Abstract
MicroRNA (miRNA) expressions in tumor biopsies have shown potential as biomarkers in cervical cancer, but suitable reference RNAs for normalization of reverse transcription quantitative polymerase chain reaction (RT-qPCR) assays in patient cohorts with different clinicopathological characteristics are not available. We aimed to identify the optimal reference miRNAs and apply these to investigate the potential of miR-9-5p as human papilloma virus (HPV) 16 biomarker and miR-210-3p as hypoxia biomarker in cervical cancer. Candidate reference miRNAs were preselected in sequencing data of 90 patients and ranked in a stability analysis by RefFinder. A selection of the most stable miRNAs was evaluated by geNorm and NormFinder analyses of RT-qPCR data of 29 patients. U6 small nuclear RNA (RNU6) was also included in the evaluation. MiR-9-5p and miR-210-3p expression was assessed by RT-qPCR in 45 and 65 patients, respectively. Nine candidates were preselected in the sequencing data after excluding those associated with clinical markers, HPV type, hypoxia status, suboptimal expression levels, and low stability. In RT-qPCR assays, the combination of miR-151-5p, miR-152-3p, and miR-423-3p was identified as the most stable normalization factor across clinical markers, HPV type, and hypoxia status. RNU6 showed poor stability. By applying the optimal reference miRNAs, higher miR-9-5p expression in HPV16- than HPV18-positive tumors and higher miR-210-3p expression in more hypoxic than less hypoxic tumors were found in accordance with the sequencing data. MiR-210-3p was associated with poor outcome by both sequencing and RT-qPCR assays. In conclusion, miR-151-5p, miR-152-3p, and miR-423-3p are suitable reference miRNAs in cervical cancer. MiR-9-5p and miR-210-3p are promising HPV16 and hypoxia biomarkers, respectively.
Introduction
MicroRNA (miRNA) expression in tumor biopsies is a novel source of biomarkers that may lead to improved tools for patient stratification and treatment decision in cervical cancer [1]. Aberrant miRNA expression has been shown to correlate with poor clinical outcome [2], [3], [4], [5], [6], [7], [8], [9], advanced tumor stage [10], [11], [12], lymph node metastases [12], [13], and infection with specific types of human papillomavirus (HPV) [14]. Moreover, miRNAs associated with tumor hypoxia, which is a well-known aggressive feature of this disease [15], have been identified in other cancer types [16] and should be evaluated in cervical cancer as well. Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) allows fast and sensitive measurement of miRNA expression that would be easy to implement in the clinic. However, the results rely on proper choice of stably expressed reference RNAs for normalization to correct nonbiological sample-to-sample variations [17]. The search for biomarkers by RT-qPCR in cervical cancer specimens poses strict requirements with regard to stability of the reference RNAs across clinicopathological characteristics that have so far only been addressed for mRNA, and not miRNA, expression assays [18].
MiRNAs belong to a different RNA class than mRNAs, with different biogenesis, size, and stability. Reference RNAs optimized for miRNA expression are therefore required [19]. The choice of candidates is challenging because current knowledge of miRNA function is scarce. Small nucleolar RNAs, such as U6 small nuclear RNA (RNU6), have been widely used in miRNA RT-qPCR studies of various cancer types, including cervical cancer [20], [21], [22], [23], [24], [25]. However, their robustness as reference RNAs has been questioned [26], [27]. Moreover, for reliable normalization of assays applied on clinical specimens, multiple RNAs are recommended [28]. In previous work, candidate miRNAs have been evaluated for their suitability to detect expression differences between normal or precancerous cervical lesions and invasive cancer [29], [30], [31]. These candidates may therefore not meet the criteria required in studies of aggressive tumor features.
In this work, we aimed to identify the optimal set of reference miRNAs for RT-qPCR studies in cervical cancer and apply these to assess the potential of miR-9-5p and miR-210-3p as HPV16 and hypoxia biomarkers, respectively. We evaluated expression level, stability, and correlation to clinical markers, HPV type, and hypoxia status for all expressed miRNAs in a cohort of 90 patients, as determined by small RNA sequencing of tumor biopsies. The most promising nine candidates together with RNU6 were further evaluated by RT-qPCR, and an optimal set of three reference miRNAs feasible for biomarker studies was identified. Our work demonstrates the importance of careful selection of references RNAs and suggests consideration of miR-210 as a novel prognostic hypoxia biomarker in cervical cancer.
Materials and Methods
Patients and Tissue Samples
In total, 110 samples from patients with locally advanced squamous cell carcinoma of the uterine cervix, enrolled for treatment at The Norwegian Radium Hospital from 2001 to 2012, were included (Supplemental Table 1). MiRNA sequencing data were utilized for 90 patients (sequencing cohort), and 29 of these patients were included in the stability evaluation analysis with RT-qPCR (RT-qPCR cohort 1; Supplemental Table 1). In the RT-qPCR validation analysis, 16 more patients were included in addition to the 29 patients from the stability evaluation analysis (RT-qPCR cohort 1 extended; Supplemental Table 1). The remaining 20 patients were included in an independent validation analysis with RT-qPCR (RT-qPCR cohort 2; Supplemental Table 1).
The study was approved by The Regional Committee for Medical and Health Research Ethics in South East of Norway (REC 2016/2179), and written informed consent was obtained from all patients. The tumor biopsies were collected before the start of treatment, immediately snap frozen, and stored at −80°C. Those with more than 50% tumor cell fraction in hematoxylin and eosin–stained histological sections were selected for analyses. All patients received external radiation followed by brachytherapy as described [32]. Concurrent cisplatin was given according to tolerance.
Clinical markers, HPV type, and hypoxia status were collected from previous work on the same patients [32], [33]. In brief, HPV type was determined based on multiplex PCR of genomic DNA using primers specific for HPV16 and HPV18 as well as two consensus primers [33]. Tumor hypoxia status was assessed as more or less hypoxia according to a 6-gene hypoxia classifier determined from Illumina gene expression profiles [32].
