Abstract
Greenberg skeletal dysplasia is an autosomal recessive, perinatal lethal disorder associated with biallelic variants affecting the lamin B receptor (LBR) gene. LBR is also associated with the autosomal recessive anadysplasia-like spondylometaphyseal dysplasia, and the autosomal dominant Pelger–Huët anomaly, a benign laminopathy characterized by anomalies in the nuclear shape of blood granulocytes. The lamin B receptor is an inner nuclear membrane protein that binds lamin B proteins (LMNB1 and LMNB2), interacts with chromatin, and exerts a sterol reductase activity. Here, we report on a novel LBR missense mutation [c.1379A>G; p.(D460R)], identified by whole exome sequencing and causing Greenberg dysplasia in two fetuses from a consanguineous Moroccan family. We revised published LBR variants to propose a genotype-phenotype correlation in LBR associated diseases. The diverse phenotypes are correlated to the functional domain affected, the heterozygous or homozygous state of the variants, and their different impact on the residual protein function. LBR represents an instructive example of one gene presenting with two different pattern of inheritance and at least three different clinical phenotypes.
Keywords: whole exome sequencing, Greenberg syndrome, fetal malformations, LBR, Pelger-Huët
INTRODUCTION
Greenberg skeletal dysplasia (GRBRD, MIM#215140) also known as Hydrops, Ectopic calcification and Moth-eaten (HEM) skeletal dysplasia, is an autosomal recessive, perinatal lethal dwarfism disorder. It belongs to the multiple congenital anomalies syndromes caused by defects in sterol metabolism, a group that includes Smith-Lemli-Opitz, Conradi-Hunermann, Congenital Hemidysplasia with Icthyosiform Nevus and Limb Defects (CHILD) syndromes, and desmosterolosis [Porter and Herman 2011]. Greenberg dysplasia is associated with biallelic variants affecting the lamin B receptor (LBR) gene [Clayton and others 2010].
LBR encodes one of the most important inner nuclear membrane proteins that binds to chromatin and lamin B proteins (LMNB1 and LMNB2). Two apparently distinct functions have been attributed to the LBR protein: the nucleoplasmatic domain has a structural function, tethers heterochromatin to the nuclear periphery, and contributes in maintaining chromatin organization and interphase nuclear shape. The transmembrane domains exhibit an enzymatic sterol reductase activity, a crucial function of the cholesterol synthesis metabolic pathway [Clayton and others 2010; Nikolakaki and others 2017], and a key activity for the viability of different human cell lines (HeLa, HEK293T and HFF) [Tsai and others 2016].
In addition to Greenberg dysplasia, homozygous or compound heterozygous LBR pathogenic variants can result in anadysplasia-like spondylometaphyseal dysplasia, a mild, spontaneously regressing bone dysplasia also known as Pelger-Huët Anomaly with mild SKeletal anomalies (PHASK, MIM# 618019) [Borovik and others 2013; Hoffmann and others 2002; Sobreira and others 2015a].
Pathogenic variants associated with GRBGD or PHASK result in a deficiency of sterol reductase activity and in elevated levels of sterol intermediates [Tsai and others 2016]. Heterozygous LBR pathogenic variants impairing its structural function, on the other hand, are associated with Pelger–Huët Anomaly (PHA, MIM#169400), a benign laminopathy characterized by abnormalities in both nuclear shape (hypolobulated nuclei) and chromatin organization of blood granulocytes [Hoffmann and others 2002]. Therefore, it was suggested that the structural role of LBR in the inner nuclear membrane was distinct from its enzymatic activity [Clayton and others 2010; Tsai and others 2016; Turner and Schlieker 2016].
Additionally, an LBR missense variant with a possible dominant-negative and cell-specific effect has been suggested in a subject with Reynolds syndrome (MIM#613471), a disease characterized by scleroderma, liver anomalies, telangiectasia, and Raynaud phenomenon [Gaudy-Marqueste and others 2010].
