Abstract
Membrane proteins play an important role in maintaining the structure and physiology of an organism. Despite their significance, spectroscopic studies involving membrane proteins remain challenging due to the difficulties in mimicking their native lipid bilayer environment. Membrane mimetic systems such as detergent micelles, liposomes, bicelles, nanodiscs, lipodisqs have improved the solubility and folding properties of the membrane proteins for structural studies, however, each mimetic system suffers from its own limitations. In this study, using three different lipid environments, vesicles were titrated with styrene-maleic acid (StMA) copolymer leading to a homogeneous SMALP system (~10 nm) at a weight ratio of 1:1.5 (vesicle: StMA solution). A combination of Dynamic Light Scattering (DLS) and Transmission Electron Microscopy (TEM) was used to characterize these SMALPs. We used a controlled synthesis mechanism to synthesize StMA based block copolymers called reversible addition-fragmentation chain transfer polymerization (RAFT) SMALPs. Incorporation of the Voltage sensor Domain of KCNQ1 (Q1-VSD) into RAFT SMALPs indicates that this is a promising application of this system to study membrane proteins using different biophysical techniques. V165C in Q1-VSD corresponding to the hydrophobic region was incorporated into the SMALP system. Continuous Wave-Electron Paramagnetic Resonance (CW-EPR) line shape analysis showed line shape broadening, exposing a lower rigid component and a faster component of the spin label.
Keywords: SMALPS, Continuous Wave-Electron Paramagnetic Resonance, Styrenemaleic Acid (StMa), Vesicles, Dynamic Light Scattering, Transmission Electron Microscopy (TEM)
1. Introduction
Membrane proteins make up almost 1/3 of total proteins and are responsible for various structural and functional properties of biological systems like transport of ions across membranes, receptors affecting down-stream signaling pathways and structure and assembly of the cells, thereby making them ideal drug targets1–3. However, the challenging task of mimicking the native membrane environment makes it extremely difficult to study membrane proteins using biophysical techniques.
Detergent micelles are the most common membrane mimetic system to solubilize membrane proteins for structural studies using high-resolution solution nuclear magnetic resonance (NMR) spectroscopy4. The structure of a membrane protein varies considerably from micelle to membrane bound environment (e.g. bicelles, liposomes, nanodiscs)5–9. Membrane mimetics, such as lipodisq nanoparticles and membrane scaffold protein (MSP)-stabilized nanodiscs, styrene-maleimide copolymer-lipid nanoparticles (SMILPs), and styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) have been shown to enhance the accuracy of biophysical studies compared to previous membrane mimetic systems8–14. However, there are advantages and drawbacks to each of these mimetic systems15. Recent studies on the modified version of styrene-maleic acid copolymers have shown ultra-stability of monodispersed lipid bilayer nanodics16. Although liposomes are a widely accepted membrane mimetic system, they tend to form heterogeneous aggregates. Protein incorporation into these proves to be extremely difficult and often leads to misfolded proteins17,18. The spherical geometry of liposomes also makes it difficult to examine the cytoplasmic regions of trans-membrane proteins19. A popular membrane mimetic system is called bicelles and is composed of long-chained phospholipids and short-chained phospholipids, such as 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC). Bicelles form a disc-shape that allows biophysical techniques to probe the cytoplasmic region, hydrophobic interior of the lipid bilayer and extracellular region of the transmembrane protein20–22. However, bicelles can be only formed using limited phospholipid combinations, which may not be able to accommodate a native environment for many membrane proteins19,23–24. Nanodiscs, composed of a scaffold protein wrapped around a bundle of phospholipids, have proven to be advantageous. Nanodiscs can result from several types of phospholipid combinations, while the scaffold protein stabilizes the lipids and protein of interest25,26. Nanodiscs allow for more accurate biophysical analysis of membrane proteins that require specific types of lipids when compared to the limited combination of lipids that bicelles offer26,27. However, formation of nanodiscs is a detergent-based technique, which may distort the structure of the protein. The stabilizing scaffold protein’s absorbance properties have also proven to interfere with the membrane protein to be studied9,25. Using polymers to form nanoparticles has proven to hold more advantages than scaffold protein nanodiscs15. Polymers such as styrene maleimide copolymer-lipid nanoparticles (SMILPs) and styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) can solubilize the protein of interest without the use of detergent. SMALPs and SMILPs form nearly homogenous nanoparticles and provide a native mimicking environment with lipids that are compatible with the protein of interest, while stabilizing the protein with minimal interference15,28.
Recent studies have used polymers to form membrane mimetic systems14,15,28 and commercial polymers have been used to characterize the formation of lipodisq nanoparticles using dynamic light scattering (DLS), 31P Solid State Nuclear Magnetic Resonance (SSNMR), and Transmission Electron Microscopy (TEM)29. Recently, our lab has shown that 3:1 styrene-maleic acid can be synthesized in a laboratory setting using reversible addition-fragmentation chain transfer polymerization (RAFT) polymerization and successfully used to characterize the structure of styrene-maleic acid copolymerlipid nanoparticles (SMALPs)15,30. RAFT polymerization is synthetically simple and provides flexibility to vary the structure of the polymer and influence the size of SMALPs15,30. SMALPs show compelling evidence that they are suitable for membrane protein incorporation15.
