Abstract
Background
Hepatocellular carcinoma (HCC) is the second leading cause of cancer mortality worldwide, however, the prognosis for HCC remains unsatisfactory. This study aimed to explore the role of miR-339-5p in HCC.
Methods
We first used quantitative real-time PCR to examine the level of miR-339-5p in HCC tissues. Then we further adopted Western blotting assay, CCK8, cell invasion assays, apoptosis detection assay, and luciferase assay to analyze how it mediate the development of HCC.
Results
We found that miR-339 is significantly decreased in primary HCC tissues. Overexpression of miR-339 in HCC cells remarkably suppressed proliferation and invasion and induced apoptosis. However, silencing miR-339 in HCC cells promoted proliferation and invasion, and reduced apoptosis. Moreover, we demonstrated that ZNF689 is a target of miR-339 and there is a negative correlation between miR-339 and ZNF689 expression in the HCC tissues. Overexpression of ZNF689 in miR-339-overexpressing HCC cells partially antagonized the inhibitory effects of miR-339.
Conclusion
Our study revealed that miR-339 inhibits HCC growth through targeting oncoprotein ZNF689 and restoration of miR-339 might be feasible therapeutic strategy for HCC treatment.
Keywords: miR-339-5p, HCC, ZNF689, treatment, proliferation
Introduction
Liver cancer is the second leading cause of cancer mortality worldwide, causing >700,000 deaths annually. Although there has been progress in treatment, the prognosis for hepatocellular carcinoma (HCC) remains unsatisfactory.1 Therefore, identifying some new biomarkers for early diagnosis of HCC and exploring the mechanisms of HCC progression might be the feasible strategy for improving the survival rate and quality of life of HCC patients.2 miRNAs are single-stranded and non-coding RNAs containing about 21 nucleotides.3
miRNAs play some important regulatory roles within cells, including posttranscriptional regulation and mRNA cleavage.4,5 Accumulating evidence shows that miRNAs coordinate many biological processes, including cell proliferation, apoptosis, migration, invasion and oncogenesis.6,7 In recent years, miR-339 has been reported to be a suppressor for various cancers, such as lung cancer, ovarian cancer, colorectal cancer (CRC) and melanoma.8–14 However, the mechanism underlying miR-399 inhibiting HCC carcinogenesis remains unclear.
Zinc finger (ZNF)-containing proteins are the transcription factors which bind to sequence-specific DNA and, therefore, regulate differentiation, metabolism, development, apoptosis, stemness maintenance and autophagy.15–19 Previous studies revealed that aberrant expression of ZNF proteins contributes to tumorigenesis. It has been reported that ZKSCAN3 (aliases ZNF306 and ZNF309) is a driver of invasive CRC progression.20 Shigematsu et al21 found that ZNF689 inhibits apoptosis of HCC cells via down-regulating Bcl-2 family members. Also, Yi et al22 showed that ZNF689 is a poor prognostic factor for HCC patients. Jen et al23 firstly verified that ZNF322A (aliases ZNF388 or ZNF489) is an oncogene which promotes lung cancer cell proliferation and metastasis. Therefore, inhibiting ZNF proteins might be a feasible strategy to suppress cancer progression.
In this study, we found that the expression of miR-339 is higher in normal liver tissues than in HCC tissues. Further study showed that miR-339 suppresses HCC cell proliferation and invasion and promotes apoptosis through inhibiting ZNF689 protein.
Materials and methods
Clinical specimens
Forty HCC tissue specimens and paracancerous tissues were surgically obtained from Zhejiang Cancer Hospital between May 2016 and May 2017. None of the patients had received chemotherapy or radiotherapy before the surgery. Also, all tissues were confirmed by pathological examination. This study was approved by Zhejiang Cancer Hospital Institutional Review Board and conforms to the Declaration of Helsinki principles. Patients who were enrolled in our study were asked to sign the informed consents.
Cell lines and cell culture
Human hepatoma cell lines HepG2, Hep3B, Bel7402, HCCLM3 and human hepatic cell line LO2 were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, P.R. China). All cell lines were cultured in DMEM supplemented with 10% FBS and 1% streptomycin and penicillin in a humidified atmosphere of 5% CO2 at 37°C.
Quantitative real-time PCR
Total RNA was extracted from the cells or tissues using E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s protocol. cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit. Next, ZNF689 cDNA was quantified with the Lightcycler 480 real-time PCR system (Roche) using SYBR Green I Master Mix (Roche). The reverse transcription of miRNA was performed with the Prime-Script miRNA cDNA Synthesis Kit (TaKaRa), and the miR-339 cDNA was detected on the Lightcycler 480 real-time PCR system (Roche) using the TaqMan MicroRNA Assay Kit (ABI, Foster City, CA, USA). GAPDH and U6 were used as the endogenous control for normalizing ZNF689 and miR-339, respectively. All primers used in this study are shown in Table S1. The sequences of miR-339 and U6 were quoted from Shen et al.24 Also, the sequences of ZNF689 and GAPDH were designed by ourselves using PubMed.
