Abstract
Purpose
To evaluate accuracy for 2 deformable image registration methods (in-house B-spline and MIM freeform) using image pairs exhibiting changes in patient orientation and lung volume and to assess the appropriateness of registration accuracy tolerances proposed by the American Association of Physicists in Medicine Task Group 132 under such challenging conditions via assessment by expert observers.
Methods and Materials
Four-dimensional computed tomography scans for 12 patients with lung cancer were acquired with patients in prone and supine positions. Tumor and organs at risk were delineated by a physician on all data sets: supine inhale (SI), supine exhale, prone inhale, and prone exhale. The SI image was registered to the other images using both registration methods. All SI contours were propagated using the resulting transformations and compared with physician delineations using Dice similarity coefficient, mean distance to agreement, and Hausdorff distance. Additionally, propagated contours were anonymized along with ground-truth contours and rated for quality by physician-observers.
Results
Averaged across all patients, the accuracy metrics investigated remained within tolerances recommended by Task Group 132 (Dice similarity coefficient >0.8, mean distance to agreement <3 mm). MIM performed better with both complex (vertebrae) and low-contrast (esophagus) structures, whereas the in-house method performed better with lungs (whole and individual lobes). Accuracy metrics worsened but remained within tolerances when propagating from supine to prone; however, the Jacobian determinant contained regions with negative values, indicating localized nonphysiologic deformations. For MIM and in-house registrations, 50% and 43.8%, respectively, of propagated contours were rated acceptable as is and 8.2% and 11.0% as clinically unacceptable.
Conclusions
The deformable image registration methods performed reliably and met recommended tolerances despite anatomically challenging cases exceeding typical interfraction variability. However, additional quality assurance measures are necessary for complex applications (eg, dose propagation). Human review rather than unsupervised implementation should always be part of the clinical registration workflow.
Introduction
Adaptive treatment techniques are becoming routine practice in radiation therapy, and the presence of advanced image registration methods in the clinic is increasing at a similar rate. The reduction in time and effort provided by image registration algorithms enables frequent plan adaptation in response to observed anatomic changes throughout the course of radiation therapy. The recent publication by the American Association of Physicists in Medicine's Task Group No. 132 (TG132) provides guidelines on the implementation of quality assurance and control for deformable image fusion and registration algorithms.1 TG132 recommends a 2-step registration evaluation. The first step is performed for a scenario in which the ground truth is available (eg, phantoms, synthetic images) and establishes essentially the best achievable performance of a deformation method through the commissioning process. The second step targets evaluation of the clinical use of deformable image registration (ie, real patient data), for which the ground truth cannot be established. Our study focuses on the latter part of the evaluation. Registration results in the form of propagated contours were also assessed qualitatively by radiation oncologists and compared with the quantitative accuracy measures.
The use of digital or physical phantoms for testing registration algorithms, as prescribed by the task group report, provides an upper bound on accuracy because of the simplicity and controlled nature of the phantom images. However, accuracy is expected to change when real patient images are used. Image sets chosen for this study ranged in difficulty of the registration problem they posed, from a relatively simple difference in respiratory phase (inhale to exhale) to a more challenging change in patient orientation (supine to prone) that exceeded the registration difficulty of cases commonly seen in oncologic clinical practice. Registration results in the form of propagated contours were also assessed qualitatively by radiation oncologists and compared with the quantitative accuracy measures.
There is a rich literature reporting on image registration errors (especially for tumors and lungs), and the overall accuracy reported is reasonable. However, there is a distribution of the image registration errors for different regions, organs, users, methods, and implementations. As such, it has been recommended that the generalization of reported accuracies be done cautiously.2 The goal of the work reported in this paper was to identify limits of accuracy for our clinic for deformable image registration of the thorax.
Methods and Materials
Image acquisition
Four-dimensional computed tomography images for 12 patients with lung cancer, obtained with patients in both prone and supine positions, were acquired under an institutional review board–approved protocol and retrospectively used to evaluate deformable image registration performance. Informed consent was obtained from all participants. Participants ranged in stage from I to IIIB and had a mean (standard deviation) age of 63 (9) years. The data sets allowed for variation of registration difficulty on an intrasubject basis by controlling the degree of respiratory change and patient orientation. The scans were performed with a Brilliance Big Bore scanner (Philips Healthcare, Cambridge, MA); a protocol with 120 kVp, 500 mAs, and 3 mm slice thickness was used. In-plane resolution ranged from 1.0 to 1.4 mm. Data were anonymized, and the primary lung tumors and normal tissue structures were delineated on the inhale and exhale phases of each scan by 1 radiation oncologist using default lung and mediastinal windows of a commercial software suite (MIM 6.6, MIM Software Inc, Cleveland, OH).
