Abstract
Cyanobacteria are dominant primary producers of various ecosystems and they colonize marine as well as freshwater and terrestrial habitats. On the basis of their oxygenic photosynthesis they are known to synthesize a high number of secondary metabolites, which makes them promising for biotechnological applications. State-of-the-art sequencing and analytical techniques and the availability of several axenic strains offer new opportunities for the understanding of the hidden metabolic potential of cyanobacteria beyond those of single model organisms. Here, we report comprehensive genomic and metabolic analyses of five non-marine cyanobacteria, that is, Nostoc sp. DSM 107007, Anabaena variabilis DSM 107003, Calothrix desertica DSM 106972, Chroococcidiopsis cubana DSM 107010, Chlorogloeopsis sp. PCC 6912, and the reference strain Synechocystis sp. PCC 6803. Five strains that are prevalently belonging to the order Nostocales represent the phylogenetic depth of clade B1, a morphologically highly diverse sister lineage of clade B2 that includes strain PCC 6803. Genome sequencing, light and scanning electron microscopy revealed the characteristics and axenicity of the analyzed strains. Phylogenetic comparisons showed the limits of the 16S rRNA gene for the classification of cyanobacteria, but documented the applicability of a multilocus sequence alignment analysis based on 43 conserved protein markers. The analysis of metabolites of the core carbon metabolism showed parts of highly conserved metabolic pathways as well as lineage specific pathways such as the glyoxylate shunt, which was acquired by cyanobacteria at least twice via horizontal gene transfer. Major metabolic changes were observed when we compared alterations between day and night samples. Furthermore, our results showed metabolic potential of cyanobacteria beyond Synechocystis sp. PCC 6803 as model organism and may encourage the cyanobacterial community to broaden their research to related organisms with higher metabolic activity in the desired pathways.
Keywords: metabolomic analysis, light microscopy, scanning electron microscopy, pigment, storage compounds, genome sequencing, phylogeny, Synechocystis sp. PCC 6803
Introduction
Cyanobacteria are found in almost any terrestrial and aquatic habitat—fresh, brackish and salt water, soil and even on desert rocks. Freshwater and marine cyanobacteria form very dense and sometimes toxic blooms (Huisman et al. 2018), and they show a rapid adaptation to rising atmospheric CO2 concentrations (Sandrini et al. 2016). An unprecedented variety of toxins is produced by cyanobacteria that threaten the quality of drinking water, influences shellfish quality via the food chain and can even cause severe health problems in humans (Dittmann et al. 2013). The unicellular genera Synechococcus and Prochlorococcus are dominant biomass producers in the oceans (Díez and Ininbergs 2014); and the development of desiccation tolerance allowed filamentous cyanobacteria to colonize inhospitable terrestrial habitats (Büdel 2011). They developed a large morphological diversity (Rippka et al. 1979). A prime example for true cellular differentiation in bacteria is the model organism Nostoc sp. PCC 7120 (synonym Anabaena; Kumar et al. 2010), where photosynthesis is carried out by vegetative cells in contrast to specialized heterocysts that are responsible for the fixation of atmospheric nitrogen.
Cyanobacteria represent early biological solar cells and their fossil footprints are preserved for hundreds of millions of years, which is documented by up to 1.9-Gyr-old stromatolite fossils (Hoffmann 1976; Brocks et al. 1999). Photosynthetic pigments are due to their exceptional chemical stability valuable molecular fossils of the geological past (Knoll et al. 2007) and the recent discovery of fossilized chlorophyll in 1.1-Gyr-old sedimentary rocks documented that the marine ecosystem was once dominated by cyanobacteria (Gueneli et al. 2018). Nowadays, cyanobacteria play an important role in the production of peptides, transfatty acids, amino acids, vitamins, and pigments (Mimouni et al. 2012). Certain carotenoid derivates, like myxol glycosides, canthaxanthin, and echinenone can be considered characteristic for cyanobacteria (Takaichi and Mochimaru 2007; Jeffrey et al. 2011), whereas other carotenoids like β-carotene and zeaxanthin are of great interest due to their antioxidant properties or anticancerous activity (Zuluaga et al. 2017; Nagy et al. 2018).
Metabolic characterization of cyanobacteria essentially links the scientific communities of researchers working on plant metabolism, bacterial metabolism and biotechnological approaches (Schwarz et al. 2013). However, most studies are focused on model organisms, mainly on the fresh water bacterium Synechocystis sp. PCC 6803 and to a lesser extent on marine Synechococcus strains such as PCC 7002. Synechocystis was previously studied with regard to photorespiration (Eisenhut, Huege, et al. 2008; Knoop et al. 2010; Orf et al. 2016), transcriptional regulators (Osanai et al. 2014; Klähn et al. 2015) carbon, iron, and nitrogen limitation (Eisenhut, Ruth, et al. 2008; Rivas-Ubach et al. 2018) and to different stress responses (Gründel et al. 2012; Narainsamy et al. 2013). For both, Synechocystis sp. PCC 6803 and different Synechococcus strains, there is a number of studies with a more biotechnological focus (Baran et al. 2013; Dempo et al. 2014; Niu et al. 2015; Kopka et al. 2017). In the same context, available metabolic models showed that the high number of isozymes in Synechocystis sp. PCC 6803 leads to more stable metabolic fluxes and most likely to lower accessibility by biochemical engineering (Jablonsky et al. 2016). Only few metabolic studies focus on other cyanobacteria and most of them used Synechocystis sp. PCC 6803 as a reference strain. A very detailed analysis showed the role of microcystins in the highlight adaptation of the more light-resistant strain Microcystis aeruginosa PCC 7806 (Meissner et al. 2015). Analyses of a microcystin-deficient strain showed slower glycolate accumulation upon highlight exposure and general stress markers accumulate such as trehalose and sucrose. Overall, the study provided detailed information on the metabolism of bloom-forming cyanobacteria and their metabolic diversity (Meissner et al. 2015). Furthermore, Arthrospira platensis as a fast-growing halophilic cyanobacterium was characterized with respect to temporal alterations under nitrate depletion showing transient increases of some amino acids during increased glycogen production (Hasunuma et al. 2013). These two publications indicated that results obtained with a model strain are not necessarily valid for other cyanobacteria. Accordingly, there is the urgent need for a taxonomically broader study of the cyanobacterial metabolism to distinguish between strain specific and generalizable traits.
The unicellular cyanobacterium Synechocystis sp. PCC 6803 that was isolated in 1968 from a fresh water lake in Oakland, CA is the most prominent model organism of this lineage, which is reflected by >2,300 scientific publications to date. It is a fast-growing, nondiazotrophic, representative of the nanophytoplankton with a comparably small genome size of 3.9 Mb (NC_000911.1; Kaneko et al. 1996). These characteristics raise the question if the insights from this model organism can be generalized for all cyanobacteria. In order to gain a holistic understanding of the effects of light and darkness on the metabolism of cyanobacteria, we investigated six non-marine representatives including the strain PCC 6803 (Rippka et al. 1979). Apart from two additional unicellular cyanobacteria, that is, Chlorogloeopsis sp. PCC 6912 and Chroococcidiopsis cubana DSM 107010 (Mitra 1950; Rippka et al. 1979), we investigated the filamentous representatives Nostoc sp. DSM 107007, Anabaena variabilis DSM 107003, and Calothrix desertica DSM 106972 (Schwabe 1960; Kenyon et al. 1972; Rippka et al. 1979).
In the current study, we established high-quality draft genomes of five cyanobacterial strains and determined their precise phylogenetic position in the tree of life. Light and electron microscopy allowed a morphological comparison with high-resolution. In a comprehensive metabolic comparison, we determined major differences between the metabolic profiles in day and night cycles and moreover between the isolates including central carbon metabolism and the use of storage compounds.
Materials and Methods
Strains and Cultivation Conditions
Nostoc sp. DSM 107007 (=SAG 25.82), A. variabilis DSM 107003 (=SAG 1403-4b), C. cubana DSM 107010 (=SAG 39.79), and C. desertica DSM 106972 (=SAG 35.79; table 1) are available at the DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). Synechocystis sp. PCC 6803 (table 1) was obtained from the Pasteur Culture Collection (PCC, Paris, France) and Chlorogloeopsis sp. PCC 6912 (=CCY9924) was kindly provided by the Culture Collection Yerseke (CCY, Royal Netherlands Institute for Sea Research [NIOZ]). Nostoc, Anabaena, Chroococcidiopsis, and Synechocystis were cultivated in 20 ml BG11+ medium (DSMZ medium 1593), Calothrix in 20 ml ES medium and Chlorogloeopsis in 20 ml BG11– medium (DSMZ medium 1592) in 25 cm2 tissue culture flasks (TPP, Trasadingen, Switzerland). All strains except for PCC 6912 were cultivated at 17 °C to represent the average temperature in their natural habitat. PCC 6912 was cultivated at 23 °C to observe sufficient growth. The six axenic cyanobacterial strains (table 1) were cultivated under low light conditions (9 µmol s−1 m−2) for 16 h (referred to as day cycle) followed by a dark period of 8 h (referred to as night cycle). All six strains were cultivated following these day–night cycles for at least 6 weeks prior to the harvesting and the final experiment was carried out in replicates: One replicate to maintain the strain in culture or for cryopreservation, one replicate for genome sequencing and four replicates each for the day and night samples, respectively.