Western blotting assay
Total cellular proteins were extracted from HCC cells which were lysed in RIPA buffer (Thermo Fisher Scientific) supplemented with protease and phosphatase inhibitors. The protein concentrations were detected with BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Forty micrograms of total cellular proteins was separated using 8% SDS-PAGE and then electrotransferred onto polyvinylidene difluoride (PVDF) membrane (EMD Millipore). Next, the PVDF membrane was incubated with 5% non-fat milk, followed by a primary antibody and a secondary antibody. Primary antibodies, including E-cadherin (1:1,000), vimen-tin (1:1,000), β-actin (1:1,000), cleaved PARP (1:1,000), cleaved caspase-3 (1:1,000) and cleaved caspase-8 (1:1,000), and secondary antibodies, including anti-mouse IgG and anti-rabbit IgG (1:5,000), were purchased from Cell Signaling Technology. The primary antibody ZNF689 (1:2,000) was obtained from Novus Biologicals. Protein bands were visualized using enhanced chemiluminescence system (Bio-Rad Clarity Western ECL; Bio-Rad Laboratories Inc.) according to the manufacturer’s protocol.
Transfection miR-339 mimics, miR-339 inhibitors and their respective negative control were purchased from GenePharma (Shanghai, P.R. China). ZNF689 overexpression plasmid (pCMV6-AC-GFP-ZNF689) and its vector pCMV6-AC-GFP were obtained from Origene Technologies Inc. (Rockville, MD, USA). Transfection was carried out using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Cell proliferation assay
HCC cells were transfected with miRNA-339 mimic, inhibitor or ZNF689 overexpression plasmid and then seeded into the 96-well plate. After that, HCC cells were treated with the Cell Counting Kit-8 (Beyotime Biotechnology, Shanghai, P.R. China). The absorbance at 450 nm was detected at indicated times (0, 24, 48 and 72 hours) using the microplate reader.
Cell invasion assays
After being transfected with miRNA-339 mimic, inhibitor or ZNF689 overexpression plasmid, HCC cells were seeded into Matrigel-coated upper well of a 24-well polycarbonate Transwell insert (Costar; Corning Incorporated, Corning, NY, USA). FBS-free medium was added to the upper well, while the complete medium was added to the lower well. After 24 hours of incubation, the cells that had invaded through the membrane were fixed and stained.
Apoptosis detection assay
After transfection, HCC cells were collected and stained with fluorescein isothiocyanate (FITC)-Annexin V and prop-idium iodide (PI) using the Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s protocol. Stained cells were subjected immediately to flow cytometry and the results were analyzed using CellQuest 3.3 software (FACScan; BD, Franklin Lakes, NJ, USA).
Luciferase assay
The 3′-UTR of ZNF689 containing the miR-339-binding sites and its mutant was cloned into pGL3-control luciferase reporter vector (Promega Corporation, Fitchburg, WI, USA). pGL3-ZNF689 or mutant pGL3-ZNF689 was co-transfected with miR-339 mimic or scramble into HCC cells by using Opti-MEM and Lipofectamine 3000. After transfection, cells were plated on a 96-well plate. The luciferase activities were evaluated by using the dual luciferase reporter analysis system (Promega Corporation). The relative firefly luciferase activity was normalized to Renilla luciferase activity 48 hours after transfection.
Statistical analyses
All statistical data were analyzed using GraphPad Prism 5.0 software. Data are expressed as the mean ± SD. Differences among groups were determined by one-way ANOVA. P<0.05 was considered as statistically significant.
Results
miR-339 is downregulated in HCC tissues
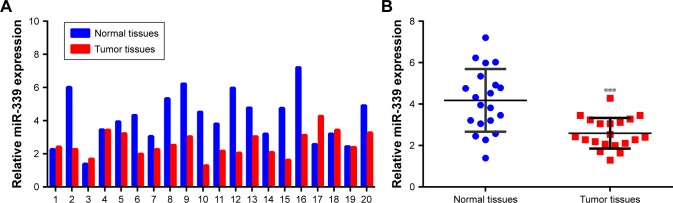
For studying the expression of miR-339 in HCC, 20 paired HCC tissues and adjacent non-tumor tissues were analyzed using quantitative PCR. As shown in Figure 1A, the expression of miR-339 was downregulated in the HCC tissues. Furthermore, miR-399 expression was markedly lower in the HCC tissues than in adjacent non-tumor tissues (Figure 1B). To investigate the clinical significance of miR-339 in HCC, we analyzed the correlation between miR-339 expression level and clinical features in these 20 HCC patients. As shown in Table S2, miR-339 expression was significantly correlated with the histological grade (P=0.008), clinical stage (P=0.025) and metastasis (P=0.001).
Figure 1.
miR-339 is downregulated in HCC tissues.
Notes: (A) The expression of miR-339 in 20 paired HCC tissues and adjacent non-tumor tissues was measured by using qPCR. (B) The expression of miR-339 was significantly higher in the HCC tissues than in adjacent non-tumor tissues. Data are shown as mean ± SD. ***P<0.001 vs the normal tissue group.
Abbreviations: HCC, hepatocellular carcinoma; qPCR, quantitative PCR.