In this study, we used SMALPs synthesized from commercial styrene-maleic acid (StMA) units. StMA was titrated with 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (POPC/POPG) vesicles and characterized using Dynamic Light Scattering (DLS), and Transmission Electron Microscopy (TEM) showing that a weight ratio of 1:1.5 resulted in a homogeneous lipid nanoparticle system. This SMALP system (1:1.5) was used to incorporate the Voltage Sensor Domain (Q1-VSD) of an integral membrane protein KCNQ1. Valine 165 in the trans-membrane segment of helix 2 of Q1-VSD was mutated to cysteine (V165C) and labeled with S-(1-oxyyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) spin labeling agent to make it EPR active for the studies using CW-EPR spectroscopy. KCNQ1 is a voltage-gated potassium channel protein with six transmembrane helices (S1–S6), involved in regulating the flow of potassium ions in the heart and composed of the voltage sensor domain, Q1-VSD (S1–S4), and the pore domain (S5–S6)31,32. CW-EPR spectra revealed higher side chain mobility in liposomes when compared to SMALPs consistent with previous studies10.
2. Materials and Methods
2.1. Typical Synthesis of a One-pot Block Copolymer of Poly(styrene-alt-maleic anhydride-b-styrene)
All materials were obtained from commercial suppliers and used as received unless otherwise specified. The synthesis of poly(styrene-alt-maleic anhydride-b-styrene) was carried out as an adaptation of a previously described procedure15,33. Briefly, Styrene (3.6608 g, 35.1 mM), Maleic Anhydride (0.98 g, 10 mM), and 2-(Dodecylthiocarbonothioylthio)propionic acid (PADTC) (0.1404 g, 0.4 mM) were combined in a 20 mL vial and dissolved in 4.64 g 1,4-dioxane. 1,1-Azobis(cyclohexanecarbonitrile) (ACHN) (0.0195 g, 0.008 mM) was added and dissolved. The contents of the vial were transferred to a 50 mL round bottom flask with a magnetic stir bar and small aliquot of solution was set aside. The flask was capped with a rubber stopper and bubbled with nitrogen for 15 minutes. The solution was heated to 90 °C for 20 hours. Once comparison by NMR with the previously set aside aliquot showed sufficient conversion (~80%), the polymer was purified by 3 sequential precipitations from tetrahydrofuran (THF) into a large excess of cold hexanes, yielding 4 g of yellow powder.
2.2. End-group Removal of Poly(styrene-alt-maleic anhydride-b-styrene)
The polymer (ca. 4 g) was dissolved in dioxane and combined with 2.4 g of benzoyl peroxide (9.9 mM) in a 50 ml round bottom flask. The flask was sealed with a rubber stopper and bubbled with nitrogen for 15 minutes. The escape needle was left in the flask and the flask was heated to 82 °C for 5 hours. Upon completion, the polymer was precipitated twice from THF into a large excess of cold hexanes, yielding a white to off-white powder.
2.3. Hydrolysis of 3:1 Styrene-maleic Anhydride
The anhydride moieties of the polymer were hydrolyzed using NaOH to their succinic acid counterparts as previously described15 with slight modifications. Briefly, equal mass of polymer and THF (1 g) were combined in a vial, and briefly heated to 95 °C while swirling to dissolve all the polymer. Then, a 4x molar excess of aqueous 2 M NaOH was added dropwise while stirring, and the resulting mixture was heated at 65 °C for 24 hours. Another 10 mL of distilled water was added, and the mixture was heated for another 24 hours. At the end of this period, the solution is stable at room temperature. The THF and excess base were removed by dialysis in 3.5 kDa cutoff tubing, using two 1 L portions of ultrapure water. The polymer was collected in a 50 mL conical tube and frozen in a −80 °C refrigerator overnight, then lyophilized, yielding a white to off-white powder. The NMR data on the hydrolyzed polymer is shown in the supporting information in Figure S1. The NMR peaks corresponding to styrene aromatic protons are observed in the 6.0 – 7.5 ppm region of the spectrum. To the best of our knowledge, precise NMR characterization of these copolymers has not been done. This is mainly due to the significant broadening observed when the maleic anhydride protons are incorporated into the polymer backbone. Hence a meaningful resolution of these protons cannot be obtained. These RAFT-synthesized copolymers are characterized by a much steeper gradient, as opposed to block copolymers.
2.4. Size Exclusion Chromatography (SEC) Procedure
5 mg of polymer was dissolved in 1.5 mL THF with 0.025% butylated hydroxy toluene (BHT). Two drops of toluene were added as a flow rate marker. The solution was then filtered through a 0.22 μm filter. Size exclusion chromatography was performed using an Agilent 1260 gel permeation chromatography system coupled with an autosampler, a guard and 2× PL Gel Mixed B columns, and a refractive index detector. The eluent was tetrahydrofuran running at 1 mL/min at 25 °C. The system was calibrated with poly(methyl methacrylate) standards in the range of 617,000 to 1010 and corrected to polystyrene using the standard Mark-Houwink parameters KMMA = 12.8, αMMA = 0.69, KSty = 11.4, αSty = 0.716. The Mn by GPC, after applying the Mark-Houwink correction for polystyrene was 7.0kDa, Mw of 9.8kDa, with a polydispersity of 1.40. The molecular weight distribution data on the polymer are also included in the supporting information in Figure S2.
