Abstract
Silver nanoparticles (Ag-NPs) have increasingly been used for coatings on various textiles and certain implants, for the treatment of wounds and burns, as a water disinfectant, and in air-freshener sprays. The wide use of Ag-NPs may have potential human health impacts. In this study, the mutagenicity of 5-nm Ag-NPs was evaluated in the mouse lymphoma assay system, and modes of action were assessed using standard alkaline and enzyme-modified Comet assays and gene expression analysis. Treatments of L5178Y/Tk+/− mouse lymphoma cells with 5-nm uncoated Ag-NPs resulted in a significant yield of mutants at doses between 3 and 6 μg/mL; the upper range was limited by toxicity. Loss of heterozygosity analysis of the Tk mutants revealed that treatments with uncoated Ag-NPs induced mainly chromosomal alterations spanning less than 34 megabase pairs on chromosome 11. Although no significant induction of DNA damage in Ag-NP-treated mouse lymphoma cells was observed in the standard Comet assay, the Ag-NP treatments induced a dose-responsive increase in oxidative DNA damage in the enzyme-modified Comet assay in which oxidative lesion-specific endonucleases were added. Gene expression analysis using an oxidative stress and antioxidant defense polymerase chain reaction (PCR) array showed that the expressions of 17 of the 59 genes on the arrays were altered in the cells treated with Ag-NPs. These genes are involved in production of reactive oxygen species, oxidative stress response, antioxidants, oxygen transporters, and DNA repair. These results suggest that 5 nm Ag-NPs are mutagenic in mouse lymphoma cells due to induction of oxidative stress by the Ag-NPs. Environ. Mol. Mutagen. 53:409419, 2012.
Keywords: silver nanoparticles, mutant frequency, chromosomal damage, oxidative stress, gene expression
INTRODUCTION
Rapid development of nanotechnology and commercialization of nanoscale products in recent years have increased human exposure to engineered nanomaterials [Nel et al., 2006; Xia et al., 2009]. In addition, the U.S. Food and Drug Administration (FDA) has already reviewed and approved some nanotechnology-based products and expects a significant increase in the use of nanomaterials in drugs, devices, biologics, cosmetics, and food [FDA, 2007]. Silver has been widely used for thousands of years for hygienic and medicinal purposes, and is used today as a broad spectrum antibacterial, antifungal, and antiviral agent for the treatment of cuts, burns, and amputation sites [Chen and Schluesener, 2008; Chaloupka et al., 2010]. Silver nanoparticles (Ag-NPs) appear to possess more potent antimicrobial activity than bulk silver and have been incorporated into wound dressings to reduce infections [Gravante et al., 2009]. They have also been used in coatings for various textiles and implants, as a water disinfectant, and in air-freshener sprays [Chen and Schluesener, 2008].
Increasing human exposure to Ag-NPs from the wide usage of Ag-NPs in medicine and related applications necessitates the generation of toxicity data. Ag-NPs have been shown to be cytotoxic and genotoxic to mammalian cell lines in vitro [Braydich-Stolle et al., 2005; Hussain et al., 2005; Arora et al., 2008; Hsin et al., 2008; AshaR-ani et al., 2009; Trickler et al., 2010; Foldbjerg et al., 2011; Li et al., 2011; Asare et al., 2012] and to rats and Zebrafish in vivo [Asharani et al., 2008; Sung et al., 2009]. However, few studies have examined the mechanisms of action of the observed toxicity. In addition, while clinical studies conducted with Ag-NP-impregnated wound dressings have showed superior prevention of infections in burn patients compared with the old silver formulation [Huang et al., 2007; Gravante et al., 2009], clinical use of Ag-NPs (15 nm)-coated wound dressings have also been associated with the development of hepa-totoxicity and argyria-like symptoms and a silver level of 107 μg/kg in plasma [Trop et al., 2006]. Therefore, additional data from studies using the standard assays with validated protocols would be useful in a weight-of-evidence evaluation of Ag-NPs for human health risk assessment.
The mouse lymphoma assay (MLA), using the Thymidine kinase (Tk) gene as the mutational target, detects a broad spectrum of genetic damage, including both point mutation and chromosomal alterations. This assay has been validated for use as a component of the genotoxic testing battery, which is used for evaluating the carcinogenic potential of chemicals [Dearfield et al., 1991; ICH, 1995; DHHS, 1997]. In this study, we assessed the mutagenic potential of Ag-NPs in mouse lymphoma cells and performed further assays to determine the mechanisms of action of the observed mutagenicity. By using loss of heterozygosity (LOH) analysis of the Tk mutants, we determined whether the mutants contained large scale loss of genetic material. To evaluate whether Ag-NPs induced oxidative stress, we also performed the oxidative DNA damage Comet assay and evaluated the gene expression changes using oxidative stress and antioxidant defense polymerase chain reaction (PCR) array.
MATERIALS AND METHODS
Materials
Uncoated Ag-NPs used in this study were obtained from Novacentrix Inc. (Austin, TX), and stored in room temperature under dark condition. Trifluorothymidine (TFT) was purchased from the Sigma Chemical Company (St. Louis, MO). Fischer’s medium was obtained from Quality Biological Inc. (Gaithersburg, MD), and all other cell culture supplies were acquired from Invitrogen Life Technologies (Carlsbad, CA). PCR Master Mix was from Promega Company (Madison, WI). The primers used for detection of LOH at the Tk locus and the D11Mit36, D11Mit20, and D11Mit74 loci were synthesized by Invitrogen Life Technologies. The RT2 First Stand Kit and the Oxidative Stress and Antioxidant Defense RT2 Profiler PCR array were obtained from Qiagen (Valencia, CA).
Nanoparticle Characterization
Transmission electron microscopy (TEM) was carried out to obtain the primary size and morphology of Ag-NPs using a field emission JEM-2100F TEM (JEOL Inc., Peabody, MA) equipped with a CCD camera. The acceleration voltage used in this study was 100 kV. Ag-NPs were measured after samples were homogeneously dispersed in water, and 5 μL of the suspension were deposited on a formvar/carbon-coated TEM grid, dried, and evacuated before analysis.
Dynamic light scattering (DLS), for characterization of hydrodynamic size and surface charge (zeta potential) of the nanoparticles in aqueous media, was performed on a Zetasizer Nano-ZS instrument (Malvern Instruments, Malvern, Worcestershire, UK). Samples were measured after dilution of Ag-NP stock solution to 50 μg/mL suspensions in water, phosphate-buffered saline (PBS), or media solution. The dilution was vortexed for 10 min, and sonicated for 30 min in ultrasonic water bath to provide a homogenous dispersion. The mixture was spun at 78g for 5 min to remove large aggregates before measuring the size [Wang et al., 2007b]. Then 1 mL of the dilute was transferred to either a 1 cm2 cuvette for dynamic size measurement or a Malvern Clear Zeta Potential cell for zeta potential measurement.
Ag-NP Preparation
Five nanometers of Ag-NPs were suspended in sterilized water at a stock concentration of 1.5%. Before each experiment, the stock solution was vortexed for 10 min, then sonicated for 30 min in an ultrasonic water bath to ensure a uniform suspension. The mixture was spun at 78g for 5 min to remove large aggregates [Wang et al., 2007b] and the supernatant was used to treat the cells at various concentrations of the Ag-NPs. The nanoparticle solution was handled under the hood to avoid bacterial contamination.