Overexpression of miR-339 inhibits HCCLM3 cells’ proliferation and invasion and induces apoptosis
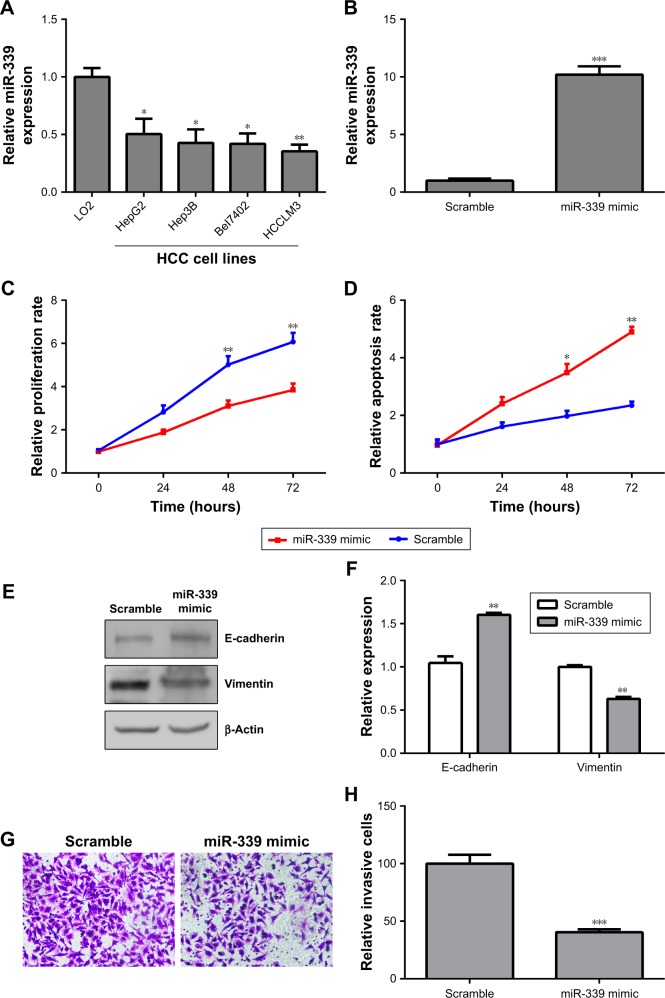
The expression of miR-339 was lower in the HCC cell lines (HepG2, Hep3B, Bel7402 and HCCLM3) than in the normal human liver cell LO2 (Figure 2A). From Figure 2A, we can see that the expression of miR-339 in HCCLM3 cells was the lowest. Therefore, we chose HCCLM3 cells for further study. miR-339 expression in the HCCLM3 cells was increased following transfection with miR-339 mimic compared with the scramble parental cells (Figure 2B). Ectopic expression of miR-339 decreased the proliferation of HCCLM3 cells (Figure 2C), but increased apoptotic cells (Figure 2D). E-cadherin and vimentin are the major markers of epithelial-to-mesenchymal transition (EMT), and EMT plays an important role in tumor metastases. Thus, we first detected whether miR-339 modulates the expression of the EMT markers. We found that E-cadherin was significantly increased in miR-339 mimic transfection cells, while vimentin was markedly decreased (Figure 2E and F). In addition, after miR-339 mimic transfection, the invasive potential of HCCLM3 cells was decreased compared with the scramble transfected cells (Figure 2G and H).
Figure 2.
Overexpression of miR-339 inhibits HCCLM3 cells’ proliferation and invasion and induces apoptosis.
Notes: (A) miR-339 expression in the HCC cell lines (HepG2, Hep3B, Bel7402 and HCCLM3) and the human normal liver cell LO2 was determined by using qPCR. (B) The expression of miR-339 in HCCLM3 cells transfected with miR-339 mimic was measured by using qPCR. (C) Overexpression of miR-339 decreased HCCLM3 cells’ proliferation. (D) Overexpression of miR-339 increased HCCLM3 cells’ apoptosis rate. (E) HCCLM3 cells were transfected with miR-339 mimic and the expression of E-cadherin and vimentin was analyzed by Western blotting. (F) Data represent the relative protein expression. (G) After transfection with miR-339 mimic, the invasive ability of HCCLM3 cells was determined by using Transwell. 200× magnification. (H) The relative amount of invasive cells was analyzed with GraphPad Prism 5.0. Data are expressed as mean ± SD, n=3. *P<0.05, **P<0.01 and ***P<0.001 vs the scramble group.
Abbreviations: HCC, hepatocellular carcinoma; qPCR, quantitative PCR.