2.5. Vesicle Preparation and Formation of SMALPs
To characterize the formation of homogeneous SMALPs, two different lipids commonly used for studying membrane proteins were used. One vesicle sample was composed of POPC, another one was composed of a molar ratio of 3:1 (POPC:POPG) and the third vesicle sample was composed of a molar ratio of 9:1 (POPC:POPG). In each of the samples, powdered lipids were dissolved in a 2-(4-(2-Hydroxyethyl)piperazin-1-yl)ethanesulfonic acid (HEPES) buffer (20 mM HEPES, 100 mM NaCl, pH 7.0) and brought to a final concentration of 25 mM. The mixture was then vortexed vigorously for one minute and followed by at least 15 freeze/sonication cycles (<30°C) to obtain a homogeneous milky solution. The vesicle solutions were then frozen with liquid nitrogen and placed in a freezer overnight (−20 °C). Dynamic light scattering was used to confirm and analyze the size of vesicles the next day.
Styrene-maleic acid was dissolved in a buffer (20mM HEPES, 100mM NaCl pH7), brought to a final concentration of 2.5% (m/v) and sonicated at 30–40 °C until the solution went clear. The vesicles (POPC, POPC/POPG) were titrated with StMA polymer by adding StMA solution dropwise obtaining the weight ratios of 1:0.5, 1:1, 1:1.25 and 1:1.5 (vesicles:StMA). The newly formed SMALP solution was mixed via slight rotation at room temperature overnight.
2.6. Preparation of Q1-VSD for CW-EPR Spectroscopic Study
The His-tag expression vector (pET-16b) containing wild type Q1-VSD (residues 100–249) was expressed and purified using Rosetta/C43(DE3) strain of E.coli as previously described4. Briefly, site-directed mutagenesis was performed to remove all the native cysteines and generate single cysteine mutants from the cys-less vector using the QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene). The mutation was confirmed by DNA sequencing and was transformed into Rosetta/C43(DE3) E.coli cells for protein overexpression.
The overexpression and purification of Rosetta/C43(DE3) E. coli cells carrying mutated Q1-VSD genes were carried out using a previously described protocol4. E. coli cells carrying mutant (V165C) was grown in an M9 minimal medium with 100 μg/mL ampicillin and 50 μg/ml chloramphenicol. The cell culture was incubated at 25 °C and 240 rpm supplemented with MEM vitamin (Mediatech) and ZnCl2 (50 μM) until the OD600 reached 0.8, at which point protein expression was induced using 1 mM isopropyl-1-thio--D-galactopyranoside (IPTG), followed by continued rotary shaking at 25°C for 24 h. Purification of Q1-VSD from inclusion bodies was carried out according to a previously described method4 and eluted using 0.1% 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) (LMPG) detergent. Protein samples were concentrated using a Microcon YM-3 (molecular weight cutoff, 3,000) filter (Amicon). The protein concentration was measured with a nanodrop and purity was confirmed by SDS-PAGE. Spin labeling, and liposomes reconstitution was carried out following a similar protocol previously described34. After purification, cysteine mutant was concentrated to 0.5 mM. Sample was then reduced with 2.5 mM Dithiothreitol (DTT), with gentle agitation at room temperature for 24 hours to ensure complete conversion to Cys-SH. MTSL spin label was added to 10 mM from a 250 mM solution in methanol into 0.5 mM Q1-VSD solution, which was then equilibrated at room temperature for 30 minutes, followed by incubation at 37 °C for 3 hours and further incubated overnight at room temperature. Sample was then buffer-exchanged into a 50 mM phosphate, 0.1% LMPG, pH 7.8. Following buffer exchange, samples were bound to Nickel resin in a column, which was then washed with 200 mL of 50 mM phosphate, 0.05% n-dodecylphosphocholine (DPC), pH 7.8 to remove excess MTSL. The spin labeled Q1-VSD was eluted using elusion buffer containing 0.5% DPC. The reconstitution of spin labeled protein into POPC/POPG (3:1) proteoliposomes was carried out via dialysis methods following a similar protocol in the literature14. The concentrated spin labeled Q1-VSD protein was mixed with stock lipid slurry (400 mM SDS, 75 mM POPC and 25 mM POPG, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 100mM imidazole (IMD), pH 7.5). The lipid slurry was pre-equilibrated to clear mixed micelles via several freeze thaw cycles. The final protein:lipid molar ratio was set to 1:250. The Q1-VSD-lipid mixture was then subjected to extensive dialysis to remove all detergent present and help form Q1-VSD/POPC/POPG vesicles.
The protein-lipid complex was incorporated into SMA-polymer following previously published protocols11,14. A 500 μl aliquot of proteoliposome-reconstituted protein sample (~30 mM POPC/POPG lipid) was added with the 2.5% of StMA polymer solution prepared in the same dialysis buffer drop-wise over 3–4 minutes at a weight ratio 1:1.5. The protein-StMA-polymer solution was equilibrated overnight at 4 °C. The resulting solution was centrifuged at 40,000xg for 30 minutes to remove the nonsolubilized protein. The supernatant was further concentrated to desired volume and concentration for CW-EPR spectroscopic measurements.
2.7. Dynamic Light Scattering (DLS) Measurements
DLS measurements were performed on a ZETASIZER NANO Series (Malvern Instruments) at 25 °C in disposable 40 μL micro cuvettes. Data were collected for 20 s and averaged for 10 scans. The size distribution in radius is shown on a log scale using Igor Pro (WaveMetrics).
2.8. CW-EPR Measurements
EPR experiments were conducted at the Ohio Advanced EPR Laboratory. CW-EPR spectra were collected at X-band on a Bruker EMX CW-EPR spectrometer using an ER041xG microwave bridge and ER4119-HS cavity coupled with a BVT 3000 nitrogen gas temperature controller. Each spin-labeled CW-EPR spectrum was acquired by signal averaging 20 42-s field scans with a central field of 3315 G and sweep width of 100 G, modulation frequency of 100 kHz, modulation amplitude of 1 G, and microwave power of 10 mW at room temperature.