Cells and Culture Conditions
The L5178Y/Tk+/−−3.7.2C mouse lymphoma cell line was utilized for the mutation assay. Cells were grown according to previously published method [Guo et al., 2011]. Briefly, the basic medium was Fischer’s medium for leukemic cells of mice with L-glutamine supplemented with pluronic F68 (0.1%), sodium pyruvate (1 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL). The treatment medium (F5p), growth medium (F10p), and cloning medium (F20p) were the basic medium supplemented with 5%, 10%, and 20% heat-inactivated horse serum, respectively. The cultures were maintained in a humidified incubator with 5% CO2 in air at 37°C.
Cell Treatment With 5 nm Ag-NPs
The Ag-NP working solution (100X) was prepared just prior to use by a series of dilutions using sterilized water. The mouse lymphoma cells were suspended in 50-mL centrifuge tubes containing 6 × 106 cells in 10 mL of treatment medium. One hundred microliters of Ag-NP working solutions was added to the cell cultures. A preliminary cytotoxicity dose range finding experiment showed that doses lower than 3 μg/mL were associated with little cytotoxicity while doses higher than 6 μg/mL with a 4 hr incubation resulted in less than 10% of relative total growth (RTG) [Moore et al., 2002, 2006]. RTG measures cytotoxicity including cell growth during treatment (4 hr), expression (2 days), and cloning (11 days). Therefore, the final concentrations of Ag-NPs in the main experiments were 3–6 μg/mL. The cells were gassed with 5% (v/v) CO2 in air and placed on a roller drum (15 rpm) in a 370C incubator for 4 hr. All experiments included an untreated control and positive control (0.1 μg/mL 4-nitroquinoline-1-oxide or methyl methanesulfonate). After treatment, the cells were centrifuged at 78g for 5 min and washed twice with fresh medium, and were then resuspended in growth medium at a density of 3 × 105 cells/mL. The culture tubes were placed on a roller drum in a 37°C incubator to begin the 2-day phenotypic expression.
Intracellular Distribution ofAg-NPs in Mouse Lymphoma Cells
The mouse lymphoma cells were cultured in F5p medium (see above) containing 5 μg/mL of the Ag-NPs for 4 hr and then washed three times with PBS to remove non-incorporated particles from the cell membrane. Five microliters of the cell suspension in PBS were placed on a glass slide and then sealed with a glass cover and clear nail polish. Images were captured using a confocal laser scanning Olympus Fluoview FV 1000 inverted microscope with an excitation of 405 nm and emission of 480 nm. Intracellular localization of the nanoparticles in cells was analyzed, and the images were created using Adobe Photoshop software.
Microwell and Soft-Agar Versions of the MLA
Following a 2-day expression period, the treated cultures were divided into two parts to perform mutant selection experiments using both the microwell and soft-agar versions of the MLA [Chen and Moore, 2004]. In the microwell version, 3 μg/mL of TFT was added to the cells in the cloning medium for mutant enumeration, and the cells were seeded into four 96-well flat-bottom microtiter plates using 200 μL per well and a final density of 2,000 cells/well. For the determination of plating efficiency, the cultures were adjusted to 8 cells/mL medium and aliquoted in 200 μL per well into two 96-well flat-bottom microtiter plates. In the soft-agar version, 1 μg/mL of TFT was added in the cloning medium to enumerate the mutants. Cells were suspended in 100 mL of cloning medium with 0.28% agar and poured onto three 100-mm diameter tissue culture dishes to give a final density of 30,000 cells/mL. Six hundred cells were suspended in 100 mL of 0.28% soft agar cloning medium for determining plating efficiency.
All 96-well plates and 100-mm tissue culture dishes were incubated at 37°C in a humidified incubator with 5% CO2 in air. After 11 days of incubation, colonies were counted, and mutant colonies were categorized as small or large [Chen and Moore, 2004]. For the microwell version, counting was performed visually and the small colonies were defined as those smaller than 25% of the diameter of the well. Mutant frequencies (MFs) were calculated using the Poisson distribution. For the soft-agar version, colony counting and sizing was performed using an automatic colony counter (Microbiology International, Frederick, MD) fitted with the capability to evaluate the size of the colonies. Mutant colonies approximately <0.6 mm in diameter were considered to be small-colony mutants. Cytotoxicity was measured using RTG that includes a measure of growth during treatment, expression, and cloning [Chen and Moore, 2004].
LOH Analysis of the Tk Mutants
Mutant clones were directly taken from TFT-selection plates. Forty-eight large and 48 small mutant colonies resulting from the treatment with 5 μg/mL Ag-NPs were analyzed. The mutant cells were washed once with PBS by centrifugation, and cell pellets were quickly frozen and stored at −20°C. Genomic DNA was extracted by digesting the cells in lysis buffer [Mei et al., 2005]. For PCR analysis of LOH at the Tk and other loci (D11Mit36, D11Mit20, and D11Mit74 loci), the amplification reactions were carried out in a total volume of 20 μL using 2× PCR Master Mix and pairs of primers described previously [Singh et al., 2005]. The thermal cycling conditions were as follows: initial incubation at 94°C for 3 min, 40 cycles of 94°C denaturation for 30 s, 55°C annealing for 30 s, and 72°C extension for 30 s, and a final extension at 72°C for 7 min. The amplification products were scored for the presence of one band (indicating LOH) or two bands (retention of heterozygosity at the given locus) after 2% agarose gel electrophoresis.
Comet Assay
The standard alkaline Comet assay was performed using established methods [Singh et al., 1988; Tice et al., 2000], with appropriate modifications. Briefly, 100 μL of the single cell suspensions after 4 hr treatment was mixed with 900 μL 1% low melting-point agarose in PBS at 37°C, and 200 μL of this suspension was applied to microscope slides (Fisher Scientific, St. Louis, MO) previously coated with 1% agarose. Cover slips were placed on the slides, and the slides were stored at 4°C for 30 min to solidify the agarose. With the cover slips gently removed, the slides were placed in freshly prepared lysis buffer (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris, with 10% dimethylsulfoxide and 1% Triton X-100 added just before use) and stored at 4°C overnight. The slides were then transferred into a chilled alkaline solution (300 mM NaOH, 1 mM EDTA, pH 13) and allowed to remain in the solution for 40 min in the dark to unwind DNA. After unwinding, electrophoresis was performed in the same solution at 4°C in the dark for 30 min at 25 V and ~ 300 mA. The slides were next washed with neutralizing buffer (0.4 M Tris, adjusted to pH 7.5 with HCl) three times for 5 min each to neutralize the remaining alkali and remove detergent; then the slides were fixed with ice cold ethanol (100%) and dried overnight. Prior to scoring, the slides were stained with SYBR Gold (Invitrogen, Carlsbad, CA) with 1:10,000 dilutions in TBE buffer. Two slides were scored from each sample, and 100 cells were selected randomly from each slide and scored using a system consisting of a Nikon 501 fluorescent microscope and Comet IV digital imaging software (Perceptive Instruments, Wiltshire, UK). The percentage of DNA in tail, defined as the fraction of DNA in the tail divided by the total amount of DNA associated with a cell multiplied by 100, was used as the parameter for DNA damage analysis.