Knockdown of miR-339 promotes HepG2 cells’ proliferation and invasion and reduces apoptosis
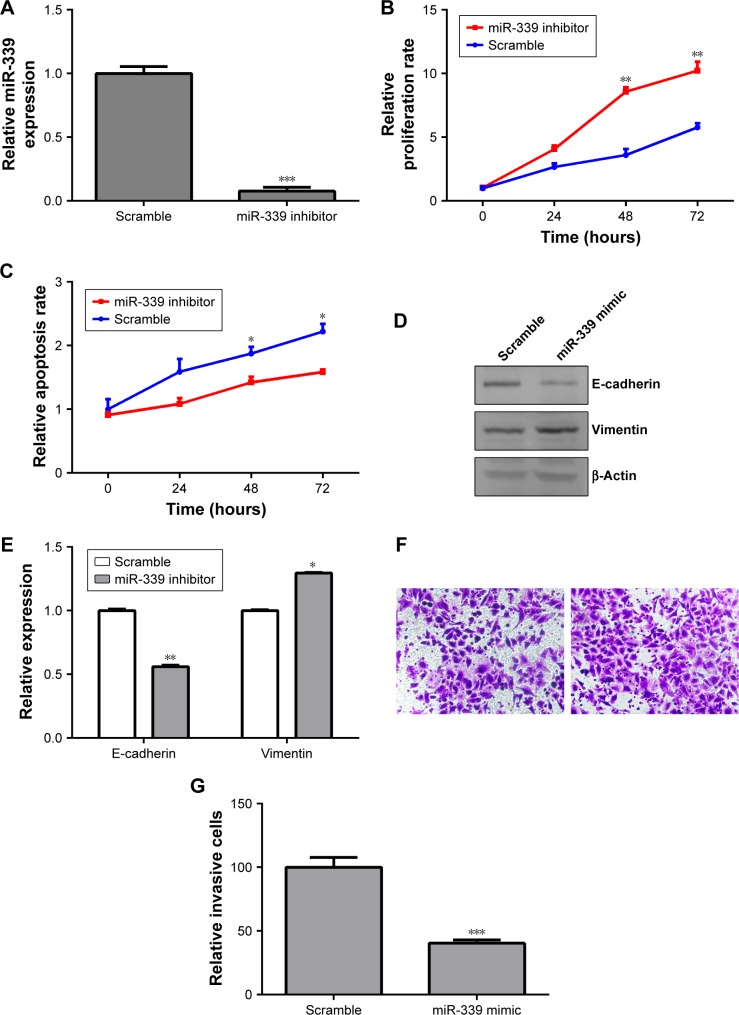
According to the result in Figure 2A, the expression of miR-339 was the highest in HepG2 cells, while it was the lowest in HCCLM3 cells among the four HCC cell lines. In order to investigate if miR-339 inhibits HCC proliferation and invasion, we used miR-339 inhibitor to silence the expression of miR-339 in HepG2 cells and miR-339 mimic to overexpress miR-339 in HCCLM3 cells. miR-339 expression in the HepG2 cells was decreased following transfection with miR-339 inhibitor compared with the scramble parental cells (Figure 3A). Silencing miR-339 increased HepG2 cells’ proliferation (Figure 3B), while it decreased the apoptosis rate of HepG2 cells (Figure 3C). We also detected the expression of E-cadherin and vimentin. It was found that E-cadherin was significantly decreased in miR-339 inhibitor transfected cells while vimentin was markedly increased (Figure 3D and E). Meanwhile, the invasive capability of HepG2 cells transfected with miR-339 inhibitor was decreased compared with the scramble transfected cells (Figure 3F and G).
Figure 3.
Knockdown of miR-339 promotes HepG2 cells’ proliferation and invasion and reduces apoptosis.
Notes: (A) HepG2 cells were treated with miR-339 inhibitor for 48 hours and the expression of miR-339 in HepG2 cells was measured by using qPCR. (B) Knockdown of miR-339 increased HepG2 cells’ proliferation. (C) Knockdown of miR-339 decreased HepG2 cells’ apoptosis rate. (D) HepG2 cells were transfected with miR-339 inhibitor and the expression of E-cadherin and vimentin was analyzed by Western blotting. (E) Data represent the relative protein expression. (F) After transfection with miR-339 inhibitor, the invasive potential of HepG2 cells was detected by using Transwell. 200× magnification. (G) The relative amount of invasive cells was analyzed with GraphPad Prism 5.0. Data are shown as mean ± SD, n=3. *P<0.05, **P<0.01 and ***P<0.001 vs the scramble group.
Abbreviation: qPCR, quantitative PCR.
miR-339 directly targets ZNF689 expression
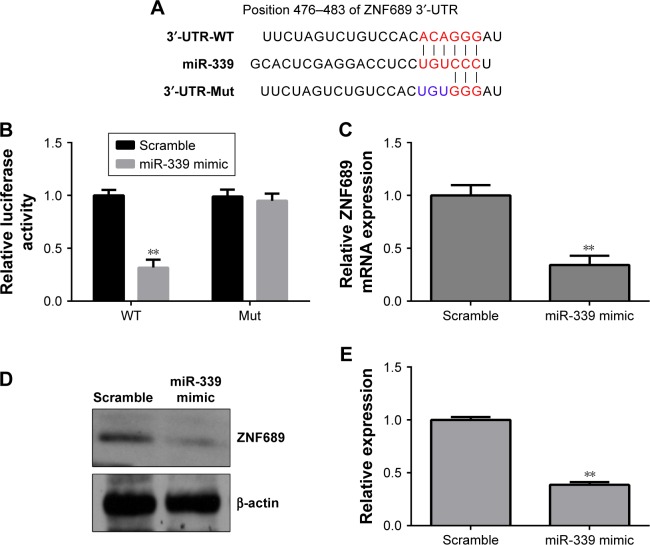
We searched the potential targets of miR-339 using the TargetScan tool and identified ZNF689 as a potential target of miR-339 (Figure 4A). As shown in Figure 4B, miR-339 overexpression inhibited the luciferase activity of the wild-type vector (ZNF689-3′-UTR-wild type [WT]), but not the mutant vector (ZNF689-3′-UTR-Mut). Moreover, overexpression of miR-339 suppressed ZNF689 mRNA and protein expression in HCCLM3 cells (Figure 4C–E).
Figure 4.
miR-339 directly targets ZNF689 expression.
Notes: (A) miR-339 and its presumptive binding sites in the 3′-UTR of ZNF689. Mutations were generated in the complementary site which binds to the seed region of miR-339. (B) miR-339 mimic and luciferase plasmids which include WT ZNF689 3′-UTR or Mut ZNF689-3′-UTR were co-transfected into HCCLM3 cells. The relative luciferase activity was determined. (C) The mRNA level of ZNF689 was measured by using qPCR after miR-339 mimic was transfected into HCCLM3 cells. (D) The protein expression of ZNF689 was detected by using Western blotting after miR-339 mimic was transfected into HCCLM3 cells. (E) Data represent the relative protein expression, shown as mean ± SD, n=3. *P<0.05, **P<0.01 vs the scramble group.
