Abstract
Non-alcoholic fatty liver disease is manifested by hepatic accumulation of triglycerides (TG) and is commonly associated with metabolic syndrome. The isoprenoid farnesol (FOH) modulates lipid metabolism and reduces hepatic TG content in rodents. This effect involves activation of at least two nuclear receptors, peroxisome proliferator-activated receptor α (PPARα) and farnesoid X receptor. We evaluated the effects of FOH (100 μM) in a cellular model of human hepatic steatosis by loading hepatocyte-like HepaRG cells with oleic acid (OA, 0.66 mM). FOH treatment decreased OA-induced TG accumulation by ~25%. Using PCR arrays, we found that FOH treatment modulated the mRNA levels of several lipid-metabolizing enzymes, both alone and when cells were loaded with OA. While FOH activated PPARα and the constitutive androstane receptor (CAR), most of the FOH-mediated effects on lipid-metabolizing genes could be attributed to activation of PPARα. In OA-loaded HepaRG cells, FOH increased fatty acid oxidation, which was accompanied by up-regulation of PPARα target genes involved in mitochondrial fatty acid oxidation, including hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase and acetyl-coenzyme A acyltransferase 2. These effects on gene expression were lost when the cells were co-treated with the PPARα antagonist, GW6471. OA treatment alone decreased the mRNA levels of the drug-metabolizing enzymes, cytochrome P450 (CYP)1A2, 2B6, and 3A4, and increased CYP2E1 expression, all of which were attenuated by FOH co-treatment. These findings show that FOH treatment increases fatty acid oxidation and decreases TG accumulation in steatotic HepaRG cells, which is likely attributable to PPARα-mediated induction of mitochondrial fatty acid oxidation.
Keywords: farnesol, HepaRG cells, steatosis, lipid metabolism, peroxisome proliferator-activated receptor
Introduction
Intrahepatic triglyceride (TG2) levels are regulated through a balance between the amounts of free fatty acids that enter hepatocytes or are synthesized de novo and the amounts that are catabolized through oxidation or secreted in very low-density lipoproteins (Koo, 2013). Any significant change in the factors mediating this balance can result in an increase in intrahepatic TG content (Anderson and Borlak, 2008; Musso et al., 2009). TG accumulation more than 5% of total liver weight is classified as steatosis (Bedogni et al., 2014). Hepatic steatosis is commonly associated with insulin resistance and the metabolic syndrome (Angulo, 2007), and in the absence of significant alcohol use, drugs, or viruses, steatosis is regarded as the hallmark of the early stages of non-alcoholic fatty liver disease (NAFLD) (Loomba and Sanyal, 2013). If left untreated, steatosis can develop into a more severe and progressive form of NAFLD known as non-alcoholic steatohepatitis (NASH), characterized by hepatic inflammation and oxidative damage (Hardy et al., 2016). Even though NAFLD has become the most common liver disease, with a worldwide prevalence ranging from 11 to 40%, there are currently no established regimens to treat NAFLD except for weight loss (Kawano and Cohen, 2013).
Farnesol (FOH) is a non-sterol isoprenoid that is produced endogenously through the dephosphorylation of farnesyl pyrophosphate (FPP), a key branch-point intermediate of the cholesterol biosynthetic pathway (Goldstein and Brown, 1990). FOH is also widely present in plants, such as peaches, tomatoes, strawberries, corn, and lemongrass (Duncan and Archer, 2008). Physiologically, FOH can function as a signaling molecule that modulates a variety of cellular processes, such as sterol biosynthesis, vasoconstriction, apoptosis, cell proliferation, and lipid metabolism (Correll et al., 1994; Edwards and Ericsson, 1999). Regarding the latter, studies in rodents have shown that FOH treatment lowers plasma and hepatic TG levels and improves metabolic abnormalities in obese animals, which is associated with lower expression of fatty acid synthesis genes and increased expression of fatty acid oxidation genes (Takahashi et al., 2002; Duncan and Archer, 2008; Goto et al., 2011). Similarly, FOH and/or its oxidative metabolites have been implicated in the TG-lowering effects produced by inhibitors of squalene synthase, the enzyme that catalyzes conversion of FPP to squalene, the first committed step in sterol biosynthesis (Hiyoshi et al., 2003). These studies suggest that FOH might be useful as a therapeutic approach to treat steatosis and alleviate progression of NAFLD.
Most of the above-described effects of FOH on lipid metabolism were shown to be mediated through nuclear receptors, such as peroxisome proliferator-activated receptors (PPARα and PPARγ), farnesoid X receptor (FXR), and retinoid X receptor β (Takahashi et al., 2002; Duncan and Archer, 2008; Goto et al., 2011). These ligand-activated transcription factors regulate expression of several lipid-metabolizing enzymes involved in fatty acid oxidation, synthesis, uptake, storage, transport, and elimination (Chawla et al., 2001; Berger and Moller, 2002; Wagner et al., 2011). While it has been demonstrated that FOH can produce beneficial effects on rodent hepatic lipid metabolism, the relevance to humans is not clear, since the activities of nuclear receptors can differ substantially across species (Gonzalez and Shah, 2008; Kiyosawa et al., 2008; Ross et al., 2010). Additionally, FOH and/or its metabolites activate the constitutive androstane receptor (CAR) in primary cultured rat and mouse hepatocytes (Kocarek and Mercer-Haines, 2002; Rondini et al., 2016). However, whether CAR activation has any role in FOH’s ability to regulate hepatic lipid metabolism has not been studied, which is important given the potential involvement of CAR in lipid metabolism and crosstalk with PPARα (Xiao et al., 2013).
HepaRG is a human hepatocellular carcinoma-derived cell line that can be differentiated in culture to cells that exhibit a hepatocyte-like phenotype, which includes expression of hepatic enzymes and transporters as well as key nuclear receptors, such as PPARα, FXR, and CAR (Aninat et al., 2006; Cerec et al., 2007; Guillouzo et al., 2007). In the current study, we therefore used oleic acid (OA)-loaded HepaRG cells as a model for human hepatic steatosis to determine the effects of FOH treatment on lipid-metabolizing pathways as well as the involvement of PPARα, FXR, and CAR in mediating the effects of FOH. We found that FOH treatment lowered cellular TG levels and increased fatty acid oxidation in the OA-loaded cells. FOH treatment also altered the mRNA expression of lipid-metabolizing genes, including several that were not previously identified in rodent studies. Further, most of the FOH-mediated changes, especially those involved in mitochondrial β-oxidation, were likely mediated by PPARα.
Materials and Methods
Materials
Trans, trans-FOH, dimethyl sulfoxide (DMSO), 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO), 3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4 isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid (GW4064), OA, hematoxylin, Oil Red O (ORO), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), triamcinolone acetonide, Triton X-100, and fatty acid-free bovine serum albumin (FF-BSA) were purchased from Sigma-Aldrich (St. Louis, MO). 2-[[4-[2-[[(Cyclohexylamino)carbonyl](4-cyclohexylbutyl)amino]ethyl]phenyl]thio]-2-methylpropanoic acid (GW7647) and N-((2S)-2-(((1Z)-1-methyl-3-oxo-3-(4-(trifluoromethyl)phenyl)prop-1-enyl)amino)-3-(4-(2-(5-methyl-2-phenyl-1,3-oxazol-4 (GW6471) were purchased from Bio-Techne (Minneapolis, MN). Williams’ Medium E, Penicillin-Streptomycin solution, and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA), and recombinant human insulin (Novolin R) from Novo Nordisk Pharmaceuticals, Inc. (Princeton, NJ). Tritiated OA ([9, 10-3H(N)]OA, 15–60Ci (0.555–2.22TBq)/mmol) was purchased from PerkinElmer (Waltham, MA). Other materials were obtained from the sources indicated below.
HepaRG cell culture and treatment
HepaRG cells were obtained from Biopredic International (Saint-Grégoire, France) under a material transfer agreement with INSERM-Transfert (Paris, France). HepaRG cells (at passage #3–4 following receipt) were plated at a density of 24,000 cells/cm2 into multi-well culture plates (6-well plates) unless otherwise indicated, and cultures were maintained in HepaRG growth medium (Williams’ Medium E supplemented with 10% FBS, 5 μg/ml insulin, 0.1 μM triamcinolone acetonide, 100 U/ml penicillin, and 100 μg/ml streptomycin) for 14 days. The confluent cells were then incubated in HepaRG differentiation medium 1 (HepaRG growth medium supplemented with 1% DMSO) for 48 hours followed by HepaRG differentiation medium 2 (HepaRG growth medium supplemented with 2% DMSO) for a further 14 days. Culture medium was renewed every 2–3 days. Upon differentiation, cells were cultured in HepaRG growth medium containing 2% FBS for 72 hours and then in HepaRG treatment medium (growth medium without serum) for 24 hours, after which treatments were performed. Synthetic nuclear receptor agonists or antagonists were added to the culture medium as concentrated stock solutions (1000X) dissolved in DMSO at the concentrations specified in the individual figure legends.