For detecting oxidative DNA damage, cells were embedded in agarose on microscope slides and stored in the lysis buffer as described earlier for the standard assay. After the slides were removed from the lysis buffer, they were washed three times for 5 min each with the enzyme buffer (40 mM HEPES, 0.1 M KCl, 0.5 mM EDTA, 0.2 mg/mL bovine serum albumin, with the solution adjusted to pH 8.0 with KOH). After the last wash, excess buffer was removed, and 200 μL of the following solutions were applied to slides prepared from each treatment group: 200 μL of enzyme buffer alone (reference slides; two slides); 200 μL of enzyme buffer containing formamidopyrimidine DNA-glycosylase (FPG) (1:1500; two slides); 200 μL of enzyme buffer containing endonuclease III (Endo III) (1:1000; two slides); and 200 μL enzyme buffer containing 8-hydroxyguanine DNA-glycosylase (hOGG1) (1:1000; two slides). The slides were kept in a moist box and incubated at 37°C for 45 min for Endo III and reference slides, 30 min for FPG and hOGG1. After the enzyme treatment, the slides were placed in a horizontal electrophoresis chamber to perform DNA unwinding and electrophoresis as described earlier for the standard assay. The slides were neutralized, dried, stained, and scored as described for the standard Comet assay.
RNA Isolation and Quality Control
Total RNA from 5 X 105 cells for three treated samples (5 μg/mL Ag-NPs for 4 hr) and three control samples was isolated using an RNeasy system (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The purity and quality of the extracted RNA were evaluated using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Rockland, DE). The integrity of RNA was assessed by analyzing both the 18 and 28 s rRNA peaks using Bio-Rad’s Experion (Bio-Rad; Hercules, CA).
Real-Time PCR Array Analysis
The first strand synthesis was performed using the RT2 PCR Array First Strand Kit and the real-time PCR was performed with the Mouse Oxidative Stress and Antioxidant Defense PCR Array (SABiosciences), according to the manufacturer’s instructions, on an ABI 7500 fast PCR machine. The thermocycler parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. There were 25 genes in the arrays that were not expressed in mouse lymphoma cells and were excluded from the analysis.
Data Analysis
The data evaluation criteria developed by the MLA Expert Workgroup of the International Workshop for Genotoxicity Tests (IWGT) were used to determine whether a specific treatment condition was positive. Positive responses are defined as those where the induced MF in one or more treated cultures exceeds the global evaluation factor of 126 mutants per 106 cells for the microwell version or 90 mutants per 106 cells for the agar version and there is also a dose-related increase with MF [Moore et al., 2002, 2006]. The LOH patterns of the mutants induced by Ag-NPs were compared using the computer program described previously [Mei et al., 2005, 2006]. The PCR data were analyzed using PCR Array Data Analysis software provided by Qiagen (Delta/delta method). The data were also input into ArrayTrack software for principal component analysis to classify the samples. Gene expression is considered significantly changed when the fold change (treatment/control) is larger than 1.5 and the P value is smaller than 0.05. Statistical analysis of data from the Comet assays was performed using SigmaStat version 3.1 (SPSS, Chicago, IL) and statistical significance was determined by oneway analysis of variance followed by the Holm-Sidak method.
RESULTS
Characterization of Ag-NPs
The primary sizes of the Ag-NPs were determined using TEM. The individual nanoparticles’ diameters were found as various sizes (Fig. 1). Over 100 nanoparticles were measured and the size distribution of the particles and aggregates were calculated. About 66% of the Ag-NPs measured had diameters in the range of 4–8 nm, while 24% of the particles had diameters of 8–12 nm. In addition, 4% of particles had diameters less than 4 nm, and 6% of the particles were above 12 nm in diameter. Because analysis of the size properties using TEM are not performed in biological environments, the clustering properties and surface charges of the Ag-NPs will not be certain due to attachment of proteins to nanoparticles or other effects in biological medium. Therefore, DLS technique was used to characterize the behavior of the Ag-NPs and the hydrodynamic sizes and surface charge were measured using Zetasizer in a biological environment (i.e., medium). The Ag-NPs were suspended in deionized water, PBS, or Fischer’s medium (F5p) for analysis of their agglomerate sizes and zeta potential. The size and surface charge characteristics of the Ag-NPs are summarized in Table I. The mean agglomerate sizes ranged from 61.2 nm (in water) to 1608.7 nm (in medium), and the surface charge ranged from —9.37 mV (in water) to —8.20 mV (in medium). Particle size observed by DLS did not coincide with the results obtained from TEM. The physical properties measured (hydrodynamic diffusion versus projected area) may partially contribute to the differences based on the techniques used in this study. The sample preparation procedure was different because the samples using TEM should be in dried condition but DLS require aqueous solution preparation. The relevance of the different characterization in the biological media could provide further information of the stability and relative size of test materials in medium. It should be noted the DLS is very sensitive to the presence of small amount of the larger nanoparticles or aggregates, which can not be avoided under the experiment condition. Furthermore, the protein binding and/or the salt in media (PBS or Fischer’s medium) may affect the physical properties of Ag-NPs in water, resulting in the different sizes or aggregation/agglomerations in different media.
Fig. 1.
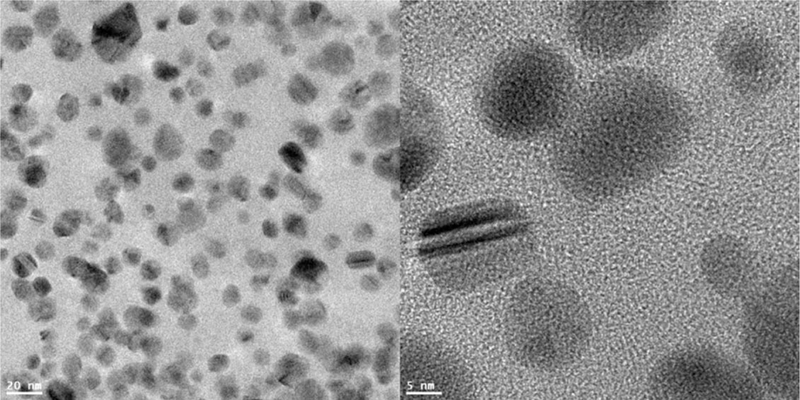
TEM characterization of Ag-NPs. Nanoparticles were deposited on formvar carbon-coated grids and dried for TEM imaging. Images were analyzed in a high resolution mode with an acceleration voltage 100 kV.
TABLE I.
Characterization of Silver Nanoparticles (Ag-NPs) in Solutionsa
| Ag-NPs solution | Z-average diameter (nm) | Zeta potential ζ (mV) |
|---|---|---|
| H2O | 61.2 ± 1.6 | −9.37 ± 0.54 |
| PBS | 1965.6 ± 284.3 | N/A |
| Fischer’s medium | 1608.7 ± 175.4 | −8.20 ± 0.26 |
Ag-NPs were dispersed in the solution described as above, vortexed for homogeneity. Dilute was transferred to a cuvette or Zeta Potential cell for dynamic size or zeta potential measurement. N/A means data were not applicable in that particular case.
Intracellular Distribution of Ag-NPs
The mouse lymphoma cells were treated with 5 nm Ag-NPs at the concentrations of 5 μg/mL for 4 hr. Figure 2 shows representative confocal images with the fluorescence representing the signal of the Ag-NPs. After 4 hr treatment, these 5 nm Ag-NPs appeared in the cytoplasm of the cells, indicating that the Ag-NPs were localized in mouse lymphoma cells.
Fig. 2.