Thoracic structures including tumor, heart, lungs, esophagus, and select vertebrae (T2, T5, and T12) were contoured. Additionally, the esophagus was segmented into upper, middle, and lower sections by evenly dividing the esophagus length into 3 parts (roughly corresponding to the cervical, thoracic, and abdominal sections) along the craniocaudal direction. The lung lobes were individually labeled.
Image registration
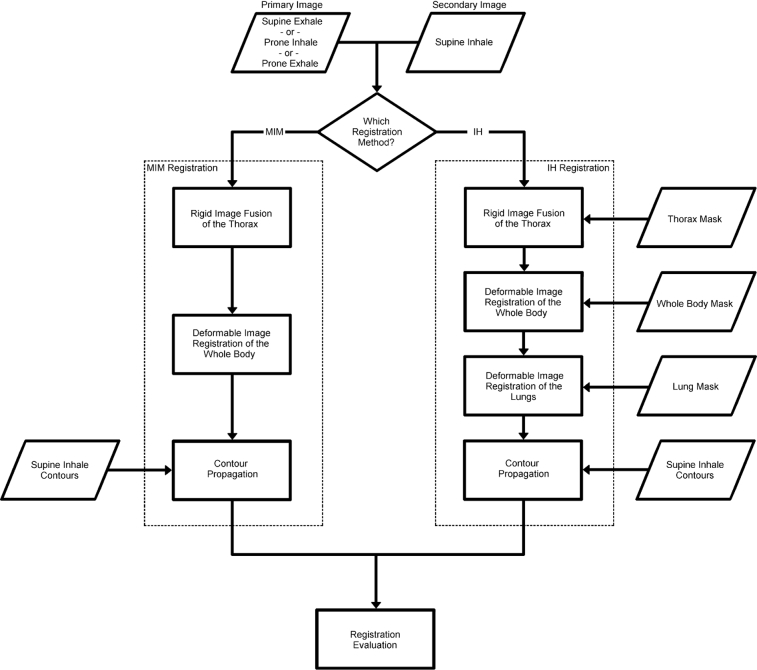
As an initial preprocessing step, prone images were rotated 180° about the z-axis, bringing prone patient anatomy into the same orientation as in the corresponding supine image. The supine inhale (SI) respiratory phase image was separately registered to the supine exhale (SE), prone inhale (PI), and prone exhale (PE) phases using both commercial software with deformable registration functionality (MIM 6.6; MIM Software Inc, Cleveland, OH) and an in-house registration algorithm (IH) based on the open-source Elastix software package, totaling 6 registrations per patient. A workflow for the image registration process is shown in Figure 1.
Figure 1.
Image registration workflow. The image registration process using the MIM and in-house (IH) algorithms is shown for a given pair of input images. The IH algorithm also requires masks of the thorax, whole body, and lung to focus the registration region at each step. Contours are propagated from the secondary image to the primary image before evaluation of results.
MIM registrations were performed using the automatic deformable image registration tool with default settings and no user adjustment. A rigid image fusion was performed as a first step. The deformable image registration tool accepted the rigid fusion results as initial input. A constrained freeform deformation model was used along with an intensity-based similarity function to perform the deformable registration. To maintain a fully automatic workflow, user input to the registrations, such as with the software's Reg Refine tool,3 was not used. In this study the term fully automatic signifies that no real-time input from the user was employed to iteratively improve or adjust the registration results.