2.9. Transmission Electron Microscopy
One drop of control POPC/POPG vesicles and StMA-lipid nanoparticle (1/1.25) samples out in a solution of 100 mM NaCl, 20 mM HEPES pH 7.0 were adsorbed to 200 mesh copper carbon-coated grids for 10 s for full absorbance. The grids were stained with two drops of 1.5% ammonium molybdate. Images were recorded using JEOL-1200EX.
3. Results and Discussion
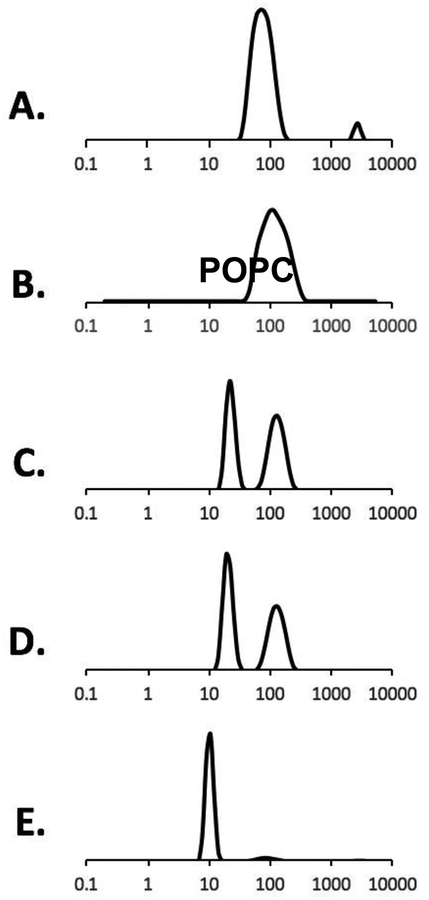
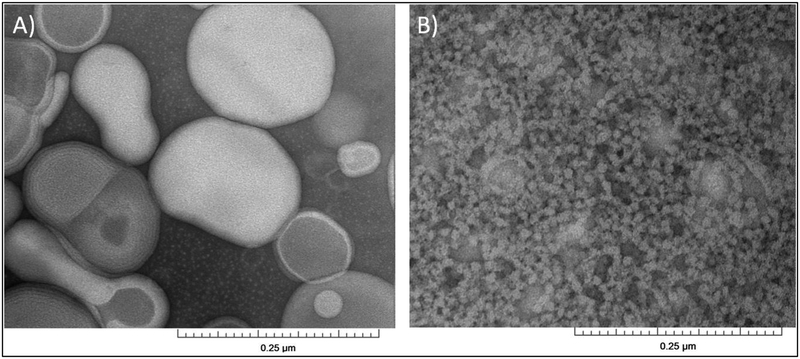
Figure 1 shows the DLS data of POPC vesicles and the addition of 2.5% (m/v) RAFT StMA polymer solution. The varying ratios of StMA can be used to control the overall particle size as shown by previously published data15 and was found to be consistent with this study. Figure 1A displays the DLS data for POPC vesicles indicating a unimodal peak with a wide size distribution representing heterogeneity in the sample. The heterogeneity of POPC vesicles was confirmed using TEM (Figure 2A). When these vesicles were titrated with a 2.5% StMA solution at a weight ratio of 1:0.5 (POPC:StMA), DLS data showed more heterogeneity as indicated by the wider size distribution as presented in Figure 1B. The addition of StMA possibly causes aggregation of the vesicle sample at this ratio. The sample at this weight ratio appeared to be mostly transparent, but some lipids remained visually undissolved. A ratio of 1:1 (POPC:StMA) shows a bimodal graph indicating two separate components in the sample as shown in Figure 1C. One component has a smaller size distribution of particles with the peak centered at 21 nm, while the other larger component showed a wider size distribution corresponding to an average particle size of 127 nm. The smaller size distribution of the first component is indicative of SMALP formation, while the larger size distribution in the second component indicates POPC vesicles in solution that have not solubilized with StMA. This mixture showed to be primarily transparent due to the solubilization of lipids by StMA. Figure 1D represents the 1:1.25 (POPC:StMA solution) ratio that shows a similar bimodal graph as Figure 1C. However, the first component with a smaller size distribution of particles in solution is depressed and slightly decreased with an average particle size of 18 nm, while the component with a larger size distribution of particles shows a decrease in the intensity, indicative of a decrease in the heterogeneity of the vesicles. The solution became visually transparent due to the solubilization of lipids. Finally, when the weight ratio of POPC to StMA solution was increased to 1:1.5, as shown in Figure 1E, DLS data showed one dominant peak with an average size distribution of 10 nm. The solution was also visually transparent at this stage. The homogeneity of the sample was confirmed using TEM as shown in Figure 2B15,29. This result is consistent with the previously published data15.
Figure 1.
DLS data showing volume graphs for 2.5 % (m/v) StMA polymer and POPC vesicles. (A) POPC vesicles (control), (B) SMALPs generated from 1:0.5 POPC:StMA, (C) SMALPs generated from 1:1 POPC:StMA, (D) SMALPs generated from1:1.25 POPC:StMA, (E) SMALPs generated from1:1.5 POPC:StMA.
Figure 2.
TEM micrographs of (A) POPC vesicles (control), and (B) SMALPs generated from 1:1.5 vesicles:StMA.