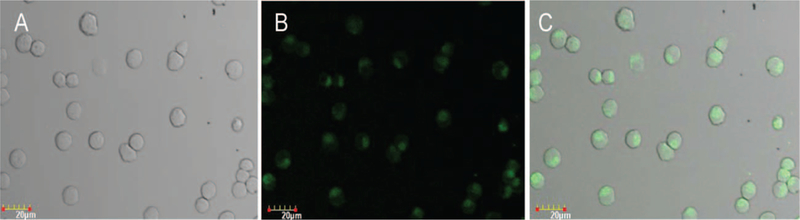
Intracellular distribution of Ag-NPs in mouse lymphoma cells. The cells were cultured in medium containing Ag-NPs (5 μg/mL) for 4 hr and then washed three times to remove the Ag-NPs bound to cell membrane. Five microliters of the cell suspension was deposited on the cover glass and then sealed on the glass slide with a clear nail polish. Images were captured with confocal laser scanning microscope with an excitation of 405 nm and emission of 480 nm. (A) Bright field image; (B) Fluorescence image; (C) Overlap image of (A) and (B).
Mutagenicity of Ag-NPs
Based on the dose range finding study, the main experiments were conducted with Ag-NPs at the dose range of 3—6 μg/mL. The cytotoxicity and Tk MFs from two experiments are presented in Figure 3 for both versions of the MLA. Both the microwell and soft-agar versions are equally acceptable MLA methodologies as determined by the MLA Expert Workgroup of IWGT [Moore et al., 2002, 2006]. The treatment of cells with the Ag-NPs resulted in increased cytotoxicity and mutagenicity. The RTGs and MFs from the two versions were similar (Fig. 3). It should be noted that the relative suspension growth during the 2-day phenotypic expression in both versions of the MLA was the same for each treatment because the same cultures were used for both versions. The MFs at the concentrations of 4 and 5 μg/mL of the Ag-NPs were 400 6 230 X 10—6 (mean ± SD) and 453 ± 171 X 10—6, respectively, while the MF for the control group was 89 ± 20 X 10—6.
Fig. 3.
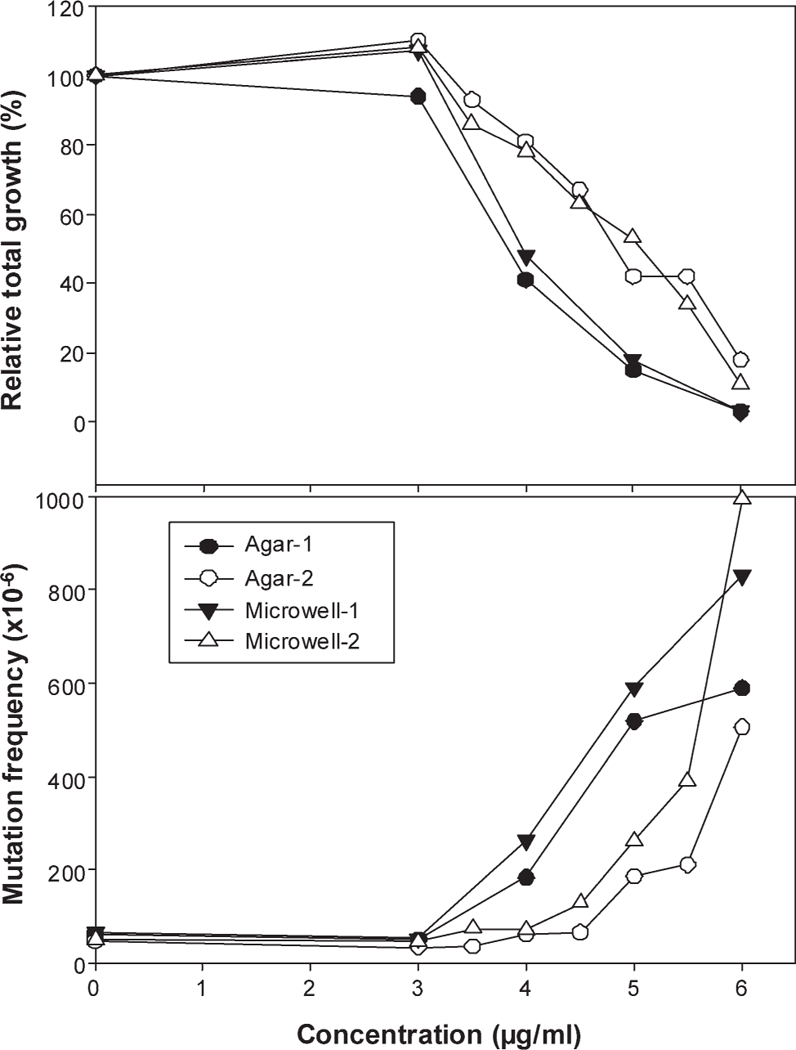
Cytotoxicity and mutagenicity of 5 nm Ag-NPs in mouse lymphoma cells. The cells were treated with Ag-NPs (3–6 μg/mL) for 4 hr, and the cytotoxicity (the top panel) and mutagenicity (the bottom panel) of 5-nm Ag-NPs were determined in two experiments (close or open circle/triangles, respectively) for both the soft-agar (circle) and microwell (triangles) versions of MLA. RTG is calculated as the product of relative suspension growth and relative plating efficiency for viability.
LOH Analysis of the Ag-NP-Induced Tk Mutants
DNA samples were isolated from 48 large and 48 small mutant colonies from the 5 μg/mL Ag-NP treatment. The LOH analysis of mutants was conducted using four microsatellite loci: Tk locus, D11Mit36, D11Mit20, and D11Mit74. These four polymorphic markers were almost evenly distributed along the full length of chromosome 11. Approximately 98% of mutants from the Ag-NP treatment lost heterozygosity at the Tk locus. The percentages of the different types of mutations in large, small, and all mutant colonies are displayed in Table II. When compared with the microsatellite mutant spectrum previously reported for the negative control [Wang et al., 2007a], the Ag-NP-induced mutational spectrum was significantly different (P < 0.0001). The most common type of mutation for the Ag-NP treatment was LOH involving only the Tk locus with 60% for large colonies and 87% for small colonies, indicating that most mutants had chromosomal damage less than approximately 34 megabase pairs (Mbp) of chromosome 11.
TABLE II.
Comparison of Mutational Types for Large, Small, and All Colonies Produced in Mouse Lymphoma Cells Treated With 5 nm Ag-NPs at the Concentration of 5 μg/mL
| Mutational type | Untreated controla |
Ag-NPs (5 ig/mL)> |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Large colonies |
Small colonies |
Total coloniesb |
Large colonies |
Small colonies |
Total coloniesb |
||||||
| N | % | N | % | % | N | % | N | % | % | ||
| non-LOH | 21 | 58.3 | 1 | 3.0 | 36.2 | 2 | 4.2 | 0 | 0 | 1.7 | |
| Tk only | 4 | 11.1 | 16 | 48.5 | 26.1 | 29 | 60.4 | 42 | 87.5 | 76.4 | |
| Tk → D11Mit36 | 6 | 16.7 | 3 | 9.1 | 13.6 | 6 | 12.5 | 3 | 6.3 | 8.8 | |
| Tk → D11Mit20 | 0 | 0 | 4 | 12.1 | 4.9 | 6 | 12.5 | 1 | 2.1 | 6.4 | |
| Tk → D11Mit74 | 5 | 13.9 | 9 | 27.3 | 19.2 | 5 | 10.4 | 2 | 4.2 | 6.7 | |
| Total mutants | 36 | 33 | 48 | 48 | |||||||
Data are from our previous study [Wang et al., 2007a].
The percentage of total colonies is the weighted sum of the number of large and small colonies considering the proportion difference of large and small colonies mutants in the MF. The “→” indicates the LOH extends from Tk locus to other 3 loci (D11Mit36, D11Mit20, and D11Mit74) on the chromosome 11, and the locus of D11Mit74 is located at the top of the chromosome 11.
