscal L, Lechuga S, Garay E. Role of tight junctions in cell proliferation and cancer. Prog Histochem Cytochem. 2007;42:1–57. doi: 10.1016/j.proghi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Lelievre SA. Tissue polarity-dependent control of mammary epithelial homeostasis and cancer development: an epigenetic perspective. J Mammary Gland Biol Neoplasia. 2010;15:49–63. doi: 10.1007/s10911-010-9168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myal Y, Blanchard A. In: Tight junctions in cancer: multifaceted players in tumorigenesis and cancer progression. Martin T, editor. Springer; New York, NY, U.S.A: 2012. [Google Scholar]
- 15.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1–deficient mice. J Cell Biol. 2002;156:1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuse M. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal R, Mori Y, Cheng Y, Jin Z, Olaru AV, Hamilton JP, David S, Selaru FM, Yang J, Abraham JM. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhawan P, Singh AB, Deane NG, No Y, Shiou S-R, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kominsky SL, Vali M, Korz D, Gabig TG, Weitzman SA, Argani P, Sukumar S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164:1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, Dissanayake SK, Earley R, Indig FE, Nickoloff BJ. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26:3846–3856. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- 22.Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, Menke A, Fensterer H, Giehl K, Lohr M. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–6271. [PubMed] [Google Scholar]
- 23.Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, An S, Yoon G, Gye MC, Yi JM. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2013;32:4873–4882. doi: 10.1038/onc.2012.505. [DOI] [PubMed] [Google Scholar]
- 24.Hou J, Wang Z, Xu H, Yang L, Yu X, Yang Z, Deng Y, Meng J, Feng Y, Guo X. Stanniocalicin 2 suppresses breast cancer cell migration and invasion via the PKC/claudin-1–mediated signaling. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14:18148–18180. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon CH, Kim MJ, Park MJ, Park IC, Hwang SG, An S, Choi YH, Yoon G, Lee SJ. Claudin-1 acts through c-Abl-protein kinase Cdelta (PKCdelta) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J Biol Chem. 2010;285:226–233. doi: 10.1074/jbc.M109.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiozaki A, Bai X-h, Shen-Tu G, Moodley S, Takeshita H, Fung S-Y, Wang Y, Keshavjee S, Liu M. Claudin 1 mediates TNFα-induced gene expression and cell migration in human lung carcinoma cells. PLoS One. 2012;7:e38049. doi: 10.1371/journal.pone.0038049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pope JL, Ahmad R, Bhat AA, Washington MK, Singh AB, Dhawan P. Claudin-1 Overexpression in intestinal epithelial cells enhances susceptibility to adenamatous polyposis coli-mediated colon tumorigenesis. Mol Cancer. 2014;13:167. doi: 10.1186/1476-4598-13-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takehara M, Nishimura T, Mima S, Hoshino T, Mizushima T. Effect of claudin expression on paracellular permeability, migration and invasion of colonic cancer cells. Biol Pharm Bull. 2009;32:825–831. doi: 10.1248/bpb.32.825. [DOI] [PubMed] [Google Scholar]
- 30.Oku N, Sasabe E, Ueta E, Yamamoto T, Osaki T. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res. 2006;66:5251–5257. doi: 10.1158/0008-5472.CAN-05-4478. [DOI] [PubMed] [Google Scholar]
- 31.Blanchard AA, Skliris GP, Watson PH, Murphy LC, Penner C, Tomes L, Young TL, Leygue E, Myal Y. Claudins 1, 3, and 4 protein expression in ER negative breast cancer correlates with markers of the basal phenotype. Virchows Arch. 2009;454:647–656. doi: 10.1007/s00428-009-0770-6. [DOI] [PubMed] [Google Scholar]
- 32.Kyuno D, Kojima T, Yamaguchi H, Ito T, Kimura Y, Imamura M, Takasawa A, Murata M, Tanaka S, Hirata K. Protein kinase Calpha inhibitor protects against downregulation of claudin-1 during epithelial-mesenchymal transition of pancreatic cancer. Carcinogenesis. 2013;34:1232–1243. doi: 10.1093/carcin/bgt057. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B, Blanchard A, Wang N, Ma X, Han J, Schroedter I, Leygue E, Myal Y. Claudin 1 promotes migration and increases sensitivity to tamoxifen and anticancer drugs in luminal-like human breast cancer cells MCF7. Cancer Investig. 2015:1–11. [PubMed] [Google Scholar]