Cell lines
HeLa and SiHa cervical cancer cell lines were used to assess miRNAs regulated by hypoxia or hypoxia combined with lactic acidosis. Culturing was performed in Dulbecco's modified Eagle medium with GlutaMAX supplemented with 10% fetal calf serum, 1% penicillin–streptomycin (Gibco, NY), and 1% L-glutamine (G7513, Sigma-Aldrich, St. Louis, MO) under 5% CO2 at 37°C. The cells were exposed to either normoxia (95% air, 5% CO2), hypoxia (0.2% O2, 5% CO2), or the combination of hypoxia (0.2% O2, 5% CO2) and lactic acidosis (10 mM lactic acid, pH 6.7) for 24 hours before they were harvested for RNA isolation. An Invivo2200 chamber (Ruskinn Technology Ltd., UK) was used to create hypoxia. Lactic acidosis was generated by adding calcium L-lactate hydrate (L4388, Sigma Aldrich) and further adjusting the culture medium to a pH of 6.7 using HCl. HEPES buffer (1 M) was used to maintain a stable pH during the experiments.
RNA Isolation
Total RNA was isolated from tissue cryotome sections and cell cultures by the miRNeasy kit (Qiagen, Hilden, Germany; sequencing cohort, cell lines) or with TRIzol Reagent (Invitrogen, Carlsbad, CA) followed by LiCl precipitation (RT-qPCR cohort 2) and dissolved in nuclease-free dH2O. RNA concentration was measured on a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). The RNA was isolated from all biopsies over a time span of 6 weeks and stored at −80°C. The RNA integrity was assessed by Bioanalyzer 2100 (Agilent Technologies, Inc. Santa Clara, CA) on all the samples within 1 year before small RNA sequencing.
Small RNA Sequencing
Small RNA preparation for sequencing was performed on samples from 90 patients (sequencing cohort) and cell cultures according to Illumina TruSeq Small RNA library protocols (Illumina, San Diego, CA). The patients included in the sequencing cohort were selected on the basis of RNA integrity of the isolated total RNA from tumor biopsies. RNA samples with an RNA integrity number (RIN) >6.5 from tumor biopsies and RIN >8 from cell cultures were considered acceptable for small RNA sequencing. Briefly, indexed 3′- and 5′-end RNA adapters were ligated to 1 μg of isolated total RNA per sample followed by reverse transcription and library amplification. The cDNA was purified on a PAGE gel by cutting out bands corresponding to 140-160 bp (length of miRNA + adapter and index sequences), followed by ethanol precipitation of pooled RNA. The Illumina HiSeq 2500 platform was used for single-end sequencing of cDNA molecules. Real-time analysis, base calling, and filtering of low quality reads were performed by Illumina's software packages (Illumina). FASTQ data were quality checked, and reads were aligned, quantified, and annotated using the miRDeep2 algorithm with miRBase v.21 for annotation of mature miRNAs [34]. Read counts were normalized as reads per million (RPM) annotated mature miRNAs. After normalization, miRNAs with read counts less than 10 across all patients were excluded from the clinical data set. Log2-transformed data were used in the analyses.
Reverse Transcription Quantitative PCR
RT-qPCR was performed on samples from 65 patients, including 45 patients in the sequencing cohort (RT-qPCR cohort 1, extended) and 20 independent patients (RT-qPCR cohort 2). The miRCURY LNA Universal RT microRNA PCR system (Exiqon Vedbaek, Denmark) was applied. cDNA was synthesized from 10 ng of total RNA using the miRCURY LNA microRNA PCR, Polyadenylation, and cDNA synthesis kit II (Exiqon). For amplification and detection of the selected miRNAs and RNU6, predeveloped microRNA LNA PCR primer sets (Supplemental Table 2) and the ExiLENT SYBR Green master mix (Exiqon) were used according to the standard protocols. Reference Dye for Quantitative PCR (1×, Sigma Aldrich R-4526) was used as passive reference. The qPCRs were run on a 7900HT Fast Real-Time PCR thermocycler (Applied Biosystems, Carlsbad, CA) using the thermal-cycling parameters recommended by the manufacturers. Amplification was analyzed using the SDS Software v.2.3 (Applied Biosystems). A no reverse transcriptase control and a no template control were included as negative controls for all reactions and showed no amplification [quantification cycle (Cq) = “undetectable”].
Primer specificity was assessed in a patient sample by melting curve analysis of the amplicons. PCR efficiency in percentage (E %) was evaluated by cDNA dilution series (80-, 320-, 1280- and 5120-fold), using the formula E% = [10(−1/slope)−1]*100. The 320-fold dilution was selected for the studies.
All samples were run in technical duplicates, and the average Cq of each sample was used in the analysis. The normalized expression level (∆Cq) of miR-9-5p and miR-210-3p was calculated by the comparative Cq method [35] using a normalization factor calculated as the average Cq values of the reference miRNAs identified in this study.
Data Analysis and Statistics
BRB-ArrayTools v.4.5 (developed by Dr. Richard Simon and the BRB-ArrayTools Development Team) [36] and SigmaPlot v12.5 (Systat Software, Inc., Chicago, IL) were used for subsetting, filtering, and statistical analyses of miRNA expression data. The web-based tool RefFinder [37] was used to rank candidates in the miRNA sequencing data based on their stability values, as assessed by the algorithms BestKeeper [38], the comparative cycle threshold (∆Ct) method [39], geNorm [40] and NormFinder [28]. Based on the stability ranking of the individual programs, a weight was given to each miRNA. The geometric mean of the weights was used for the final ranking in RefFinder. GeNorm and NormFinder were further used to evaluate the selected candidates in RT-qPCR experiments. The geNorm algorithm calculates a stability value (M) for each candidate based on the average pairwise variation for each candidate with all other candidates. GeNorm excludes the least stable candidate (highest M) in a stepwise process until the most stable gene pair is left, i.e., the pair with the lowest average standard deviation (SD) of expression ratios between the candidates. GeNorm also calculates a normalization factor (NF) which determines the number of genes required for reliable normalization. Starting with the two most stable miRNAs (lowest M values), geNorm continuously adds another gene and recalculates the NF. The optimal number of reference genes is attained when the pairvise variation value (VNF) is below the recommended cutoff value of 0.15 [40]. NormFinder is an ANOVA-based algorithm used to estimate overall variation of candidate reference genes across subgroups based on both their intergroup and intragroup variation. The calculations are combined into a stability value (SV) for each candidate, where a low SV indicates higher stability [28].