FOH stock solutions (1000X) were prepared in ethanol (EtOH). For FOH treatments, the required volume of FOH stock was added to 8.8% FF-BSA (w/v) prepared in HepaRG treatment medium and allowed to form complex for 30 min. EtOH (0.1%) diluted in FF-BSA was used as a vehicle control for FOH treatment experiments. For OA treatment, OA was first complexed with FF-BSA at a 6:1 molar ratio (OA:BSA). Briefly, an 8 mM OA stock was prepared by adding 2.26 mg of OA to 1 ml of a sterile 8.8% FF-BSA solution in HepaRG treatment medium. The stock solution was then diluted in HepaRG treatment medium to obtain the desired FOH, OA,and OA + FOH concentrations, as described in the individual figure legends. In all groups, the final content of FF-BSA in the cell culture medium was 0.7% (w/v).
Measurement of cell viability
The MTT assay was used to assess FOH treatment effects on cell viability, essentially as described previously (Mosmann, 1983). Briefly, MTT was dissolved at a concentration of 1 mg/mL in phosphate-buffered saline (PBS) and sterile-filtered prior to use. Cells were cultured in 12-well plates. At the selected times following treatment, culture medium was replaced with 0.3 mL of MTT solution, and cells were further incubated for 30 minutes at 37° C. Following incubation, cells were washed with PBS, and MTT formazan was extracted with 0.3 ml 100% isopropanol. Aliquots of the isopropanol extracts were diluted 1:5 in isopropanol and absorbance was measured at 560 nm using a SpectraMax Plus Absorbance Microplate Reader (Molecular Devices, Sunnyvale, CA). Each sample was measured in triplicate, and each experiment was repeated in 3 independent HepaRG cultures.
Measurement of neutral lipids and TG
ORO staining was used to determine treatment effects on the cellular levels of neutral lipids, as previously described (Ramirez-Zacarias et al., 1992). Briefly, ORO was dissolved at a concentration of 3.5 mg/ml in isopropanol and then filtered with Whatman paper. For each experiment, a working solution of ORO was prepared fresh by mixing 6 ml of ORO stock with 4 ml of water (0.21% w/v), and the solution was filtered again after incubating at room temperature for 20 min. To visualize cellular lipids, 24 hours after the last treatment HepaRG cells were washed with PBS and then fixed in 10% (v/v) neutral-buffered formalin (pH 7.4) for 1 hour at room temperature. Fixed cells were washed 3 times with double-distilled water and then dehydrated in isopropanol for 10 minutes at room temperature, after which the isopropanol was removed from the wells and cells allowed to dry. The cells were then incubated for 30 minutes at room temperature in ORO solution, followed by extensive washing with double-distilled water to remove non-specific staining. Nuclei were then counterstained with hematoxylin, and ORO-stained lipid vesicles were visualized at 200X using a phase-contrast microscope equipped with a digital camera (Olympus; Waltham, MA). To quantify differences in neutral lipid accumulation, ORO was extracted from the cells with isopropanol and absorbance read at 510 nm using isopropanol as blank. The experiment was repeated in 2 independent HepaRG experiments.
To measure TG levels directly, HepaRG cells were collected by scraping (2 wells/sample) and then pelleted by centrifugation at 1000 x g for 5 minutes. Pellets were then resuspended in ice-cold PBS containing 1% Triton X-100. The suspension was sonicated and then centrifuged at 13,200 x g for 10 minutes at 4°C to remove cellular debris. Supernatants were collected and 10 μl of sample was used to measure TG concentrations using the Serum Triglyceride Quantification Kit (Colorimetric) from Cell Biolabs (San Diego, CA) according to the manufacturer’s instructions. Total cellular TG levels were normalized to protein content in each sample, which was determined using the bicinchoninic acid assay (Sigma-Aldrich). All assays were performed in triplicate, and each experiment repeated 3 times.
Total RNA isolation and quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Twenty-four hours following the last treatment, HepaRG cells were harvested in Ambion lysis buffer and total RNA was extracted and column purified using the Purelink RNA isolation kit (Ambion; Carlsbad, CA). Complementary DNA (cDNA) was then synthesized using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies; Grand Island, NY), following the manufacturer’s instructions. Relative changes in gene expression were determined using a StepOnePlus Real Time PCR System (Applied Biosystems, Foster City, CA). Reactions were prepared using 10 μL of either SYBR Green or TaqMan Gene Expression Master Mix (Applied Biosystems), 50 ng RNA equivalents, and gene-specific primers. TaqMan Gene Expression Assays (Applied Biosystems, Supplemental Table 1) were used for initial studies demonstrating treatment effects on nuclear receptor target genes [CYP2B6 for CAR, perilipin 2 (PLIN2) for PPAR, and small heterodimer partner (SHP) for FXR]. All other RT-qPCR assays were conducted using SYBR Green Master Mix with primer pairs purchased from Integrated DNA Technologies (Coralville, IA). The primers were designed using the Primer-BLAST software tool (National Center for Biotechnology Information website), and the sequences are shown in Supplemental Table 1. For all assays, commercially available primer sets to detect TATA-box binding protein (P/N 4326322E from Applied Biosystems; Hs.PT.58v.39858774 from IDT) were purchased and used as the endogenous control. Relative fold-changes in mRNA levels were calculated using the comparative CT (ΔΔCT) method. All assays were performed in duplicate, and each experiment was repeated in at least 3 independent HepaRG cultures.
Measurement of hepatic lipid-metabolizing gene expression using customized PCR arrays
Custom-designed RT2 Profiler PCR Array plates (96-wells) were purchased from Qiagen (Carlsbad, CA), which mostly contained primer sets to detect transcripts for proteins involved in fatty acid, triglyceride, and sterol metabolism. Only lipid-metabolizing genes shown to be expressed in human liver and to regulate critical steps in hepatic lipid metabolism were included on the array. Primers to detect CAR, PPAR, and FXR transcripts were also included on the array, as were primers to detect prototypical targets of these receptors (CYP2B6, PLIN2, and SHP, respectively), as positive controls. A small number of genes involved in carbohydrate metabolism and inflammation were also included, as were 4 reference genes as internal controls. A list of genes with descriptions is provided in Supplemental Table 2 [in addition to those listed, ELOVL2, ELOVL3, ELOVL4, GPAT2, and TNF were included on the array but were excluded from analysis due to their undetectable or high CT values (>34)]. Following treatment, HepaRG cells were harvested and total RNA isolated using the Purelink RNA isolation kit. cDNA was then prepared using the RT2 First Strand Synthesis Kit (Qiagen) and PCR conducted using the RT2 SYBR Green qPCR Master Mix according to the manufacturer’s (Qiagen) recommendations. Relative fold changes in gene expression were determined using the RT2 Profiler PCR Array Data Analysis system (Qiagen), which is based on the ΔΔCT method. Raw CT values for the target transcripts were normalized to corresponding CT values for the TBP reference gene. Treatment-induced changes in mRNA levels were then normalized to vehicle-treated controls. A heat map was generated using Morpheus matrix visualization and analysis software (https://software.broadinstitute.org/morpheus). Treatments were performed on 2 independent HepaRG cultures, and values in the heat map represent average fold-changes relative to control for the 2 biological replicates.
Measurement of fatty acid oxidation
Treatment effects on fatty acid oxidation were measured in intact cells cultured on 12-well plates, as described previously (Zhang et al., 2012), using [9,10-3H(N)]OA as substrate and tritiated water as the end-product of oxidation. Briefly, a complex of FF-BSA:OA (8 mM OA) was prepared as described above. [9,10-3H(N)]OA was included to achieve a final radiolabel concentration of 1 μCi/mL in the treatment medium. One mL of medium containing [3H]OA was added to each well and the cells incubated at 37 °C for 3 hours, after which the medium was collected and protein precipitated by adding 100% trichloroacetic acid to a final concentration of 9% (v/v) and centrifuging. The supernatants were alkalinized by adding 6 N NaOH and then loaded onto columns containing 0.2 g/mL Dowex 1×2–400 resin (Sigma-Aldrich) diluted in water. Tritiated water was eluted from the columns using 1.5 mL water into scintillation vials containing 10 mL Ultima-Flo™ M scintillation cocktail (PerkinElmer; Waltham, MA) and radioactivity was measured using a Beckman-Coulter liquid scintillation counter (model LS3801; Indianapolis, IN). Results were corrected for nonspecific radioactivity obtained from medium containing [3H]OA that was not added to HepaRG cultures. Etomoxir (10 μM; Sigma-Aldrich), an inhibitor of carnitine palmitoyltransferase 1, was used as a negative control in 2 of the experiments. Experiments were conducted in quadruplicate and repeated in 3 independent HepaRG cultures.