Two independent mouse strains carrying heterozygous loss-of function variants in the Lbr gene recapitulate the Pelger-Huët anomaly blood phenotype [Green and others 1975; Hoffmann and others 2002]. Moreover, mice homozygous for deleterious Lbr variants show a PHA-like blood phenotype and develop other abnormalities, including alopecia, variable expression of syndactyly, and hydrocephalus [Shultz and others 2003]. Finally a homozygous knock-out mouse model carrying a frameshift insertion in the Lbr gene and resulting in a C-terminally truncated protein, exhibits embryonic lethality with incomplete penetrance, shortened postnatal life span, hydrocephaly, and syndactyly, as well as chromatin atypia in neutrophils [Cohen and others 2008].
Here, we report on a novel homozygous Greenberg skeletal dysplasia-associated missense change [c.1379A>G; p.(D460R)] identified in two fetuses fulfilling GRBRD clinical diagnostic criteria. Starting from this variant, we re-analyzed published LBR variants and their related phenotypes, to speculate on the genotype-phenotype correlation of LBR-associated diseases.
MATERALS AND METHODS
Genetic analyses
The study was performed with the approval of the Internal Review Board, and informed consents were obtained by patients’ legal representatives. Genetic analyses were performed on DNA extracted from cartilage sample of aborted fetus (Figure 1A, II-1 and II-4) or from blood (Figure 1A, I-1 and I-2 and II-5).
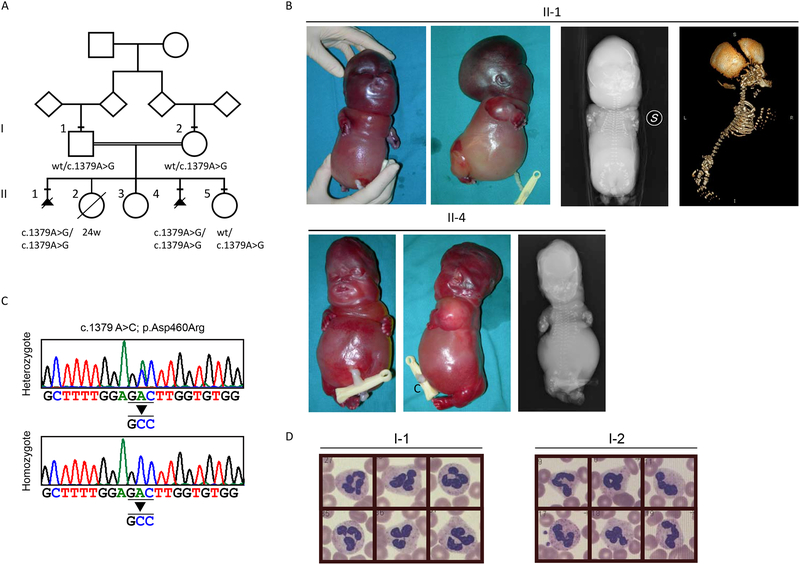
Figure 1.
Genetics and phenotype. A) Pedigree of the family. Squares indicate males, circles indicate females, and a rhombus indicate that the gender is unknown. Black triangles indicate a termination of pregnancy. Subjects whose DNA was available are marked with a bar above the symbol. The variants identified are reported below each tested subject. B) Fetus II-1 external examination (upper left panels) shows severe hydrops fetalis micromelia, dysmorfic features and ambiguous genitalia. Radiological exams (upper right panels) shows the skeletal dysplasia deficient ossification and hypoplastic and skewed bones in X-rays and 3D-CT exams. Fetus II-4 external examination (lower left panels) shows severe hydrops fetalis micromelia and dysmorfic features. The X-ray exam (lower right panel) shows skeletal dysplasia (short ribs, platyspondyly, skewed long bones). C) Sequence electropherograms showing the c.1379A>C mutation in the heterozygous father (I-1) and an homozygous affected fetus (II-1). D) Absence of the pelgeroid change (hyposegmented nucleus of neutrophils) in patients’ parents.