The miRNA information database miRBase v.22 [41] was used to assess miRNA family affiliations. MiRNA expression was compared between patient groups by Mann-Whitney U test or Student's t test. Pearson's and Spearman's correlation analyses were used to correlate the expression level between miR-28 family members and correlation between Cq-values of the miRNAs versus age of biopsy, respectively. Cox proportional hazards univariate analysis was used to search for associations between miRNAs and survival, with progression-free survival or disease-specific survival with a maximum follow-up time of 5 years as clinical end point. For progression-free survival and disease-specific survival, time from diagnosis to the first event of recurrence or disease-related death was used, respectively. Kaplan-Meier analysis was performed by comparing survival curves for patients with high and low miR-210-3p expression using log-rank test.
Results
Preselection from Small RNA Sequencing Data
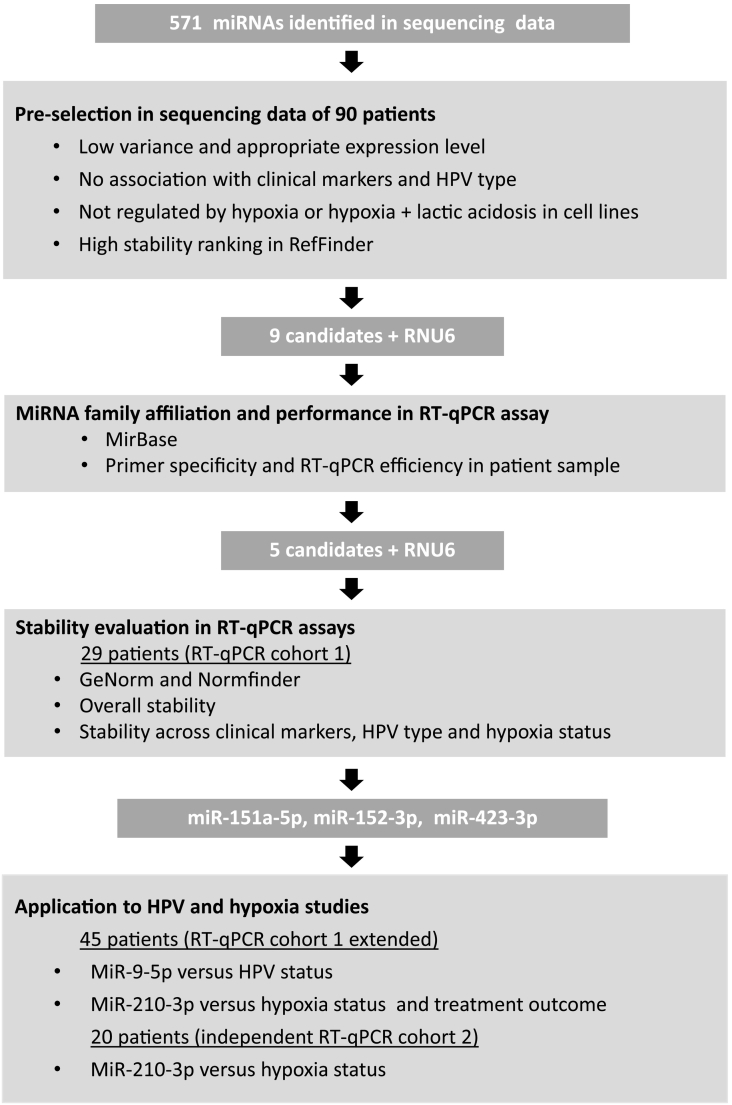
A total of 571 different miRNAs detected by small RNA sequencing of tumor samples from 90 cervical cancer patients were the basis of our selection of candidate reference miRNAs (Figure 1). To be selected, we required that the miRNA was expressed in all samples and exhibited low expression variance across the patients, and included only the 25% least variable miRNAs. In addition, to ensure an appropriate expression level within the range of most other miRNAs [42], those with a median log2-transformed expression ranging from 4 to 12 rpm were included. Only 5% of the miRNAs had an expression above 12 rpm, and those with expression below 4 rpm might be difficult to detect in RT-qPCR experiments. These two criteria excluded 477 miRNAs (Supplemental Table 3).
Figure 1.
Schematic presentation of the study workflow.
To optimize our list of candidates for prognostic biomarker studies, we excluded miRNAs that were responsive to hypoxia or hypoxia combined with lactic acidosis in the cervical cancer cell lines HeLa and SiHa using a threshold of 1.5-fold change to define the responsive ones (Supplemental Table 3). We further required that the candidate showed no differences in expression between patient groups exhibiting different clinical variables, including age, tumor stage, tumor volume, lymph node status, and HPV type, or was associated with survival. After the selection and filtering criteria were met, the list was reduced to 20 different miRNAs (Supplemental Table 3).
Stability ranking of the remaining 20 candidates by RefFinder showed good accordance between the 4 integrated algorithms (Supplemental Table 4). The nine top recommended miRNAs were selected for evaluation in RT-qPCR assays. They were among the nine most stable candidates by all algorithms, except for miR-151b which was ranked as number 10 by NormFinder. In addition, we included RNU6 to compare its performance with the candidate miRNAs.
MiRNA Family Affiliation and RT-qPCR Assay Performance
To avoid inclusion of highly co-regulated miRNAs, family affiliation analysis was carried out on the nine selected candidates. Three miRNAs, miR-151a-5p, miR-151b, and miR-28-5p, were identified to belong to the same miR-28 family (Supplemental Table 5) and thus likely to be co-regulated. MiR-151b is identical to miR-151a-5p, except for three extra bases on the 3′ terminus in miR-151a-5p (Supplemental Figure 1A). The sequencing data of the two miRNAs were identical (Supplemental Figure 1B). MiR-151b was therefore excluded from the further analysis, as it was ranked lower than miR-151a-5p in the RefFinder analysis (Supplemental Table 4). Before excluding either miR-28-5p or miR-151a-5p, which also showed correlation in expression (Supplementary Figure 1C), their PCR efficiency was analyzed together with the remaining candidates.