Statistical analysis
Results are presented as mean ± SEM. Significance testing was performed using GraphPad Prism version 6.00 (GraphPad Software; San Diego, CA). As appropriate, treatment groups were compared using one- or two-way analysis of variance with the Newman-Keuls test for multiple comparisons, unpaired t-tests, or one-sample t-tests against the hypothetical value of 1. P <0.05 was considered to be statistically significant.
Results
FOH suppresses OA-mediated TG accumulation in HepaRG cells
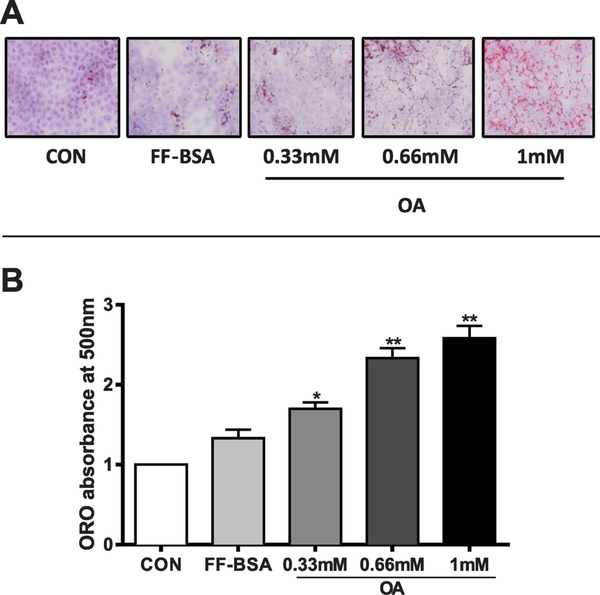
HepaRG cells have been previously shown to be a suitable in vitro system for studying hepatic lipid metabolic processes (Samanez et al., 2012; Brown et al., 2013). Here, we used HepaRG cells as a model to evaluate the effects of FOH treatment on human hepatic steatosis. HepaRG cells were treated for 48 hours with OA (0.33, 0.66, or 1 mM), a known inducer of TG accumulation in human hepatic cells, including HepaRG cells (Ricchi et al., 2009; Anthérieu et al., 2011). ORO staining revealed an OA concentration-dependent increase of intracytoplasmic lipid droplets (stained in red, Figure 1A). Quantification of the staining showed that absorbance was significantly increased by 1.3-, 1.7-, and 1.9- fold at 0.33, 0.66, and 1 mM OA, respectively, compared to cultures incubated with medium containing FF-BSA vehicle (Figure 1B).
Figure 1. Determination of neutral lipid accumulation by ORO staining in OA-loaded HepaRG cells.
HepaRG cells were incubated for 48 hours in HepaRG treatment medium alone (CON) or containing 0.7% FF-BSA, either alone or complexed with 0.33 mM, 0.66 mM, or 1 mM OA. Treatment medium was replaced once after 24 hours. (A) ORO was used to stain neutral lipids, and cells were photographed under phase-contrast optics at 200X magnification. Neutral lipids are stained red, hepatocyte nuclei are purple/blue. (B) ORO was extracted from the cells and absorbance measured at 510 nm. Each bar represents the mean ± SEM (n=4 wells per group, derived from combining the data from 2 independent HepaRG experiments with duplicate treatments). *Significantly different from vehicle control (FF-BSA), P<0.05, **P<0.01.
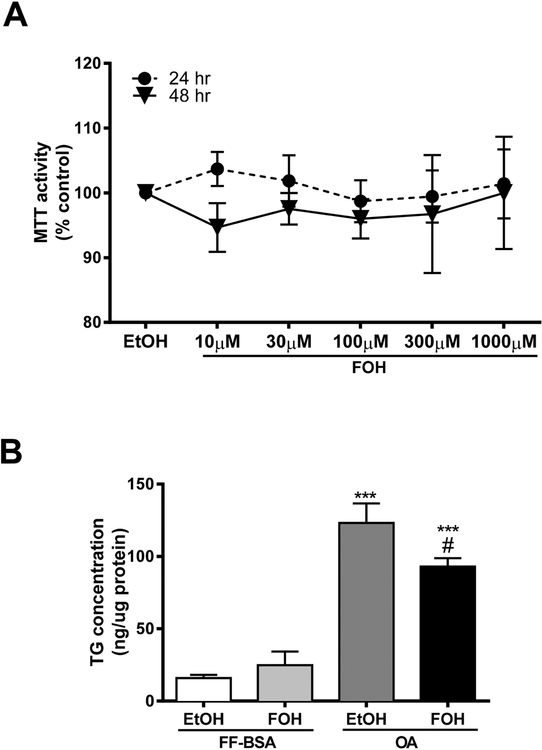
We next determined whether FOH treatment caused any cytotoxicity in HepaRG cultures. HepaRG cells were treated with increasing concentrations of FOH (10, 30, 100, 300 and 1000 μM) for 24 or 48 hours, and cell viability was measured by the MTT assay. FOH treatment had no significant effect on MTT formazan absorbance at any of the concentrations or treatment times that were tested, although greater variability in MTT activity was noted at the 2 highest FOH concentrations (Figure 2A). Also, no changes in cellular morphology were noted when the cells were observed under phase-contrast microscopy (data not shown). Therefore, we selected 100 μM FOH for further study. To determine the effect of FOH on intracellular TG accumulation in OA-loaded HepaRG cells, HepaRG cell cultures were pre-incubated with either FF-BSA or 0.66 mM OA for 24 hours followed by treatment with either vehicle or 100 μM FOH in the absence or presence of OA for another 48 hours, and intracellular TG levels were measured (Figure 2B). FOH treatment had no effect on TG levels under control conditions. Exposure to OA increased the amount of intracellular TG by 7.7–fold in HepaRG cells when compared to FF-BSA-treated control. This OA-induced increase in TG levels was significantly reduced (by 24.4%) when the HepaRG cells were co-treated with 100 μM FOH.
Figure 2. Effects of FOH on HepaRG viability and TG content.
(A) HepaRG cells were cultured for 24 or 48 hours in HepaRG treatment medium containing either vehicle control [FFBSA+0.1% EtOH (EtOH)] alone or FF-BSA complexed with 10 – 1000 μM FOH. Cell viability was determined using the MTT assay, as described in Materials and Methods. Each bar represents the mean MTT formazan absorbance ± SEM from three independent experiments (n=3). Values were normalized to those of the EtOH-treated group. (B) HepaRG cells were pre-treated for 24 hours in HepaRG treatment medium containing FF-BSA, either alone or complexed with 0.66 mM OA. The cells were then incubated for 48 hours in pre-treatment medium (i.e., FF-BSA or OA) containing either 0.1% EtOH or 100 μM FOH and harvested for TG measurement, as described in Materials and Methods. The amount of TG was normalized to protein content for each sample. Each bar is the mean ± SEM from 3 independent experiments (n=3). ***Significantly different from FF-BSA:EtOH group, P<0.001.#Significantly different from OA:EtOH group, P<0.05.
FOH activates CAR and PPARα, but not FXR, in HepaRG cells
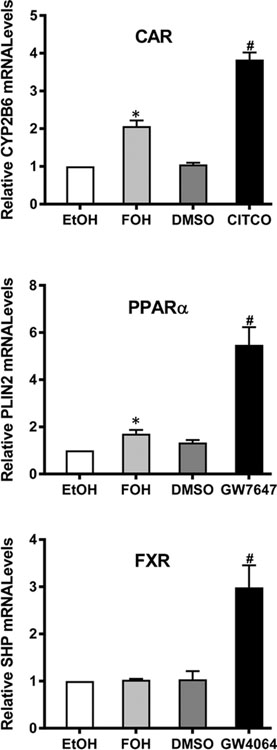
Since FOH has been shown to regulate cellular processes through activation of CAR, PPARα, and FXR, we hypothesized that one or more of these receptors might mediate the observed reduction of TG by FOH in our human hepatic steatosis model. Therefore, we first determined whether FOH treatment affected the activity of CAR, PPARα, or FXR in HepaRG cells. HepaRG cultures were treated with either FOH or a known synthetic agonist of each receptor for 48 hours, and mRNA levels of target genes were measured by RT-qPCR (Figure 3). Treatment with the CAR agonist CITCO (0.1 μM), PPARα agonist GW7647 (10 μM), or FXR agonist GW4064 (1 μM) significantly increased the mRNA levels of CYP2B6 (3.8-fold), PLIN2 (5.5-fold), and SHP (2.9-fold), respectively, compared to vehicle-treated control. Treatment with 100 μM FOH significantly increased CYP2B6 mRNA levels by 2.1-fold and PLIN2 mRNA levels by 1.7-fold. However, FOH treatment had no effect on SHP mRNA content. These findings indicate that FOH modulates the activity of CAR and PPARα, but not FXR, in HepaRG cells under the experimental conditions that were used, although FOH’s effects on CAR and PPARα were somewhat weaker than those produced by the synthetic agonists.