- 34.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dias K, Dvorkin-Gheva A, Hallett RM, Wu Y, Hassell J, Pond GR, Levine M, Whelan T, Bane AL. Claudin-low breast cancer; clinical & pathological characteristics. PLoS One. 2017;12 doi: 10.1371/journal.pone.0168669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabatier R, Finetti P, Guille A, Adelaide J, Chaffanet M, Viens P, Birnbaum D, Bertucci F. Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization. Mol Cancer. 2014;13:228. doi: 10.1186/1476-4598-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchard AA, Ma X, Dueck KJ, Penner C, Cooper SC, Mulhall D, Murphy LC, Leygue E, Myal Y. Claudin 1 expression in basal-like breast cancer is related to patient age. BMC Cancer. 2013;13:268. doi: 10.1186/1471-2407-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myal Y, Leygue E, Blanchard AA. Claudin 1 in breast tumorigenesis: revelation of a possible novel "claudin high" subset of breast cancers. J Biomed Biotechnol. 2010;2010:956897. doi: 10.1155/2010/956897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu S, Singh K, Mangray S, Tavares R, Noble L, Resnick MB, Yakirevich E. Claudin expression in high-grade invasive ductal carcinoma of the breast: correlation with the molecular subtype. Mod Pathol. 2013;26:485–495. doi: 10.1038/modpathol.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heerma van Voss MR, van Diest PJ, Smolders YH, Bart J, van der Wall E, van der Groep P. Distinct claudin expression characterizes BRCA1-related breast cancer. Histopathology. 2014;65:814–827. doi: 10.1111/his.12490. [DOI] [PubMed] [Google Scholar]
- 41.Palmer RH, Ridden J, Parker PJ. Identification of multiple, novel, protein kinase C-related gene products. FEBS Lett. 1994;356:5–8. doi: 10.1016/0014-5793(94)01202-4. [DOI] [PubMed] [Google Scholar]
- 42.Webb BL, Hirst SJ, Giembycz MA. Protein kinase C isoenzymes: a review of their structure, regulation and role in regulating airways smooth muscle tone and mitogenesis. Br J Pharmacol. 2000;130:1433–1452. doi: 10.1038/sj.bjp.0703452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koizumi J, Kojima T, Ogasawara N, Kamekura R, Kurose M, Go M, Harimaya A, Murata M, Osanai M, Chiba H. Protein kinase C enhances tight junction barrier function of human nasal epithelial cells in primary culture by transcriptional regulation. Mol Pharmacol. 2008;74:432–442. doi: 10.1124/mol.107.043711. [DOI] [PubMed] [Google Scholar]
- 44.Allen-Petersen BL, Miller MR, Neville MC, Anderson SM, Nakayama KI, Reyland ME. Loss of protein kinase C delta alters mammary gland development and apoptosis. Cell Death Dis. 2010;1:e17. doi: 10.1038/cddis.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masso-Weich PA, Verstovsek G, Ip MM. Alterations in the expression and localization of protein kinase C isoforms during mammary gland differentiation. Eur J Cell Biol. 1999;78:497–510. doi: 10.1016/s0171-9335(99)80076-4. [DOI] [PubMed] [Google Scholar]
- 46.Gonzales-Mariscal L, Garay E, Quiros M. Vol. 65. Elsevier Inc.; Burlington, MA, U.S.A.: 2010. Regulation of Claudins by Posttranslational Modifications and Cell-Signaling Cascades; pp. 113–150. [Google Scholar]
- 47.Way KJ, Chou E, King GL. Identification of PKC-isoform-specific biological actions using pharmacological approaches. Trends Pharmacol Sci. 2000;21:181–187. doi: 10.1016/s0165-6147(00)01468-1. [DOI] [PubMed] [Google Scholar]
- 48.Parker PJ, Murray-Rust J. PKC at a glance. J Cell Sci. 2004;117:131–132. doi: 10.1242/jcs.00982. [DOI] [PubMed] [Google Scholar]
- 49.Besson A, Davy A, Robbins SM, Yong VW. Differential activation of ERKs to focal adhesions by PKC epsilon is required for PMA-induced adhesion and migration of human glioma cells. Oncogene. 2001;20:7398–7407. doi: 10.1038/sj.onc.1204899. [DOI] [PubMed] [Google Scholar]
- 50.Miyata Y, Sato T, Yano M, Ito A. Activation of protein kinase C betaII/epsilon-c-Jun NH2-terminal kinase pathway and inhibition of mitogen-activated protein/extracellular signal-regulated kinase 1/2 phosphorylation in antitumor invasive activity induced by the polymethoxy flavonoid, nobiletin. Mol Cancer Ther. 2004;3:839–847. [PubMed] [Google Scholar]
- 51.Urtreger AJ, Kazanietz MG, Bal de Kier Joffe ED. Contribution of individual PKC isoforms to breast cancer progression. IUBMB Life. 2012;64:18–26. doi: 10.1002/iub.574. [DOI] [PubMed] [Google Scholar]