Melting curve analysis of the amplicons in RT-qPCR showed the presence of a single peak for all primer sets (Supplemental Figure 2). PCR efficiencies were in the satisfactory range of 93% (miR-652-3p) to 108% (RNU6), except for miR-532-5p (70% efficiency) and miR-28-5p (111% efficiency), which were excluded (Supplemental Table 6). MiR-671-3p showed Cq values >35 in the least diluted samples (×80) and was excluded since accurate quantification could be difficult, leaving five candidates and RNU6 for the subsequent analyses.
Stability Evaluation in RT-qPCR Assays
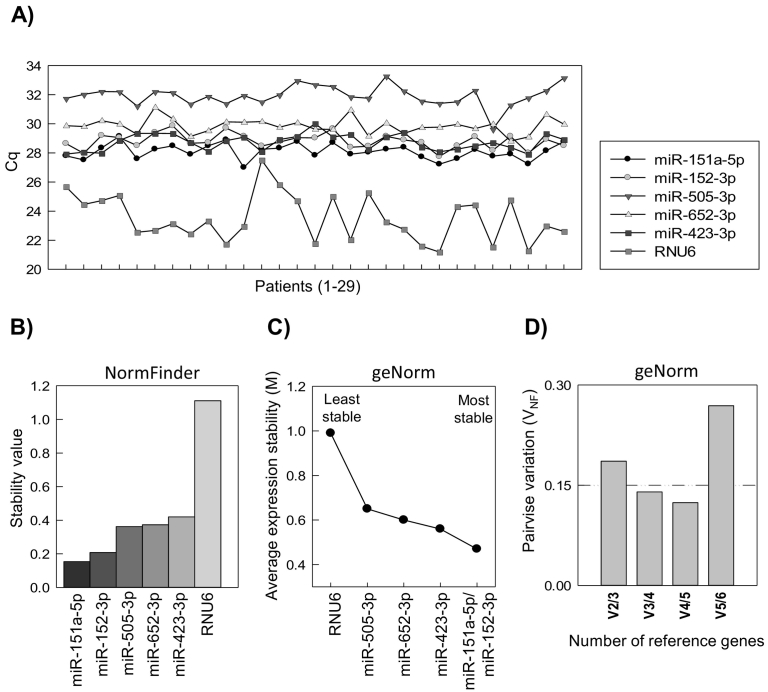
To visualize expression level and variation of the candidates, raw Cq values were plotted for 29 patients in the sequencing cohort (RT-qPCR cohort 1). The candidates displayed mean ± SD Cq values in the range from 23.5 ± 1.6 (RNU6) to 31.9 ± 0.7 (miR-505-3p), where RNU6 clearly showed high expression and variance across the samples compared to the other candidates (Figure 2A, Supplemental Table 7). No relationship was found between the Cq value and year of biopsy sampling for the miRNAs evaluated with RT-qPCR (Supplemental Figure 3), indicating no degradation of the investigated miRNAs during storage time.
Figure 2.
Overall stability analysis of 6 candidate reference RNAs by RT-qPCR in 29 patients (RT-qPCR cohort 1). (A) Quantification cycle (Cq) of candidate reference miRNAs and RNU6 across the cohort. (B) Overall stability value of candidate reference miRNAs and RNU6 by NormFinder, where a lower value represents higher stability. (C) Average expression stability (M) of candidate reference miRNAs and RNU6 after stepwise exclusion of the least stable RNA in geNorm. The x-axis indicates ranking of the candidates according to their expression stability with increasing stability from left to right. (D) geNorm pairwise variation analysis of number of reference RNAs required in the normalization factor. VNF indicates the pairwise variation between two sequential normalization factors (NFs) containing an increasing number of RNAs and starting with the most stable pair. The thin line represents the cutoff value for VNF of 0.15, for which inclusion of more reference RNAs has no significant effect on the normalization factor.
In the overall stability analysis, NormFinder ranked miR-151a-5p as the most stable candidate, followed by miR-152-3p. NormFinder ranked RNU6 as the least stable RNA (Figure 2B). Similarly, miR-151a-5p and miR-152-3p were the most stable candidates in the geNorm ranking, as shown by the M value of 0.47 (Figure 2C). This is below the expected M value range of 0.5-1 when evaluating reference genes in cancer biopsies, and where M < 1 is required for suitability as reference genes [43]. In accordance with NormFinder, geNorm ranked RNU6 as the least stable RNA. The M value was reduced from 0.99 to 0.65 after stepwise removal of RNU6 (Figure 2C).
To determine the optimal number of reference miRNAs required for normalization, geNorm's recommended cutoff value of 0.15 for pairwise variation between normalization factors (VNF) was used. The first VNF value below 0.15 appeared at V3/4, suggesting that the top three stable miRNAs, miR-151a-5p, miR-152-3p, and miR-423-5p, were sufficient (Figure 2D). The high V5/6 value (Figure 2D) was consistent with the steep decrease in M value obtained by the stepwise exclusion of the lowest rated candidate, RNU6 (Figure 2C).