Table 1.
Morphologic, Biochemical, Genetic, and Biogeographic Characteristics of Six Non-Marine Cyanobacteria
| Nostoc sp. | Anabaena variabilis | Calothrix desertica | Chlorgloeopsis sp. | Chroococcidiopsis cubana | Synechocystis sp. | |
|---|---|---|---|---|---|---|
| Morphology | ||||||
| Cells | Filamentous | Filamentous | Filamentous | Unicellular (tetrade) | Unicellular (tetrade) | Unicellular |
| Cell shape | Barrel to oval | Spherical | Disc or isodiametric | Oval deformed | Oval | Spherical to oval |
| Hormogonia | – | – | + | – | – | – |
| Sheet | + | + | + | (+) | + | – |
| Branching | – | – | – | – | – | – |
| Heterocysts | Terminal or intercalary | Terminal or intercalary | Terminal | – | – | – |
| Cell length width (µm) | 5.05 (± 0.78) 4.22 (± 0.36) | 4.52 (± 0.82) 4.55 (± 0.46) | 4.23 (± 1.59) 7.53 (± 2.02) | 5.51 (± 0.72) 4.50 (± 0.82) | 5.83 (± 0.99) 4.93 (± 0.82) | 2.65 (± 0.22) 2.30 (± 0.33) |
| Chain length (µm) | >200 | >1000 | >500 | – | – | – |
| Phylogeny | ||||||
| Cladea | B1 | B1 | B1 | B1 | B1 | B2 |
| Order | Nostocales | Nostocales | Nostocales | Nostocales | Chroococci diopsidiales | Synecho coccales |
| Family | Nostocaceae | Nostocaceae | Nostocaceae | Chlorogloe opsidaceae | Chroococci diopsidaceae | Merismo pediaceae |
| Genome | ||||||
| Status | Draftb/complete | Draft | Draft | Draft | Draft | Complete |
| Contigs | 139/7 | 54 | 110 | 84 | 295 | 5 |
| Size (Mb) | 7.26/7.21 | 6.12 | 11.62 | 7.92 | 8.29 | 3.95 |
| G+C | 41.24/41.22 | 39.31 | 38.56 | 41.48 | 44.51 | 47.35 |
| Accession number | RSCN00000000/NC_003272.1 | RSCM00000000 | RSCL00000000 | RSCJ00000000 | RSCK00000000 | NC_000911.1 |
| Isolation | ||||||
| Origin | x | Utrecht, NL | Antofagasta desert, Chile | Allahabad, India | Pinar del Rio, Cuba | Oakland, CA |
| Habitat | Fresh water | Fresh water | Soil | Garden soil | Dry soil | Freshwater |
| Scientist | Haselkorn | Pringsheim | Schwabe | Mitra | Hindák | Kunisawa |
| Year | Before 1982 | Before 1954 | 1958 | Before 1950 | 1966 | 1968 |
| First Publication | Rippka et al. (1979) | Kenyon et al. (1972) | Schwabe (1960) | Mitra (1950) | Rippka et al. (1979) | Stanier and Cohen (1977) |
| Collection ID | ||||||
| DSM | 107007 | 107003 | 106972 | — | 107010 | — |
| SAG | 25.82 | 1403-4b | 35.79 | 1411-1a | 39.79 | — |
| PCC | 7120 | 6309 | 7102 | 6912 | 7433 | 6803 |
| ATCC | 27893 | 29211 | 27901 | 27193 | 29381 | 27184 |
| UTEX | 2576 | 377 | — | — | — | — |
| CCAP | — | 1403/4B | — | 1411/A1 | — | — |
Current study.
Cells for pigment, glycogen and metabolome analysis were harvested by centrifugation (4 °C, 12,000 × g, 5–10 min) at the end of the day or night cycle, respectively. Small LED balls with a diameter of 1.4 cm were adjusted to 9 µmol s−1 m−2 and added to the light samples during the harvesting. Cultures were washed with cold water (ES medium) or isotonic sodium chloride solution (0.1 g l−1 for BG11– and 1 g l−1 for BG11+ medium) and suspensions were transferred into weighted reaction tubes. After centrifugation, the supernatant was discarded, the amount of wet biomass was determined and the pelleted cells were immediately frozen in liquid nitrogen and stored at –80 °C prior to extraction.
Cryopreservation
Culture flasks with cyanobacterial suspensions were cooled for 10 min by placing the flasks on ice. DMSO was slowly added under shaking to the suspension in the culture flasks to a final percentage of 9% DMSO. Immediately after addition of the DMSO, aliquots of 1.5 ml of the cyanobacterial suspension were filled in a 1.8 ml cryovial and were kept on ice for 20 min. The cryovials were subsequently placed into a CoolCell device (CoolCell LX Freezing Container, BioCision, CA) and incubated in a refrigerator at –80 °C. After 4 h of incubation, the vials were cooled in liquid nitrogen thereby avoiding direct contact with the top part of the vials. Vials were stored in the gas phase of the liquid nitrogen tank.
For recovery, cryovials were thawed in sterile water at 40 °C for 3 min. The suspension of each cryovial was transferred into a standard culture flask with 10 ml growth medium. The cyanobacterial suspension in the culture flask was incubated for 24 h in the dark and then exposed to standard growth conditions. All cultures but Nostoc sp. DSM 107007 showed growth and were thus successfully cryopreserved.
Light and Electron Microscopy
Cyanobacteria were harvested after 16 h (day cycle) and after 8 h (night cycle), and they were fixed in 1% glutaraldehyde at 4 °C overnight (ON). Subsequently, cells were washed three times with PBS and were analyzed by light and scanning electron microscopy.
Bright-field (BF) and fluorescence z-stacks were taken using a Nikon N Plan Apochromat λ x100/1.45 oil objective and the Nikon DS-Ri2 or ORCA FLASH 4.0 HAMAMATSU camera, respectively. Autofluorescence images were detected using GFP (485/20 nm) excitation and Cy5 (705/72 nm) emission filters. Specimens were immobilized in MatTek glass bottom dishes (35 mm, No. 1.5) employing a 1% agarose cushion (Boedeker et al. 2017). To determine the cell size, the width and lengths of at least 100 representative cells were determined manually using the Nikon NIS-Elements software (Version 4.3). Fluorescence images were postprocessed using the 3D Landweber deconvolution algorithm.
Scanning electron microscopy (SEM) was performed as previously described (Rast et al. 2017). In short, samples were placed onto poly-l-lysine coated cover slips (12 mm) for 10 min, then fixed with 2% glutaraldehyde in TE buffer (10 mM TRIS, 1 mM EDTA, pH 6.9) and dehydrated with a graded series of acetone (10%, 30%, 50%, 70%, 90%, 100%) on ice 10 min for each step. After critical point drying with CO2 samples were mounted onto aluminum stubs with adhesive tape, sputter coated with gold-palladium and examined in a Zeiss Merlin field emission scanning electron microscope. Images were taken with the SEM software version 5.05 at an acceleration voltage of 5 kV with the Inlens SE-detector and HESE2 SE-detector in a 75:25 ratio.
Genome Sequencing
DNA was extracted using Qiagen’s Blood and Tissue Kit (Qiagen, Hilden, Germany) following instructions of the manufacturer. The protocol was optimized by incubating the cyanobacteria with glass beads for 30 min, at 37 °C on a shaker, following the enzymatic lysis step. Illumina libraries were prepared using the NEBNext Ultra DNA Library Prep Kit (New England Biolabs, Frankfurt, Germany) as per the instructions of the manufacturer. DNA was sheared using a Covaris S220 sonication device (Covaris Inc; MA) with the following settings: 50 s, 105 W, 5% duty factor, 200 cycles of burst. Size selection and an enrichment PCR of seven cycles were performed according to the protocol; 300-bp paired-end sequencing of the libraries was performed on the Illumina MiSeq system using the v3 chemistry (600 cycles) following standard protocol. Quality control and adapter clipping of the sequences was done using fastq-mcf tool of ea-utils (Aronesty 2011). DNA-Seq reads were de novo assembled with VELVET 1.2.10 (Zerbino and Birney 2008).
Genome Quality and Phylogenetic Analyses
Sequences of 207 cyanobacterial genomes were downloaded from NCBI. Together with the five genomes in this study, all 212 genomes were evaluated for their completeness and contamination with the program CheckM and the option lineage_wf with default parameters (Parks et al. 2015). For phylogeny, the intermediate concatenated alignment of 43 marked genes was further analyzed with raxmlHPC (parameters: -f a -# 20 -m PROTGAMMAAUTO -p 12345 -x 12345). In addition, all genomes were annotated with DFAST with default parameters (Tanizawa et al. 2018) with certain manual corrections. The COG categories were extracted from the resulting gbk files (Tatusov et al. 2003).
The quality of five established cyanobacterial draft genome sequences and >200 reference genomes from the public databases were investigated with CheckM (Parks et al. 2015). This program allows to determine the completeness of a genome and to detect possible contaminations based on a reference data set of 43 universal protein encoding single copy genes. The absence of additional copies provides very strong and independent evidence for the axenic status of a bacterium, which is the prerequisite for meaningful metabolome analyses performed in the current study.