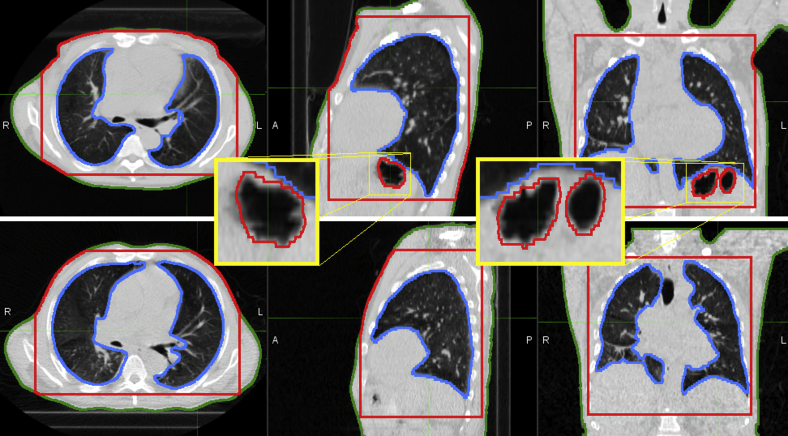
Thoracic image registration is made challenging by a number of issues, such as sliding interfaces, respiratory motion and associated density changes of lung parenchyma, and cardiac motion. In an attempt to overcome these challenges, particularly by providing realistic deformation in the presence of sliding pleural interfaces,4, 5, 6 an in-house B-splines-based Elastix7, 8 registration method was developed that involved 3 steps. First, a rigid registration of the thorax was performed using the normalized cross-correlation similarity metric. Second, a deformable image registration of the entire patient was performed. Finally, a deformable registration of the lungs was added to improve vessel and bronchus alignment. The deformable registrations used a sum of squared intensity differences similarity metric and a bending energy regularization penalty on the transformation. The registrations required contours as additional input: whole-body patient anatomy minus extremities and large air pockets (eg, in the stomach and bowel), thorax region, and lung parenchyma, as shown in Figure 2. Removal of air pockets (ie, exclusion of large cavities of gas within the stomach in both the whole body and thorax masks) was found to be necessary to exclude any dramatic changes in topology, primarily between prone and supine orientations, because this was a registration problem that was not the focus of this study. Except for the creation of additional contours to mask portions of the image from the algorithm, no user intervention was required.
Figure 2.
Registration contours for one image pair. Body (green), thorax (red), and lung (blue) contours required for registration are shown for the inhale prone (top) and inhale supine (bottom) images of one patient. Note the large air pockets that have been excluded from both the whole-body and thorax masks in the sagittal and coronal views of the inhale prone image (magnified), which are not present in the supine image. (A color version of this figure is available at http://doi.dx.10.1016/j.adro.2018.08.023.)
Registration evaluation
Physician-specified contours of the SI phase were propagated to all other images using the resulting transformation of each registration. Agreement between registration-propagated contours and ground-truth physician contours was assessed with multiple metrics to quantify registration accuracy: Dice similarity coefficient (DSC),9 mean distance to agreement (MDA),10 and Hausdorff distance (HD).11 DSC quantifies agreement between contours according to the degree of overlap of their volumes. Accuracy of contours can also be measured by calculating the distance of closest approach of each point on the surface of one contour to all points on the surface of the other contour. MDA and HD are the mean and maximum distances of closest approaches, respectively. The spatial Jacobian and deformation vector field images were used to assess the plausibility of the resulting transformations. Negative spatial Jacobian values and irregularities in the deformation field indicated regions of nonphysical deformation and localized registration failure. Visual inspection of the deformed images was also performed by physicians to detect any large, noticeable issues with the transformation. The accuracy of each registration method was compared against recommendations set forth in the TG132 report, which recommends DSC of >0.8 to 0.9 and MDA <2 to 3 mm, depending on the treatment site.12, 13
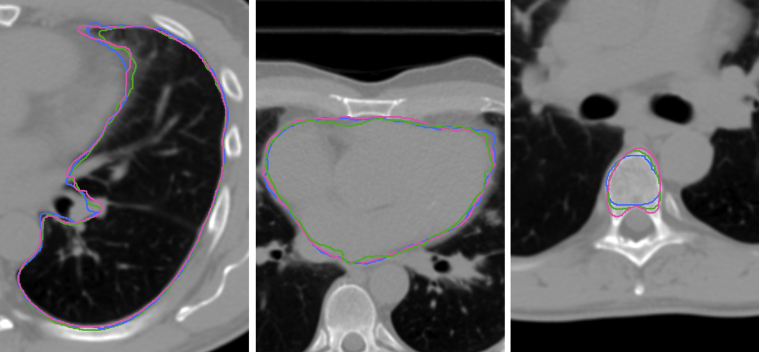
The accuracy assessment resulted in 3 contours per structure for the SE, PI, and PE images: physician drawn, MIM propagated, and IH propagated. Physician-observers (n = 3) were tasked with rating each of the contours in the set for quality and choosing the highest quality delineation of the 3. A rating of “A” meant the contour was clinically acceptable as is, a rating of “B” signified minor changes were necessary, and a “C” rating was given to contours deemed unacceptable to use for treatment planning purposes without major revision. The contours were anonymized and randomized such that the observers were blinded to their mode of creation. Examples of anonymized contours for several structures are given in Figure 3. Physician assessments for each registration method were compared against the accuracy metrics recommended by the task group report.