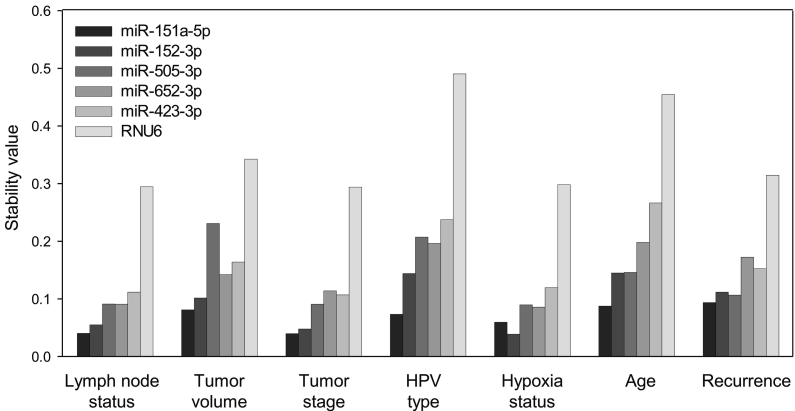
NormFinder was further used for stability evaluation across clinical subgroups of the 29 patients. Stability ranking based on inter- and intragroup variation for lymph node status, tumor volume, tumor stage, hypoxia status, HPV type, age, and cancer recurrence showed that miR-151a-5p was the most stable candidate in all groups, except for hypoxia status where miR-152-3p was rated as the most stable (Figure 3). RNU6 was the least stable candidate in all analyses, with a predominantly high inter- and intragroup variation (Supplemental Figure 4).
Figure 3.
Stability analysis of 6 candidate reference RNAs by RT-qPCR across subgroups of 29 patients (RT-qPCR cohort 1). Average stability value of candidate reference miRNAs and RNU6 by NormFinder based on the inter- and intragroup variation for lymph node status, tumor volume, tumor stage, HPV type, hypoxia status, age, and recurrence. Lower value represents higher stability. Subgroup characteristics and number of samples are described in Supplemental Table 1.
The best combination of candidates was miR-151a-5p and miR-152-3p for all markers, except age, in the NormFinder analysis (Table 1). These two miRNAs therefore appeared as the overall best combination for a normalization factor. However, for application of the normalization factor in HPV and hypoxia studies, the third most stable candidate in the geNorm ranking, miR-423-5p, was also included based on the pairwise variation analysis described above (Figure 2D).
Table 1.
Optimal Combination of Candidate Reference miRNAs by NormFinder
| Group | Best miRNA Combination | Stability |
|---|---|---|
| Lymph node status | miR-151a-5p + miR-152-3p | 0.035 |
| Tumor volume | miR-151a-5p + miR-152-3p | 0.076 |
| Tumor stage | miR-151a-5p + miR-152-3p | 0.032 |
| HPV type | miR-151a-5p + miR-152-3p | 0.091 |
| Hypoxia status | miR-151a-5p + miR-152-3p | 0.040 |
| Age | miR-151a-5p + miR-505-3p | 0.094 |
| Recurrence | miR-151a-5p + miR-152-3p | 0.055 |
Application in HPV and Hypoxia Studies
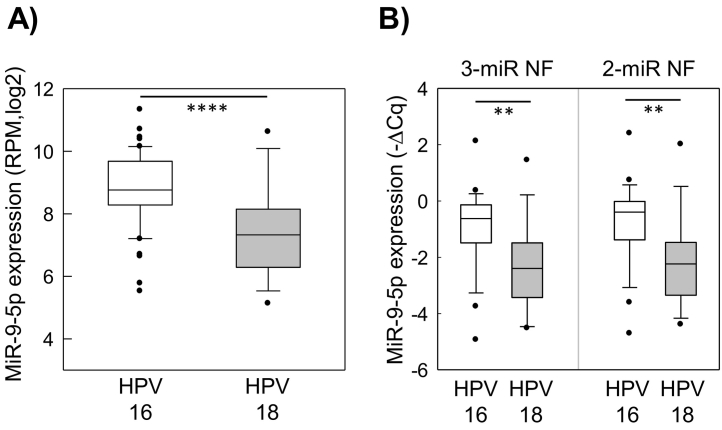
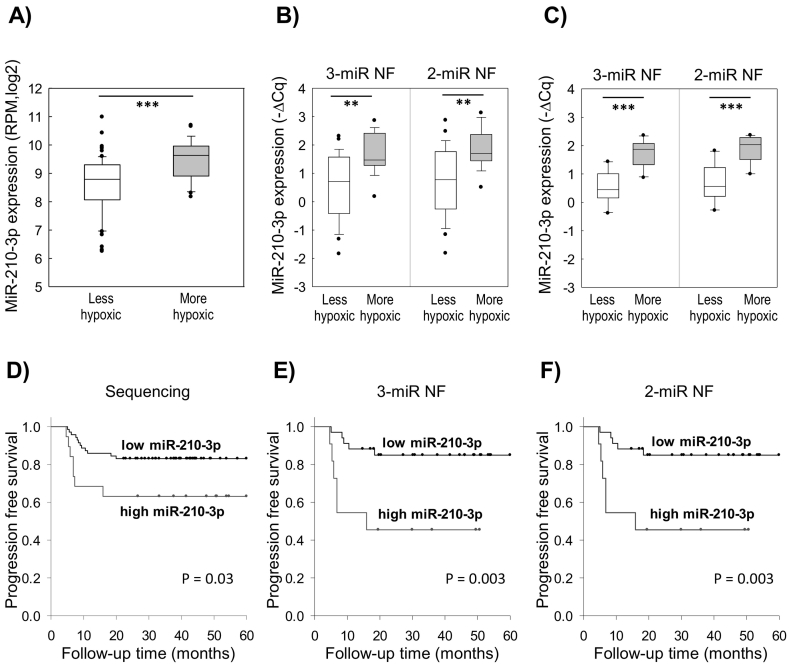
A normalization factor based on miR-151a-5p, miR-152-3p, and miR-423-5p or on only miR-151a-5p and miR-152-3p was evaluated and compared in studies of miR-9-5p and miR-210-3p in the extended RT-qPCR cohort 1 of 45 patients. The importance of these miRNAs in cervical cancer pathogenesis was first demonstrated in our sequencing data of 90 patients, showing a higher miR-9-5p expression in HPV16- than HPV18-positive tumors (Figure 4A) and a higher miR-210-3p expression in more hypoxic than less hypoxic tumors (Figure 5A). By RT-qPCR, we found that the normalization factors based on three miRNAs (miR-151a-5p, miR-152-3p, miR-423-5p) and two miRNAs (miR-151a-5p, miR-152-3p) were both suitable for distinguishing miR-9-5p expression between HPV16- and HPV18-positive tumors (Figure 4B). Moreover, the difference in expression between the two groups was almost equal for the two normalization factors. Similarly, a significant difference in miR-210-3p expression between less and more hypoxic tumors that was almost equal for both normalization factors was found (Figure 5B). In addition, a larger difference between less and more hypoxic tumors was detected in both cases in the independent RT-qPCR cohort 2, which included an equal number of 10 highly hypoxic tumors and 10 tumors with minor hypoxia (Figure 5C).