Whole exome sequencing (WES), including library construction, exome capture, sequencing, and data analysis were performed by the Baylor-Hopkins Center for Mendelian Genomics (www.mendeliangenomics.org) in the sample from the female fetus using Illumina HiSeq 2000 (Illumina Inc., San Diego, CA, USA). Variant filtering was performed using the PhenoDB Variant Analysis tool [Sobreira and others 2015b]. We captured the CCDS exonic regions and flanking intronic regions totaling ~51 Mb by using the Agilent SureSelect XT kit (Agilent, Santa Clara, CA, USA) and generated paired end 100 bp reads with the Illumina HiSeq2500 platform. We aligned each read to the HG19/GRCh37 human genome reference with the Burrows-Wheeler Alignment (BWA) version 0.5.10-tpx [Li and Durbin 2009]. Local realignment around indels and base call quality score recalibration were performed using the Genome Analysis Toolkit (GATK) [DePristo and others 2011] version 2.3–9-ge5ebf34. Variant filtering was done using the Variant Quality Score Recalibration (VQSR) method [Choi and others 2012; DePristo and others 2011].
Sequence validation and segregation analyses were performed by Sanger sequencing using an ABI 3130XL and the ABI BigDye Terminator Sequencing Kit V.3.1 (Thermo Fisher Scientific, Wlatham, MA, USA) and the SeqScape v2.6 Software (Thermo Fisher Scientific).
RESULTS
Clinical Report
A 22-year-old Moroccan woman was referred for a genetic counseling to evaluate her familial recurrence risk for a severe form of skeletal dysplasia. She and her husband were healthy and first-degree cousins. No parental blood group incompatibility and no family history of skeletal dysplasia were reported. In the proband’s obstetric history, there were two therapeutic terminations of pregnancy. In each fetus, ultrasonography showed hydrops fetalis and severe micromelia. In both cases the fetuses were autopsied (Figure 1A, II-1 and II-4). A third pathologic pregnancy ended at 24 weeks for premature rupture of membranes (fetal ultrasounds were reported to be normal at 20 weeks of gestation). Finally, two other uneventful pregnancies of the couple gave birth to two healthy daughters (Figure 1A, II-3 and II-5).
Fetus 1 (Figure 1B, II-1). The pregnancy was uncomplicated and there was no exposure to alcohol or drugs. Prenatal karyotype, performed on amniotic fluid, was 46,XY. Fetal ultrasound measurements of the femur, tibia, humerus, ulna and radius were all below the 5th centile for gestational age at 20 weeks. Induced abortion was carried out at 21 weeks.
At autopsy, the fetus presented ambiguous genitalia, severe hydrops fetalis, severe tetramicromelia and dysmorphic features (frontal bossing, micrognathia, severe nasal and midface hypoplasia, and low-set ears). The chest was narrow and the abdomen was prominent with marked ascites. Hands were broad and fingers were short with postaxial bilateral hexadactyly. Internal examination revealed mild hypoplastic lungs and heart, and micronodular surface of the liver.
Radiological examination of the fetus showed a globally delayed and skewed ossification (particularly severe in the skull, cervical and first thoracic vertebrae, and ischiatic-iliac-pubic bones) costal hypoplasia, and shortened, skewed and misaligned long bones.
Fetus 2 (Figure 1B, II-4). The pregnancy was uncomplicated and there was no exposure to alcohol or medications during pregnancy. Induced abortion was carried out at 20 weeks. At autopsy, the fetus presented female genitalia, severe hydrops fetalis, thorax hypoplasia, severe tetramicromelia without polydactyly, severe nasal and midface hypoplasia and low-set dysmorphic ears. The internal examination didn’t reveal any anomaly.
Radiological examination showed global hypomineralized ossification, sparse ossification of the membranous part of the skull, small maxillary and mandibular bones, midface hypoplasia with depressed nasal bridge, narrow chest with short and hypoplastic ribs, deficient ossification of the vertebral column with platyspondyly, and skewed long bones with severely shortened and curved diaphysis. Histological examination of the bones and cartilages showed a severely disordered chondro-osseous development.
Genetic studies
Due to the similarities between the phenotype of the two fetuses and the consanguinity of our patient with her husband, we suspected an autosomal recessive inheritance of the skeletal dysplasia.