CheckM was used to retrieve the set of the 43 universal proteins from the selected genomes and to generate an alignment of the concatenated protein sequences. This alignment was manually refined using the Edit-option of the MUST-package (Philippe 1993). G-blocks was applied to eliminate ambiguously aligned amino acid positions, with the following parameters (conserved position: half identical; flanking position: three-fourths identical; maximum number of contiguous nonconserved positions: 2; Gap positions kept if less than half; Talavera and Castresana 2007). A phylogenetic maximum likelihood (ML) tree of the data set was generated with RAxML v8.2.10 (Stamatakis 2014) using the GTR + 4Γ model and parallel computation with 14 PThreads. To estimate the statistical support of the nodes 100 bootstrap-replicates were performed under the same conditions with the “rapid bootstrap analysis”-option of RAxML.
Pigment Extraction and Analyses
For the analysis of chlorophyll a (Chl a) and carotenoids, the cell pellets were resuspended in methanol (10 µl per 1 mg wet weight) and treated with three cycles of freezing in liquid nitrogen and thawing (4 °C; Sartory and Grobbelaar 1984; Henriques et al. 2007). Samples were incubated overnight in the dark at 4 °C and centrifuged for 10 min (10,000 × g). Pigments in the supernatant were separated by HPLC (Agilent 1260 Series, Agilent Technologies, Santa Clara, CA) equipped with a reverse phase C18 column (2.7 µm, 4.6 × 125 mm, Agilent Poroshell 120 EC-C18) and a diode array detector at 37 °C. The absorption spectra (428 nm Chl a, 450 nm β,β-carotene, 470 nm carotenoids) were collected from four independent samples for each cyanobacterium. The mobile phase, at a constant flow rate of 1 ml min−1, consisted of a mixture 0.5 M ammonium acetate (pH 7.2):methanol (20:80, v/v) for 5 min, followed by a gradient of 10 min towards 100% methanol and an isocratic run of 100% methanol for 25 min. Chl a and carotenoid pigments were identified by the retention times and by absorption spectra in comparison with commercial references. These standards, that is, canthaxanthin, β,β-carotene, Chl a, echinenone, myxoxanthophyll, and zeaxanthin were purchased from DHI LAB Products.
Phycobiliproteins were extracted from harvested biomass in 0.1 M phosphate buffer (pH 7.0) (10 µl per 1 mg wet weight), employing freezing (–20 °C, 1 h), and thawing (room temperature), for three times and a final sonication step in an ultrasonic bath filled with ice slurry for 5 min (Lawrenz et al. 2011). The samples were stored in the dark for 48 h, after which supernatants were obtained (4 °C, dark) by centrifugation (10,000 × g, 10 min). Analyses were carried out using a double-beam spectrophotometer at room temperature (PerkinElmer Lambda 25 UV/VIS). Continuous spectra of absorbance (300–700 nm) were collected from four biological replicates. The concentration of phycobiliproteins, phycocyanin (PC) and allophycocyanin (APC) were determined using the equation given by Bennett and Bogorad (1973). All results were referred to 1 mg of wet weight.
Metabolome Analyses
Metabolome extraction and analysis were performed as described previously (Zech et al. 2009; Krall et al. 2009) with some modifications: Cell pellets were resuspended in methanol (10 µl per 1 mg wet weight) followed by cell lysis in an ultrasonic bath for 10 min at room temperature. H2O (10 µl per 1 mg wet weight) was added to the suspension followed by vigorous shaking for 1 min. Subsequently, 500 µl of chloroform were added and vigorously shaken for 1 min followed by a centrifugation for 5 min at 4 °C and 12,000 × g. 50–200 µl of the polar phase were transferred into vials, spiked with 13C-ribitol as internal standard and dried in a vacuum concentrator for 1 h. For determination of extracellular metabolites, 10 µl of the supernatant were spiked with ribitol and dried for 1 h. Metabolome analysis was performed on an Agilent GC-MSD system (7890B coupled to a 5977 GC) equipped with a high-efficiency source (HES) and a PAL RTC system. A two-step derivatization with a methoxyamine hydrochloride solution (20 mg ml−1 in pyridine) and N-methyl-N-(trimethylsilyl)-trifluoracetamide was automatically performed with the PAL RTC system. One microliter of the sample was injected into a multimode inlet in pulsed splitless mode (30 psi until 0.75 min, 50 ml min−1 at 1 min). For quantification of the compatible solutes the samples were additionally injected in pulsed split mode with a split ratio of 10:1 (split flow of 12 ml min−1). Separation was conducted on an Agilent VF-5ms column with a helium flow of 1.2 ml min−1. The oven temperature was hold at 70 °C for 6 min and then linearly increased with 6 °C min−1 up to 325 °C. Ions were detected in scan mode from 70 to 700 m/z with 2.3 scans s−1.
Data analysis of intracellular metabolites was performed as previously described (Neumann-Schaal et al. 2015; Hofmann et al. 2018). The Wilcoxon–Mann–Whitney test using Benjamini–Hochberg correction was used for statistical analyses (Steglich et al. 2018). The concentration of lactate in the supernatants was corrected by the lactate concentration measured in blanks.
Ethanol and acetate were extracted and measured as previously described (Neumann-Schaal et al. 2015) with some minor modifications: 80 µl sulfuric acid and 200 µl o-cresol as internal standard were added to 800 µl supernatant. The volatile compounds were extracted with 150 µl tert-methylbutylether.
Determination of Glycogen Content
Cell pellets were resuspended in 150 µl 1× PBS followed by cell lysis in an ultrasonic bath filled with ice slurry for 10 min. Cells were centrifuged for 5 min and the glycogen content in 50 µl of the supernatant was determined with the colorimetric glycogen assay kit MET-5022 according to the instructions of the manufacturer (Cell Biolabs, Inc., San Diego).
Results
Genome Sequencing of Five Cyanobacteria
Draft genome sequencing of the five investigated cyanobacteria served as a quality control of the authenticity of the strains deposited at the DSMZ (table 1), and it provided the basis for the interpretation of the metabolic data established in the current study. The annotated draft genomes with an at least 114-fold average coverage were deposited at GenBank with the following accession numbers: Nostoc sp. DSM 107007 (147-fold; RSCN00000000); A. variabilis DSM 107003 (217-fold; RSCM00000000); C. desertica DSM 106972 (132-fold; RSCL00000000); Chlorogloeopsis sp. PCC 6912 (114-fold; RSCJ00000000); C. cubana DSM 107010 (173-fold; RSCK00000000). The CheckM analysis documented the absence of additional single copy genes in our newly established draft genomes of Nostoc and Anabaena (supplementary table S1, Supplementary Material online), thus confirming their axenic state, which was independently documented by light and electron microscopy (Figs. 1 and 2). The CheckM analyses revealed putative “contamination” levels between 1.59% and 1.87% in the axenic cultures of Chroococcidiopsis, Chlorogloeopsis, and Calothrix reflecting the occurrence of additional core gene copies in the predefined reference set. However, the comparison of all 91 completely sequenced cyanobacteria, which were investigated in the current study (supplementary table S1, Supplementary Material online), showed that only 45% of the strains (41/91) have a CheckM-based “contamination” level of 0%. 15% of these axenic strains (14/91) exhibit alleged “contaminations” of >1% and the finished genome of Acaryochloris marina MBIC 11017, which contains nine single-copy plasmids (Swingley et al. 2008), even exhibits a value of 5% (table 2). The detected pseudocontaminations reflect a low number of authentic core gene homologs, which might correlate with the considerable number of extrachromosomal elements in cyanobacteria. On the basis of our comparative genome analyses, we conclude that CheckM can be used as a reliable in silico tool to validate the axenicity of genome-sequenced cyanobacteria. We recommend the utilization of three stringent quality criteria, that is, 1) a “contamination” level below 2%, 2) a genome “completeness” of at least 98%, and 3) at least a 100-fold genome coverage.
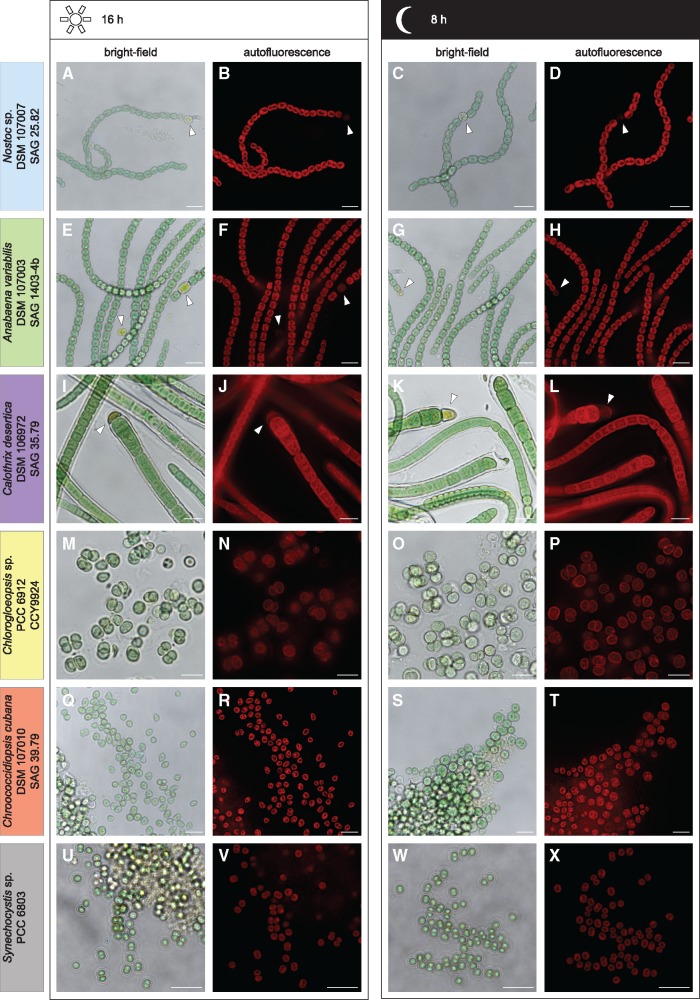
Fig. 1.