Figure 4.
MiR-9-5p expression in relation to HPV type. (A) MiR-9-5p expression in HPV16- and HPV18-positive tumors based on sequencing data of 90 patients (sequencing cohort). Values are given as log2 transformed RPM. (B) MiR-9-5p expression in HPV16- and HPV18-positive tumors by RT-qPCR in 45 patients (extended RT-qPCR cohort 1). Values are given as relative expression calculated by the ∆Cq method using the 3-miR NF (miR-151a-5p, miR-152-3p, and miR-423-3p) or the 2-miR NF (miR-151a-5p and miR-152-3p). Boxes represent lower and upper quartiles with median as horizontal line; whiskers depict the 10-90 percentiles; circles mark outliers. P values calculated by the Student's t test (two-sided) are shown. **P < .01, ****P < .0001.
Figure 5.
MiR-210-3p expression in relation to hypoxia status and treatment outcome. (A) MiR-210-3p expression in less and more hypoxic tumors based on sequencing data of 90 patients (sequencing cohort). (B, C) MiR-210-3p expression in less and more hypoxic tumors by RT-qPCR in 45 patients (extended RT-qPCR cohort 1; B) and 20 independent patients (RT-qPCR cohort 2; C). (A-C) Boxes represent lower and upper quartiles, with median as horizontal line; whiskers depict the 10-90 percentiles; circles mark outliers. P values were calculated by the Student's t test (two-sided) or the Mann-Whitney U test. (D, E, F) Kaplan-Meier curves of progression-free survival illustrating difference in recurrence for patients with high (n = 19) and low (n = 71) miR-210-3p expression by sequencing (sequencing cohort, 90 patients) (D) and high (n = 15) and low (n = 30) miR-210-3p expression by RT-qPCR (extended RT-qPCR cohort 1, 45 patients; E, F). P values by log-rank test are shown. The analyses were based on log2 transformed RPM (A, D) or relative expression calculated by the ∆Cq method using the 3-miR NF (miR-151a-5p, miR-152-3p, and miR-423-3p) or the 2-miR NF (miR-151a-5p and miR-152-3p) (B, C, E, F). **P < .005, ***P < .001.
To investigate the feasibility of the reference miRNAs in the search for prognostic biomarkers, we first assessed the relationship between miR-210-3p expression and treatment outcome in the sequencing cohort. The patient group classified with high miR-210-3p expression showed significantly increased risk of disease progression (Figure 5D), suggesting that miR-210-3p expression could serve as a prognostic biomarker in cervical cancer. The relationship to outcome was validated by RT-qPCR, and the significance was not altered by any of the two normalization factors (Figure 5E and F). Altogether, it seems that our reference miRNAs were suitable for biomarker studies in cervical cancer and that there was no benefit of including miR-423-5p in addition to miR-151a-5p and miR-152-3p in calculation of the normalization factor.
Discussion and Conclusions
Evaluation of normalized miRNA sequencing data of a large patient cohort made it possible to identify all potentially suitable reference miRNAs for testing in RT-qPCR assays. The preselected candidates were particularly promising for biomarker studies, with an appropriate expression level that was neither associated with clinical markers, treatment outcome, or HPV type nor responsive to hypoxia or hypoxia combined with lactic acidosis. Among the excluded candidates were miR-191-5p and miR-23a-3p, which have been suggested as reference miRNAs in studies comparing normal cervix, precancerous lesions, and cancer [30], [31]. In our study, miR-191-5p had a higher expression level than most other miRNAs, and miR-23a-3p expression was associated with treatment outcome, demonstrating the need for optimizing reference RNAs for each study purpose. Further stability analysis by RefFinder based on the sequencing data enabled selection of nine top-ranked candidates for testing in RT-qPCR assays. The accuracy of the RefFinder ranking has been questioned, as bias due to differences in assay efficiency may occur when the input is noncorrected, raw Cq values from RT-qPCR [44]. However, our analysis was based on normalized sequencing data, making the ranking more robust.
After excluding four miRNAs based on family affiliation, poor efficiency, and expression level in RT-qPCR assays, five preselected candidates, together with RNU6, remained for a rigorous evaluation by geNorm and NormFinder analyses on RT-qPCR data. The output of geNorm and NormFinder is a stability value per gene and per combination of genes. In addition, geNorm provides guidelines for suitable stability values, and M values in the range of 0.5-1 are acceptable when evaluating candidate reference genes in heterogeneous samples like cancer biopsies [43]. In our study, the M values of the preselected miRNA candidates ranged from of 0.65 using all five candidates to 0.47 using the two most stable ones. NormFinder provides no similar guidelines for stability values. However, the consistency in candidate ranking between NormFinder and geNorm ensured that the selected candidates are suitable as reference miRNAs.
RNU6 showed a high expression level and the lowest stability in all analyses by both geNorm and NormFinder. The M value increased to 0.99 if RNU6 was included, which is close to the recommended M value limit of 1. Also, including RNU6 in the normalization factor increased the VNF value above the recommended cutoff of 0.15. Hence, RNU6 was found to be less suitable as reference RNA, in accordance with other reports [26], [27]. Of particular importance for biomarker studies, RNU6 had the highest inter- and intragroup variation across clinical markers, HPV type, and hypoxia status. Consequently, the ability to minimize technical variation between samples would be reduced when using RNU6 as normalization factor, and small changes in miRNA expression could be difficult to detect. RNU6 is a longer RNA specie of 106 bp and has different physicochemical properties, isolation efficiency, and stability in solutions than miRNAs [27]. The poor performance of RNU6 could therefore be due to partly degradation or suboptimal RT-qPCR conditions.