Figure 3. Effects of FOH on nuclear receptor (CAR, PPARα, and FXR) activities in HepaRG cells.
HepaRG cells were cultured in HepaRG treatment medium containing FF-BSA + 0.1% ethanol (EtOH), FF-BSA + 100 μM FOH, 0.1% DMSO, or one of the following nuclear receptor agonists: 0.1 μM CITCO (CAR), 10 μM GW7647 (PPARα), or 1 μM GW4064 (FXR). Treatment medium was replaced once after 24 hours. After 48 hours of treatment, cells were harvested for RNA isolation, and CYP2B6, PLIN2, and SHP mRNA levels were quantified by RT-qPCR, as described in Materials and Methods. Each bar represents the mean ± SEM of normalized mRNA values from 3 (PLIN2, SHP; n=3) or 4 (CYP2B6; n=4) independent experiments, and within each experiment data were normalized to the EtOH-treated control group (i.e., EtOH=1). *Significantly different from 1, P<0.05.#Significantly different from DMSO-treated group, P<0.05.
FOH treatment regulates the expression of lipid-metabolizing genes in HepaRG cells
To investigate the mechanism(s) underlying the FOH-mediated reduction of TG content in OA-loaded HepaRG cells, we used a customized PCR array to profile treatment-induced changes in the expression of genes involved in various human hepatic lipid-metabolizing pathways, including those associated with fatty acid uptake and transport, fatty acid oxidation, fatty acid synthesis, TG biosynthesis, lipoprotein secretion, and cholesterol biosynthesis. HepaRG cells were treated either with FOH or OA alone, OA and FOH in combination (i.e., effects of FOH in OA-loaded cells), or a control agonist for CAR, PPARα, or FXR. The treatment effects on gene expression (fold changes in mRNA levels relative to control) are shown in Supplemental Table 2.
The PCR array data confirmed the effects of the positive control nuclear receptor agonist treatments that were observed by RT-qPCR (Figure 3), namely increased expression of CYP2B6 (9.9-fold increase in mRNA levels), SHP (5.0-fold), and PLIN2 (3.8-fold) by treatment with CITCO (CAR), GW4064 (FXR), or GW7647 (PPARα), respectively. Other than CYP2B6 induction, treatment with the CAR agonist CITCO had little effect on expression of other genes on the array, and the few CITCO-mediated changes were not focused on any particular lipid metabolism pathway. Treatment of control (i.e., not OA-loaded) HepaRG cells with the FXR agonist GW4064 changed the expression of several lipid-metabolizing genes and had a major suppressive effect on phosphoenolpyruvate carboxykinase 1, the main control point for gluconeogenesis, as well as on several CYPs including CYP2B6, CYP2E1, and CYP411. GW4064 treatment of OA-loaded HepaRG cells reduced the mRNA levels for most of the genes on the array. The PPARα agonist GW7647 changed the expression of numerous genes. While GW7647 suppressed the expression of four genes, apolipoprotein-4 (APOC4), interleulin-6 (IL-6), SHP, and CAR, all other genes profiled that changed by at least 1.3-fold were upregulated by GW7647 treatment. The major pathways affected by PPARα activation were fatty oxidation, particularly in mitochondria, uptake, and transport.
The PCR array data shown in Supplemental Table 2 also confirmed FOH’s ability to activate CAR (as indicated by increased CYP2B6 mRNA levels) and PPARα (PLIN2) but not FXR (SHP). The magnitude of the FOH-mediated increase in CYP2B6 mRNA content determined on the arrays was the same as that determined by RT-qPCR (2.1-fold) while the increase in PLIN2 mRNA levels was somewhat lower on the arrays (1.3-fold) than it was by RT-qPCR (1.7-fold). In contrast to the RT-qPCR analysis, which indicated that FOH had no effect on FXR activity, the array data suggested a modest suppressive effect (1.3-fold decrease in SHP mRNA content).
FOH treatment of control HepaRG cells changed the expression of 27 genes on the array (using the criterion of an at least 1.3-fold change in mRNA level), and the affected genes were distributed among the various functional categories represented on the array. By this criterion, treatment of HepaRG cells with OA alone affected the expression of 24 genes. Of the genes regulated by FOH or OA treatment alone, only 9 were regulated in the same direction (i.e., either increased or decreased) by both treatments. FOH treatment of OA-loaded HepaRG cells changed the mRNA levels of 37 genes by at least 1.3-fold. The most notable change in affected genes occurred within the mitochondrial β-oxidation category, where only 3 or 4 of the 18 genes on the array were regulated by FOH or OA treatment alone, but 11 of those genes were regulated by the combination of FOH and OA. Ten of those 11 genes were upregulated by FOH treatment, and those 10 genes were also upregulated by the PPARα agonist GW7647 (Supplemental Table 2).
Although FOH and OA co-treatment produced changes of 1.3-fold or more in the mRNA levels of other categories of lipid-metabolizing genes that were included on the array (Supplemental Table 2), in general a smaller fraction of the genes in those other categories was regulated than was seen for the mitochondrial β-oxidation genes. Of the 6 fatty acid uptake/transport genes that were evaluated, only the mRNA level of SLC27A1 (Gene Symbol, see Supplemental Table 2 for Gene Name) was changed by FOH+OA treatment (1.3-fold increase). Likewise, FOH+OA only affected expression of one peroxisomal β-oxidation gene, ACOX1 (1.3-fold increase in mRNA level). Four of 13 fatty acid biosynthesis genes were regulated by FOH+OA: ELOVL6 (1.4-fold increase), FASN (1.3-fold increase), SCD (1.4-fold decrease), and SCD5 (1.3-fold decrease). Three of 6 genes that were classified as fatty acid metabolism (other) were changed: ACOT1 (1.3-fold increase), ACOT6 (1.3-fold decrease), and ACOT12 (3.9-fold increase). Four of 11 triglyceride biosynthesis genes were regulated by FOH+OA: LPIN1 (1.3-fold increase), AGPAT1 (1.3-fold decrease), AGPAT6 (1.4-fold increase), and DGAT2 (1.3-fold increase). Four of 11 cholesterol pathway genes were changed: FNTA (1.3-fold increase), HMGCR (1.4-fold increase), RABGGTA (1.4-fold decrease), and RABGGTB (1.3-fold decrease).
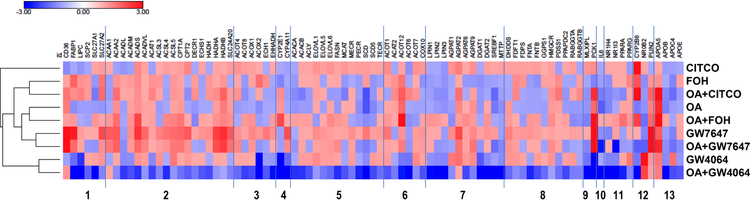
Figure 4 shows a heat map of the PCR array results, in which the genes are presented in the same order (and in the same categories) that is shown in Supplemental Table 2, and the treatment groups are clustered to indicate how similarly they affected the gene expression profile. The heat map shows that the gene expression profile produced by CITCO treatment was distinct from the other treatments. The groups treated with GW4064 (alone or in the presence of OA) clustered together, and apart from the other groups. The groups treated with GW7647 (alone or with OA) clustered together and the groups treated with OA alone or OA+FOH clustered together, and all four of these treatment groups showed similarities in their gene expression profiles, likely indicating the dominant influence of PPARα activation on gene expression in these 4 treatment groups. Finally, the group treated with FOH alone clustered most closely with the group treated with OA+CITCO, suggesting that the effect of FOH was largely a combination of CAR and PPARα activation.
Figure 4. Heat map showing effects of FOH, OA, and nuclear receptor agonists on lipidmetabolizing gene expression.
HepaRG cells were pre-treated for 24 hours in HepaRG treatment medium containing FF-BSA alone or complexed with 0.66 mM OA. The cells were then incubated for 48 hours (media change after 24 hours) in pre-treatment medium (i.e., FFBSA or OA) containing 0.1% EtOH, 100 μM FOH, 0.1% DMSO, 0.1 μM CITCO, 10 μM GW7647, or 1 μM GW4064. Cells were then harvested, and mRNA levels of 96 genes were measured using a customized PCR Array, as described in Materials and Methods. Effects of treatments normalized to their respective vehicle controls (EtOH for farnesol, DMSO for nuclear receptor agonists) are presented as a heat map, where positive log-ratios (log2-fold change) for upregulated genes are shown in red and negative log-ratios for downregulated genes are shown in blue. Treatment groups are hierarchically clustered using one minus Pearson correlation clustering. Functional classifications of genes are (1) fatty acid uptake and transport, (2) mitochondrial fatty acid oxidation, (3) peroxisomal β-oxidation, (4) microsomal ω-oxidation, (5) fatty acid biosynthesis, (6) fatty acid metabolism (other), (7) TG biosynthesis, (8) cholesterol metabolism, (9) carbohydrate metabolism, (10) inflammation, (11) nuclear receptors, (12) nuclear receptor targets, and (13) apolipoproteins.