Figure 3.
Examples of randomized contours. Physician-drawn and registration-propagated contours for 3 structures are shown: left lung (left), heart (center), and T5 thoracic vertebra.
Results
Registration accuracy
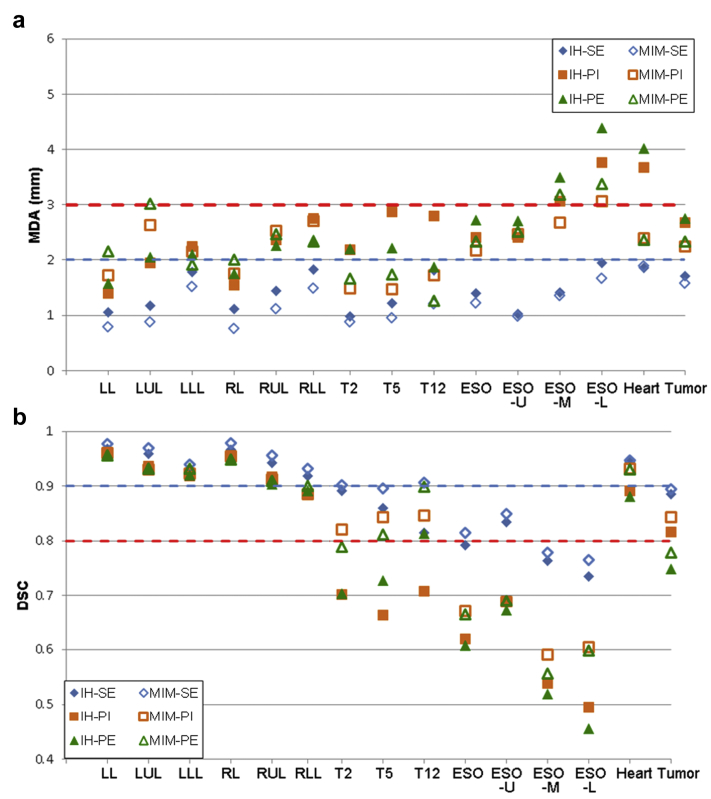
Across all patients and registrations, the mean (standard deviation) of accuracy metrics were DSC 0.8 (0.1), MDA 2.1 (1.4) mm, and HD 22.6 (17.7) mm for MIM; and DSC 0.8 (0.2), MDA 2.3 (1.4) mm, and HD 22.5 (17.2) mm for IH. Results for the contour-based accuracy assessment are provided in Figure 4. MIM performed better with both complex structures (eg, vertebrae) and low-contrast organs (eg, esophagus), whereas IH performed better with lungs (whole and individual lobes). Accuracy metrics worsened for both methods when propagating from supine to prone patient orientation, but most structures remained within recommended tolerances. Esophagus accuracy was often less than suggested limits across all registrations, likely because of poor contrast and high delineation uncertainty of the structure, particularly in the mid and lower third of the esophagus.
Figure 4.
Registration accuracy comparisons. Results are shown for 2 metrics, averaged over 12 patients: (a) mean distance to agreement (MDA) and (b) Dice similarity coefficient (DSC). Dashed horizontal lines represent the general accuracy limits reported in the literature. Abbreviations: ESO = esophagus; ESO-L = lower esophagus; ESO-M = middle esophagus; ESO-U = upper esophagus; IH = in-house B-splines registration; LL = left lung; LLL = left lower lobe; LUL = left upper lobe; MIM = freeform MIM registration; PE = supine inhale to prone exhale; PI = supine inhale to prone inhale; RL = right lung; RLL = right lower lobe; RUL = right upper lobe; SE = supine inhale to supine exhale; T2 = thoracic vertebra 2; T5 = thoracic vertebra 5; T12 = thoracic vertebra 12.
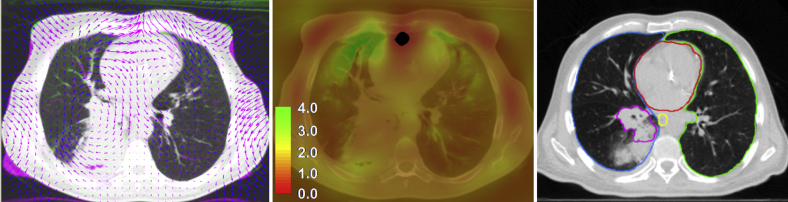
Most contours met the TG132 accuracy constraints of DSC >0.8 and MDA <3 mm; however, the spatial Jacobian images often contained regions with negative values, indicating nonphysical deformation (folding of space). An example of a nonphysical transformation region is shown in Figure 5.