Both geNorm and NormFinder suggested miR-151a-5p and miR-152-3p to be the best pair of reference miRNAs. These miRNAs were ranked as the most stable ones based on overall and inter- and intragroup stability across clinical markers, treatment outcome, HPV type, and hypoxia status, strongly indicating suitability in biomarker studies. One exception was NormFinder's analysis of the age groups, where a combination of miR-151a-5p and miR-505-3p was found to be more stable. However, the individual stability values for miR-152-3p and miR-505-3p were similar. The ranking of miR-505-3p above miR-152-3p could therefore be due to the lower intergroup variability of miR-505-3p. Since this discrepancy between NormFinder and geNorm was minor, it seems reasonable to select miR-151a-5p and miR-152-3p as the optimal pair of miRNAs. MiR-423-3p was selected as the third reference miRNA since this candidate was ranked as number three in the geNorm analysis and the overall stability increased by adding miR-423-3p to the combination of miR-151a-5p and miR-152-3p. Moreover, miR-423 has been identified as a suitable reference miRNA in cervical tissues in previous work [29].
Our studies of miR-9-5p and miR-210-3p in relation to HPV type and hypoxia status, respectively, showed that the three-miRNA combination (miR-151a-5p, -152-3p, -423-3p) and two-miRNA combination (miR-151a-5p, -152-3p) performed equally well as normalization factor. In particular, both combinations led to a significant association between miR-210-3p expression and treatment outcome. Moreover, the association between miR-210-3p and hypoxia status was validated in an independent patient cohort in both cases. These results strongly support the suitability of the two sets of reference miRNAs in studies of prognostic biomarkers. To increase robustness of the normalization factor, it is recommended to use at least three reference RNAs [40]. However, the benefit of using the three-miRNA rather than two-miRNA combination seems to be small and should be evaluated against the added cost.
In conclusion, miR-151-5p, miR-152-3p, and miR-423-3p are suitable reference miRNAs in RT-qPCR analyses of cervical cancer biopsies. Application of the reference miRNAs in biomarker studies showed a higher miR-9-5p expression in HPV16- than HPV18-positive tumors, supporting our results based on miRNA sequencing data and previous findings in cervical cancer [14]. Moreover, a higher miR-210-3p expression in more hypoxic than less hypoxic tumors and its association to poor treatment outcome were demonstrated, supporting findings in other cancer types [16] and representing novel results for cervical cancer. These results encourage development of HPV16 and hypoxia biomarkers based on miR-9-5p and miR-210-3p expression in cervical cancer. Moreover, RT-qPCR with the reference miRNAs identified here would probably be a feasible assay for assessment of such biomarkers.
The following are the supplementary data related to this article.
Supplementary material
Exclusion of miRNAs and Preselection of Candidate Reference miRNAs
Footnotes
Funding: This work was supported by The Norwegian Cancer Society (grant no. 107438-PR-2007-0179).
Declaration of interest: None.
References
- 1.Li J, Liu Q, Clark LH, Qiu H, Bae-Jump VL, Zhou C. Deregulated miRNAs in human cervical cancer: functional importance and potential clinical use. Future Oncol. 2017;13(8):743–753. doi: 10.2217/fon-2016-0328. [DOI] [PubMed] [Google Scholar]
- 2.Fang H, Shuang D, Yi Z, Sheng H, Liu Y. Up-regulated microRNA-155 expression is associated with poor prognosis in cervical cancer patients. Biomed Pharmacother. 2016;83:64–69. doi: 10.1016/j.biopha.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 3.How C, Pintilie M, Bruce JP, Hui AB, Clarke BA, Wong P, Yin S, Yan R, Waggott D, Boutros PC. Developing a prognostic micro-RNA signature for human cervical carcinoma. PLoS One. 2015;10(4):e0123946. doi: 10.1371/journal.pone.0123946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu X, Schwarz JK, Lewis JS, Jr., Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70(4):1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang B, Li Y, Wang T. A three miRNAs signature predicts survival in cervical cancer using bioinformatics analysis. Sci Rep. 2017;7(1):5624. doi: 10.1038/s41598-017-06032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo M, Shen D, Wang W, Xian J. Aberrant expression of microRNA-26b and its prognostic potential in human cervical cancer. Int J Clin Exp Pathol. 2015;8(5):5542–5548. [ http://www.ncbi.nlm.nih.gov/pubmed/26191262] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Jiang T. MicroRNA-335 represents an independent prognostic marker in cervical cancer. Tumour Biol. 2015;36(8):5825–5830. doi: 10.1007/s13277-015-3252-2. [DOI] [PubMed] [Google Scholar]
- 8.Wang N, Zhou Y, Zheng L, Li H. MiR-31 is an independent prognostic factor and functions as an oncomir in cervical cancer via targeting ARID1A. Gynecol Oncol. 2014;134(1):129–137. doi: 10.1016/j.ygyno.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 9.Yin ZL, Wang YL, Ge SF, Guo TT, Wang L, Zheng XM, Liu J. Reduced expression of miR-503 is associated with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci. 2015;19(21):4081–4085. [ http://www.ncbi.nlm.nih.gov/pubmed/26592830] [PubMed] [Google Scholar]
- 10.Gocze K, Gombos K, Juhasz K, Kovacs K, Kajtar B, Benczik M, Gocze P, Patczai B, Arany I, Ember I. Unique microRNA expression profiles in cervical cancer. Anticancer Res. 2013;33(6):2561–2567. [ http://www.ncbi.nlm.nih.gov/pubmed/23749909] [PubMed] [Google Scholar]
- 11.Ye C, Sun NX, Ma Y, Zhao Q, Zhang Q, Xu C, Wang SB, Sun SH, Wang F, Li W. MicroRNA-145 contributes to enhancing radiosensitivity of cervical cancer cells. FEBS Lett. 2015;589(6):702–709. doi: 10.1016/j.febslet.2015.01.037. [DOI] [PubMed] [Google Scholar]