FOH treatment increases fatty acid oxidation gene expression and activity in OA-loaded HepaRG cells
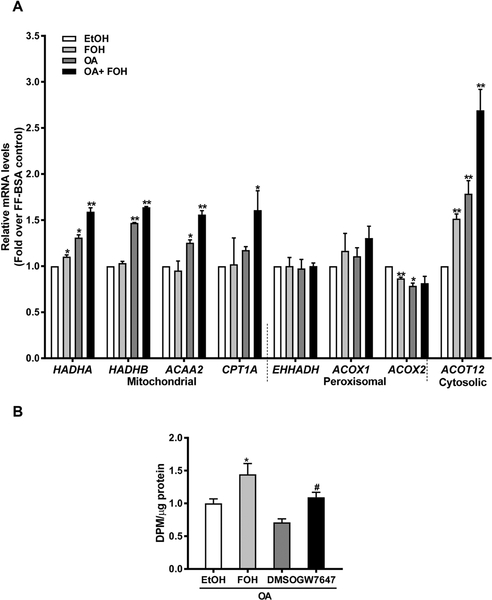
We validated results from our microarray experiments by RT-qPCR, using primer pairs that were different from those used in the PCR arrays (Supplemental Table 1). Since fatty acid oxidation was a major pathway modulated by FOH treatment of OA-loaded HepaRG cells, we measured the mRNA levels of several mitochondrial and peroxisomal β-oxidation genes in additional batches of HepaRG cells that were treated the same as those used for the array experiments. Under control conditions, treatment with FOH alone had no significant effect on the expression of mitochondrial β-oxidation genes except for hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit (HADHA) (Figure 5A). OA treatment significantly increased expression of HADHA (~1.5-fold), hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit (HADHB, 1.5-fold), and acetyl-coenzyme A acyltransferase 2 (ACAA2, 1.3-fold). Co-treatment with FOH and OA further increased the expression of HADHA (1.2-fold), HADHB (1.2-fold), and ACAA2 (1.3-fold), when compared to the OA-only treated samples. Carnitine palmitoyltransferase 1A (CPT1A) expression was not changed by FOH or OA treatment alone but was significantly increased by co-treatment with OA and FOH (1.7-fold over EtOH-treated control). In contrast, neither FOH nor OA had any effect on the expression of peroxisomal β-oxidation genes except for ACOX2, which was suppressed by both FOH (1.2-fold) and OA (1.3-fold). Additionally, expression of the cytosolic thioesterase, acyl-CoA thioesterase 12 (ACOT12), increased by 1.5-fold over control in response to FOH treatment and 1.8-fold by OA treatment. Co-treatment with FOH further increased the OA-mediated increase in ACOT12 expression by 1.5-fold, in agreement with the data from the PCR arrays (Supplemental Table 2).
Figure 5. Effects of FOH on fatty acid oxidation gene expression and activity in HepaRG cells.
(A) HepaRG cells were pre-treated for 24 hours in HepaRG treatment medium containing FF-BSA alone or complexed with 0.66 mM OA. The cells were then incubated for 48 hours (media change after 24 hours) in pre-treatment medium containing either 0.1% EtOH or 100 μM FOH. Cells were then harvested, and mRNA levels of genes regulating fatty acid oxidation were quantified by RT-qPCR. Each bar represents the mean ± SEM of normalized mRNA levels from 3 independent experiments (n=3), and within each experiment data were normalized to the EtOH-treated control group (i.e., EtOH=1). *Significantly different from 1, P<0.05, **P<0.01. (B) HepaRG cells were pre-treated for 24 hours with HepaRG treatment medium containing FFBSA complexed with 0.66 mM OA. The cells were then treated for 48 hours (media change after 24 hours) with medium containing FF-BSA+OA and either 0.1% EtOH, 100 μM FOH, 0.1% DMSO, or 10 μM GW7647. Following treatment, cells were processed to measure fatty acid oxidation, as described in Materials and Methods. Tritiated water DPM were normalized to cellular protein content and all treatment groups were normalized to the EtOH-treated control group. Each bar represents the mean ± SEM of normalized values from 3 independent experiments (n=3). *Significantly different from EtOH-treated group, P<0.05.#Significantly different from DMSO-treated group, P<0.05.
The panel of genes changed following FOH co-treatment with OA suggested a main effect on mitochondrial β-oxidation, although the individual gene expression changes were generally small. Therefore, to determine the integrated impact of FOH treatment on fatty acid oxidation in intact HepaRG cells, we measured the release of tritiated water from OA (Figure 5B). In OA-loaded HepaRG cells, FOH significantly increased fatty acid oxidation by 1.5-fold compared to treatment-matched controls. Fatty acid oxidation increased to a similar extent following treatment with the PPARα agonist GW7647, whereas treatment with the carnitine palmitoyltransferase 1 inhibitor etomoxir reduced mitochondrial β-oxidation by 46% (data not shown). Taken together, our findings indicate that under steatogenic conditions, FOH treatment increases mitochondrial fatty acid oxidation but has little effect on expression of peroxisomal fatty acid oxidation genes.
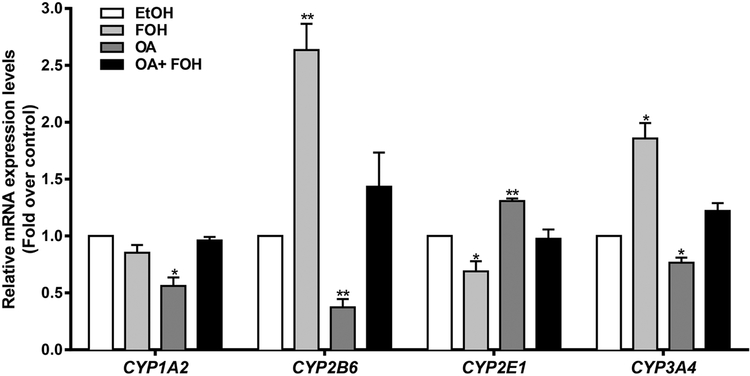
FOH treatment modulates expression of CYPs in HepaRG cells
Since FOH has been reported to be a substrate and inhibitor of certain CYPs (Raner et al., 2002), the effects of FOH treatment on CYP expression in control and OA-loaded HepaRG cells were evaluated. As shown in Figure 6, OA treatment significantly decreased the mRNA levels of CYP1A2 (1.8-fold), CYP2B6 (2.6-fold), and CYP3A4 (1.2-fold) and increased CYP2E1 mRNA levels (1.3-fold). Treatment with FOH alone significantly increased CYP2B6 (2.6-fold) and CYP3A4 (1.9-fold) mRNA levels in control HepaRG cells and reversed the OA-mediated decreases of CYP1A2, CYP2B6, and CYP3A4 as well as the OA-mediated increase of CYP2E1.
Figure 6. Effects of FOH on CYP expression in HepaRG cells.
HepaRG cells were pre-treated for 24 hours with HepaRG treatment medium containing FF-BSA alone or complexed with 0.66 mM OA. The cells were then incubated for 48 hours (media change after 24 hours) in pre-treatment medium (i.e., FF-BSA or OA) containing either 0.1% EtOH or 100 μM FOH. Cells were then harvested, and CYP mRNA levels were quantified by RT-qPCR, as described in Materials and Methods. Each bar represents the mean ± SEM of normalized mRNA levels from 3 independent experiments (n=3), and within each experiment data were normalized to the EtOH-treated control group (i.e., EtOH=1). *Significantly different from 1, P<0.05, **P<0.01.