—Light microscopic analysis of the day–night cycle. Micrographs of the strains Nostoc sp. DSM 107007 (A–D), Anabaena variabilis DSM 107003 (E–H), Calothrix desertica DSM 106972 (I–L), Chlorogloeopsis sp. PCC 6912 (M–P), Chroococcidiopsis cubana DSM 107010 (Q–T), and Synechocystis sp. PCC 6803 (U–X). Cells were either analyzed after 16 h (day) or after 8 h (night). No change in the autofluorescence intensity could be observed during the day–night cycle (B&D, F&H, J&L, N&P, and R&T). While Nostoc sp. (A–D), A. variabilis (E–H), and C. desertica (I–L) are filamentous and form heterocysts (arrow heads), Chlorogloeopsis sp. (M–P), C. cubana (Q–T), and Synechocystis sp. (U–X) show multicellular aggregates or a unicellular form. Synechocystis sp. shows a higher amount of aggregates during the day (arrow head), which are hardly to find after 8 h of darkness (U–X). Scale bar 10 µm.
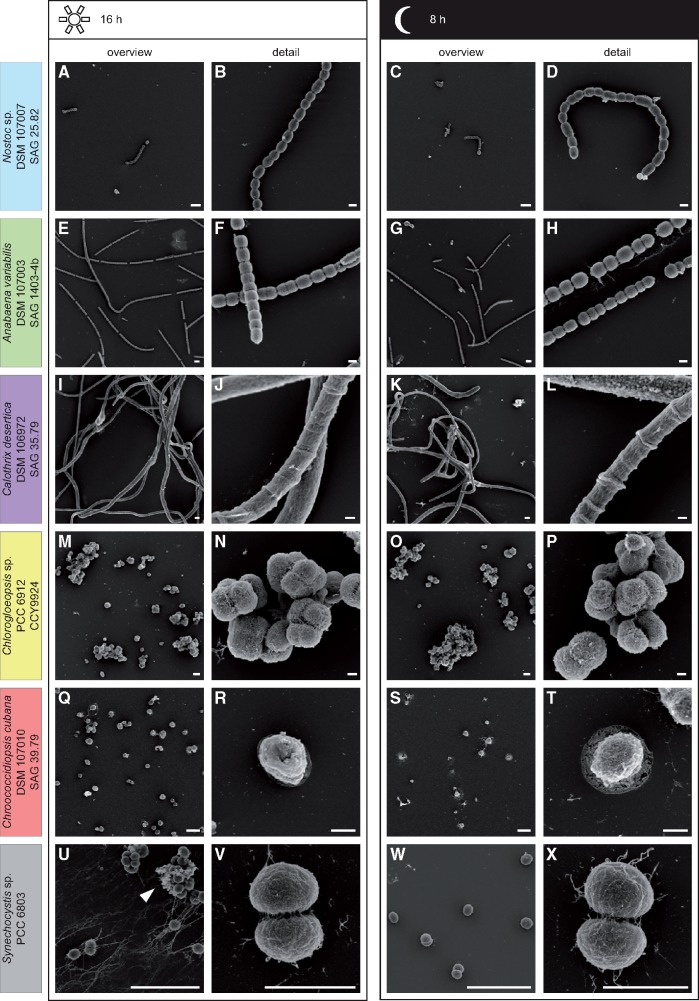
Fig. 2.
—Scanning electron microscopy analysis of the day–night cycle. Micrographs of the strains Nostoc sp. DSM 107007 (A–D), Anabaena variabilis DSM 107003 (E–H), Calothrix desertica DSM 106972 (I–L), Chlorogloeopsis sp. PCC 6912 (M–P), Chroococcidiopsis cubana DSM 107010 (Q–T), and Synechocystis sp. PCC 6803 (U–X). Cells were either analyzed after 16 h (day) or after 8 h (night) and are depicted as overview (A&C, E&G, I&K, M&O, Q&S, and U&W) and detailed (B&D, F&H, J&L, N&P, R&T, and V&X) illustration. No change in morphology could be observed during the day–night cycle (A–T). However, Synechocystis sp. (U–X) appears to form more aggregates after 16 h of light (arrow head). Scale bar overview: 10 µm and detail: 2 µm.
Table 2.
Genome Characteristics and CheckM Results of (A) Five Newly Established High-Quality Draft Genomes and (B) 20 Complete Reference Genomes from Axenic Cyanobacteria
| (A) High-Quality Draft Genomes | |||||
| Contigs | 23S, 16S, 5Sa | tRNAs | “Completeness” | “Contamination” | |
| Nostoc sp. DSM 107007 | 139 | 1, 1, 1 | 63 | 98.74 | 0.00 |
| Anabaena variabilis DSM 107003 | 54 | 1, 1b, 1 | 37 | 99.67 | 0.00 |
| Calothrix desertica DSM 106972 | 110 | 1, 1, 1 | 69 | 99.27 | 1.87 |
| Chlorogloeopsis sp. PCC 6912 | 84 | 1, 1, 1 | 38 | 99.76 | 1.77 |
| Chroococcidiopsis cubana DSM 107010 | 295 | 1, 1, 1 | 40 | 99.48 | 1.59 |
| (B) Complete Genomes | |||||
| Chr + ECRs | 23S, 16S, 5S | tRNAs | “Completeness” | “Contamination” | |
| Synechocystis sp. PCC 6803 | 1 + 4 | 2, 2, 2 | 41 | 99.78 | 0.00 |
| Synechococcus elongatus PCC 7942 | 1 + 1 | 2, 2, 2 | 44 | 100.00 | 0.00 |
| Prochlorococcus marinus CCMP 1375 | 1 + 0 | 1, 1, 1 | 40 | 100.00 | 0.00 |
| Calothrix sp. 336/3 | 1 + 3 | 1, 1, 1 | 60 | 100.00 | 0.00 |
| Crinalium epipsammum PCC 9333 | 1 + 8 | 4, 4, 4 | 43 | 99.48 | 0.00 |
| Cyanothece sp. PCC 7424 | 1 + 6 | 3, 3, 3 | 44 | 99.71 | 0.00 |
| Calothrix sp. NIES-4071 | 1 + 8 | 5, 5, 5 | 63 | 99.04 | 1.14 |
| Gloeobacter kilaueensis JS1 | 1 + 0 | 1, 1, 1 | 49 | 98.29 | 1.14 |
| Halomicronema hongdechloris C2206 | 1 + 0 | 2, 2, 2 | 45 | 98.82 | 1.18 |
| Leptolyngbya boryana IAM M-101 | 1 + 3 | 3, 3, 3 | 67 | 99.41 | 1.18 |
| Nostoc sp. N6 | 1 + 10 | 4, 4, 4 | 93 | 99.44 | 1.19 |
| Rivularia sp. PCC 7116 | 1 + 2 | 3, 3, 3 | 51 | 99.78 | 1.22 |
| Synechococcus sp. JA-3-3Ab | 1 + 0 | 2, 2, 2 | 46 | 100.00 | 1.32 |
| Moorea producens PAL-8-15-08-1 | 1 + 1 | 2, 2, 2 | 55 | 99.26 | 1.37 |
| Chroococcidiopsis thermalis PCC 7203 | 1 + 2 | 3, 3, 3 | 46 | 99.63 | 1.44 |
| Microcystis aeruginosa PCC 7806SL | 1 + 0 | 2, 2, 2 | 42 | 99.67 | 1.45 |
| Leptolyngbya sp. O-77 | 1 + 0 | 2, 2, 2 | 44 | 98.70 | 1.53 |
| Chamaesiphon minutus PCC 6605 | 1 + 2 | 3, 3, 3 | 55 | 99.48 | 2.11 |
| Nostoc flagelliforme CCNUN1 | 1 + 8 | 6, 6, 5 | 65 | 100.00 | 3.07 |
| Acaryochloris marina MBIC 11017 | 1 + 9 | 2, 2, 2 | 67 | 99.53 | 5.07 |
Note.—Putative CheckM-based “contamination” levels above 1% are highlighted in bold.
Coverage differences indicate the presence of more than one rRNA operon per strain.
Partial 16S-rRNA gene located on two contigs; Chr, chromosome; ECRs, extrachromosomal elements. The complete CheckM analysis is shown in supplementary table S1, Supplementary Material online.