Using WES and subsequent Sanger sequencing, we identified a homozygous c.1379A>G (p.D460R) missense change in the LBR gene shared by both fetuses (Figure 1A and C, II-1 and II-4). This change was predicted as a class 4 variant (likely pathogenic) based on the American College of Medical Genetics and Genomics (ACMG) rules, and predicted pathogenic by several in silico bioinformatics tools (Supplementary table 1) [Richards and others 2015]. The variant was not present in gnomAD (http://gnomad.broadinstitute.org/) [Lek and others 2016]. Both parents resulted heterozygotes, as well as the available healthy sister (Figure 1A and C, II-5). We evaluated the presence of the Pelger–Huët anomaly in the heterozygous parents (Figure 1A, I-1 and I-2), but granulocytes had a normal nuclear shape (Figure 1D).
Evaluation of genotype-phenotype correlation
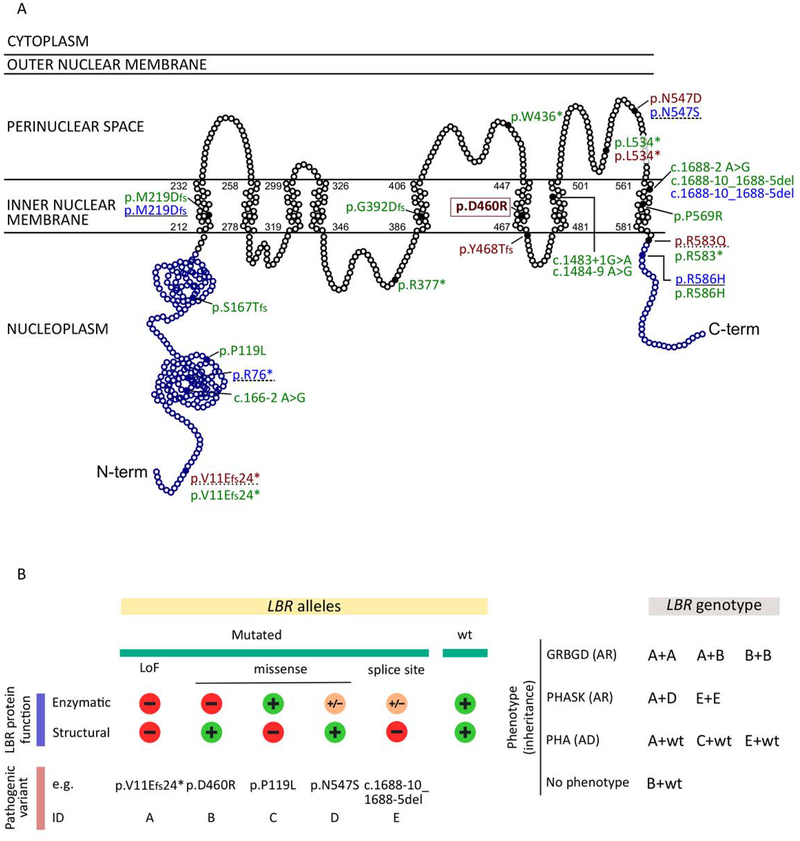
Using available literature data, we summarized all cases with at least one LBR variant including ours, and collected available clinical data (HGMD Professional 2018.2). We could map a total of 23 different variants on the LBR sequence (Figure 2A). Sixteen were nonsense variants (stop gain, frameshift, splicing variants), and seven were missense. Using clinical and laboratory data, we classified cases into one of the following four categories: Greenberg dysplasia, PHA with mild skeletal anomalies, isolated PHA, and no phenotype. Biallelic loss-of-function variants, either nonsense or missense affecting the enzymatic sterolreductase domain, were always associated with Greenberg dysplasia (Figure 2A and 2B). PHASK was reported in two subjects who are compound heterozygous for one nonsense and one missense variant, and in one homozygous subject carrying a c.1688–10_1688–5del (Figure 2A and 2B) [Hoffmann and others 2002; Mattout-Drubezki and Gruenbaum 2003]. Finally, either nonsense or missense monoallelic variants affecting LBR structural function were associated with PHA (Figure 2A and 2B).
Figure 2.
LBR variants and genotype-phenotype correlation. A) Schematic representation of the lamin B receptor protein: nucleoplasmatic (blue) and transmembrane (black) domains are reported. Each dot represents one amino acid. All published pathogenic variants are shown (full dots). In green, pathogenic changes associated with PHA (autosomal dominant). Homozygous or compound heterozygous pathogenic changes associated with GRBGD and PHASK (autosomal recessive) are shown in red and in light blue, respectively. The couple of variants identified in compound heterozygous subjects are indicated by a continuous or hyphened underlining, respecitvely. The c.1379 A>G p.(D460R) variant identified in our patient is boxed.