Abbreviations: Mut, mutant type; qPCR, quantitative PCR; WT, wild type.
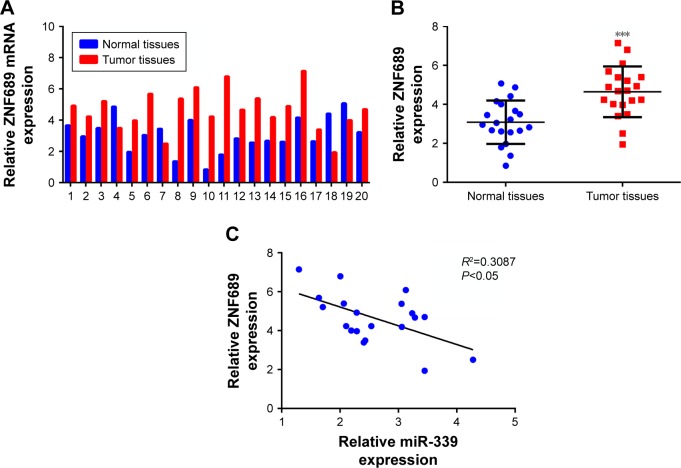
ZNF689 is upregulated in HCC tissues
As shown in the Figure 5A, the expression of ZNF689 was upregulated in the HCC tissues. Furthermore, ZNF689 expression was significantly higher in the HCC tissues than in adjacent non-tumor tissues (Figure 5B). In addition, we verified that there was a significant inverse correlation between miR-339 and ZNF689 expression in the HCC tissues (Figure 5C).
Figure 5.
ZNF689 is upregulated in HCC tissues.
Notes: (A) The mRNA level of ZNF689 in the 20 paired HCC tissues and adjacent non-tumor tissues was measured by using qPCR. (B) The mRNA expression of ZNF689 was significantly higher in the HCC tissues than in adjacent non-tumor tissues. (C) The relationship between miR-339 and ZNF689 expression was analyzed by using GraphPad Prism 5.0 software. Data are shown as mean ± SD. ***P<0.001 vs the normal tissue group.
Abbreviations: HCC, hepatocellular carcinoma; qPCR, quantitative PCR.
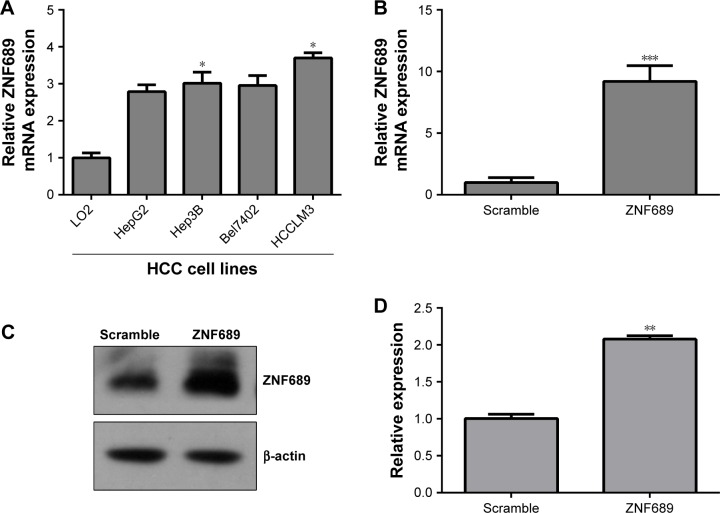
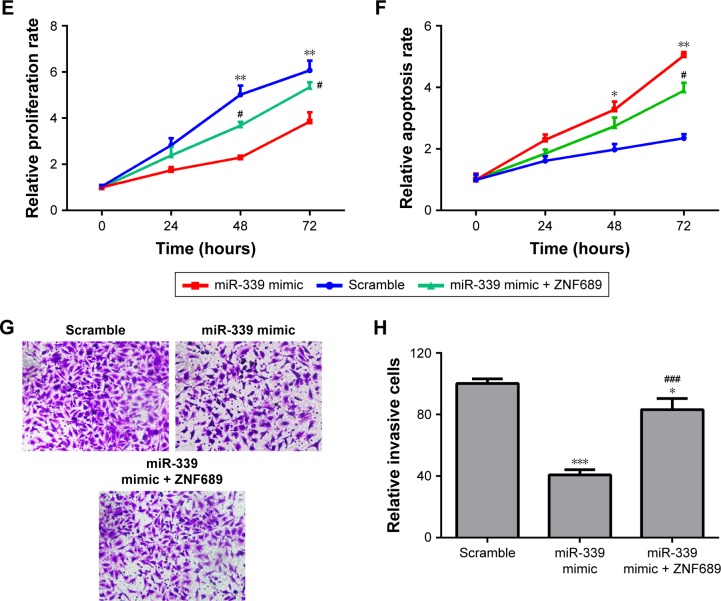
Overexpression of ZNF689 antagonizes miR-339 inhibitory effect on HCCLM3 cells
ZNF689 expression was higher in the HCC cell lines (HepG2, Hep3B, Bel7402 and HCCLM3) than in the human normal liver cell LO2 (Figure 6A). The mRNA and protein expression of ZNF689 in HCCLM3 cells was increased following transfection with ZNF689 overexpression plasmid (Figure 6B–D). Next, ZNF689 overexpression plasmid was co-transfected with miR-339 mimic into HCCLM3 cells. Also, we found that the proliferation and invasion potential of miR-339–overexpressing cells was partially increased after treatment with ZNF689 overexpression plasmid (Figure 6E–H). Furthermore, ectopic expression of ZNF689 suppressed apoptosis of HCCLM3 cells overexpressing miR-339 (Figure 6F). After ZNF689 overexpression plasmid was co-transfected with miR-339 mimic into HCCLM3 cells, the protein expression of cleaved PARP, cleaved caspase-3 and cleaved caspase-8 was decreased at 72 hours (Figure S1).