DNA Damage Measured by the Comet Assay
The Comet assay was also performed using the same treatment procedure as that for MLA. After the 4 hr treatment, cells were fixed for the assay. All experiments were independently performed at least three times. The results are shown in Figure 4. In the standard Comet assay, there was no significant induction of DNA damage although the percentage of DNA in the tail increased slightly with increasing dose. The sensitivity and specificity of the Comet assay for oxidative DNA adducts can be improved by incubating the lysed cells with lesion-specific endonucleases that recognize particular oxidized bases and create additional breaks. Three enzymes, FPG, EndoIII, and hOOGl, were used for the modified Comet assays in this study. FPG can specifically excise oxidized purines, including 8-oxo-7,8-dihydroguanine (8-oxoGua), 2,6-dia- mino-4-hydroxy-5-formamidopyrimidine (FaPyGua), and 4,6-diamino-5-formamidopyrimidine (FaPyAde), as well as other ring-opened purines. hOGGl can also recognize oxidized bases, such as 8-oxoGua, but only when they are paired with cytosine. EndoIII recognizes oxidized pyrimidines, including thymine glycol and uracil glycol [David and Williams, 1998]. In the oxidative DNA damage comet assays, addition of lesion-specific endonucleases resulted in significant induction of DNA breaks in a dose-dependent manner (Fig. 4).
Fig. 4.
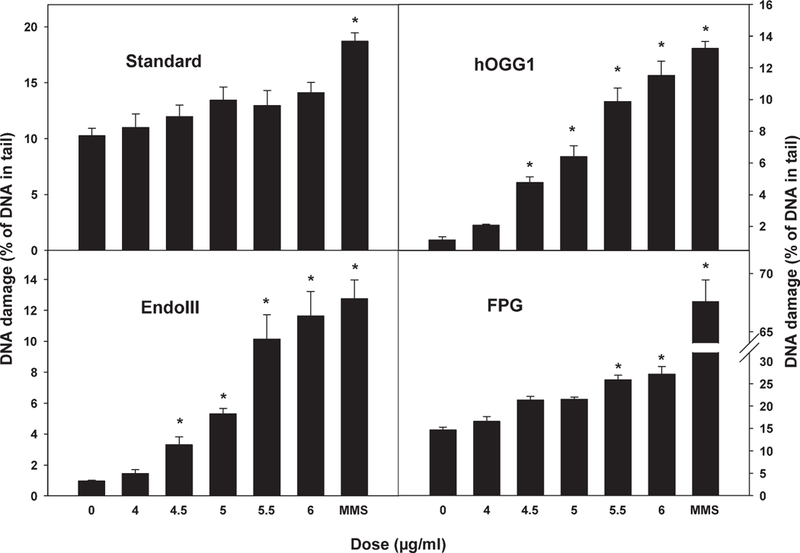
DNA damage induced by 5-nm Ag-NPs in mouse lymphoma cells. The Comet assay was performed with the standard procedure (top left panel), and addition of lesion-specific endonucleases: human 8-hydroxyguanine DNA-glycosylase (hOGG1, top right panel), endonuclease III (EndoIII, bottom left panel) and formamidopyrimidine DNA-glycosylase (FPG, bottom right panel) following treatment of L5178Y cells with different doses of 5 nm Ag-NPs. Methyl methanesulfonate (MMS) was used as the positive control. The data points represent the mean 6 1 standard errors for three independent experiments. Asterisk indicates P < 0.01 when the treatment group was compared with the concurrent control.
Gene Expression Analysis of the Ag-NP-Induced Oxidative Stress
To investigate the potential role of oxidative stress in the Ag-NP-induced genotoxicity, a pathway-specific PCR array was utilized for gene expression analysis. The Mouse Oxidative Stress and Antioxidant Defense RT2 Profile PCR Array can measure the expression of 84 genes related to oxidative stress. Six vehicle control and six Ag-NP (5 μg/mL) treated cell samples were used for the gene expression study. Among the 84 genes, 59 genes were actively expressed in mouse lymphoma cells. The expressed genes were analyzed by principal component analysis according to their expression intensities (Fig. 5). A separation between the control and Ag-NP-treated groups was clearly observed, suggesting that the expression pattern of the genes related to oxidative stress was significantly altered by the Ag-NP treatment.
Fig. 5.
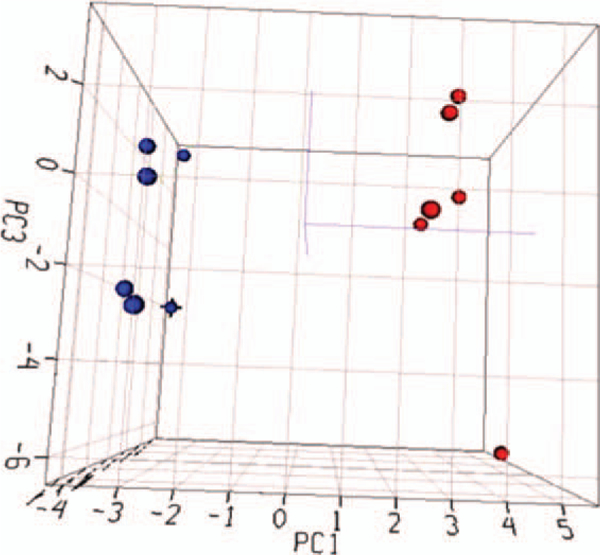
Principal component analysis of gene expression profiles from control and Ag-NP-treated cell samples. No specific cut-off was applied and the intensity of gene expression data for the expressed 59 genes in the PCR array was used for the analysis. The blue and red dots indicate the control and Ag-NP-treated samples, respectively.
To select differentially expressed genes, minimum requirements were established as both a 1.5-fold change in the gene expression compared with untreated controls and a p-value less than 0.05. A total of 17 genes satisfied this requirement with nine genes up-regulated and eight down-regulated by the Ag-NP treatments (Table III). The functions of these differentially expressed genes are involved in the production of reactive oxygen species (ROS), antioxidants, oxidative stress response, oxygen transporters, and DNA repair.
Table III.
List of Genes Whose Expressions Were Significantly Altered by Ag-NP Treatment (P < 0.05 and fold change ≥ 1.5)
| Gene name | Gene description | P | Fold change | Gene function |
|---|---|---|---|---|
| Ncf2 | Neutrophil cytosolic factor 2 | 0.000l | 5.2 | Production of reactive oxygen species |
| Nqol | NAD(P)H dehydrogenase, quinone 1 | 0.000l | 2.6 | Oxidative stress response |
| Prdx6 | Peroxiredoxin 6 | 0.000l | 2.3 | Antioxidants; oxidative stress response |
| Ppp1r15b | Protein phosphatase 1, regulatory (inhibitor) subunit 15b | 0.0004 | 2.2 | Oxidative stress response |
| Prdx6-rs1 | Peroxiredoxin 6, related sequence 1 | 0.006 | 2.l | Antioxidants |
| Cat | Catalase | 0.0002 | 2.0 | Antioxidants; oxidative stress response |
| Txnip | Thioredoxin interacting protein | 0.0005 | l.9 | Oxidative stress response |
| Txnrdl | Thioredoxin reductase 1 | 0.006 | l.9 | Oxidative stress and oxygen transporters |
| Sodl | Superoxide dismutase 1, soluble | 0.04 | l.5 | Antioxidants; production of reactive oxygen species; oxidative stress response |
| Idhl | Isocitrate dehydrogenase 1 (NADP+), soluble | 0.02 | −1.5 | Oxidative stress response |
| Xpa | Xeroderma pigmentosum, complementation group A | 0.0l | −1.5 | DNA repair |
| Prdx5 | Peroxiredoxin 5 | 0.04 | −1.5 | Antioxidants |
| Txnrd2 | Thioredoxin reductase 2 | 0.02 | −1.6 | Antioxidants; oxidative stress response |
| Kif9 | Kinesin family member 9 | 0.02 | −1.6 | Antioxidants |
| Ercc2 | Excision repair cross-complementing rodent repair deficiency, complementation group 2 | 0.04 | −1.7 | DNA repair |
| Recql4 | RecQ protein-like 4 | 0.009 | −1.9 | Production of reactive oxygen species |
| Duoxl | Dual oxidase 1 | 0.007 | −2.7 | Oxidative stress response |
DISCUSSION
There is little doubt that Ag-NPs can be highly toxic to cells, as it has long been known that bulk silver possesses antibacterial properties and these properties are retained when silver is synthesized in nanoscale products. As such, most potential applications of Ag-NPs are related to their capacity to kill bacteria that can cause diseases through the contamination of food, water, and wounds [Faunce and Watal, 2010]. In addition, Ag-NPs have been used in the manufacture of clothing and various food contact materials. Therefore, exposure to Ag-NPs in the body is becoming increasingly widespread. Thus, Ag-NPs have gained an increasing chance to access to tissues, cells, and biological molecules within the human body [Chen and Schluesener, 2008].