The comparison of the newly established draft genome sequences with deposited 16S rRNA sequences, the draft genome of C. cubana CCALA 043 (=DSM 107010) and the complete genome of Nostoc sp. PCC 7120 validated the authenticity of the strains. Illumina MiSeq sequencing and de novo assembly resulted in high-quality draft genomes sensu (Bowers et al. 2017) with >99% genome completeness harboring between 54 and 295 contigs (tables 1 and 2, supplementary table S1, Supplementary Material online). The at first sight rather disappointing number of 295 contigs for Chroococcidiopsis probably reflects a higher number of repetitive sequences within the genome; this view is supported by the first draft genome of this cyanobacterium deposited at GenBank that even contains 926 contigs (NZ_PVWM00000000.1). The estimated genome sizes of the five newly established high-quality draft genomes range between 6.12 Mb for Anabaena and 11.62 Mb for Calothrix. The latter contains a nearly 3-fold higher number of genes compared with the unicellular reference strain Synechocystis with a small genome of only 3.95 Mb (table 1). Furthermore, C. desertica has the third largest cyanobacterial genome sequenced to date, besides two closely related strains namely Scytonema hofmannii PCC 7110 and Calothrix sp. NIES-4071 with genome sizes of 12.29 Mb and 12.05 Mb, respectively (NZ_ANNX00000000.2, NZ_AP018255; fig. 3).
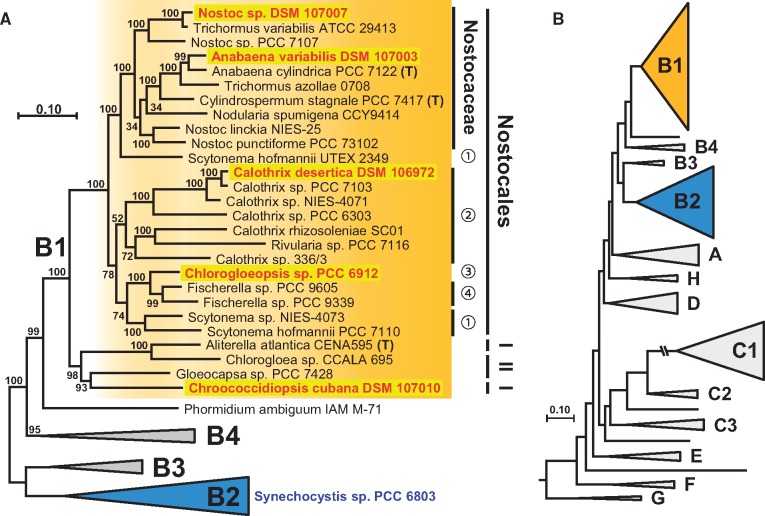
Fig. 3.
—Phylogenetic maximum likelihood tree of 45 cyanobacteria. (A) The MLSA phylogeny was inferred by RAxML under the GTR + 4Γ model based on a concatenated alignment of 43 proteins with 6,566 amino acid positions. Bootstrap support of 100 replicates is indicated. Twenty-seven cyanobacterial taxa of subclade B1 are shown in detail; subclade B2 that contains the reference strain Synechocystis sp. PCC 6803, B3 and B4 are shown as triangles. I, Chroococcidiopsidales; II, Chroococcales; numbers represent families of the prevalent order Nostocales: 1, Scytonemaceae; 2, Rivulariaceae; 3, Chlorogloeopsidaceae; 4, Haplosiphonaceae. (B) Schematic overview of the cyanobacterial diversity (see MLSA tree of 208 taxa; supplementary fig. S1, Supplementary Material online).
Phylogenetic Analyses
Selection of Cyanobacterial Reference Taxa for Phylogenetic Analyses
One aim of the current study was the establishment of a comprehensive phylogenetic tree of genome sequenced cyanobacteria based on a multilocus sequence alignment (MLSA) of 43 universal proteins. The phylogenetically broad taxon sampling based on the reference study of Shih et al. (2013) and comprehensive BLAST searches in the public databases with the beta subunit of the DNA-dependent RNA polymerase (RpoB), which is with ∼1,100 amino acid positions a well-suited and comparably large phylogenetic marker (Case et al. 2007; Petersen and Wagner-Döbler 2017). The sampled taxa should represent the phylogenetic diversity of the phylum cyanobacteria and thus only a comparably small subset of, for example, >600 available Prochlorococcus genomes was included in the analysis. We established a set of 207 cyanobacterial reference genomes in addition to the five high-quality draft genomes that were sequenced in the current study (see above). The quality of the sequence data varied according to the NCBI criteria between complete, nearly complete and draft genomes with up to 300 contigs, and some genomes from nonaxenic strains even originated in meta-genome binning approaches (Cornet, Bertrand, et al. 2018). Accordingly, and based on a recent report revealing comparably high contamination levels in some cyanobacterial genomes (Cornet, Meunier, et al. 2018), we investigated all genomes regarding their completeness and possible contaminations. CheckM analyses of the 207 reference genomes resulted in the removal of four strains (supplementary table S1, Supplementary Material online). Calothrix sp. NIES-4101 was eliminated, because it displayed a considerable contamination level of 46%, and three strains were deleted due to their estimated completeness of <90%. All five newly sequenced cyanobacterial strains passed our strict CheckM-based quality criteria, which moreover ensured that the underlying data set of 43 universal marker proteins for the phylogenetic analyses was with 99.23% data coverage nearly complete.
Phylogenetic MLSA Tree of the Phylum Cyanobacteria Represented by 208 Strains
The complete phylogenetic MLSA tree with 208 taxa that is based on the concatenated alignment of 43 proteins with 6,458 amino acid positions are shown in supplementary figure S1, Supplementary Material online. The tree was rooted by two genome sequenced Gloeobacter species, which ensures the comparability with the study of Shih et al. (2013), even if the actual positioning of this “violet” lineage is still a matter of debate (Szöllosi et al. 2012; Saw et al. 2013). The division into higher order groups designated as clade A to G corresponds to the MLSA tree of Shih et al. (2013), which is based on high bootstrap support in a comparable protein phylogeny of 31 universal core genes, originally introduced by Wu and Eisen (2008). The general structure of our tree is irrespective of almost twice the number of taxa comparable to those of Shih et al. (2013). All distinct lineages, including subclades B1–B3 and C1–C3, were recovered and the improved taxon sampling allowed the detection of novel monophyletic lineages such as clade H located between clade A and D (supplementary fig. S1, Supplementary Material online).
Subclade B1 that is shown on top of the tree comprises all five genome sequenced strains, whose metabolomes were analyzed in the current study (supplementary fig. S1, Supplementary Material online). The reference strain Synechocystis sp. PCC 6803 is located in subclade B2. The monophyly of clade B is in contrast to clade A and D highly supported (97% Bootstrap proportion [BP]), and subclades B1 and B2 are based on their phylogenetic diversity and complexity the probably best sampled lineages among cyanobacteria. About one third of the 208 taxa in our analysis (69 taxa) belong to subclade B1 and 20% (41 taxa) are placed in subclade B2. Its slowly evolving sistergroup B3 comprises only four taxa. Crinalium epipsammum PCC 9333 and Chamaesiphon minutus PCC 6605, whose phylogenetic positioning was previously unresolved (Shih et al. 2013), form together with Merismopedia glauca CCAP 1448/3 a well-supported subclade (99% BP) that was designated B4. Finally, the newly analyzed strain Phormidium ambiguum IAM M71 represents a connecting link between subclades B1 and B4, and it is due to its statistically well-supported distinct positioning within the tree probably the first representative of a fifth subclade.
The current study recovered the clades and branching pattern of the reference study (Shih et al. 2013), but the doubling of the taxa resulted in a clear reduction of the statistical support, especially of the backbone of the tree connecting the distinct clades (supplementary fig. S1, Supplementary Material online). The lower support correlates with the inclusion of new and deeply diverging taxa that likely represent the first members of novel (sub)clades. Specifically, and apart from P. ambiguum IAM M71 in clade B, it is Prochlorothrix hollandica PCC 9006 (Schyns et al. 1997) and Limnothrix sp. PR1529 representing two novel lineages within the clade C, as well as Gloeomargarita lithophora Alchichica-D10 CCAP 1437/1 (Moreira et al. 2017) that was recently identified as closest free-living relative of chloroplasts (Ponce-Toledo et al. 2017; de Vries and Archibald 2017).
The by far fastest evolving lineage in supplementary figure S1, Supplementary Material online is subclade C1 that mainly comprises marine Synechococcus and Prochlorococcus strains with strongly reduced genomes usually with sizes below 3.0 and 2.0 Mb, respectively. It’s very long basal branch (100% BP), which reflects a phase of pronounced accelerated evolution, is shown with a third of its original length. C1 is with >600 available single-cell genomes just for the genus Prochlorococcus (Kashtan et al. 2014) a very intensely studied lineage of cyanobacteria. In contrast, C2 is only represented by the three highly similar Synechococcus elongatus strains PCC 7942, UTEX2973 and PCC 6301 (not shown).