- 52.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 53.Ainsworth PD, Winstanley JH, Pearson JM, Bishop HM, Garrod DR. Protein kinase C alpha expression in normal breast, ductal carcinoma in situ and invasive ductal carcinoma. Eur J Cancer. 2004;40:2269–2273. doi: 10.1016/j.ejca.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 54.Kerfoot C, Huang W, Rotenberg SA. Immunohistochemical analysis of advanced human breast carcinomas reveals downregulation of protein kinase C alpha. J Histochem Cytochem. 2004;52:419–422. doi: 10.1177/002215540405200314. [DOI] [PubMed] [Google Scholar]
- 55.Ways DK, Kukoly CA, deVente J, Hooker JL, Bryant WO, Posekany KJ, Fletcher DJ, Cook PP, Parker PJ. MCF-7 breast cancer cells transfected with protein kinase C-alpha exhibit altered expression of other protein kinase C isoforms and display a more aggressive neoplastic phenotype. J Clin Invest. 1995;95:1906–1915. doi: 10.1172/JCI117872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banan A, Zhang LJ, Shaikh M, Fields JZ, Choudhary S, Forsyth CB, Farhadi A, Keshavarzian A. theta Isoform of protein kinase C alters barrier function in intestinal epithelium through modulation of distinct claudin isotypes: a novel mechanism for regulation of permeability. J Pharmacol Exp Ther. 2005;313:962–982. doi: 10.1124/jpet.104.083428. [DOI] [PubMed] [Google Scholar]
- 57.D'Souza T, Agarwal R, Morin PJ. Phosphorylation of claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. J Biol Chem. 2005;280:26233–26240. doi: 10.1074/jbc.M502003200. [DOI] [PubMed] [Google Scholar]
- 58.Lippoldt A, Liebner S, Andbjer B, Kalbacher H, Wolburg H, Haller H, Fuxe K. Organization of choroid plexus epithelial and endothelial cell tight junctions and regulation of claudin-1, -2 and -5 expression by protein kinase C. Neuroreport. 2000;11:1427–1431. doi: 10.1097/00001756-200005150-00015. [DOI] [PubMed] [Google Scholar]
- 59.French AD, Fiori JL, Camilli TC, Leotlela PD, O'Connell MP, Frank BP, Subaran S, Indig FE, Taub DD, Weeraratna AT. PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int J Med Sci. 2009;6:93–101. doi: 10.7150/ijms.6.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Skliris GP, Hube F, Gheorghiu I, Mutawe MM, Penner C, Watson PH, Murphy LC, Leygue E, Myal Y. Expression of small breast epithelial mucin (SBEM) protein in tissue microarrays (TMAs) of primary invasive breast cancers. Histopathology. 2008;52:355–369. doi: 10.1111/j.1365-2559.2007.02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leitges M, Plomann M, Standaert ML, Bandyopadhyay G, Sajan MP, Kanoh Y, Farese RV. Knockout of PKC alpha enhances insulin signaling through PI3K. Mol Endocrinol. 2002;16:847–858. doi: 10.1210/mend.16.4.0809. [DOI] [PubMed] [Google Scholar]
- 63.Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteomics. 2007;6:1855–1867. doi: 10.1074/mcp.M700183-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Yan Y, Penner CC, Skliris GP, Cooper C, Nugent Z, Blanchard A, Hamedani MK, Wang X, Myal Y, Murphy LC. Steroid receptor RNA activator protein (SRAP) expression as a prognostic factor in ER+ human breast tumors. J Cancer Res Clin Oncol. 2013;139:1637–1647. doi: 10.1007/s00432-013-1485-2. [DOI] [PubMed] [Google Scholar]
- 65.Fortino V, Torricelli C, Capurro E, Sacchi G, Valacchi G, Maioli E. Antiproliferative and survival properties of PMA in MCF-7 breast cancer cell. Cancer Investig. 2008;26:13–21. doi: 10.1080/07357900701637949. [DOI] [PubMed] [Google Scholar]
- 66.Dumont JA, Bitonti AJ. Modulation of human melanoma cell metastasis and adhesion may involve integrin phosphorylation mediated through protein kinase C. Biochem Biophys Res Commun. 1994;204:264–272. doi: 10.1006/bbrc.1994.2454. [DOI] [PubMed] [Google Scholar]
- 67.Gopalakrishna R, Barsky SH. Tumor promoter-induced membrane-bound protein kinase C regulates hematogenous metastasis. Proc Natl Acad Sci U S A. 1988;85:612–616. doi: 10.1073/pnas.85.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the β-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12:469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 69.Awadelkarim KD, Callens C, Rosse C, Susini A, Vacher S, Rouleau E, Lidereau R, Bieche I. Quantification of PKC family genes in sporadic breast cancer by qRT-PCR: evidence that PKCiota/lambda overexpression is an independent prognostic factor. Int J Cancer. 2012;131:2852–2862. doi: 10.1002/ijc.27600. [DOI] [PubMed] [Google Scholar]