Figure 5.
Example of an ill-behaved transformation. The plausibility of the resulting transformations was evaluated by examining the spatial Jacobian of the transformation and by visualizing the deformable vector field. Additionally, the deformed images were visually inspected by physicians for noticeable registration errors. Shown is an ill-behaved region of the transformation resulting from deformable image registration using the in-house algorithm. Folding of space by the transformation occurs at the anterior edge of the heart in the axial slices shown, as illustrated in the complementary color overlay of the prone inhale and deformed supine inhale images and overlaid deformation vector field (DVF) (left). The spatial Jacobian of the transformation is shown (center). Values greater than 1.0 indicate expansion, values less than 1.0 indicate compression, and negative values (shown in black) represent nonphysical transformation. Contours propagated from prone inhale to supine inhale are shown for the left lung (green), right side of the lung (blue), heart (red), esophagus (yellow), and tumor (magenta) using the in-house registration result (right). Atelectasis (posterior to the tumor) was contoured but excluded from the analysis. Despite the issues with the transformation, contour propagation using the transformation still yields reasonable accuracy. (A color version of this figure is available at http://doi.dx.10.1016/j.adro.2018.08.023.)
Contour quality assessment
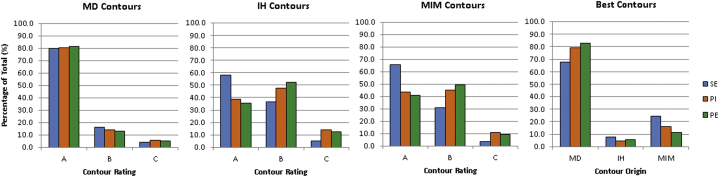
Combined results of the contour assessment performed by 3 physicians are shown in Figure 6. Across all 3 registrations, 50.0%, 41.8%, and 8.2% of contours propagated via MIM registration were rated A, B, and C, respectively. For IH-propagated contours across all registrations, 43.8%, 45.2%, and 11.0% were scored as A, B, and C, respectively. Of the physician-drawn contours, 80.6%, 14.5%, and 4.9% were rated A, B, and C, respectively. The latter set of values can be considered a control for physician variability of delineation and assessment combined.
Figure 6.
Summary of observer contour assessment. Organ delineations were created on all images, and the SI image contours were transferred to the supine exhale (SE), prone inhale (PI), and prone exhale (PE) images via 2 deformable image registrations. Physician-specified (MD), in-house-propagated (IH), and MIM-propagated (MIM) contour sets on the SE, PI, and PE images were rated for quality by physician observers blinded to the origin of the contours. A rating of “A” signified a clinically acceptable delineation, whereas a rating of “B” indicated minor revisions were necessary. A rating of “C” meant the contour contained major errors and was not suitable for clinical use. Additionally, observers were asked to choose which of the 3 structures was of the highest quality.
Mean distance contours were ranked the highest quality, followed by MIM-propagated and then IH-propagated structures. The results of the contour assessment were in agreement with the registration accuracy metrics. SI-PI registrations had lower contour quality compared with SI-SE registrations, whereas SI-PE registrations had the lowest ratings with the least number of A-ranked contours and greatest number of B-ranked contours. The number of contours considered unacceptable for clinical use increased from 4.2% with the SI-SE registration to 9.0% to 10.3% for the registrations involving a change in patient orientation.
Discussion
The accuracy of 2 very different deformable image registration methods was investigated for challenging cases using clinical images in the context of recent TG132 recommendations. The transformation model of MIM’s deformable registration tool is freeform with no practical limit on the degrees of freedom available. The MIM registration method has the advantage of being implemented in a popular software suite that is already integrated into the clinical workflow at many institutions. The in-house registration method differs from MIM in the transformation model and input required, using a B-spline basis and needing a set of additional contours instead. These additional contours were necessary only to create masks of the registration region so that tissue outside this region of interest (ie, thorax) was ignored. The masks did not impart any information regarding structure boundaries or organ surfaces. Although Elastix registration must be performed outside the standard clinical software, the framework allows for far-superior flexibility via input parameter files so that the registration algorithm design can be customized on a site-specific basis, allowing for a highly accurate registration in challenging clinical scenarios (eg, resolution of atelectasis). Despite their differences, both registration algorithms performed well according to accuracy metrics based on propagated contours under scenarios in which anatomic variations were larger than those typically encountered on a fraction-to-fraction basis.