Figure 6.
Overexpression of ZNF689 antagonizes miR-339 inhibitory effect on HCCLM3 cells.
Notes: (A) ZNF689 mRNA expression in the HCC cell lines (HepG2, Hep3B, Bel7402 and HCCLM3) and the human normal liver cell LO2 was determined by using qPCR. (B) The mRNA expression of ZNF689 in HCCLM3 cells transfected with ZNF689 overexpression plasmid was measured by using qPCR. (C) The protein level of ZNF689 in HCCLM3 cells transfected with ZNF689 overexpression plasmid was measured by using Western blotting. (D) Data represent the relative protein expression. (E–G) ZNF689 overexpression plasmid was co-transfected with miR-339 mimic into HCCLM3 cells. (E) The cell proliferation was measured by using MTT. (F) Apoptotic cells were determined by using Annexin V-FITC/PI Apoptosis Detection Kit. (G) The cell invasive capacity was measured by using Transwell. 200× magnification. (H) The relative amount of invasive cells was analyzed with GraphPad Prism 5.0. Data are presented as mean ± SD, n=3. *P<0.05, **P<0.01 and ***P<0.001 vs the scramble group. #P<0.05 and ###P<0.001 vs the miR-339 mimic group.
Abbreviations: FITC, fluorescein isothiocyanate; HCC, hepatocellular carcinoma; PI, propidium iodide; qPCR, quantitative PCR.
Discussion
miR-339 has been confirmed as a tumor suppressor in many types of cancer. The reduced miR-339-5p expression is associated with an increase in metastatic rate and high clinical stages of breast cancer.25 Weber et al13 showed that miR-339-3p is differentially expressed in melanoma cells and healthy melanocytes. Further study determined that miR-339-3p functions as a strong inhibitor of melanoma and high levels of miR-339-3p decrease the invasive aggressiveness of melanoma cells. It was observed that miR-339-5p was significantly downregulated in Taxol-A549 cells compared with A549 cells, and this study suggested that miR-339-5p downregulation contributes to Taxol resistance in small-cell lung cancer.26 In ovarian cancer, miR-339-5p is speculated to be a biomarker for inhibiting metastasis.12 Zhou et al14 demonstrated that the low-level expression of miR-339-3p was significantly associated with lymph node metastasis in patients with CRC. Higher expression of miR-339-5p indicates low metastatic potential in the non-small-cell lung cancer (NSCLC) cells.27 Wang et al28 showed that miR-339-5p expression level was significantly lower in HCC tissues compared with non-cancerous liver tissues, and patients with lower miR-339-5p expression level were associated with a poorer overall survival.
In this study, we found that the expression of miR-339 was lower in HCC tissues than in the adjacent non-tumor tissues. Moreover, the expression of miR-339 was significantly lower in patients with metastases. Decreased expression of miR-339 was associated with clinical stage (P=0.025), histological grade (P=0.008) and tumor metastasis (P=0.001). Downregulation of miR-339 facilitated metastasis of HCC, indicating poorer prognosis. This is consistent with previous studies, implying the potential of miR-339 as a positive marker for cancer metastasis and prognosis. Further studies demonstrated that overexpression of miR-339 inhibited HCC cells’ proliferation and invasion, but induced apoptosis. However, knockdown of miR-339 promoted HCC cells’ proliferation and invasion and reduced apoptosis, suggesting the tumor suppressor role of miR-339 in HCC.
Previous studies have shown the various oncogenes involved in the mechanisms of miR-339 inhibiting cancer cells’ proliferation, migration and invasion and inducing apop-tosis. Ren et al29 proved that miR-339 targets the 3′-UTR of Skp2 mRNA to inhibit lung cancer cells’ proliferation. Shan et al12 demonstrated that miR-339-5p suppresses migration and invasion of ovarian cancer cell lines through targeting NACC1 and BCL6. miR-339-5p was reported to inhibit the EMT via targeting B-cell chronic lymphocytic leukemia/ BCL6 and, therefore, restrain cell migration and invasion in NSCLC.10 In CRC cells, miR-339-5p functions as a tumor suppressor which suppresses cell growth, colony formation and metastasis by downregulating the expression of PRL-1.30 A study by Jansson et al31 demonstrated that miR-339-5p downregulates MDM2 expression through directly targeting the 3′-UTR of MDM2 mRNA, and thus promotes p53 function in breast cancer cell MCF-7 and CRC cell HCT-116.