B) Schematic representation of the impact of LBR mutations on protein function, and LBR-associated phenotypes. Left: loss-of-function (LoF) mutations (nonsense / frameshift changes) cause the impairing of both enzymatic and structural function of the protein (mutation ID: A). Missense changes may have different effects on the protein: they can affect exclusively the enzymatic activity (mutation ID: B) or the structural function (mutation ID: C), or they can have only some impact on the enzymatic function preserving the structural one (mutation ID: D). Finally, splice site mutations can completely impair the structural function but preserve a residual enzymatic activity (mutation ID: E). For each ID category, an example of pathogenic variant is reported.
Right: LBR-related phenotypes and the pattern of heritance are shown. For each phenotype all the possible associated genotypes are shown. Genotypes not shown have not been reported in literature.
GRBGD: Greenberg dysplasia; PHA: Pelger–Huët anomaly; PHASK: Pelger-Huët Anomaly with mild SKeletal anomalies; AR: autosomal recessive phenotype; AD: autosomal dominant phenotype. Minus sign: variants completely impairing the relative function; “+”: variants preserving the relative function; Plus/minus sign: variants partially impairing the relative function.
DISCUSSION
Greenberg skeletal dysplasia is a very rare fetal anomaly described in approximately ten cases worldwide; causative variants in the LBR gene have been identified in five of them [Clayton and others 2010; Konstantinidou and others 2008; Waterham and others 2003]. We solved a familial case with two affected fetuses from consanguineous parents of Moroccan descent. Using WES, we identified a novel homozygous c.1379A>G; p.(D460R) missense variant in the enzymatic domain of the LBR gene, predicted to be pathogenic. Both healthy parents were confirmed to be carriers of the variant but they did not present the PHA cellular phenotype, which should behave as autosomal dominant. Whereas heterozygous loss-of-function variants in LBR are always associated with PHA, it is clearly not the case for missense changes, as showed by our family and reported in literature for the p.N547D and the p.R583Q [Clayton and others 2010]. This observation prompted us to review literature data on LBR variants (Figure 2). PHA is reported to be associated with monoallelic loss-of-function variants, or with missense changes affecting the structural role of the protein (p.P119L; p.P569R; p.R586H; Figure 2). As expected, two out of the three missense variants are located in the nucleoplasmatic domain. Interestingly, the p.Pro569Arg change, located in the transmembrane domain of the protein, also seems to impair the LBR structural function [Nikolakaki and others 2017], causing PHA. On the other hand, two amino acid changes (p.N547D, and p.R583Q) are reported affecting the enzymatic activity but not the structural function. As the p.D460R here described, they do not cause nuclear shape alteration in heterozygosis, but are associated with Greenberg dysplasia when in homozygous or compound heterozygous state (Figure 2). Therefore, we suggest that it is not recommended to exclude a Greenberg dysplasia diagnosis in a fetus when parents do not show the PHA anomaly in blood granulocytes.
Other missense variants causing p.N547S and p.R586H are associated with the PHASK phenotype when in compound heterozygous state with a loss-of-function mutation [Borovik and others 2013; Sobreira and others 2015a]. Here, the mild skeletal phenotype is due to the co-occurrence of a loss-of-function variant completely impairing LBR function and a missense variant likely maintaining residual enzymatic function. The combination is therefore not sufficient to cause the Greenberg dysplasia phenotype (Figure 2). Supporting this hypothesis, a homozygous splicing variant c.1688–10_1688–5del causing the skipping of exon 13 and resulting in an in-frame deletion (p.H522R-L523_C562del) is also associated with PHASK. As expected, a small amount of wild-type protein has been detected in this patient, likely preserving residual enzymatic activity [Hoffmann and others 2002]. Overall, these findings suggest a spectrum in LBR-associated phenotypes, ranging from Greenberg skeletal dysplasia to isolated PHA (Figure 2). Such variability, also reported as one gene - multiple phenotypes phenomenon, is common in Mendelian diseases (1,239 genes with ≥2 phenotypes at 31st August, 2018; https://www.omim.org/statistics/geneMap), and can be explained by at least four different molecular mechanisms [Zhu and others 2014].