The demonstration of registration accuracy for any finite data set of image pairs cannot ensure that accuracy is maintained in all possible clinical scenarios. The images chosen for this study were limited to changes in thoracic anatomy caused by respiration and the transition from prone to supine orientation. These factors caused substantial deformations in the anatomy of the thorax comparable to or exceeding deformations that may occur on an intrafraction basis as a result of patient repositioning. Longitudinal imaging throughout the course of radiation therapy can reveal anatomic changes from addition or loss of tissue (eg, tumor regression, edema, weight loss, etc.).14, 15, 16, 17 Such cases remain challenges for most deformable image registration algorithms and are the subject of ongoing research.18, 19 Because the prone and supine acquisitions of this study were performed during the same imaging session, anatomy was consistent between images used for registration. It was therefore expected that deformable image registration algorithms should register these cases with acceptable accuracy.
The 3 image registration cases used in this study varied in difficulty, with the easiest being registration of inhale to exhale supine images. The SI to PE registration posed the greatest challenge, presenting a registration problem more difficult than images containing no change in patient orientation. The switch from supine to prone orientation induces significant deformations in the abdomen and thorax and is further complicated by lung volume differences between inhale and exhale. The accuracy of MIM’s deformable image registration tool based on contour assessment has been estimated previously as ranging from 1.1 to 1.4 mm.20 Based on the accuracy assessment of this work, the SI to SE registrations were in agreement, with MDA of 1.27 (0.836) mm. A slightly higher registration error of 2.51 (1.6) mm was obtained for the SI to PE registrations, reflecting the difficulty of the anatomic variations used for this investigation. The in-house deformable image registration algorithm had a performance similar to that of MIM and can therefore be assumed to have similar accuracy. Deformable image registration accuracy of thoracic registration for the purpose of contour propagation has been reported to be on the order of 1 to 3 mm for other algorithms, on par with the registration accuracy found in this study.21, 22 Therefore both algorithms met the accuracy requirements of TG132 for actual patient data.
The accuracy of propagated contours in this study was in agreement with findings in the literature. In an interobserver contouring investigation, DSC for thoracic structures was reported to range between 0.74 for esophagus and 0.96 for lungs, with the esophagus generating the largest interobserver variability.13 Other studies report esophagus DSC ranging from 0.7 to 0.8 and HD of approximately 10 mm.23, 24, 25 Studies reporting lung contour propagation accuracy have found MDA, HD, and DSC values ranging from 1.5 to 2 mm, 20 to 25 mm, and 0.91 to 0.96, respectively.4, 23, 24, 26, 27 For lung, a DSC >0.9 is expected in instances of acceptable agreement.26 Automatic segmentation of vertebrae has been found to result in contours with DSC values between 0.8 and 0.95 compared with ground-truth contours, depending on the segmentation or propagation method used.28, 29 The cord, heart, and lung tumor DSC values using segmentation or propagation have also been reported to range between 0.8 and 0.9 for clinically acceptable results.23, 24 Values for the quantitative accuracy metrics in Figure 4 of the present study were in agreement with these previously reported findings.
Qualitative contour assessment was previously performed in a similar study wherein propagated contours were rated as clinically acceptable as is (82%), requiring minor editing (14%), or unacceptable (3%).24 Although different deformable image registration algorithms were used, the results of the present study indicated a decrease in “A”-ranked contours, attributed to the increased difficulty (prone to supine, inhale to exhale) of the registrations used. High accuracy according to contour-based metrics was not always indicative of a clinically acceptable registration. Across all observers and registrations, 7.8% of contours were given a “C” rating, indicating the propagated contour could not be used in a clinical treatment plan. Some variability in observer judgment was present as well, with 4.9% of physician-drawn contours ranked as “C.” Although the majority of image registrations obtained in this study were suitable for contour propagation, human review should always be part of the clinical workflow despite an algorithm meeting quantitative accuracy metrics.
The lungs are usually treated as 2 organs, right and left, and are not segmented on a lobe-wise basis during standard clinical workflow. Because of the large volume of the lungs, accuracy at the high-contrast lung boundaries may not be indicative of accuracy within the parenchyma away from the pleural surfaces. To investigate accuracy within the lung volume, this study also used lung lobes for accuracy quantification. Accuracy was only slightly decreased for individual lobes compared with that of whole lungs as shown in Figure 4. Lobe contour propagation performance is quite satisfactory, considering that the lobes deform independently with changes in lung volume and that fissures that do not appear with sufficient contrast in computed tomography images. This provides some confidence that contour propagation accuracy is maintained through the volume of interest for registration.