FOH regulates mitochondrial β-oxidation genes through PPARα
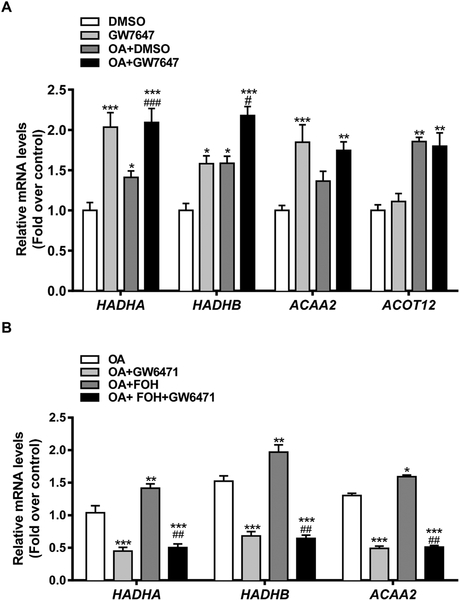
To establish the role of PPARα in the regulation of mitochondrial β-oxidation gene expression in control and OA-loaded HepaRG cells, differentiated cells were treated with 10 μM GW7647, and changes in gene expression were measured by RT-qPCR (Figure 7A). In control cells, PPARα activation significantly increased HADHA (2.0-fold), HADHB (1.6-fold), and ACAA2 (1.8-fold) mRNA levels. As seen in the above-described experiment (Figure 5A), OA treatment also upregulated HADHA (1.4-fold), HADHB (1.6-fold), and ACAA2 (1.4-fold), and the OA-mediated increases were increased further by GW7647 co-treatment (by 1.5-fold, 1.4-fold, and 1.3-fold, respectively, for HADHA, HADHB, and ACAA2 mRNA). In contrast to these 3 genes encoding mitochondrial β-oxidation proteins, although OA treatment increased cytosolic ACOT12 mRNA content (1.9-fold), treatment with GW7647 did not increase ACOT12 expression in control cells and did not enhance OA-induced ACOT12 expression. Next, OA-loaded HepaRG cells were treated with FOH in the presence or absence of the PPARα antagonist GW6471 (10 μM) for 48 hours, and expression of HADHA, HADHB, and ACAA2 was measured using RT-qPCR (Figure 7B). GW6471 treatment significantly reduced HADHA, HADHB, and ACAA2 expression in OA-loaded cells and antagonized the FOH-mediated increases, indicating that under steatogenic conditions, FOH treatment increases the expression of mitochondrial β-oxidation genes through activation of PPARα in HepaRG cells.
Figure 7. Role of PPARα in FOH-mediated regulation of mitochondrial fatty acid β-oxidation genes in HepaRG cells.
(A) HepaRG cells were cultured in HepaRG treatment medium containing either 0.1% DMSO, 10 μM GW7647, FF-BSA complexed with 0.66 mM OA+DMSO, or 0.66 mM OA+10 μM GW7647 for 48 hours. Treatments were replaced once after 24 hours. Cells were then harvested, and mRNA levels of fatty acid oxidation genes were quantified by RT-qPCR. Each bar represents the mean ± SEM of mRNA levels normalized to the DMSO-treated control group from 3 (HADHB, ACAA2; n=3) or 4 (HADHA, ACOT12; n=4) independent experiments. *Significantly different from DMSO-treated group, P<0.05, **P<0.01, ***P<0.001.#Significantly different from OA+DMSO-treated group, P<0.05,###P<0.001. (B) HepaRG cells were pre-treated for 24 hours with HepaRG treatment medium containing FF-BSA complexed with 0.66 mM OA, followed by treatment for 48 hours (media change after 24 hours) with medium containing 0.66 mM OA, either alone or in combination with 100 μM FOH, 10 μM GW6471, or FOH+GW6471. Cells were then harvested, and mRNA levels of mitochondrial fatty acid β-oxidation genes were quantified by RT-qPCR. Each bar represents the mean ± SEM of mRNA levels normalized to the OA-treated group from 3 (HADHA, ACAA2; n=3) or 4 (HADHB; n=4) independent experiments. *Significantly different from OA-treated group, P<0.05, **P<0.01, ***P<0.001.##Significantly different from OA+FOH-treated group, P<0.01.
Discussion
The pathogenesis of NAFLD involves dysregulation of lipid metabolic pathways, many of which are regulated by nuclear receptors, such as the PPARs (Sambasiva Rao and Reddy, 2004; Tailleux et al., 2012), FXR (Sinal et al., 2000), CAR (Yamazaki et al., 2007), pregnane X receptor (Sookoian et al., 2010), and liver X receptor (Ducheix et al., 2013). Therefore, pharmacological manipulations of these nuclear receptors using synthetic modulators are regarded as potential approaches for NAFLD treatment (López-Velázquez et al., 2012; Machado and Cortez-Pinto, 2014). In addition to synthetic activators, these nuclear receptors are targeted by several classes of endogenous compounds, including bile salts (FXR) (Wang et al., 1999) and lipoproteins, eicosanoids, fatty acids, and prostaglandins (PPARs) (Kliewer et al., 1997; Schupp and Lazar, 2010). Here, we studied FOH, an endogenous isoprenoid that has previously been shown to suppress hepatic TG accumulation in rodents through activation of PPARα, PPARγ, and/or FXR (Forman et al., 1995; Takahashi et al., 2002; Hiyoshi et al., 2003; Goto et al., 2011). However, extrapolation of FOH’s effects on lipid metabolism from rodents to human is complicated by interspecies differences in the activities and functions of these receptors; particularly for PPARα, which in known to regulate diverse sets of lipid-metabolizing genes in mouse and human hepatocytes (Rakhshandehroo et al., 2009), possibly due to differences in ligand affinity, the peroxisome proliferator response elements (PPREs) in target genes, and the transcriptional co-regulators that are recruited (Ammerschlaeger et al., 2004; Gonzalez and Shah, 2008; Yang et al., 2008). In view of the above-described differences, we evaluated whether FOH treatment could suppress TG accumulation under steatogenic conditions in human hepatic cells, using OA-treated HepaRG cells as our model. FOH treatment lowered TG levels by nearly 25% in OA-loaded HepaRG cells, suggesting that FOH can potentially reduce steatosis in human hepatocytes.
FOH-mediated activation of human CAR, PPARα, and FXR has not yet been adequately defined. In transfection assays conducted in CV1 cells, FOH treatment activated a chimeric receptor containing the human PPARα ligand-binding domain and GAL4 DNA-binding domain as well as the full-length receptor (Takahashi et al., 2002). Also, in the same study, FOH increased expression of PPARα target genes in HepG2 cells engineered to overexpress human PPARα. In our initial experiments, we found that FOH treatment increased expression of the PPARα target gene PLIN2, showing that FOH can activate endogenous PPARα in HepaRG cells. FOH treatment also increased expression of CYP2B6, indicating that FOH can modulate human CAR-mediated responses. FOH was previously shown to activate FXR in cultured rat hepatocytes (Goto et al., 2011). However, FOH treatment did not change expression of the FXR target gene SHP in our study, even though FXR was robustly activated by the positive control agonist GW4064. This failure to increase SHP expression might be attributable to FOH being a relatively weak FXR agonist (Forman et al., 1995) or to species differences in FXR activation. In addition, while SHP is a well-known FXR-target gene, we noted that SHP was regulated not only by FXR (GW4064 treatment) but also by the PPARα agonist GW7647, which produced a suppressive effect on SHP expression (Supplemental Table 2). It therefore seems possible that the net lack of effect of farnesol treatment on SHP expression in HepaRG cells could have been due, at least in part, to a negation of any FXR-mediated activating effect by a PPARα-mediated suppressive effect.
FOH upregulates genes involved in fatty acid oxidation in cultured rat hepatocytes (Duncan and Archer, 2008) and the livers of high fat-fed mice (Goto et al., 2011). The results from our customized PCR array and RT-qPCR analyses indicated that FOH treatment of OA-loaded HepaRG cells increased the expression of several genes that are responsible for hepatic fatty acid oxidation, which translated to increased fatty acid oxidation activity. FOH treatment has also been reported to suppress expression of the lipogenic genes SREBF1 and FAS in obese mice (Goto et al., 2011) and FAS but not SREBF1 in cultured clone 9 rat hepatic cells (Duncan and Archer, 2008). Our PCR array data indicated that FOH treatment did not suppress expression of SREBF1 or FAS in HepaRG cells, further suggesting that FOH-mediated regulation of hepatic lipid-metabolizing gene expression differs across species.
OA-treated HepaRG cells have been reported to have lower mRNA expression of lipogenic genes and increased expression of genes regulating fatty acid oxidation and lipid droplet formation (Anthérieu et al., 2011). When OA-loaded HepaRG cells were treated with PPARα or dual PPARα/γ agonists, TG accumulation was suppressed, which was accompanied by increases in fatty acid oxidation, up-regulation of genes regulating fatty acid oxidation, and downregulation of lipogenic genes (Rogue et al., 2014), suggesting that activation of PPARs in OA-treated HepaRG cells can reverse steatosis. In the current study, FOH treatment of control or OA-loaded HepaRG cells increased expression of several lipid-metabolizing genes that were also upregulated following treatment with the synthetic PPARα agonist GW7647. Treatment of control and OA-loaded HepaRG cells with GW7647 increased expression of the mitochondrial β-oxidation genes, HAHDA, HADHB, and ACAA2, and FOH-mediated up-regulation of these genes was attenuated by the PPARα antagonist GW6471 in OA-loaded HepaRG cells, demonstrating that FOH’s effects on these mitochondrial β-oxidation genes are mediated through PPARα.