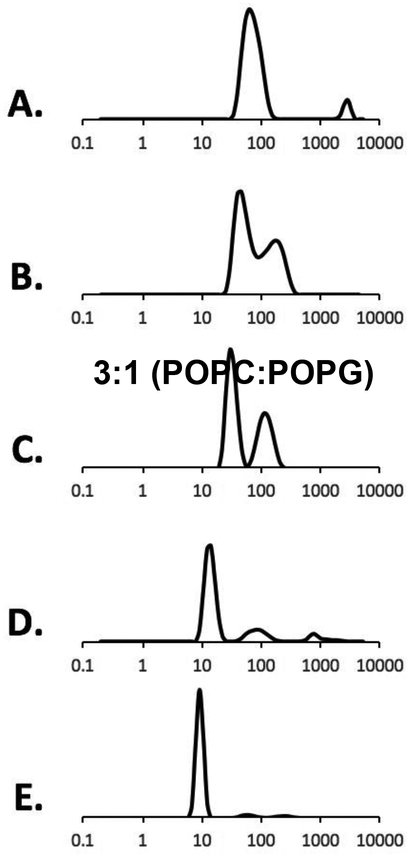
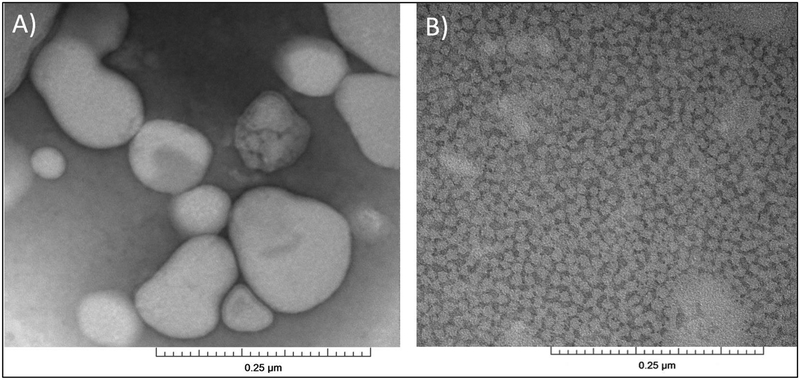
Figure 3 shows the DLS data of 3:1 (POPC:POPG) titrated with the same 2.5% (m/v) StMA solution. Similar heterogeneity (reported above) was observed for 3:1 (POPC:POPG) vesicle sample as shown in Figure 3A with a relatively large size and the finding was confirmed using TEM15,29 (Figure 4A). Vesicles were first titrated with the StMA at weight ratio of 1:0.5 (Figure 3B), resulting in a bimodal graph by DLS and a partially clear solution with some visible aggregation was still present. Although it showed a wide size distribution, it resulted in a 2-component peak. The more intense peak indicated SMALP formation due to the lower size distribution, while the less-intense peak replicates the unsolubilized vesicles shown in Figure 3A. Figure 3C reveals a weight ratio (vesicles:StMA) of 1:1 and showed improvements from the previous ratio of 1:0.5 with respect to the homogeneity and the clarity of the sample. At the weight ratio of 1:1, the solution became visually transparent. The data indicate two components; one with a relatively low size distribution of particles with the peak centered at 30 nm indicating that the StMA is beginning to solubilize the vesicles to form SMALPs; the component with a larger size distribution of particles similar to vesicles (Figure 3A) remained in solution and indicates some nonsolubilized vesicles. As the 3:1 (POPC:POPG) vesicles were further titrated to a weight ratio of 1:1.25 (Figure 3D), StMA solubilizes the majority of the vesicles present as shown in Figure 3C and showed a visually transparent solution. The component with a smaller size distribution of the particles also decreased with an average particle size distribution of 30 nm. It can be inferred that as more polymer is added to the vesicles, the SMALP formation caused reduction; both in size itself and the size distribution and agrees with previous literature studies15,29. This is also observed as the vesicles are further titrated with 1:1.5 (vesicles:StMA solution) as shown in Figure 3E. At this ratio, the entire solution was clear and has fully solubilized the vesicles to form SMALPs with a decreased size distribution as indicated by the peak centered at 10 nm. The homogeneity of the SMALPs is further confirmed using TEM (Figure 4B).
Figure 3.
DLS data showing volume graphs for 2.5 % (m/v) StMA polymer and 3:1 (POPC:POPG) vesicles. (A) POPC:POPG, 3:1 vesicles (control), (B) SMALPs generated from1:0.5 vesicles:StMA, (C) SMALPs generated from 1:1 vesicles:StMA, (D) SMALPs generated from1:1.25 vesicles:StMA, (E) SMALPs generated from1:1.5 vesicles:StMA.
Figure 4.
TEM micrographs of (A) POPC:POPG (3:1) vesicles (control), and (B) SMALPs generated from 1:1.5 vesicles:StMA.