MIM provides tools for user-guided, iterative improvement of deformable registration results. Using the Reg Refine tool, the automatic deformable image registration results may be substantially improved for many scenarios. However, the addition of user input enables the introduction of errors as well. For example, if the user were to lock an incorrect point using the tool, the algorithm would be prevented from accurately registering the tissue near that location. Interuser variability of registration accuracy when using the semiautomatic workflow was avoided in this study by only considering accuracy of the fully automatic registration but could be the subject of future investigation.
Although contour-based metrics suggested no issues with the registrations, inspection of the resulting transformations uncovered major issues in the form of nonphysical transformations within subvolumes of the anatomy of interest. As shown in Figure 5, the volume with negative Jacobian values (black voxels) occurs at the boundaries of the right lung, left lung, and heart, though contour appearance and propagation accuracy for these organs were largely unaffected. This finding suggests that contour propagation is largely insensitive to localized errors in the deformation. However, when using deformable image registration for voxel-based applications such as dose accumulations, these issues with the transformation will likely translate to errors in summed dose.
Conclusions
For the 2 deformable registration algorithms tested on clinical data sets, most contour-based accuracy limits recommended by TG132 were met for thoracic structures, but irregularities in the transforms and negative values in the spatial Jacobian images were identified, indicating registration failure in localized regions. Despite algorithm accuracy within tolerance limits, a substantial number of propagated contours resulting from the registration were not of clinically acceptable quality. Therefore the accuracy limits were sufficient for contour propagation where careful review by an observer is possible but not for more complex tasks such as dose accumulation.
Acknowledgments
The authors would like to thank William Sleeman IV for his extensive contribution to the curation of patient data used in this work.
Footnotes
Sources of support: This work had no specific funding.
Conflicts of interest: Dr Weiss reports grants from the National Institutes of Health, grants from Varian, nonfinancial support from Philips, and royalties from UpToDate outside the submitted work. All other authors have nothing to disclose.
References
- 1.Brock K.K., Mutic S., McNutt T.R., Li H., Kessler M.L. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys. 2017;44:e43–e76. doi: 10.1002/mp.12256. [DOI] [PubMed] [Google Scholar]
- 2.Kashani R., Hub M., Balter J.M. Objective assessment of deformable image registration in radiotherapy: A multi-institution study: Multi-institution assessment of image registration. Med Phys. 2008;35:5944–5953. doi: 10.1118/1.3013563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson P.B., Padgett K.R., Chen K.L., Dogan N. Evaluation of the tool “Reg Refine” for user-guided deformable image registration. J Appl Clin Med Phys. 2016;17:158–170. doi: 10.1120/jacmp.v17i3.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z., Rietzel E., Boldea V., Sarrut D., Sharp G.C. Evaluation of deformable registration of patient lung 4DCT with subanatomical region segmentations: Evaluation of deformable registration of 4DCT with segmentations. Med Phys. 2008;35:775–781. doi: 10.1118/1.2828378. [DOI] [PubMed] [Google Scholar]
- 5.Pace D.F., Enquobahrie A., Yang H., Aylward S.R., Niethammer M. Deformable image registration of sliding organs using anisotropic diffusive regularization. Proc IEEE Int Symp Biomed Imaging. 2011:407–413. doi: 10.1109/ISBI.2011.5872434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delmon V., Rit S., Pinho R., Sarrut D. Registration of sliding objects using direction dependent B-splines decomposition. Phys Med Biol. 2013;58:1303–1314. doi: 10.1088/0031-9155/58/5/1303. [DOI] [PubMed] [Google Scholar]
- 7.Klein S., Staring M., Murphy K., Viergever M.A., Pluim J. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 8.Shamonin D. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease. Front Neuroinform. 2013;7 doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 10.Chalana V., Kim Y. A methodology for evaluation of boundary detection algorithms on medical images. IEEE Trans Med Imaging. 1997;16:642–652. doi: 10.1109/42.640755. [DOI] [PubMed] [Google Scholar]