In the wcurrent study, we used TargetScan Human tool to predict the target of miR-339, and the result showed that miR-339 directly targeted the 3′-UTR of ZNF689 and there was a significant inverse correlation between miR-339 and ZNF689 expression in the HCC tissues. ZNF689, a C2H2-type ZNF, is a putative transcription regulating factor. A previous study reported that downregulated ZNF689 inhibits cancer cells’ proliferation.32 Also, it has been verified that ZNF689 suppresses apoptosis of HCC cell lines by decreasing the expression of the proapoptotic factors.21
Conclusion
In our study, overexpressing ZNF689 antagonized the inhibitory effect of miR-339 on HCC, providing the first clue that miR-339 mechanistically acts by suppressing ZNF689. All these results including ours suggest that miR-339 is associated with inhibition of tumorigenesis via targeting multiple oncoproteins in tumors and, therefore, may be developed as a therapeutic target in HCC.
Supplementary materials
Overexpression of ZNF689 antagonizes the apoptotic effect induced by miR-339 in HCCLM3 cells.
Note: After ZNF689 overexpression plasmid was co-transfected with miR-339 mimic into HCCLM3 cells, the protein expression of cleaved-PARP, cleaved-caspase3 and cleaved-caspase8 was detected at 72 h by using Western blotting.
Table S1.
Primer sequences used in qPCR assay
| Gene | Forward (5′-3′) Reverse (5′-3′) |
|---|---|
|
| |
| miR-339 | GGGTCCCTGTCCTCCA |
| TGCGTGTCGTGGAGTC | |
| U6 | GCTTCGGCAGCACATATACTAAAAT |
| CGCTTCACGAATTTGCGTGTCAT | |
| ZNF689 | TGGAACGAAACACCGATGACT |
| CCATTCTTCTTTCTGGTTCTGCT | |
| GAPDH | TGCACCACCAACTGCTTAGC |
| GGCATGGACTGTGGTCATGAG | |
Abbreviation: qPCR, quantitative PCR.
Table S2.
Correlation between miR-339 levels and the 20 HCC patients’ clinicopathological parameters
| Parameters | MiR-339 expression
|
P-value | |
|---|---|---|---|
| High (n=9) | Low (n=11) | ||
|
| |||
| Age | 0.964 | ||
| ≤60 | 5 | 6 | |
| >60 | 4 | 5 | |
| Gender | 0.653 | ||
| Men | 5 | 5 | |
| Women | 4 | 6 | |
| Histological grade | 0.008 | ||
| Well | 7 | 2 | |
| Moderate or poor | 2 | 9 | |
| Clinical stage | 0.025 | ||
| I, II | 7 | 3 | |
| III, IV | 2 | 8 | |
| Metastasis | 0.001 | ||
| Yes | 1 | 10 | |
| No | 8 | 1 | |
Note: P-value was calculated with a two-sided Pearson chi-square test.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21(37):10573–10583. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5(5):396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 6.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 7.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 8.Afgar A, Fard-Esfahani P, Mehrtash A, et al. miR-339 and especially miR-766 reactivate the expression of tumor suppressor genes in colorectal cancer cell lines through DNA methyltransferase 3B gene inhibition. Cancer Biol Ther. 2016;17(11):1126–1138. doi: 10.1080/15384047.2016.1235657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P, Liu H, Li Y, Wang Y, Zhao L, Wang H. miR-339-5p inhibits lung adenocarcinoma invasion and migration by directly targeting BCL6. Oncol Lett. 2018;16(5):5785–5790. doi: 10.3892/ol.2018.9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Zhang X, Yang Z, Li Y, Han B, Chen LA. miR-339-5p inhibits metastasis of non-small cell lung cancer by regulating the epithelial-to-mesenchymal transition. Oncol Lett. 2018;15(2):2508–2514. doi: 10.3892/ol.2017.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren H, Zhang Y, Zhu H. miR-339 depresses cell proliferation via directly targeting S-phase kinase-associated protein 2 mRNA in lung cancer. Thorac Cancer. 2018;9(3):408–414. doi: 10.1111/1759-7714.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan W, Li J, Bai Y, Lu X. miR-339-5p inhibits migration and invasion in ovarian cancer cell lines by targeting NACC1 and BCL6. Tumour Biol. 2016;37(4):5203–5211. doi: 10.1007/s13277-015-4390-2. [DOI] [PubMed] [Google Scholar]
- 13.Weber CE, Luo C, Hotz-Wagenblatt A, et al. miR-339-3p is a tumor suppressor in melanoma. Cancer Res. 2016;76(12):3562–3571. doi: 10.1158/0008-5472.CAN-15-2932. [DOI] [PubMed] [Google Scholar]
- 14.Zhou C, Lu Y, Li X. miR-339-3p inhibits proliferation and metastasis of colorectal cancer. Oncol Lett. 2015;10(5):2842–2848. doi: 10.3892/ol.2015.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong CJ, Park H, Yu SW. Autophagy for the quality control of adult hippocampal neural stem cells. Brain Res. 2016;1649(Pt B):166–172. doi: 10.1016/j.brainres.2016.02.048. [DOI] [PubMed] [Google Scholar]