- 70.Lonne GK, Cornmark L, Zahirovic IO, Landberg G, Jirstrom K, Larsson C. PKCalpha expression is a marker for breast cancer aggressiveness. Mol Cancer. 2010;9:76. doi: 10.1186/1476-4598-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pal D, Outram SP, Basu A. Upregulation of PKCeta by PKCepsilon and PDK1 involves two distinct mechanisms and promotes breast cancer cell survival. Biochim Biophys Acta. 2013;1830:4040–4045. doi: 10.1016/j.bbagen.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain K, Basu A. Protein kinase C-epsilon promotes EMT in breast cancer. Breast Cancer (Auckl) 2014;8:61–67. doi: 10.4137/BCBCR.S13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brennan K, Offiah G, McSherry EA, Hopkins AM. Tight junctions: a barrier to the initiation and progression of breast cancer? J Biomed Biotechnol. 2010;2010:460607. doi: 10.1155/2010/460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yano K, Imaeda T, Niimi T. Transcriptional activation of the human claudin-18 gene promoter through two AP-1 motifs in PMA-stimulated MKN45 gastric cancer cells. Am J Physiol Gastrointest Liver Physiol. 2008;294:G336–G343. doi: 10.1152/ajpgi.00328.2007. [DOI] [PubMed] [Google Scholar]
- 75.Inai T, Kobayashi J, Shibata Y. Claudin-1 contributes to the epithelial barrier function in MDCK cells. Eur J Cell Biol. 1999;78:849–855. doi: 10.1016/S0171-9335(99)80086-7. [DOI] [PubMed] [Google Scholar]
- 76.Kim J, Choi S, Kim JO, Kim KK. Autophagy-mediated upregulation of cytoplasmic claudin 1 stimulates the degradation of SQSTM1/p62 under starvation. Biochem Biophys Res Commun. 2018;496:159–166. doi: 10.1016/j.bbrc.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 77.Tanaka M, Kamata R, Sakai R. Phosphorylation of ephrin-B1 via the interaction with claudin following cell-cell contact formation. EMBO J. 2005;24:3700–3711. doi: 10.1038/sj.emboj.7600831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dukes JD, Fish L, Richardson JD, Blaikley E, Burns S, Caunt CJ, Chalmers AD, Whitley P. Functional ESCRT machinery is required for constitutive recycling of claudin-1 and maintenance of polarity in vertebrate epithelial cells. Mol Biol Cell. 2011;22:3192–3205. doi: 10.1091/mbc.E11-04-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu CJ, Mannan P, Lu M, Udey MC. Epithelial cell adhesion molecule (EpCAM) regulates claudin dynamics and tight junctions. J Biol Chem. 2013;288:12253–12268. doi: 10.1074/jbc.M113.457499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Assender JW, Gee JM, Lewis I, Ellis IO, Robertson JF, Nicholson RI. Protein kinase C isoform expression as a predictor of disease outcome on endocrine therapy in breast cancer. J Clin Pathol. 2007;60:1216–1221. doi: 10.1136/jcp.2006.041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tonetti DA, Morrow M, Kidwai N, Gupta A, Badve S. Elevated protein kinase C alpha expression may be predictive of tamoxifen treatment failure. Br J Cancer. 2003;88:1400–1402. doi: 10.1038/sj.bjc.6600923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsons M, Keppler MD, Kline A, Messent A, Humphries MJ, Gilchrist R, Hart IR, Quittau-Prevostel C, Hughes WE, Parker PJ. Site-directed perturbation of protein kinase C–integrin interaction blocks carcinoma cell chemotaxis. Mol Cell Biol. 2002;22:5897–5911. doi: 10.1128/MCB.22.16.5897-5911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pan Q, Bao LW, Kleer CG, Sabel MS, Griffith KA, Teknos TN, Merajver SD. Protein kinase C epsilon is a predictive biomarker of aggressive breast cancer and a validated target for RNA interference anticancer therapy. Cancer Res. 2005;65:8366–8371. doi: 10.1158/0008-5472.CAN-05-0553. [DOI] [PubMed] [Google Scholar]
- 84.Platet N, Prevostel C, Derocq D, Joubert D, Rochefort H, Garcia M. Breast cancer cell invasiveness: correlation with protein kinase C activity and differential regulation by phorbol ester in estrogen receptor-positive and -negative cells. Int J Cancer. 1998;75:750–756. doi: 10.1002/(sici)1097-0215(19980302)75:5<750::aid-ijc14>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 85.Sjo A, Magnusson KE, Peterson KH. Protein kinase C activation has distinct effects on the localization, phosphorylation and detergent solubility of the claudin protein family in tight and leaky epithelial cells. J Membr Biol. 2010;236:181–189. doi: 10.1007/s00232-010-9289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]