- 11.Huttenlocher D.P., Klanderman G.A., Rucklidge W.J. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell. 1993;15:850–863. [Google Scholar]
- 12.Kumarasiri A., Siddiqui F., Liu C. Deformable image registration based automatic CT-to-CT contour propagation for head and neck adaptive radiotherapy in the routine clinical setting. Med Phys. 2014;41:12. doi: 10.1118/1.4901409. [DOI] [PubMed] [Google Scholar]
- 13.McCall R., MacLennan G., Taylor M. Anatomical contouring variability in thoracic organs at risk. Med Dosim. 2016;41:344–350. doi: 10.1016/j.meddos.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Møller D.S., Khalil A.A., Knap M.M., Hoffmann L. Adaptive radiotherapy of lung cancer patients with pleural effusion or atelectasis. Radiother Oncol. 2014;110:517–522. doi: 10.1016/j.radonc.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Van Zwienen M., van Beek S., Belderbos J. Anatomical changes during radiotherapy of lung cancer patients. Int J Radiat Oncol Biol Phys. 2008;72:S111. [Google Scholar]
- 16.Kwint M., Conijn S., Schaake E. Intra thoracic anatomical changes in lung cancer patients during the course of radiotherapy. Radiother Oncol. 2014;113:392–397. doi: 10.1016/j.radonc.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M.L., Hoffmann L., Kandi M., Møller D.S., Poulsen P.R. Dosimetric impact of respiratory motion, interfraction baseline shifts, and anatomical changes in radiotherapy of non-small cell lung cancer. Acta Oncol. 2013;52:1490–1496. doi: 10.3109/0284186X.2013.815798. [DOI] [PubMed] [Google Scholar]
- 18.Guy C.L., Weiss E., Jan N., Reshko L.B., Christensen G.E., Hugo G.D. Effect of atelectasis changes on tissue mass and dose during lung radiotherapy. Med Phys. 2016;43:6109–6117. doi: 10.1118/1.4965807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guy C.L., Weiss E., Christensen G.E., Jan N., Hugo G.D. CALIPER: A deformable image registration algorithm for large geometric changes during radiotherapy for locally advanced non-small cell lung cancer. Med Phys. 2018;45:2498–2508. doi: 10.1002/mp.12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piper J. Evaluation of an intensity-based free-form deformable registration algorithm. Med Phys. 2007;34:2353–2354. [Google Scholar]
- 21.Kadoya N., Fujita Y., Katsuta Y. Evaluation of various deformable image registration algorithms for thoracic images. J Radiat Res. 2014;55:175–182. doi: 10.1093/jrr/rrt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wijesooriya K., Weiss E., Dill V. Quantifying the accuracy of automated structure segmentation in 4D CT images using a deformable image registration algorithm: Automatic segmentation using deformable image registration. Med Phys. 2008;35:1251–1260. doi: 10.1118/1.2839120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peroni M., Spadea M.F., Riboldi M. Validation of automatic contour propagation for 4D treatment planning using multiple metrics. Technol Cancer Res Treat. 2013;12:501–510. doi: 10.7785/tcrt.2012.500347. [DOI] [PubMed] [Google Scholar]
- 24.Hardcastle N., Van Elmpt W., De Ruysscher D., Bzdusek K., Tomé W.A. Accuracy of deformable image registration for contour propagation in adaptive lung radiotherapy. Radiat Oncol. 2013;8:243. doi: 10.1186/1748-717X-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sila Kurugol. Machine Learning and Model Based 3D Segmentation Algorithms for Challenging Medical Imaging Problems, Ph.D. Dissertation, Department of Electrical and Computer Engineering, Northeastern University, Boston Massachusetts, 2011.
- 26.van Rikxoort E.M., de Hoop B., Viergever M.A., Prokop M., van Ginneken B. Automatic lung segmentation from thoracic computed tomography scans using a hybrid approach with error detection: Automatic lung segmentation from thoracic CT scans. Med Phys. 2009;36:2934–2947. doi: 10.1118/1.3147146. [DOI] [PubMed] [Google Scholar]
- 27.Ross J.C., Kindlmann G.L., Okajima Y. Pulmonary lobe segmentation based on ridge surface sampling and shape model fitting. Med Phys. 2013;40:121903. doi: 10.1118/1.4828782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egger J, Nimsky C. Cellular automata segmentation of the boundary between the compacta of vertebral bodies and surrounding structures. Poster presented at: SPIE Medical Imaging Conference. February 27–March 3, 2016; San Diego, CA.
- 29.Yao J., Burns J.E., Forsberg D. A multi-center milestone study of clinical vertebral CT segmentation. Comput Med Imaging Graph. 2016;49:16–28. doi: 10.1016/j.compmedimag.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]