Variants affecting one or more diverse functional domains can result in different isolated or combined phenotypes as for the LBR gene.
Variants with qualitatively different effects, such as happens for loss-of-function vs. gain-of-function changes in the sodium voltage-gated channel alpha subunit 9 (SCN9A; OMIM*603415) gene [Cox and others 2006; Yang and others 2004].
Variants which impact on the amount of protein, being associated with a mild to severe phenotype as for the transmembrane protein-67 gene (TMEM67; MIM*609884) in nephronophtisis type 11 (MIM#613550)[Brancati and others 2009; Otto and others 2009].
Finally, we recently showed an uncommon one gene – multiple phenotype phenomenon due to variants affecting tissue-specific splicing isoforms in the proteome of centriole protein 1A gene (POC1A, MIM*614783) [Giorgio and others 2017].
In conclusion, LBR is an instructive example of a gene in which variants are associated with a continuum of phenotypes ranging from no clinical phenotype, benign Pelger-Huët anomaly, mild skeletal abnormalities with bilobed neutrophil nuclei, to perinatal lethal Greenberg skeletal dysplasia. The diverse phenotypes are associated with different functional domains affected, the heterozygous or homozygous state of the variants, and their different impact on the residual protein function.
Supplementary Material
ACKNOWLEDGMENTS
Sequencing and analysis was provided by the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) and was funded by the National Human Genome Research Institute (grant 1U54HG006542, David Valle and James Lupski), the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 to Daniel MacArthur and Heidi Rehm. The work was supported by the Fondazione Umberto Veronesi (post-doctoral fellowship 2017 to EG). This research received funding specifically appointed to Department of Medical Sciences from the Italian Ministry for Education, University and Research (Ministero dell’Istruzione, dell’Università e della Ricerca - MIUR) under the program “Dipartimenti di Eccellenza 2018 – 2022” Project D15D18000410001.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- Badens C, Lacoste C, Philip N, Martini N, Courrier S, Giuliano F, Verloes A, Munnich A, Leheup B, Burglen L, Odent S, Van Esch H, Levy N. 2006. Mutations in PHD-like domain of the ATRX gene correlate with severe psychomotor impairment and severe urogenital abnormalities in patients with ATRX syndrome. Clin Genet 70(1):57–62. [DOI] [PubMed] [Google Scholar]
- Borovik L, Modaff P, Waterham HR, Krentz AD, Pauli RM. 2013. Pelger-huet anomaly and a mild skeletal phenotype secondary to mutations in LBR. Am J Med Genet A 161A(8):2066–2073. [DOI] [PubMed] [Google Scholar]
- Brancati F, Iannicelli M, Travaglini L, Mazzotta A, Bertini E, Boltshauser E, D’Arrigo S, Emma F, Fazzi E, Gallizzi R, Gentile M, Loncarevic D, Mejaski-Bosnjak V, Pantaleoni C, Rigoli L, Salpietro CD, Signorini S, Stringini GR, Verloes A, Zabloka D, Dallapiccola B, Gleeson JG, Valente EM. 2009. MKS3/TMEM67 mutations are a major cause of COACH Syndrome, a Joubert Syndrome related disorder with liver involvement. Hum Mutat 30(2):E432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7(10):e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton P, Fischer B, Mann A, Mansour S, Rossier E, Veen M, Lang C, Baasanjav S, Kieslich M, Brossuleit K, Gravemann S, Schnipper N, Karbasyian M, Demuth I, Zwerger M, Vaya A, Utermann G, Mundlos S, Stricker S, Sperling K, Hoffmann K. 2010. Mutations causing Greenberg dysplasia but not Pelger anomaly uncouple enzymatic from structural functions of a nuclear membrane protein. Nucleus 1(4):354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TV, Klarmann KD, Sakchaisri K, Cooper JP, Kuhns D, Anver M, Johnson PF, Williams SC, Keller JR, Stewart CL. 2008. The lamin B receptor under transcriptional control of C/EBPepsilon is required for morphological but not functional maturation of neutrophils. Hum Mol Genet 17(19):2921–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al-Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM, Woods CG. 2006. An SCN9A channelopathy causes congenital inability to experience pain. Nature 444(7121):894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudy-Marqueste C, Roll P, Esteves-Vieira V, Weiller PJ, Grob JJ, Cau P, Levy N, De Sandre-Giovannoli A. 2010. LBR mutation and nuclear envelope defects in a patient affected with Reynolds syndrome. J Med Genet 47(6):361–370. [DOI] [PubMed] [Google Scholar]