Several research groups have investigated the toxicity of Ag-NPs in mammalian cell lines. It has been reported that Ag-NPs (7–20 nm) induce a dose-dependent cytotoxicity with IC50 values of 10.6 μg/mL in HT-1080 human fibrosarcoma cells and 11.6 μg/mL in A431 human skin carcinoma cells [Arora et al., 2008]. When IMR-90 human lung fibroblast cells and U251 human glioblastoma cells were treated with Ag-NPs (6–20 nm), extensive and dose-dependent DNA damage was observed using the Comet assay [AshaRani et al., 2009]. After A549 human lung carcinoma cells were exposed to Ag-NPs (30–50 nm), dose-dependent cellular toxicity was demonstrated by the MTT and annexin V/propidium iodide assays [Foldbjerg et al., 2011]. In addition, Ag-NPs (10–20 nm) have been demonstrated to be cytotoxic based on the MTT assay and the lactase dehydrogenase leakage assay in mouse germline stem cells [Braydich-Stolle et al., 2005; Asare et al., 2012], BRL 3A rat liver cells [Hussain et al., 2005], and human hepatoma HepG2 cells [Kim et al., 2009]. In our previous study, Ag-NPs did not induce mutations in the Ames test, but displayed dose-dependent cytotoxicity and genotoxicity in the human lymphoblast TK6 cell micronucleus assay [Li et al., 2011]. It is also worth noting that ionic silver is cytotoxic but not genotoxic in several different genotoxicity assays. Silver iodide was evaluated for mutagenicity in the Ames/micro-some test and in the Sister Chromatid Exchanges assay, and was found to be negative in both assays even at toxic doses [Eliopoulos and Mourelatos, 1998]. Silver nitrate was also negative in the Drosophila wing somatic mutation assay while Ag-NPs were positive in this assay [Demir et al., 2010].
In this study, 5-nm Ag-NPs induced cytoxicity in mouse lymphoma cells and mutagenicity in both the microwell and soft-agar versions of the MLA. The dose-related increases in MFs were associated with dose-related increases in cytotoxicity (Fig. 3). The mean MF for treatment with 5 μg/mL Ag-NPs was about sevenfold higher than the control. In the MLA, clastogens tend to result in a relatively higher proportion of small colony of Tk mutants and predominantly LOH types of mutations, whereas chemical compounds that induce point mutations result in a relatively higher proportion of large colony mutants and less LOH [Applegate et al., 1990]. In this study, LOH at the Tk locus occurred in 95% of large colony mutants and 100% of small colony mutants from the 5 μg/mL Ag-NP treatment (Table II), and the most common type of mutation for the Ag-NP treatment was LOH involving only the Tk locus (76%). Based on the severity of DNA damage of the Tk mutants, LOH can also occur at other loci along chromosome 11 in addition to the Tk gene. Because the four polymorphic markers used in this study are about evenly distributed along the full length of chromosome 11 and the microsatellite locus of D11Mit 36 is located at about 34 Mbp from the Tk locus, the major type of mutation for the Ag-NP treatment was LOH involving < 34 Mbp of chromosome 11. LOH can result from any of several mechanisms, including large deletions, mitotic recombination, and whole chromosome loss [Honma et al., 2001], and LOH is an important mutational event in tumor-igenesis. The results from this study indicate that the mutagenicity of Ag-NPs results from a clastogenic mode-of- action. Previous research also demonstrated the clastogenic effect of Ag-NPs by the chromosomal breaks as significant numbers of micronuclei in cultured human lung fibroblast cells and glioblastoma cells using cytokinesis-blocked micronucleus assay [AshaRani et al., 2009], and by the chromosomal aberrations in human mesenchymal stem cells [Hackenberg et al., 2011]. The clastogenic effect of Ag-NPs could result from oxidative stress. Previous studies with mouse lymphoma cells treated with chemicals that cause oxidative stress [Harrington-Brock et al., 2003; Singh et al., 2005; Moore and Chen, 2006] have shown patterns for the ratio of large colonies vs. small colonies and LOH percentage similar to those found in this study.
Currently, the best-developed paradigm for nanoparticle toxicity is that several characteristics of nanoparticles can culminate in ROS generation [Nel et al., 2006]. After Ag-NP exposure, the level of bulky DNA adducts was strongly correlated with the cellular ROS levels [Foldb-jerg et al., 2011]. To determine whether the mutagenicity of Ag-NPs in the mouse lymphoma cells resulted from an oxidative stress mechanism, DNA breakage was measured with the standard Comet assay together with oxidative stress Comet assays. While the Ag-NP treatment of the mouse lymphoma cells did not increase DNA breaks in the standard assay, DNA breaks were induced in the oxidative stress Comet assay (Fig. 4). The induction of DNA breaks by the addition of FPG, EndoIII, and hOGG1 indicates that the DNA damage resulting from the Ag-NP treatment was mainly oxidized nucleotides.