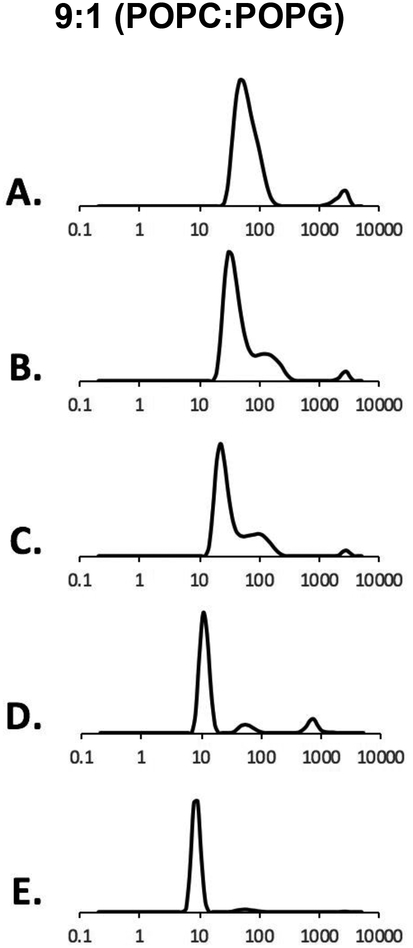
Figure 5 shows the DLS data of 9:1 (POPC:POPG) vesicles. Vesicles were synthesized with a molar ratio of 9:1 (POPC:POPG) and titrated with 2.5% (m/v) StMA in buffer (20mM HEPES, 100mM NaCl pH7). Figure 5A presents the DLS data for 9:1 (POPC:POPG) vesicles. The vesicles show consistent data by providing a heterogeneous solution which is further confirmed using TEM as shown in Figure 6A. As StMA is further titrated with the 9:1 (POPC:POPG) vesicles to a 1:0.5 weight ratio (vesicles:StMA) as displayed in Figure 5B, a significant amount of StMA solubilizes the vesicles, but shows a small amount of aggregation. This is reflected in the DLS data as the dominant peak in the bimodal graph is further depressed and narrows down, which is a consistent trend of SMALP formation15. However, a portion of the solution showed a small peak indicative of the unsolubilized vesicles at a larger distance. Figure 5C shows the titration data of 1:1 (vesicles:StMA) with a slight improvement from the previous titration ratio of 1:0.5 and showed to be nonsolubilized. Although DLS generated a similar graph to that of Figure 5B, the dominant peak showed a downward shift as well as a decrease in size distribution, indicative that StMA is still solubilized with the vesicles to form a homogeneous SMALP system. As more StMA is added to a ratio of 1:1.25 (vesicles:StMA) as shown in Figure 5D, a majority of vesicles are solubilized by StMA to form homogeneous SMALPs as shown by the dominant peak with a size distribution of 10 nm, only a small portion of vesicles have not solubilized with StMA even though the sample remains visually transparent. Finally, a weight ratio of 1:1.5 (vesicles:StMA) was also completely clear and found to give the best data with the average size distribution of ~9 nm for completely homogeneous SMALPs and is presented in Figure 5E. The finding and size distribution were further confirmed using TEM as shown in Figure 6B.
Figure 5.
DLS data showing volume graphs for 2.5 % (m/v) StMA polymer and 9:1 (POPC:POPG) vesicles. (A) POPC:POPG 9:1 vesicles (control), (B) SMALPs generated from1:0.5 vesicles:StMA, (C) SMALPs generated from 1:1 vesicles:StMA, (D) SMALPs generated from1:1.25 vesicles:StMA, (E) SMALPs generated from 1:1.5 vesicles:StMA.
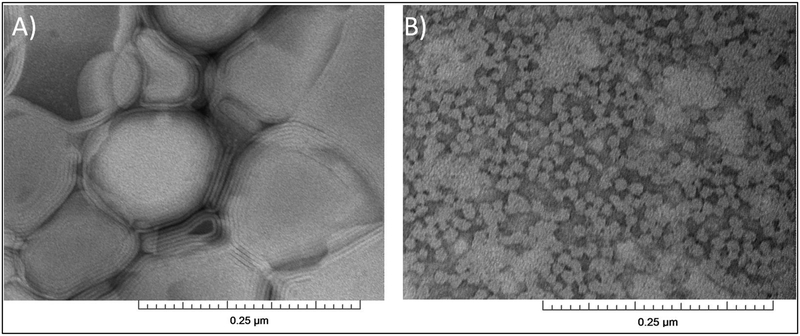
Figure 6.
TEM micrographs of (A) POPC:POPG (9:1) vesicles (control), and (B) SMALPs generated from 1:1.5 vesicles:StMA.
As stated above, TEM was used to further confirm the data obtained from DLS experiments as DLS tends to be biased towards larger particle sizes15,29. Our TEM data is in good agreement with the DLS data obtained for different ratios of vesicles to StMA and clearly shows that at 2.5% StMA and a ratio of 1:1.5 (vesicles:StMA) we were able to obtain homogeneous SMALPs. Using the three titrations presented Figures 1, 3, and 5, SMALP systems were successfully characterized in different lipid environments at a critical weight ratio of 1:1.5. Table 1 presents the size distributions for each of the three titrations presented above. The data clearly indicates that the formation of SMALPs clearly works for a variety of phospholipids. Further investigation of StMA in the presence of other lipids will progress the study of membrane proteins in different lipid environments. For all the liposome samples studied the addition of more StMA (upto 1:2) did not significantly reduce the size of the SMALPs.
Table 1.