- 16.Krebs CJ, Zhang D, Yin L, Robins DM. The KRAB zinc finger protein RSL1 modulates sex-biased gene expression in liver and adipose tissue to maintain metabolic homeostasis. Mol Cell Biol. 2014;34(2):221–232. doi: 10.1128/MCB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X, Huang M, Wang Z, Liu B, Zhu Z, Li C. ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J Cancer. 2016;7(1):60–68. doi: 10.7150/jca.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauhan S, Goodwin JG, Chauhan S, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50(1):16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai KP, Chen J, He M, et al. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135(8):1790–1799. doi: 10.1002/ijc.28819. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Hamilton SR, Sood A, et al. The previously undescribed ZKSCAN3 (ZNF306) is a novel “driver” of colorectal cancer progression. Cancer Res. 2008;68(11):4321–4330. doi: 10.1158/0008-5472.CAN-08-0407. [DOI] [PubMed] [Google Scholar]
- 21.Shigematsu S, Fukuda S, Nakayama H, et al. ZNF689 suppresses apop-tosis of hepatocellular carcinoma cells through the down-regulation of Bcl-2 family members. Exp Cell Res. 2011;317(13):1851–1859. doi: 10.1016/j.yexcr.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Yi PS, Wu B, Deng DW, Zhang GN, Li JS. Positive expression of ZNF689 indicates poor prognosis of hepatocellular carcinoma. Oncol Lett. 2018;16(4):5122–5130. doi: 10.3892/ol.2018.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jen J, Lin LL, Chen HT, et al. Oncoprotein ZNF322A transcriptionally deregulates alpha-adducin, cyclin D1 and p53 to promote tumor growth and metastasis in lung cancer. Oncogene. 2016;35(18):2357–2369. doi: 10.1038/onc.2015.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen B, Zhang Y, Yu S, et al. MicroRNA-339, an epigenetic modulating target is involved in human gastric carcinogenesis through targeting NOVA1. FEBS Lett. 2015;589(20 Pt B):3205–3211. doi: 10.1016/j.febslet.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Wu ZS, Wu Q, Wang CQ, et al. miR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer. 2010;10:542. doi: 10.1186/1471-2407-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan CZ, Li G, Luo QS, Li HM. miR-339-5p downregulation contributes to Taxol resistance in small-cell lung cancer by targeting α1,2-fucosyltransferase 1. IUBMB Life. 2017;69(11):841–849. doi: 10.1002/iub.1679. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Zhao W, Bao P, et al. miR-339-5p inhibits cell migration and invasion in vitro and may be associated with the tumor-node-metastasis staging and lymph node metastasis of non-small cell lung cancer. Oncol Lett. 2014;8(2):719–725. doi: 10.3892/ol.2014.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YL, Chen CM, Wang XM, Wang L. Effects of miR-339-5p on invasion and prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016;40(1):51–56. doi: 10.1016/j.clinre.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Ren H, Zhang Y, Zhu H. miR-339 depresses cell proliferation via directly targeting S-phase kinase-associated protein 2 mRNA in lung cancer. Thorac Cancer. 2018;9(3):408–414. doi: 10.1111/1759-7714.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C, Liu G, Wang L, et al. miR-339-5p regulates the growth, colony formation and metastasis of colorectal cancer cells by targeting PRL-1. PLoS One. 2013;8(5):e63142. doi: 10.1371/journal.pone.0063142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansson MD, Damas ND, Lees M, Jacobsen A, Lund AH. miR-339-5p regulates the p53 tumor-suppressor pathway by targeting Mdm2. Oncogene. 2015;34(15):1908–1918. doi: 10.1038/onc.2014.130. [DOI] [PubMed] [Google Scholar]
- 32.Silva FP, Hamamoto R, Furukawa Y, Nakamura Y. TIPUH1 encodes a novel KRAB zinc-finger protein highly expressed in human hepatocellular carcinomas. Oncogene. 2006;25(36):5063–5070. doi: 10.1038/sj.onc.1209517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression of ZNF689 antagonizes the apoptotic effect induced by miR-339 in HCCLM3 cells.
Note: After ZNF689 overexpression plasmid was co-transfected with miR-339 mimic into HCCLM3 cells, the protein expression of cleaved-PARP, cleaved-caspase3 and cleaved-caspase8 was detected at 72 h by using Western blotting.
Table S1.
Primer sequences used in qPCR assay
| Gene | Forward (5′-3′) Reverse (5′-3′) |
|---|---|
|
| |
| miR-339 | GGGTCCCTGTCCTCCA |
| TGCGTGTCGTGGAGTC | |
| U6 | GCTTCGGCAGCACATATACTAAAAT |
| CGCTTCACGAATTTGCGTGTCAT | |
| ZNF689 | TGGAACGAAACACCGATGACT |
| CCATTCTTCTTTCTGGTTCTGCT | |
| GAPDH | TGCACCACCAACTGCTTAGC |
| GGCATGGACTGTGGTCATGAG | |
Abbreviation: qPCR, quantitative PCR.
Table S2.
Correlation between miR-339 levels and the 20 HCC patients’ clinicopathological parameters
| Parameters | MiR-339 expression
|
P-value | |
|---|---|---|---|
| High (n=9) | Low (n=11) | ||
|
| |||
| Age | 0.964 | ||
| ≤60 | 5 | 6 | |
| >60 | 4 | 5 | |
| Gender | 0.653 | ||
| Men | 5 | 5 | |
| Women | 4 | 6 | |
| Histological grade | 0.008 | ||
| Well | 7 | 2 | |
| Moderate or poor | 2 | 9 | |
| Clinical stage | 0.025 | ||
| I, II | 7 | 3 | |
| III, IV | 2 | 8 | |
| Metastasis | 0.001 | ||
| Yes | 1 | 10 | |
| No | 8 | 1 | |
Note: P-value was calculated with a two-sided Pearson chi-square test.