Subclade B1 and the Classification of Cyanobacteria
The MLSA tree in figure 3, which is based on a largely reduced taxon sampling of clade B (45 taxa in fig. 3 vs 117 taxa in supplementary fig. S1, Supplementary Material online), shows that the five newly sequenced cyanobacteria cover the phylogenetic depth of subclade B1. The position of the subclades B1–B4 with respect to the entire cyanobacterial diversity is depicted in figure 3B. Subclade B1, which is highlighted in orange throughout the manuscript, harbors the cyanobacterial taxa with the largest genomes (see above) and represents from a morphological perspective with unicellular, tetrade forming and filamentous strains the most versatile lineage among cyanobacteria (table 1; see also microscopic images in Figs. 1 and 2). B1 contains three different cyanobacterial orders, that is, a monophyletic group with 23 Nostocales (100% BP) and a paraphyletic assemblage comprising two Chroococcales and two Chroococcidiopsidales including the unicellular strain C. cubana DSM 107010 (98% BP). The considerable statistical support of subclade B1 revealed several taxonomic incongruencies of the studied cyanobacteria. One example is the weakly supported family Rivulariaceae (52% BP), where the genus Calothrix displays a greater phylogenetic depth than the family Nostocaceae (fig. 3). A similar situation is encountered in the genus Nostoc, which represents the entire diversity of the family, encompassing several other genera (Anabaena, Cylindrospermum, Nodularia, and Trichormus). Finally, members of the family Scytonemaceae are located in two highly distinct subtrees, suggesting a so far cryptic new family (supplementary fig. S1, Supplementary Material online). A prime example of misclassification on species level is the occurrence of a S. hofmannii strain in both subtrees. Strain UTEX2349 is a close relative of Tolypothrix sp. NIES-4075 in the comprehensive tree with 208 taxa, whereas strain PCC 7110 forms a separate branch together with Scytonema sp. NIES-4073 and Mastigocladopsis repens PCC 10914 (supplementary fig. S1, Supplementary Material online). These examples document the actual dilemma of the cyanobacterial classification with paraphyletic groups at different taxonomic levels.
Comparison of MLSA, RpoB, and 16S rRNA Phylogenies
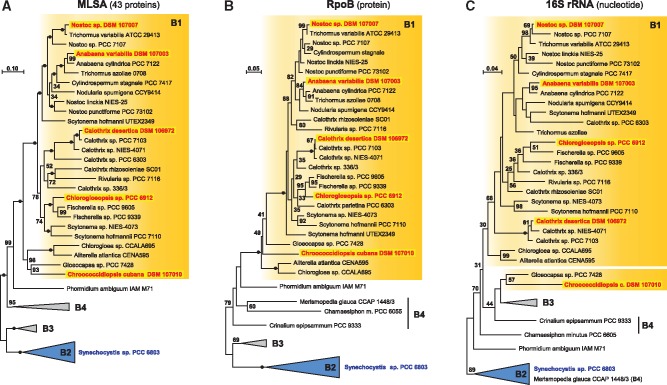
The MLSA analysis in figure 3 that is based on 43 universally conserved marker proteins provided a solid approximation of the evolutionary relationship of cyanobacteria in subclade B1. Figure 4 compares the resolving power of the MLSA, RpoB, and 16S rRNA phylogenies based on a comparable taxon sampling. The two former protein phylogenies were calculated based on 6,566 and 1,082 amino acid position, respectively, the 16S rRNA gene tree comprised 1,481 nucleotide positions. The 16S rRNA showed the by far lowest resolution, which is reflected by only a single subtree that is supported by maximal statistical support comprising three closely related taxa of the genus Calothrix. In contrast, 100% BP was obtained for 5/31 and 19/31 nodes in the RpoB and MLSA phylogeny, respectively. If we use the less stringent cut-off value of 70% BP, considered by some authors as significant, the picture is similar, but the dominance of the protein data sets is with 28/31 (MLSA) and 14/31 (RpoB) versus 6/31 (16S rRNA) less pronounced. According to its resolving power, the 16S rRNA phylogeny did not even recover the monophyly of subclade B1 (fig. 4C), which was supported by 100% BP in both protein trees (fig. 4A and B). The same observation was made for the monophyly of the Nostocaceae that was not shown by the 16S rRNA, but supported by 82% (RpoB) and 100% BP (MLSA) in the protein trees. The sistergroup positioning of S. hofmannii UTEX2349 and Nostocaceae could not be recovered in the RpoB tree, but the correct positioning was not statistically contradicted, which reflects typical limits of comparably small data sets. The MLSA analysis shows in comparison to the two single gene/protein phylogenies the by far best resolution with solid statistical support for the internal structure of subclade B1. However, the monophyly of Rivulariaceae is only weakly supported with 52% BP and the actual relationships between 1) Nodularia spumigena, 2) two distinct Nostoc strains, and 3) the Anabaena, Trichormus, and Cylindrospermus subtree remains unresolved.
Fig. 4.
—Comparison of MLSA, RpoB, and 16S rRNA phylogenies. Three phylogenetic maximum likelihood analyses of clade B with the same taxon sampling (45 taxa). Subclade B1 is highlighted in orange, subclades B2–B4 are depending on their monopyly collapsed to triangles. MLSA and RpoB trees were midpoint rooted by the outgroup (B2, B3), the 16S rRNA gene tree was rooted with B2. 100% BP is indicated by a black dot. The scale bar represents the mean number of inferred nonsynonymous substitutions per amino acid position and in case of the 16S rDNA the number of substitutions inferred by the model.
Phylogenetic RAxML Analyses of the Isocitrate Lyase, Malate Synthase, and PEP Carboxykinase
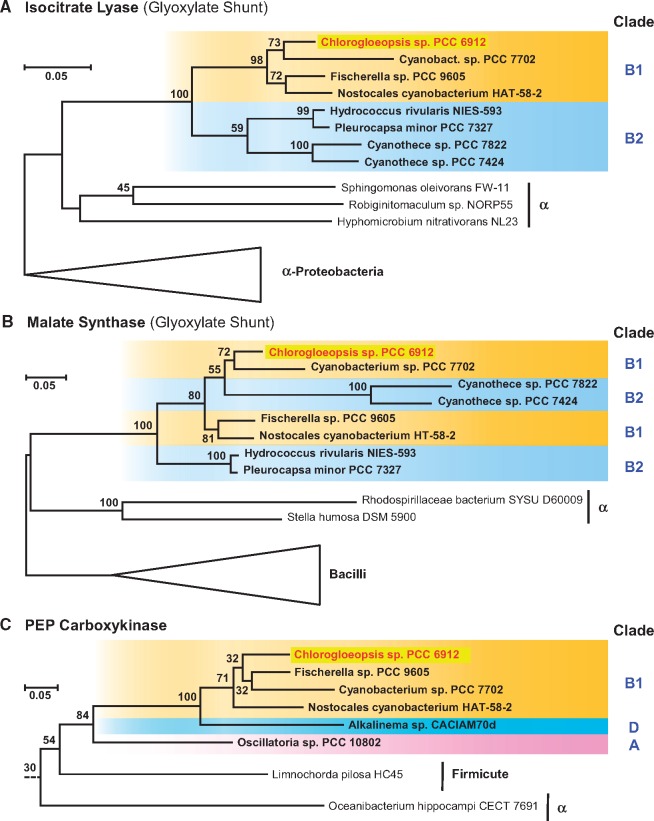
The reconstruction of their central carbon metabolism showed the occurrence of lineage-specific pathways among the six cyanobacteria investigated in the current study (see below). Prime examples are the enzymes of the glyoxylate shunt (isocitrate lyase [ICL, EC 4.1.3.1], malate synthase [MLS, EC 2.3.3.9]) and the phosphoenolpyruvate carboxykinase (PEPCK, EC 4.1.1.49), which have exclusively been detected in Chlorogloeopsis. Comprehensive BLASTP searches in the nonredundant (NR) NCBI protein database with hundreds of cyanobacterial (draft-) genomes revealed only eight cyanobacterial hits for orthologous enzymes of the glyoxylate shunt and six specific hits for the PEPCK (fig. 5, supplementary fig. S2, Supplementary Material online).
Fig. 5.
—Phylogenetic RAxML analysis of the (A) isocitrate lyase (ICL), (B) malate synthase (MLS), and (C) PEP carboxykinase (PEPCK). Cyanobacterial sequences are highlighted in bold and the color code of their phylogenetic affiliation corresponds to the MLSA tree presented in supplementary figure S1, Supplementary Material online. Outgroup sequences are indicated by triangles or a dotted line. The complete trees including sequence accession numbers are presented in supplementary figure S2, Supplementary Material online.
The phylogenetic RAxML analysis of the ICL shows a common branching of the four strains from subclade B1, that is, Chlorogloeopsis sp. PCC 6912, Cyanobacterium sp. PCC 7702, Fischerella sp. PCC 9605 and Nostocales cyanobacterium HAT-58-2, as well as the four strains from subclade B2, that is, Hydrococcus rivularis NIES-593, Pleurocapsa minor PCC 7327 and two Cyanothece strains (PCC 7822, PCC 7424). The common branching of subclades B1 and B2 (100% BP) and their positioning among bacterial reference sequences clearly show that cyanobacteria recruited the ICL via a single horizontal gene transfer (HGT) probably from an alphaproteobacterium (fig. 5). The distinct positioning of the cyanobacterial strains is indicative of a second HGT between subclade B1 and B2 (supplementary fig. S1, Supplementary Material online). The actual localization of ICL-containing cyanobacteria in the MLSA-tree provides evidence for at least two secondary losses of the ICL gene. The most parsimonious scenario, which is shown in supplementary figure S1, Supplementary Material online, predicts one loss in the common ancestor of the Fischerella clade ranging from strain PCC 9431 to PCC 7521 (subclade B1) and a second loss in the common ancestor of Aphanothece hegewaldii and Microcystis aeruginosa (subclade B2).