The regulation of ACOT12 was unusual in that its expression was increased by FOH and/or OA treatment, but GW7647 treatment had less (as determined by PCR arrays) or no (as determined by RT-qPCR) effect. ACOT12 is a cytoplasmic enzyme that catalyzes the hydrolysis of acetyl-CoA to acetate and CoA, and the enzyme’s activity is regulated by ADP/ATP levels (Swarbrick et al., 2014). ACOT12 in rats has a PPRE motif in its promotor region (Suematsu and Isohashi, 2006), which is consistent with the observed increase in ACOT12 activity upon treatment with the PPARα agonist clofibrate in rat hepatocytes (Ebisuno et al., 1988). However, the PPRE motif on the human ACOT12 gene is located outside the putative promoter region (Suematsu and Isohashi, 2006) and its responsiveness to PPAR activators is not known. Nonetheless, the up-regulation of ACOT12 by OA and FOH observed in this study suggests a role for this protein in fatty acid metabolism under steatotic conditions.
OA treatment decreased the expression of CYP1A2, CYP2B6, and CYP3A4 and increased CYP2E1 expression. Changes in the expression of various CYPs are routinely seen in the livers of NAFLD patients, resulting in altered drug metabolism (Merrell and Cherrington, 2011). CYP2E1 is also directly involved in NAFLD pathogenesis because of its role in lipid peroxidation that increases oxidative stress, one of the factors responsible for NAFLD progression (Chalasani et al., 2003). All OA-induced changes in CYP expression were attenuated by FOH co-treatment. FOH has previously been reported to modulate expression of rat CYPs and to interact with mammalian CYPs as a substrate and/or inhibitor. Specifically, FOH has been shown to increase CYP2B mRNA levels in primary cultured rat hepatocytes (Kocarek and Mercer-Haines, 2002), induce several monooxygenase activities in rat liver (Horn et al., 2005), inhibit monooxygenase activities in rabbit liver microsomes (Raner et al., 2002b), and be a substrate for ω-hydroxylation by rabbit and human CYP2E1 (DeBarber et al., 2004). Our findings suggest that FOH can potentially normalize fatty acid-induced changes in the hepatic expression of several human drug-metabolizing enzymes.
In conclusion, our results indicate that FOH can exert potentially beneficial effects on human hepatic lipid and drug metabolism. FOH reversed fatty acid-induced steatosis in human hepatic cells by increasing the expression of genes responsible for mitochondrial β-oxidation, primarily through activation of the nuclear receptor PPARα. FOH’s ability to normalize expression of human CYPs might be an effective approach to ameliorate altered metabolism of drugs during treatment of NAFLD, but this possibility requires further investigation. Additionally, FOH has been shown to function as a dual human PPARα/γ activator (Takahashi et al., 2002), and farnesol or a metabolite has been shown to regulate adipocyte functions through PPARγ activation (Takahashi et al., 2002; Goto et al., 2011; Torabi and Mo, 2016). Although we did not include PPARγ in this study, it would be interesting to determine whether PPARγ contributes to any of FOH’s effects using PPARγ-deficient hepatocytes or PPARγ selective antagonists, such as GW9662 (Zhang et al., 2015).
Supplementary Material
Highlights.
Farnesol treatment decreased triglyceride levels in steatotic HepaRG cells
Farnesol activated nuclear receptors CAR and PPARα in HepaRG cells
Farnesol-mediated PPARα activation increased mitochondrial fatty acid oxidation
Farnesol normalized oleic acid-induced changes in cytochrome P450 expression
Acknowledgements
This research was supported by grants from the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01 HL050710] and the National Institutes of Health National Institute of Environmental Health Sciences [Center Grant P30 ES020957]. Asmita Pant was supported, in part, through the Thomas C. Rumble fellowship awarded through Wayne State University. The authors thank Drs. Zofia Duniec-Dmuchowski and Hailin Fang for their technical guidance during these experiments, and thank the developers of the HepaRG cell line at the Institut National de la Santé et de la Recherche Médicale (INSERM) for providing us with the cell line.
Abbreviations
- ACAA2
acetyl-coenzyme A acyltransferase 2
- ACOT12
acyl-CoA thioesterase 12
- CAR
constitutive androstane receptor (NR1I3)
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime
- CoA
coenzyme A
- cDNA
complementary DNA
- CT
cycle threshold
- CYP
cytochrome P450
- DMSO
dimethyl sulfoxide
- DPM
disintegrations per minute
- EtOH
ethanol
- FOH
farnesol
- FF-BSA
fatty acid-free bovine serum albumin
- FPP
farnesyl pyrophosphate
- FXR
farnesoid X receptor (NR1H4)
- FBS
fetal bovine serum
- GW4064
3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid
- GW6471
N-((2S)-2-(((1Z)-1-methyl-3-oxo-3-(4-(trifluoromethyl)phenyl)prop-1-enyl)amino)-3-(4-(2-(5-methyl-2-phenyl-1,3-oxazol-4 yl)ethoxy) phenyl)propyl)propanamide
- GW7647
2-[[4-[2-[[(cyclohexylamino)carbonyl](4-cyclohexylbutyl)amino]ethyl]phenyl]thio]-2-methylpropanoic acid
- HADHA
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit
- HADHB
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NAFLD
non-alcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- OA
oleic acid
- ORO
Oil Red O
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PLIN2
perilipin 2
- PPARα
peroxisome proliferator-activated receptor α (NR1C1)
- PPARγ
peroxisome proliferator-activated receptor γ (NR1C3)
- PPRE
peroxisome proliferator response element
- PXR
pregnane X receptor (NR1I2)
- RT-qPCR
quantitative reverse transcription-polymerase chain reaction
- SHP
small heterodimer partner (NR0B2)
- SREBP
sterol regulatory element-binding protein
- TG
triglyceride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no actual or potential conflicts of interest.
References
- Ammerschlaeger M, Beigel J, Klein K-U, Mueller SO, 2004. Characterization of the Species-Specificity of Peroxisome Proliferators in Rat and Human Hepatocytes. Toxicol Sci 78, 229–240. [DOI] [PubMed] [Google Scholar]
- Anderson N, Borlak J, 2008. Molecular Mechanisms and Therapeutic Targets in Steatosis and Steatohepatitis. Pharmacol Rev 60, 311–357. [DOI] [PubMed] [Google Scholar]
- Angulo P, 2007. GI epidemiology: nonalcoholic fatty liver disease. Aliment Pharmacol Ther 25, 883–889. [DOI] [PubMed] [Google Scholar]
- Aninat C, Piton A, Glaise D, Le Charpentier T, Langouet S, Morel F, Guguen-Guillouzo C, Guillouzo A, 2006. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34, 75–83. [DOI] [PubMed] [Google Scholar]
- Anthérieu S, Rogue A, Fromenty B, Guillouzo A, Robin M-A, 2011. Induction of vesicular steatosis by amiodarone and tetracycline is associated with up-regulation of lipogenic genes in heparg cells. Hepatology 53, 1895–1905. [DOI] [PubMed] [Google Scholar]
- Bedogni G, Nobili V, Tiribelli C, 2014. Epidemiology of fatty liver: An update. World J Gastroenterol 20, 9050–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Moller DE, 2002. The Mechanisms of Action of PPARs. Annu Rev Med 53, 409–435. [DOI] [PubMed] [Google Scholar]
- Brown MV, Compton SA, Milburn MV, Lawton KA, Cheatham B, 2013. Metabolomic signatures in lipid-loaded HepaRGs reveal pathways involved in steatotic progression. Obesity 21, 10.1002/oby.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerec V, Glaise D, Garnier D, Morosan S, Turlin B, Drenou B, Gripon P, Kremsdorf D, Guguen-Guillouzo C, Corlu A, 2007. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology 45, 957–967. [DOI] [PubMed] [Google Scholar]
- Chalasani N, Gorski JC, Asghar MS, Asghar A, Foresman B, Hall SD, Crabb DW, 2003. Hepatic cytochrome P450 2E1 activity in nondiabetic patients with nonalcoholic steatohepatitis. Hepatology 37, 544–550. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ, 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866–1870. [DOI] [PubMed] [Google Scholar]
- Correll CC, Ng L, Edwards PA, 1994. Identification of farnesol as the non-sterol derivative of mevalonic acid required for the accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem 269, 17390–17393. [PubMed] [Google Scholar]
- DeBarber AE, Bleyle LA, Roullet JB, Koop DR, 2004. Omega-hydroxylation of farnesol by mammalian cytochromes p450. Biochim Biophys Acta 1682, 18–27. [DOI] [PubMed] [Google Scholar]
- Ducheix S, Montagner A, Theodorou V, Ferrier L, Guillou H, 2013. The liver X receptor: a master regulator of the gut-liver axis and a target for non alcoholic fatty liver disease. Biochem Pharmacol 86, 96–105. [DOI] [PubMed] [Google Scholar]
- Duncan RE, Archer MC, 2008. Farnesol decreases serum triglycerides in rats: identification of mechanisms including up-regulation of PPARalpha and down-regulation of fatty acid synthase in hepatocytes. Lipids 43, 619–627. [DOI] [PubMed] [Google Scholar]