ROS have been documented as intrinsic signaling molecules that modulate multiple cellular responses and cause alteration of gene expression. Thus, expression alteration of the genes related to oxidative stress may suggest possible oxidative stress. Therefore, we further determined the alteration of gene expression related to oxidative stress using a PCR array. The expression of many of these genes was markedly affected by the Ag-NP treatment in the mouse lymphoma cells (Fig. 5). The unsupervised principal component analysis of gene expression in the Mouse Oxidative Stress and Antioxidant Defense PCR Array profiles in mouse lymphoma cells clearly demonstrate that a large number of these genes related to oxidative stress were altered by Ag-NPs. Nine genes were upregulated about 1.5–5.2-fold, and eight were downregulated by 1.5–2.7-fold in response to Ag-NP treatments (Table III). The Ncf2 gene was the most upregulated gene. This gene is involved in the production of ROS because it encodes p67phox, an essential component of the multi-protein NADPH oxidase enzyme, the major source of superoxide [Gauss et al., 2006]. The Duox1 gene was the most down-regulated gene. DUOX1 and DUOX2 are identified as H2O2-producing enzymes, and it has been reported that the expression of Duoxl and Duox2 is often suppressed in lung cancer cells by hypermethylation of the CpG-rich promoter regions in both Duox genes [Luxen et al., 2008]. Some genes such as Xpa and Ercc2 are related to DNA repair. Their dysregulation suggest that there is an impact of the oxidative stress induced by the Ag-NPs on DNA repair genes [Jaiswal, 2000]. Nqol, one of the NADPH-depend-ent enzymes, was also upregulated by 2.6-fold. This gene has protective roles in detoxification of xenobiotic carbonyls and quinones [Jaiswal, 2000]. Upregulation of this downstream effector gene can stimulate NADPH synthesis and increase levels of glutathione regeneration. In addition, the gene expression analysis of oxidative stress-related genes also suggested that the Ag-NP treatment resulted in the stimulation of several canonical pathways, including the Nrf2-mediated oxidative stress response and mitochondrial dysfunction pathways according to our pathway analyses (data not shown). It has been reported that Ag-NP exposure (~ 60 nm, 10 μg/mL) dysregulated many genes associated with detoxification, SOS response, oxidative/redox stress, drug resistance/sensitivity, and protein stress in Escherichia coli [Gou et al., 2010]. Ag-NPs also increased superoxide dismutase 1 (Sod1) expression levels in human hepatoma cells [Kim et al., 2009], which is consistent with our results (Table III). Several genes that were downregulated are involved in DNA repair such as Xpa and Ercc2 and antioxidants such as Prdx5 and Kif9. Down-regulation of these genes may result in more DNA damage and less DNA repair, thus increasing gene mutations and chromosomal alterations.
In summary, 5 nm Ag-NPs induced dose-dependent cytotoxicity and mutagenicity in mouse lymphoma cells in both the microwell and soft-agar versions of the MLA. The major type of mutations induced by Ag-NPs was LOH involving <34 Mbp chromosomal alterations in chromosome 11, indicating a clastogenic mode of action, possibly via oxidative stress. The DNA damage resulting from the induction of oxidative stress by Ag-NPs was confirmed by the Comet assay and gene expression analysis of the oxidative stress-related genes. These results suggest that 5 nm Ag-NPs are mutagenic in mouse lymphoma cells and that the mutation induction results from the Ag-NP-induced oxidative stress; thus, this study provides insights into the mechanisms underlying toxicity and genotoxicity of Ag-NPs.
ACKNOWLEDGMENTS
N.M. and X.G. performed the MLA and LOH analysis, and prepared the cells for other experiments. Y.C. performed the real-time PCR array analysis. W.D. performed the Comet assay. Y.Z. and A.S.B. performed the nanoparticle characterization. S.F.A., P.R., and M.M.M. helped with planning the experiments and editing the manuscript. T.C. designed the study. N.M., Y.Z., and T.C. prepared the manuscript. All authors approved the final version of manuscript.
Footnotes
The views presented in this paper do not necessarily reflect those of the U.S. Food and Drug Administration, but express the opinions of the authors.
REFERENCES
- Applegate ML, Moore MM, Broder CB, Burrell A, Juhn G, Kasweck KL, Lin PF, Wadhams A, Hozier JC. 1990. Molecular dissection of mutations at the heterozygous thymidine kinase locus in mouse lymphoma cells. Proc Natl Acad Sci USA 87:51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Jain J, Rajwade JM, Paknikar KM. 2008. Cellular responses induced by silver nanoparticles: In vitro studies. Toxicol Lett 179:93–100. [DOI] [PubMed] [Google Scholar]
- Asare N, Instanes C, Sandberg WJ, Refsnes M, Schwarze P, Kruszewski M, Brunborg G. 2012. Cytotoxic and genotoxic effects of silver nanoparticles in testicular cells. Toxicology 291:65–72. [DOI] [PubMed] [Google Scholar]
- AshaRani PV, Low Kah Mun G, Hande MP, Valiyaveettil S. 2009. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 3:279–290. [DOI] [PubMed] [Google Scholar]
- Asharani PV, Wu YL, Gong Z, Valiyaveettil S. 2008. Toxicity of silver nanoparticles in Zebrafish models. Nanotechnology 19:1–8. [DOI] [PubMed] [Google Scholar]
- Braydich-Stolle L, Hussain S, Schlager JJ, Hofmann MC. 2005. In vitro cytotoxicity of nanoparticles in mammalian germline stem cells. Toxicol Sci 88:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaloupka K, Malam Y, Seifalian AM. 2010. Nanosilver as a new generation of nanoproduct in biomedical applications. Trends Bio-technol 28:580–588. [DOI] [PubMed] [Google Scholar]
- Chen T, Moore MM. 2004Screening for chemical mutagens using the mouse lymphoma assay In: Yan Z, Caldwell GW, editors. Optimization in Drug Discovery: In-vitro Methods. Totowa, NJ:Hu-mana Press; pp337–352. [Google Scholar]
- Chen X, Schluesener HJ. 2008. Nanosilver: a nanoproduct in medical application. Toxicol Lett 176:1–12. [DOI] [PubMed] [Google Scholar]
- David SS, Williams SD. 1998. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev 98:1221–1262. [DOI] [PubMed] [Google Scholar]
- Dearfield KL, Auletta AE, Cimino MC, Moore MM. 1991. Considerations in the U.S. environmental protection agency’s testing approach for mutagenicity. Mutat Res 258:259–283. [DOI] [PubMed] [Google Scholar]
- Demir E, Vales G, Kaya B, Creus A, Marcos R. 2010. Genotoxic analysis of silver nanoparticles in Drosophila. Nanotoxicology 5:417–424. [DOI] [PubMed] [Google Scholar]
- DHHS. 1997. Department of Health and Human Services, Genotoxicity: A Standard Battery for Genotoxicity Testing of Pharmaceuticals, International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use, Food and Drug Administration, Rockville, MD. [Google Scholar]
- Eliopoulos P, Mourelatos D. 1998. Lack of genotoxicity of silver iodide in the SCE assay in vitro, in vivo, and in the Ames/microsome test. Teratog Carcinog Mutagen 18:303–308. [DOI] [PubMed] [Google Scholar]
- Faunce T, Watal A. 2010. Nanosilver and global public health: international regulatory issues. Nanomedicine (Lond) 5:617–632. [DOI] [PubMed] [Google Scholar]
- FDA. 2007. Nanotechnology task force report 2007. USFDA, http://www.fda.gov/downloads/ScienceResearch/SpecialTopics/Nanotechnology/ucm110856.pdf.