The MLS-phylogeny is largely consistent with the ICL-tree, since it comprises the same cyanobacterial taxa and shows a comparable branching pattern. The sole difference is the nested positioning of the two Cyanothece sequences (B2) within subclade B1. The very long branch might either indicate an additional HGT from subclade B1 combined with an accelerated evolution rate or it reflects a phylogenetic misplacement associated with a rather low bootstrap support. Nevertheless, a synchronous evolution of these marker enzymes of the glyoxylate shunt is strongly supported by the characteristic tandem organization of both genes (MLS-ICL), which has been observed in all eight cyanobacteria harboring this metabolic bypass of the tricarboxylic acid (TCA) cycle. A comparable genetic organization of this mini-pathway has also been reported for Escherichia coli (Ahn et al. 2016), but the presence of ∼300 nucleotides in the noncoding area between both genes argues against a current function as a bicistronic operon. However, the tight spatial coupling and the synchronous evolution provides strong evidence that this pathway has been horizontally transferred en bloc at least in cyanobacterial evolution.
The PEPCK phylogeny with six cyanobacterial sequences documents an independent recruitment from a heterotrophic bacterium (fig. 5C). The most parsimonious scenario for the current distribution of PEPCK genes in clade A, D and subclade B1 proposes one HGT into cyanobacteria followed by two additional internal transfers. The distribution in subclade B1 is noteworthy, because the PEPCK was also specifically found in those four taxa that contain the glyoxylate shunt. A possible co-evolution of the currently noncoupled PEPCK and MLS-ICL genes remains speculative, because the internal PEPCK branching pattern in B1 is not resolved. However, the PEPCK gene is absent in the genomes of 66 other cyanobacteria all located in subtree B1 (supplementary fig. S1, Supplementary Material online), which might indicate a beneficial metabolic linkage with the glyoxylate shunt.
Light and Electron Scanning Microscopy
To verify the axenicity and to describe and compare the morphology of the strains Nostoc, Anabaena, Calothrix, Chroococcidiopsis, Chlorogloeopsis, and Synechocystis depending on the day–night cycle, we performed light- and scanning electron microscopy (figs. 1 and 2). While the filamentous strains Nostoc, Anabaena, and Calothrix formed trichomes up to 1 mm in length ((figs. 1 and 2A–L), the strains Chlorogloeopsis, Chroococcidiopsis, and Synechocystis were unicellular or form multicellular aggregates ((figs. 1 and 2M–X). Filamentous strains can contain heterocysts that appeared yellowish (fig. 2, white arrowhead) and showed a lower autofluorescence than vegetative cells. However, no microbial contamination by other bacteria or fungi could be observed. Cell morphologies appeared as described before (Rippka et al. 1979; Gonzalez-Esquer et al. 2016). Morphological traits and individual cell sizes are summarized in table 1. By comparing cells during the day and night cycle, only Synechocystis showed a morphological difference. Cells exposed to light, formed more aggregates and exopolysaccharides. However, after the lack of light for 8 h, cells are looser and showed hardly any aggregates ((figs. 1 and 2U–X).
Photosynthetic Pigments
The primary photosynthetic pigments in cyanobacteria are Chl a, various carotenoids and phycobilins (Jeffrey et al. 2011). The pigment composition of Chl a to the main carotenoids (fig. 6A) and the abundances of phycobiliproteins in the day and night cycles (fig. 6B) were compared studying the effect of low light/dark cycling.
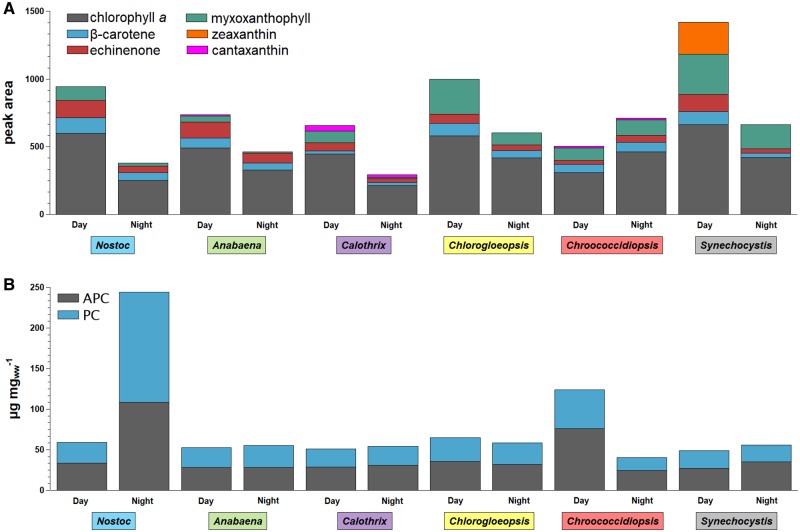
Fig. 6.
—Comparison of pigments at the end of the day and night cycle. Shown are peak areas for Chl a and carotenoids (A) and phycobiliprotein concentration PC and APC [µg mg−1] (B) for Nostoc sp. DSM 107007 (blue), Anabaena variabilis DSM 107003 (green), Calothrix desertica DSM 106972 (purple), Chlorogloeopsis sp. PCC 6912 (yellow), Chroococcidiopsis cubana DSM 107010 (red), and Synechocystis sp. PCC 6803 (gray) in comparison of day and night. PC, C-phycocyanin; APC, allophycocyanin.
In the day cycle, Chl a was most abundant in Synechocystis, whereas in the night cycle, Chl a was most abundant in Chroococcidiopsis. Chroococcidiopsis was the sole cyanobacterium studied that showed a higher Chl a content (1.5-fold higher) in the night cycle than in the day cycle. The overall lowest Chl a content was found in Calothrix at the end of the night cycle (fig. 6A).
In cyanobacteria the most abundant carotenoids are carotenes (e.g., β-carotene) and xanthophylls (e.g., canthaxanthin, echinenone, myxoxanthophyll, zeaxanthin), which are oxygenated derivatives of the carotenes (Takaichi and Mochimaru 2007). The studied carotenoids were selected according to previously published data. Under the studied conditions Synechocystis was the sole cyanobacterium in this study that contained zeaxanthin (supplementary table S2, Supplementary Material online), whereas trace amounts previously described for Nostoc could not be detected (Takaichi et al. 2001, 2005). Anabaena, Calothrix, Chlorogloeopsis, and Chroococcidiopsis contained the characteristic cyanobacterial carotenoids β-carotene, echinenone, and myxoxanthophyll. Canthaxanthin could exclusively be detected in Calothrix and Chlorogloeopsis.
When comparing the combined carotenoid concentration with the Chl a concentration, all ratios increased in the day cycle (supplementary table S2, Supplementary Material online). This increase is lowest for Chroococcidiopsis (1.16-fold) and highest for Synechocystis (1.98-fold). However, in the combined carotenoid concentration, only the five specifically analyzed pigments were considered.
Cyanobacteria were named according to their characteristic color, which is caused by phycobiliproteins. These form sophisticated antenna complexes for light harvesting called phycobilisomes. All cyanobacterial species contain APC and almost all species contain C-PC (Stadnichuk et al. 2015).
After the night cycle Nostoc showed with 4.16-fold the highest increase concomitant with the highest measured phycobiliprotein concentration (244 µg mg−1; supplementary table S3, Supplementary Material online). In contrast, Chroococcidiopsis showed a high decrease in PC-APC concentration by 3.07-fold in the night cycle (fig. 6B).
