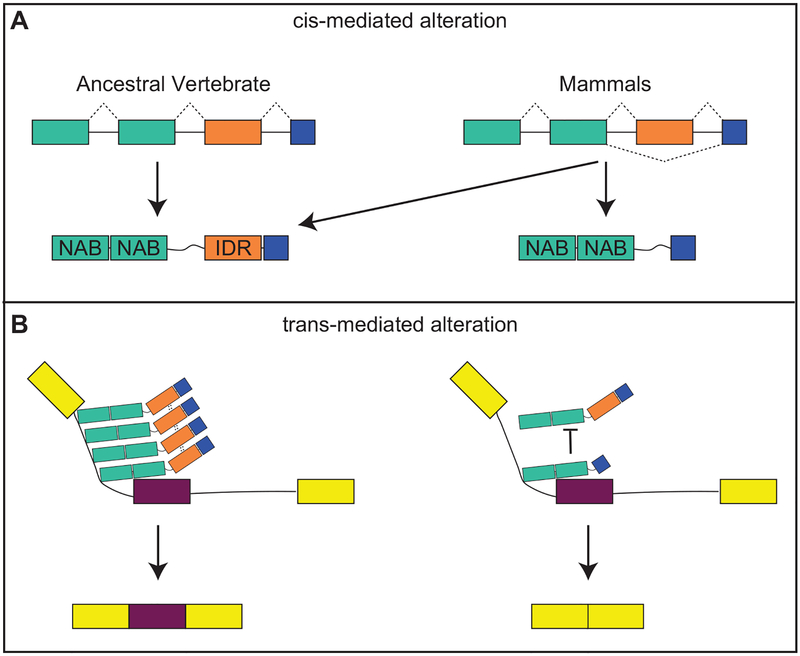
Figure 3: Mechanisms contributing to the evolution of splicing patterns.
Both A) cis-mediated and B) trans-mediated alterations contribute to the evolution of splicing patterns. A) Mutations affecting splice recognition sites contribute to increased exon skipping in mammals. Such mutations are enriched around exons containing intrinsic disordered regions (IDR) and under-represented around nuclei acid binding domains (NAB). In this example, mammals are able to produce two protein isoforms from the same mRNA, one containing and IDR and one lacking the IDR. B) IDRs contribute to protein-protein interactions. A protein complex assembled among IDR-containing RNPs may promote exon inclusion (left). In the absence of IDRs, such complexes are not formed and exon skipping occurs (right). Image modeled after Gueroussov et al. 2017.