- Foldbjerg R, Dang DA, Autrup H. 2011. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch Toxicol 85:743–750. [DOI] [PubMed] [Google Scholar]
- Gauss KA, Bunger PL, Crawford MA, McDermott BE, Swearingen R, Nelson-Overton LK, Siemsen DW, Kobayashi SD, Deleo FR, Quinn MT. 2006. Variants of the 5’-untranslated region of human NCF2: expression and translational efficiency. Gene 366:169–179. [DOI] [PubMed] [Google Scholar]
- Gou N, Onnis-Hayden A, Gu AZ. 2010. Mechanistic toxicity assessment of nanomaterials by whole-cell-array stress genes expression analysis. Environ Sci Technol 44:5964–5970. [DOI] [PubMed] [Google Scholar]
- Gravante G, Caruso R, Sorge R, Nicoli F, Gentile P, Cervelli V. 2009. Nanocrystalline silver: a systematic review of randomized trials conducted on burned patients and an evidence-based assessment of potential advantages over older silver formulations. Ann Plast Surg 63:201–205. [DOI] [PubMed] [Google Scholar]
- Guo X, Verkler TL, Chen Y, Richter PA, Polzin GM, Moore MM, Mei N. 2011. Mutagenicity of 11 cigarette smoke condensates in two versions of the mouse lymphoma assay. Mutagenesis 26:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg S, Scherzed A, Kessler M, Hummel S, Technau A, Froelich K, Ginzkey C, Koehler C, Hagen R, Kleinsasser N. 2011. Silver nanoparticles: evaluation of DNA damage, toxicity and functional impairment in human mesenchymal stem cells. Toxicol Lett 201:27–33. [DOI] [PubMed] [Google Scholar]
- Harrington-Brock K, Collard DD, Chen T. 2003. Bromate induces loss of heterozygosity in the thymidine kinase gene of L5178Y/Tk(+/−)-3.7.2C mouse lymphoma cells. Mutat Res 537:21–28. [DOI] [PubMed] [Google Scholar]
- Honma M, Momose M, Sakamoto H, Sofuni T, Hayashi M. 2001. Spindle poisons induce allelic loss in mouse lymphoma cells through mitotic non-disjunction. Mutat Res 493:101–114. [DOI] [PubMed] [Google Scholar]
- Hsin YH, Chen CF, Huang S, Shih TS, Lai PS, Chueh PJ. 2008. The ap-optotic effect of nanosilver is mediated by a ROS- and JNK-dependent mechanism involving the mitochondrial pathway in NIH3T3 cells. Toxicol Lett 179:130–139. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li X, Liao Z, Zhang G, Liu Q, Tang J, Peng Y, Liu X, Luo Q. 2007. A randomized comparative trial between Acticoat and SD-Ag in the treatment of residual burn wounds, including safety analysis. Burns 33:161–166. [DOI] [PubMed] [Google Scholar]
- Hussain SM, Hess KL, Gearhart JM, Geiss KT, Schlager JJ. 2005. In vitro toxicity of nanoparticles in BRL 3A rat liver cells. Toxicol In Vitro 19:975–983. [DOI] [PubMed] [Google Scholar]
- ICH. 1995. Topic S2A Genotoxicity: Guidance on Specific Aspects of Regulatory Genotoxicity Tests for Pharmaceuticals, International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use, Harmonised Tripartite Guideline CPMP/ICH/141/95 Approved September 1995 Available at: http://www.ifpma.org/ich1991.html [Google Scholar]
- Jaiswal AK. 2000. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic Biol Med 29:254–262. [DOI] [PubMed] [Google Scholar]
- Kim S, Choi JE, Choi J, Chung KH, Park K, Yi J, Ryu DY. 2009. Oxidative stress-dependent toxicity of silver nanoparticles in human hepatoma cells. Toxicol In Vitro 23:1076–1084. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen DH, Yan J, Chen Y, Mittelstaedt RA, Zhang Y, Biris AS, Heflich RH, Chen T. 2011. Genotoxicity of silver nanoparticles evaluated using the Ames test and in vitro micronucleus assay. Mutat Res [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Luxen S, Belinsky SA, Knaus UG. 2008. Silencing of DUOX NADPH oxidases by promoter hypermethylation in lung cancer. Cancer Res 68:1037–1045. [DOI] [PubMed] [Google Scholar]
- Mei N, Xia Q, Chen L, Moore MM, Chen T, Fu PP. 2006. Photomutagenicity of anhydroretinol and 5,6-epoxyretinyl palmitate in mouse lymphoma cells. Chem Res Toxicol 19:1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Xia Q, Chen L, Moore MM, Fu PP, Chen T. 2005. Photomutagenicity of retinyl palmitate by ultraviolet a irradiation in mouse lymphoma cells. Toxicol Sci 88:142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MM, Chen T. 2006. Mutagenicity of bromate: implications for cancer risk assessment. Toxicology 221:190–196. [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Bolcsfoldi G, Burlinson B, Cifone M, Clarke J, Delongchamp R, Durward R, Fellows M, Gollapudi B, Hou S, Jenkinson P, Lloyd M, Majeska J, Myhr B, O’Donovan M, Omori T, Riach C, San R, Stankowski LF Jr, Thakur AK, Van Goethem F, Wakuri S, Yoshimura I. 2006. Mouse lymphoma thymidine kinase gene mutation assay: follow- up meeting of the International Workshop on Genotoxicity Testing—Aberdeen, Scotland, 2003—Assay acceptance criteria, positive controls, and data evaluation. Environ Mol Mutagen 47:1–5. [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Harrington-Brock K, Awogi T, Bolcsfoldi G, Cifone M, Collard D, Fellows M, Flanders K, Gol- lapudi B, Jenkinson P, Kirby P, Kirchner S, Kraycer J, McEnaney S, Muster W, Myhr B, O’Donovan M, Oliver J, Ouldelhkim MC, Pant K, Preston R, Riach C, San R, Shimada H, Stankowski LF Jr. 2002. Mouse lymphoma thymidine kinase gene mutation assay: follow-up International Workshop on Genotoxicity Test Procedures, New Orleans, Louisiana, April 2000. Environ Mol Mutagen 40:292–299. [DOI] [PubMed] [Google Scholar]
- Nel A, Xia T, Madler L, Li N. 2006. Toxic potential of materials at the nanolevel. Science 311:622–627. [DOI] [PubMed] [Google Scholar]
- Singh SP, Chen T, Chen L, Mei N, McLain E, Samokyszyn V, Thaden JJ, Moore MM, Zimniak P. 2005. Mutagenic effects of 4-hydroxynonenal triacetate, a chemically protected form of the lipid peroxidation product 4-hydroxynonenal, as assayed in L5178Y/Tk+/− mouse lymphoma cells. J Pharmacol Exp Ther 313:855–861. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. 1988. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 175:184–191. [DOI] [PubMed] [Google Scholar]
- Sung JH, Ji JH, Park JD, Yoon JU, Kim DS, Jeon KS, Song MY, Jeong J, Han BS, Han JH, Chung YH, Chang HK, Lee JH, Cho MH, Kelman BJ, Yu IJ. 2009. Subchronic inhalation toxicity of silver nanoparticles. Toxicol Sci 108:452–461. [DOI] [PubMed] [Google Scholar]
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. 2000. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen 35:206–221. [DOI] [PubMed] [Google Scholar]
- Trickler WJ, Lantz SM, Murdock RC, Schrand AM, Robinson BL, Newport GD, Schlager JJ, Oldenburg SJ, Paule MG, Slikker W Jr, Hussain SM, Ali SF. 2010. Silver nanoparticle induced blood-brain barrier inflammation and increased permeability in primary rat brain microvessel endothelial cells. Toxicol Sci 118:160–170. [DOI] [PubMed] [Google Scholar]
- Trop M, Novak M, Rodl S, Hellbom B, Kroell W, Goessler W. 2006. Silver-coated dressing acticoat caused raised liver enzymes and argyria-like symptoms in burn patient. J Trauma 60:648–652. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen T, Honma M, Chen L, Moore MM. 2007a. 3’-azido-3’- deoxythymidine induces deletions in L5178Y mouse lymphoma cells. Environ Mol Mutagen 48:248–257. [DOI] [PubMed] [Google Scholar]
- Wang JJ, Sanderson BJ, Wang H. 2007b. Cytotoxicity and genotoxicity of ultrafine crystalline SiO2 particulate in cultured human lym- phoblastoid cells. Environ Mol Mutagen 48:151–157. [DOI] [PubMed] [Google Scholar]
- Xia T, Li N, Nel AE. 2009. Potential health impact of nanoparticles. Annu Rev Public Health 30:137–150. [DOI] [PubMed] [Google Scholar]