- Giorgio E, Rubino E, Bruselles A, Pizzi S, Rainero I, Duca S, Sirchia F, Pasini B, Tartaglia M, Brusco A. 2017. A syndromic extreme insulin resistance caused by biallelic POC1A mutations in exon 10. Eur J Endocrinol 177(5):K21–K27. [DOI] [PubMed] [Google Scholar]
- Green MC, Shultz LD, Nedzi LA. 1975. Abnormal nuclear morphology of leukocytes in the mouse mutant ichthyosis. Transplantation 20(2):172–175. [PubMed] [Google Scholar]
- Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, Karl H, Kaps R, Muller D, Vaya A, Aznar J, Ware RE, Sotelo Cruz N, Lindner TH, Herrmann H, Reis A, Sperling K. 2002. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly). Nat Genet 31(4):410–414. [DOI] [PubMed] [Google Scholar]
- Konstantinidou A, Karadimas C, Waterham HR, Superti-Furga A, Kaminopetros P, Grigoriadou M, Kokotas H, Agrogiannis G, Giannoulia-Karantana A, Patsouris E, Petersen MB. 2008. Pathologic, radiographic and molecular findings in three fetuses diagnosed with HEM/Greenberg skeletal dysplasia. Prenat Diagn 28(4):309–312. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout-Drubezki A, Gruenbaum Y. 2003. Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell Mol Life Sci 60(10):2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E, Mylonis I, Giannakouros T. 2017. Lamin B Receptor: Interplay between Structure, Function and Localization. Cells 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Tory K, Attanasio M, Zhou W, Chaki M, Paruchuri Y, Wise EL, Wolf MT, Utsch B, Becker C, Nurnberg G, Nurnberg P, Nayir A, Saunier S, Antignac C, Hildebrandt F. 2009. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11). J Med Genet 46(10):663–670. [DOI] [PubMed] [Google Scholar]
- Porter FD, Herman GE. 2011. Malformation syndromes caused by disorders of cholesterol synthesis. J Lipid Res 52(1):6–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics 17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, Dreger C, Herrmann H, Kalscheuer V, Olins AL, Olins DE, Sperling K, Hoffmann K. 2003. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet 12(1):61–69. [DOI] [PubMed] [Google Scholar]
- Sobreira N, Modaff P, Steel G, You J, Nanda S, Hoover-Fong J, Valle D, Pauli RM. 2015a. An anadysplasia-like, spontaneously remitting spondylometaphyseal dysplasia secondary to lamin B receptor (LBR) gene mutations: further definition of the phenotypic heterogeneity of LBR-bone dysplasias. Am J Med Genet A 167A(1):159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. 2015b. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat 36(4):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PL, Zhao C, Turner E, Schlieker C. 2016. The Lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EM, Schlieker C. 2016. Pelger-Huet anomaly and Greenberg skeletal dysplasia: LBR-associated diseases of cholesterol metabolism. Rare Dis 4(1):e1241363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, Mooyer P, Noort Gv G, Kelley RI, Wilcox WR, Wanders RJ, Hennekam RC, Oosterwijk JC. 2003. Autosomal recessive HEM/Greenberg skeletal dysplasia is caused by 3 beta-hydroxysterol delta 14-reductase deficiency due to mutations in the lamin B receptor gene. Am J Hum Genet 72(4):1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Li S, Xu Z, Li H, Ma L, Fan J, Bu D, Liu B, Fan Z, Wu G, Jin J, Ding B, Zhu X, Shen Y. 2004. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet 41(3):171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Need AC, Petrovski S, Goldstein DB. 2014. One gene, many neuropsychiatric disorders: lessons from Mendelian diseases. Nature neuroscience 17(6):773–781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.