- Ebisuno S, Isohashi F, Nakanishi Y, Sakamoto Y, 1988. Acetyl-CoA hydrolase: relation between activity and cholesterol metabolism in rat. Am J Physiol 255, R724–730. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J, 1999. Sterols and Isoprenoids: Signaling Molecules Derived from the Cholesterol Biosynthetic Pathway. Annu Rev Biochem 68, 157–185. [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C, 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81, 687–693. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS, 1990. Regulation of the mevalonate pathway. Nature 343, 425–430. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Shah YM, 2008. PPARα: Mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology 246, 2–8. [DOI] [PubMed] [Google Scholar]
- Goto T, Kim YI, Funakoshi K, Teraminami A, Uemura T, Hirai S, Lee JY, Makishima M, Nakata R, Inoue H, Senju H, Matsunaga M, Horio F, Takahashi N, Kawada T, 2011. Farnesol, an isoprenoid, improves metabolic abnormalities in mice via both PPARalpha-dependent and -independent pathways. Am J Physiol Endocrinol Metab 301, E1022–1032. [DOI] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C, 2007. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168, 66–73. [DOI] [PubMed] [Google Scholar]
- Hardy T, Oakley F, Anstee QM, Day CP, 2016. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu Rev Pathol. [DOI] [PubMed] [Google Scholar]
- Hiyoshi H, Yanagimachi M, Ito M, Yasuda N, Okada T, Ikuta H, Shinmyo D, Tanaka K, Kurusu N, Yoshida I, Abe S, Saeki T, Tanaka H, 2003. Squalene synthase inhibitors suppress triglyceride biosynthesis through the farnesol pathway in rat hepatocytes. J Lipid Res 44, 128–135. [DOI] [PubMed] [Google Scholar]
- Horn TL, Long L, Cwik MJ, Morrissey RL, Kapetanovic IM, McCormick DL, 2005. Modulation of hepatic and renal drug metabolizing enzyme activities in rats by subchronic administration of farnesol. Chem Biol Interact 152, 79–99. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Cohen DE, 2013. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol 48, 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa N, Kwekel JC, Burgoon LD, Dere E, Williams KJ, Tashiro C, Chittim B, Zacharewski TR, 2008. Species-specific regulation of PXR/CAR/ER-target genes in the mouse and rat liver elicited by o, p’-DDT. BMC Genomics 9, 487–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM, 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci USA 94, 4318–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocarek TA, Mercer-Haines NA, 2002. Squalestatin 1-Inducible Expression of Rat CYP2B: Evidence That an Endogenous Isoprenoid Is an Activator of the Constitutive Androstane Receptor. Mol Pharmacol 62, 1177–1186. [DOI] [PubMed] [Google Scholar]
- Koo SH, 2013. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clin Mol Hepatol 19, 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomba R, Sanyal AJ, 2013. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol 10, 686–690. [DOI] [PubMed] [Google Scholar]
- López-Velázquez JA, Carrillo-Córdova LD, Chávez-Tapia NC, Uribe M, Méndez-Sánchez N, 2012. Nuclear Receptors in Nonalcoholic Fatty Liver Disease. J Lipids 2012, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado MV, Cortez-Pinto H, 2014. Nuclear receptors: how do they position in non-alcoholic fatty liver disease treatment? Liver Int 34, 1291–1294. [DOI] [PubMed] [Google Scholar]
- Merrell MD, Cherrington NJ, 2011. Drug metabolism alterations in nonalcoholic fatty liver disease. Drug Metab Rev 43, 317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T, 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65, 55–63. [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M, 2009. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog Lipid Res 48, 1–26. [DOI] [PubMed] [Google Scholar]
- Rakhshandehroo M, Hooiveld G, Müller M, Kersten S, 2009. Comparative Analysis of Gene Regulation by the Transcription Factor PPARα between Mouse and Human. PLoS ONE 4, e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W, 1992. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 97, 493–497. [DOI] [PubMed] [Google Scholar]
- Raner GM, Muir AQ, Lowry CW, Davis BA, 2002a. Farnesol as an inhibitor and substrate for rabbit liver microsomal P450 enzymes. Biochem Biophys Res Commun 293, 1–6. [DOI] [PubMed] [Google Scholar]
- Raner GM, Muir AQ, Lowry CW, Davis BA, 2002b. Farnesol as an inhibitor and substrate for rabbit liver microsomal P450 enzymes. Biochem Biophys Res Commun 293, 1–6. [DOI] [PubMed] [Google Scholar]
- Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, Banni S, Lonardo A, Carulli N, Loria P, 2009. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol 24, 830–840. [DOI] [PubMed] [Google Scholar]
- Rogue A, Anthérieu S, Vluggens A, Umbdenstock T, Claude N, de la Moureyre-Spire C, Weaver RJ, Guillouzo A, 2014. PPAR agonists reduce steatosis in oleic acid-overloaded HepaRG cells. Toxicol Appl Pharmacol 276, 73–81. [DOI] [PubMed] [Google Scholar]
- Rondini EA, Duniec-Dmuchowski Z, Kocarek TA, 2016. Nonsterol Isoprenoids Activate Human Constitutive Androstane Receptor in an Isoform-Selective Manner in Primary Cultured Mouse Hepatocytes. Drug Metab Dispos 44, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Plummer SM, Rode A, Scheer N, Bower CC, Vogel O, Henderson CJ, Wolf CR, Elcombe CR, 2010. Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol Sci 116, 452–466. [DOI] [PubMed] [Google Scholar]
- Samanez CH, Caron S, Briand O, Dehondt H, Duplan I, Kuipers F, Hennuyer N, Clavey V, Staels B, 2012. The human hepatocyte cell lines IHH and HepaRG: models to study glucose, lipid and lipoprotein metabolism. Arch Physiol Biochem 118, 102–111. [DOI] [PubMed] [Google Scholar]
- Sambasiva Rao M, Reddy JK, 2004. PPARα in the pathogenesis of fatty liver disease. Hepatology 40, 783–786. [DOI] [PubMed] [Google Scholar]
- Schupp M, Lazar MA, 2010. Endogenous Ligands for Nuclear Receptors: Digging Deeper. The J Biol Chem 285, 40409–40415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ, 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ, 2010. The nuclear receptor PXR gene variants are associated with liver injury in nonalcoholic fatty liver disease. Pharmacogenet Genomics 20, 1–8. [DOI] [PubMed] [Google Scholar]
- Suematsu N, Isohashi F, 2006. Molecular cloning and functional expression of human cytosolic acetyl-CoA hydrolase. Acta Biochim Pol 53, 553–561. [PubMed] [Google Scholar]
- Swarbrick CMD, Roman N, Cowieson N, Patterson EI, Nanson J, Siponen MI, Berglund H, Lehtiö L, Forwood JK, 2014. Structural Basis for Regulation of the Human Acetyl-CoA Thioesterase 12 and Interactions with the Steroidogenic Acute Regulatory Protein-related Lipid Transfer (START) Domain. J Biol Chem 289, 24263–24274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailleux A, Wouters K, Staels B, 2012. Roles of PPARs in NAFLD: Potential therapeutic targets. Biochim Biophys Acta 1821, 809–818. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kawada T, Goto T, Yamamoto T, Taimatsu A, Matsui N, Kimura K, Saito M, Hosokawa M, Miyashita K, Fushiki T, 2002. Dual action of isoprenols from herbal medicines on both PPARγ and PPARα in 3T3-L1 adipocytes and HepG2 hepatocytes. FEBS Lett 514, 315–322. [DOI] [PubMed] [Google Scholar]
- Wagner M, Zollner G, Trauner M, 2011. Nuclear receptors in liver disease. Hepatology 53, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM, 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 3, 543–553. [DOI] [PubMed] [Google Scholar]
- Xiao L, Wang J, Jiang M, Xie W, Zhai Y, 2013. The emerging role of constitutive androstane receptor and its cross talk with liver X receptors and peroxisome proliferator-activated receptor A in lipid metabolism. Vitam Horm 91, 243–258. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Kakizaki S, Horiguchi N, Sohara N, Sato K, Takagi H, Mori M, Negishi M, 2007. The role of the nuclear receptor constitutive androstane receptor in the pathogenesis of non-alcoholic steatohepatitis. Gut 56, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Nagano T, Shah Y, Cheung C, Ito S, Gonzalez FJ, 2008. The PPARα-Humanized Mouse: A Model to Investigate Species Differences in Liver Toxicity Mediated by PPARα. Toxicol Sci 101, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cui Y, Wang X-L, Shang X, Qi Z-G, Xue J, Zhao X, Deng M, Xie M-L, 2015. PPARα/γ agonists and antagonists differently affect hepatic lipid metabolism, oxidative stress and inflammatory cytokine production in steatohepatitic rats. Cytokine 75, 127–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.