Average particle size distribution of SMALPs resulting from different ratios of vesicles:StMA
| Particle Size (nm) | |||||
|---|---|---|---|---|---|
| Name |
1:0 (Control vesicles) |
1:0.5 | 1:1 | 1:1.25 | 1:1.5 |
| POPC | 90 | 120 | 21 | 18 | 10 |
| 3:1 (POPC:POPG) | 60 | 45 | 30 | 15 | 10 |
| 9:1 (POPC:POPG) | 55 | 30 | 21 | 10 | 9 |
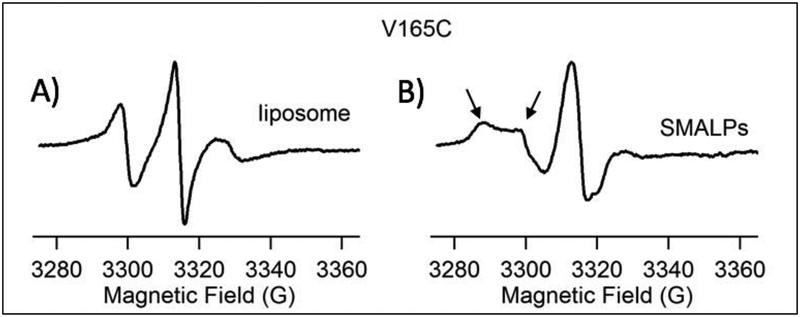
Successful characterization of SMALPs in a 3:1 (POPC:POPG) environment was tested with the integral membrane protein Q1-VSD. Q1-VSD is composed of four trans-membrane helices4. To test the compatibility of the SMALP system and membrane proteins, residue V165C was spin-labeled and incorporated into 3:1 (POPC:POPG) vesicles and StMa solution was added until a weight ratio of the protein sample to StMA solution was 1:1.5. For all the liposome samples studied, the addition of more StMA (up to 1:2) did not significantly reduce the size the SMALPs. V165C is an amino acid mutant embedded in the second helix within the lipid bilayer of Q1-VSD. Spin labeled residue in a SMALP system versus spin labeled residue in a 3:1 vesicle environment (liposome) were compared using CW-EPR line shape analysis as displayed in Figure 7. The spin-labeled Q1-VSD V165C in the SMALP system showed line shape broadening, revealing two motional components signified by the two arrows. The left arrow shows the slow and more rigid component, while the right arrow points to the faster motional component of the spin label. These two components, however, are not present in the liposome. This data is consistent with previous studies of characterizing the membrane protein KCNE1 in the presence of lipodisq nanoparticles10 clearly suggesting that the SMALP system is compatible with membrane proteins and allows for the analysis of membrane proteins using biophysical technique, such as CW-EPR spectroscopy.
Figure 7.
CW-EPR data on V165C-Q1-VSD mutant in (A) POPC:POPG (3:1) liposomes as control, and (B) SMALPs formed from 1:1.5 (vesicles:StMA), Left arrow shows the slower, rigid limit component of the spin label while the right arrow indicates a faster component.
4. Conclusion
Knowledge and research on SMALP systems is exponentially growing10,15,29. In this study, SMALPs were characterized by titrating different ratios of POPC:POPG with the RAFT synthesized StMA. The resulting homogeneous SMALP solution was found to have an average size of 10 nm. This newly synthesized RAFT SMALP system was used as a new membrane mimetic system to show successful incorporation of Q1-VSD membrane protein and analyzed using CW-EPR spectroscopy. Overall, our data suggests a promising future for this membrane mimetic system with a great potential for studies using biophysical techniques. RAFT SMALPs offer great potential in being synthetically simple, and economical alternative to other current membrane mimetic systems. It provides flexibility with the structure of the polymer and therefore better control on the size of SMALPs15.
Highlights:
The size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) were characterized using Dynamic Light Scattering.
EPR spectra showed the incorporation of a membrane protein into the nanoparticles and liposomes.
TEM images show the size and distribution of the SMALPs.
Acknowledgements
This work was generously supported by the NIGMS/NIH Maximizing Investigator’s Research Award (MIRA) R35 GM126935 award and a NSF CHE-1807131 grant. The pulsed EPR spectrometer was purchased through funding provided by the NSF (MRI-1725502), the Ohio Board of Reagents, and Miami University. Gary A. Lorigan would also like to acknowledge support from the John W. Steube Professorship.
Funding Sources
National Institutes of Health Grant R35 GM126935
National Science Foundation Grant CHE-1807131
National Science Foundation Grant MRI-1725502
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest
References
- 1.Hang Y, and Flynn AD, 2016. Drugging membrane protein interactions. Annu Rev Biomed 18, 51–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull SC, and Doig AJ, 2015. Properties of protein drug target classes. PLoS One, 10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overington JP, Al-Lazikani B, and Hopkins AL, 2006. How many drug targets are there? Nat Rev Drug Discovery 5, 993–996. [DOI] [PubMed] [Google Scholar]
- 4.Peng D, Kim JH, Kroncke BM, Law CL, Xia Y, Droege KD, Van Horn WD, Vanoye CG and Sanders CR 2014. Purification and structural study of the voltage-sensor domain of the human KCNQ1 potassium ion channel. Biochemistry 53, 2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coey AT, Sahu ID, Gunasekera TS, Troxel KR, Hawn JM, Wickenheiser MR, Welch RC, Vanoye CG, Kang C, Sanders CR, and Lorigan GA, 2011. Reconstitution of KCNE1 into Lipid Bilayers: Structural and Dynamic Differences in Micelle and Vesicle Environments. Biochemistry 50, 10851–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu L, Wang W, Ling S, Liu S, Xiao L, Xin Y, Lai C, Xiong Y, Zhang L, and Tian C, 2015. CW-EPR studies revealed different motional properties and oligomeric states of the integrin β1a transmembrane domain in detergent micelles or liposomes. Scientific Reports 5, 7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seddon AM, Curnow P, and Booth PJ, 2004. Membrane proteins, lipids, and detergents: not just a soap opera. Biochim Biophys Acta 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- 8.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, and Opella SJ, 2004. High resolution NMR spectroscopy of membrane proteins in aligned bicelles. J Am Chem Soc 126, 15340–1341. [DOI] [PubMed] [Google Scholar]