- 12.Zhao S, Yao D, Chen J, Ding N, Ren F. MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14(9):2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Gao G, Hu X, Wang Y, Schwarz JK, Chen JJ, Grigsby PW, Wang X. Activation of miR-9 by human papillomavirus in cervical cancer. Oncotarget. 2014;5(22):11620–11630. doi: 10.18632/oncotarget.2599. [ http://www.ncbi.nlm.nih.gov/pubmed/25344913] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyng H, Malinen E. Hypoxia in cervical cancer: from biology to imaging. Clinical Transl Imaging. 2017;5(4):373–388. doi: 10.1007/s40336-017-0238-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid Redox Signal. 2014;21(8):1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data Normalization Strategies for MicroRNA Quantification. Clin Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fjeldbo CS, Aarnes EK, Malinen E, Kristensen GB, Lyng H. Identification and validation of reference genes for RT-qPCR studies of hypoxia in squamous cervical cancer patients. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0156259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14(5):844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azizmohammadi S, Safari A, Azizmohammadi S, Kaghazian M, Sadrkhanlo M, Yahaghi E, Farshgar R, Seifoleslami M. Molecular identification of miR-145 and miR-9 expression level as prognostic biomarkers for early-stage cervical cancer detection. QJM. 2017;110(1):11–15. doi: 10.1093/qjmed/hcw101. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Zhang S, Zhao Z, Mao X, Huang J, Wu Z, Zheng L, Wang Q. MicroRNA-27b up-regulated by human papillomavirus 16 E7 promotes proliferation and suppresses apoptosis by targeting polo-like kinase2 in cervical cancer. Oncotarget. 2016;7(15):19666–19679. doi: 10.18632/oncotarget.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Ni S. Association between genetic polymorphisms in the promoters of let-7 and risk of cervical squamous cell carcinoma. Gene. 2018;642:256–260. doi: 10.1016/j.gene.2017.11.038. [DOI] [PubMed] [Google Scholar]
- 23.Sun P, Shen Y, Gong JM, Zhou LL, Sheng JH, Duan FJ. A New MicroRNA expression signature for cervical cancer. Int J Gynecol Cancer. 2017;27(2):339–343. doi: 10.1097/IGC.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Wang F, Xu J, Wang X, Ye F, Xie X. Micro ribonucleic acid-93 promotes oncogenesis of cervical cancer by targeting RAB11 family interacting protein 1. J Obstet Gynaecol Res. 2016;42(9):1168–1179. doi: 10.1111/jog.13027. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Han LR, Zhou YX, Li Y. MiR-195 suppresses cervical cancer migration and invasion through targeting Smad3. Int J Gynecol Cancer. 2016;26(5):817–824. doi: 10.1097/IGC.0000000000000686. [DOI] [PubMed] [Google Scholar]
- 26.Lamba V, Ghodke-Puranik Y, Guan W, Lamba JK. Identification of suitable reference genes for hepatic microRNA quantitation. BMC Res Notes. 2014;7:129. doi: 10.1186/1756-0500-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wotschofsky Z, Meyer HA, Jung M, Fendler A, Wagner I, Stephan C, Busch J, Erbersdobler A, Disch AC, Mollenkopf HJ. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal Biochem. 2011;417(2):233–241. doi: 10.1016/j.ab.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 29.Babion I, Snoek BC, van de Wiel MA, Wilting SM, Steenbergen RDM. A strategy to find suitable reference genes for miRNA quantitative PCR analysis and its application to cervical specimens. J Mol Diagn. 2017;19(5):625–637. doi: 10.1016/j.jmoldx.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Leitao Mda C, Coimbra EC, de Lima Rde C, Guimaraes Mde L, Heraclio Sde A, Silva Neto Jda C, de Freitas AC. Quantifying mRNA and microRNA with qPCR in cervical carcinogenesis: a validation of reference genes to ensure accurate data. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0111021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y, Li Y, Ye F, Wang F, Wan X, Lu W, Xie X. Identification of miR-23a as a novel microRNA normalizer for relative quantification in human uterine cervical tissues. Exp Mol Med. 2011;43(6):358–366. doi: 10.3858/emm.2011.43.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fjeldbo CS, Julin CH, Lando M, Forsberg MF, Aarnes EK, Alsner J, Kristensen GB, Malinen E, Lyng H. Integrative analysis of DCE-MRI and gene expression profiles in construction of a gene classifier for assessment of hypoxia-related risk of chemoradiotherapy failure in cervical cancer. Clin Cancer Res. 2016;22(16):4067–4076. doi: 10.1158/1078-0432.CCR-15-2322. [DOI] [PubMed] [Google Scholar]
- 33.Lando M, Wilting SM, Snipstad K, Clancy T, Bierkens M, Aarnes EK, Holden M, Stokke T, Sundfor K, Holm R. Identification of eight candidate target genes of the recurrent 3p12-p14 loss in cervical cancer by integrative genomic profiling. J Pathol. 2013;230(1):59–69. doi: 10.1002/path.4168. [DOI] [PubMed] [Google Scholar]
- 34.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Informat. 2007;3:11–17. [ http://www.ncbi.nlm.nih.gov/pubmed/19455231] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012 doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [ http://www.ncbi.nlm.nih.gov/pubmed/15127793] [DOI] [PubMed] [Google Scholar]
- 39.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:7. doi: 10.1186/gb-2002-3-7-research0034. [RESEARCH0034, http://www.ncbi.nlm.nih.gov/pubmed/12184808] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, Borghi L, Verbrugghe P, Kalaydjieva L, Bleuler S, Laule O. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 2011;12(156) doi: 10.1186/1471-2164-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellemans J, Vandesompele J. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol Biol. 2014;1160:19–26. doi: 10.1007/978-1-4939-0733-5_3. [DOI] [PubMed] [Google Scholar]
- 44.De Spiegelaere W, Dern-Wieloch J, Weigel R, Schumacher V, Schorle H, Nettersheim D, Bergmann M, Brehm R, Kliesch S, Vandekerckhove L. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Exclusion of miRNAs and Preselection of Candidate Reference miRNAs