- 9.Bayburt TH, and Sligar SG, 2003. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci 12, 2476–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu ID, Zhang R, Dungan MM, Craig AF, Lorigan GA, 2017. Characterization of KCNE1 inside lipodisq nanoparticles for EPR spectroscopic studies of membrane proteins. J Phys Chem, 121, 5312–5321. [DOI] [PubMed] [Google Scholar]
- 11.Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Grobner G, and Watts A, 2012. Detergent free formation and physicochemical characterization of nanosized lipids-polymer complexes: lipodisq. Angew Chem, Int Ed. 51, 4653–4657. [DOI] [PubMed] [Google Scholar]
- 12.Park SH, Opella SJ, 2012. TritonX-100 as the “short-chain lipid” improves the magnetic alignment and stability of membrane proteins in phosphatidylcholine bilayers for oriented-sample solid-state NMR spectroscopy. J. Am. Chem. Soc 132 (36) 12552–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Angelis AA, Opella SJ, 2007. Bicelle samples for solid-state NMR of membrane proteins. Nat. Protoc 2 (10), 2332–2338. [DOI] [PubMed] [Google Scholar]
- 14.Sahu ID, McCarrick RM, Troxel KR, Zhang R, Smith HJ, Dunagan MM, Swartz MS, Rajan PV, Kroncke BM, Sanders CR, Lorigan GA, 2013. DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles. Biochemistry 52 (38), 6627–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig AF, Clark EE, Sahu ID, Zhang R, Frantz ND, Al-Abdul-Wahid S, Dabney-Smith C, Konkolewicz D, Lorigan GA, 2016. Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim. Biophys. Acta 1858, 2931–2939. [DOI] [PubMed] [Google Scholar]
- 16.Ravula T, Hardin NZ, Ramadugu SK, Cox SJ, Ramamoorthy A, 2018. Formation of pH-Resistant Monodispersed Polymer-Lipid Nanodiscs. Angew Chem, Int Ed. 57 (5), 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 17.Geertsma ER, Mahmood NABN, Schuurman-Wolters GK, and Poolman B, 2008. Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc 3 (2), 256–266. [DOI] [PubMed] [Google Scholar]
- 18.Fang G, Friesen R, Lanfermeijer F, Hagting A, Poolman B, Konings WN, 1999. Manipulation of activity and orientation of membrane-reconstituted di-tripeptide transport protein DtpT of Lactococcus lactis. Mol. Membr. Biol 16 (4), 297–304. [DOI] [PubMed] [Google Scholar]
- 19.Raschle T, Hiller S, Etzkorn M, Wagner G, 2010. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr. Opin. Struct. Biol 20 (4), 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau T-L, Kim C, Ginsberg MH, Ulmer TS, 2009. The structure of the integrin alpha IIb beta 3 transmembrane complex explains integrin transmembrane signaling. EMBO J. 28 (9), 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D, Walter KFA, Brueckner A-K, Hilty C, Becker S, Griesinger C, 2008. Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J. Am. Chem. Soc 130 (42), 13822–13823. [DOI] [PubMed] [Google Scholar]
- 22.Sanders CR, Prosser RS, 1998. Bicelles: a model membrane system for all seasons? Structure 6 (10), 1227–1234. [DOI] [PubMed] [Google Scholar]
- 23.Vold RR, Prosser RS, 1996. Magnetically oriented phospholipid bilayered micelles for structural studies of polypeptides. Does the ideal bicelle exist? J. Magn. Reson. Ser. B 113 (3), 267–271. [Google Scholar]
- 24.Duerr UHN, Gildenberg M, Ramamoorthy A, 2012. The magic of bicelles lights up membrane protein structure. Chem. Rev 112 (11), 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayburt TH, Sligar SG, 2010. Membrane protein assembly into nanodiscs. FEBS Lett. 584 (9), 1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borch J, Hamann T, 2009. The nanodisc: a novel tool for membrane protein studies. Biol. Chem 390 (8), 805–814. [DOI] [PubMed] [Google Scholar]
- 27.Hagn F, Etzkorn M, Raschle T, Wagner G, 2013. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc 135 (5), 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall SCL, Tognoloni C, Charlton J, Bragginton ÉC, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Elder KJ, Dafforn TA, 2018. An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles. Nanoscale (10), 10609–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Sahu ID, Liu L, Osatuke A, Comer RG, Dabney-Smith C, Lorigan GA, 2015. Characterizing the structure of lipodisq nanoparticles for membrane spectroscopic studies. Biochem. Biophys. Acta 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AAA, Autzen HE, Laursen T, Wu V, Yen M, Hall A, Hansen SD, Cheng Y, and Xu T, 2017. Controlling styrene maleic acid lipid particles through RAFT. Biomacromolecules, 18, 3706–3713. [DOI] [PubMed] [Google Scholar]
- 31.Jespersen T, Grunnet M, Olesen SP, 2005. The KCNQ1 potassium channel: From gene to physiological function. Physiology, 20, 408–416. [DOI] [PubMed] [Google Scholar]
- 32.Sun J, and MacKinnon R, 2017. Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell 169, 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bapat AP, Ray JG, Savin DA, Hoff EA, Patton DL, Sumerlin BS, 2012. Dynamic-covalent nanostructures prepared by Diels-Alder reactions of styrene-maleic anhydride-derived copolymers obtained by one-step cascade block copolymerization. Polym. Chem-Uk 3 (11), 3112–3120. [Google Scholar]
- 34.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR, 2012. The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol. Science, 336 (6085) [DOI] [PMC free article] [PubMed] [Google Scholar]