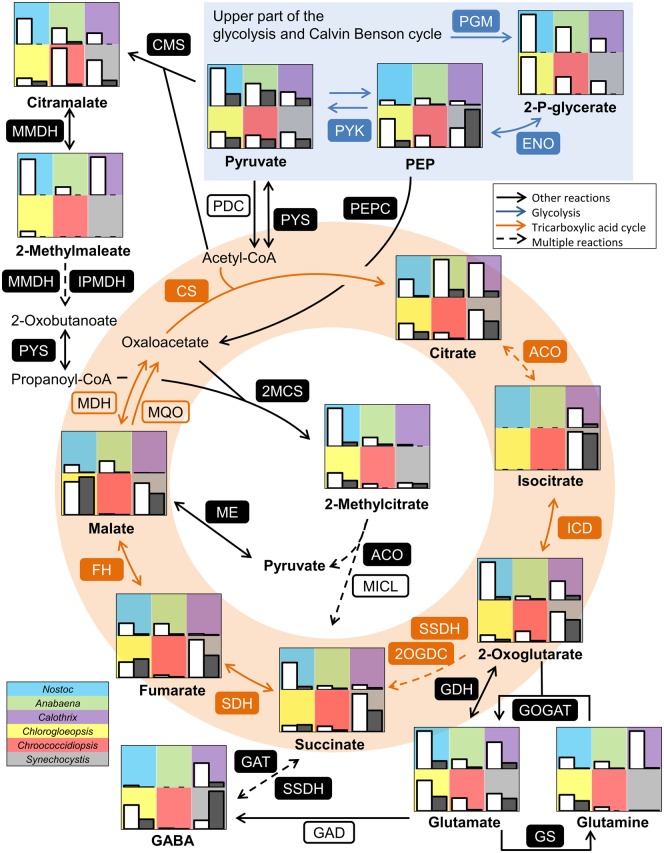
Reconstruction of the Central Carbon Metabolism and Metabolic Changes during Day–Night Cycles
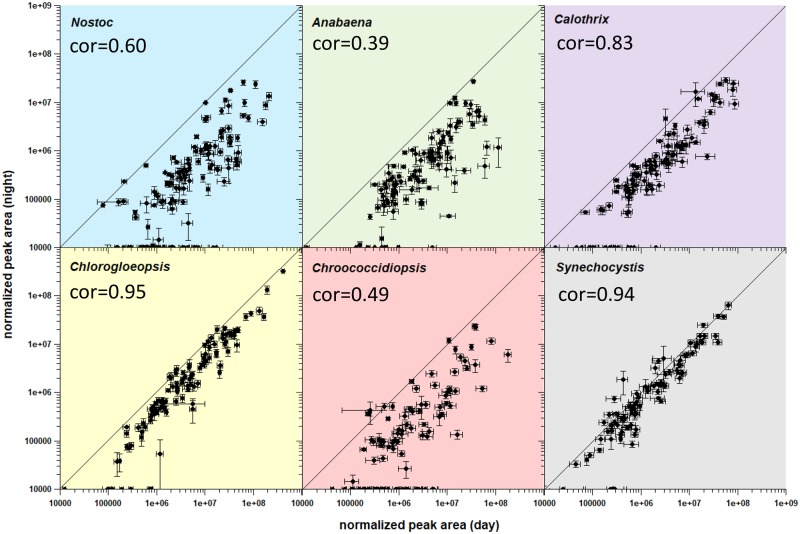
Overall, we identified 92–126 metabolites in cells after 16 h of light exposure and 67–100 metabolites in cells after 8 h in the dark (fig. 7). The concentration of almost all metabolites was drastically reduced after the night cycle. Five to 33 metabolites were not detectable in the night samples. We analyzed extracellularly the relative abundance of three putative fermentation products lactate, ethanol, and acetate (supplementary fig. S3, Supplementary Material online). Lactate, ethanol, and acetate were only detectable in very low amounts or even below the detection limit in the culture supernatant and were not increased in the night cycle. Thus, we assume that the cultures were not anoxygenic during the night cycle. The difference between day and night was lowest for Synechocystis: Actually, only 4-aminobutanoate (γ-amino butyric acid [GABA]), glycerol-3-phosphate, and phosphoenolpyruvate were nonsignificantly increased in this strain. Furthermore, the decreasing effects were lower for Chlorogloeopsis compared with Nostoc, Anabaena, Calothrix, and Chroococcidiopsis. Metabolites of Chlorogloeopsis and Synechocystis showed a roughly positive linear correlation between the day and night (Pearson correlation of 0.95 or 0.94, respectively).
Fig. 7.
—Comparison of logarithmized normalized peak areas of day and night cycle. Shown are metabolites for Nostoc sp. DSM 107007 (blue), Anabaena variabilis DSM 107003 (green), Calothrix desertica DSM 106972 (purple), Chlorogloeopsis sp. PCC 6912 (yellow), Chroococcidiopsis cubana DSM 107010 (red), and Synechocystis sp. PCC 6803 (gray) in comparison of day and night. Error bars represent the standard error. For detailed values see supplementary table S4, Supplementary Material online. The Pearson correlation coefficient (cor) was determined by using the R function “cor.test().”
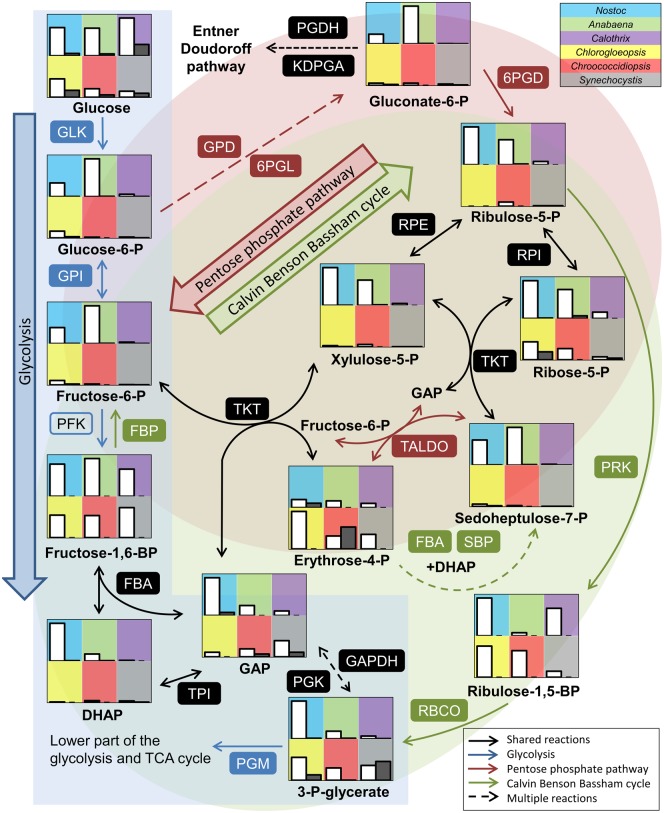
We detected a majority of the metabolites that can be assigned to the Calvin Benson Bassham cycle (CBB cycle) and the oxidative pentose phosphate pathway (OPP pathway; fig. 8). On the genomic level, both pathways were reported to be conserved in all cyanobacteria (Beck et al. 2012). All metabolites were more abundant in the day cycle than in the night cycle. The presence of 6-phosphogluconate indicated an active OPP pathway. 6-Phosphogluconate was most abundant in Anabaena and intermediates of the CBB cycle were lower abundant compared with the other strains which might indicate a major role of the OPP pathway and a minor role of the CBB cycle in Anabaena. Furthermore, intermediates of the OPP pathway and CBB cycle were less abundant in Synechocystis, which showed a higher abundance of metabolites of the TCA cycle. Calothrix showed also an active CBB cycle but intermediates of the OPP pathway were only detectable in small amounts. Interestingly, intermediates of the glycolysis had a low abundance even though this strain showed the highest intracellular glucose level. Chroococcidiopsis showed higher levels of erythrose-4-phosphate at the end of the night cycle compared with the day, whereas all other metabolites of the central carbon metabolism were less abundant at night.
Fig. 8.
—Comparison of the Calvin Benson Bassham cycle, glycolysis/gluconeogenesis and pentose phosphate pathway. Shown are relative normalized peak areas for Nostoc sp. DSM 107007 (blue), Anabaena variabilis DSM 107003 (green), Calothrix desertica DSM 106972 (purple), Chlorogloeopsis sp. PCC 6912 (yellow), Chroococcidiopsis cubana DSM 107010 (red), and Synechocystis sp. PCC 6803 (gray) at day (white bars) and night (gray bars). For detailed values see supplementary table S4, Supplementary Material online. Enzymes present in all strains are labeled with filled boxes. Descriptions of the abbreviations for the enzymes and the corresponding gene locus for each strain are listed in supplementary table S5, Supplementary Material online.
Previous studies on genomic level showed that other parts of the central carbon metabolism including the glycolysis and the TCA cycle are less conserved among cyanobacteria (Beck et al. 2012). Most enzymes of the classical glycolysis are present in the annotation of all six strains analyzed, but neither the phosphofructokinase (PFK, EC 2.7.1.11) nor orthologs of alternative enzymes for this reaction (EC 2.7.1.90, EC 2.7.1.46) could be identified in Calothrix (supplementary table S5, Supplementary Material online). This observation, which based on in silico analyses of the high-quality draft genome (GenBank file RSCL00000000; supplementary table S1, Supplementary Material online), was independently supported by the low abundance of the respective metabolites. The Entner–Doudoroff pathway (ED pathway) is described as alternative glucose degradation pathway in cyanobacteria (Chen et al. 2016). We could find homologous genes in all strains (supplementary table S5, Supplementary Material online). However, the key metabolite 2-keto-3-deoxy-6-phosphogluconate (KDPG) was not detectable in the analyzed strains. Consequently, we conclude that Calothrix is the sole strain with a nonfunctional glycolysis and hence degrades sugars and sugar polymers rather via the OPP pathway than the ED pathway. We observed higher levels of glucose-6-phosphate and fructose-6-phosphate in Anabaena and Nostoc, whereas C3 intermediates were enriched only in Nostoc (fig. 8). The lower part of the glycolytic pathway showed a shift, as these intermediates became more abundant in Chlorogloeopsis, Chroococcidiopsis, and Synechocystis, whereas pyruvate as the entry metabolite of the TCA cycle and several biosynthetic pathways showed a rather homogenous abundance in all strains (fig. 9).
Fig. 9.
—Comparison of the lower part of the glycolysis and TCA cycle. Shown are relative normalized peak areas for Nostoc sp. DSM 107007 (blue), Anabaena variabilis DSM 107003 (green), Calothrix desertica DSM 106972 (purple), Chlorogloeopsis sp. PCC 6912 (yellow), Chroococcidiopsis cubana DSM 107010 (red), and Synechocystis sp. PCC 6803 (gray) at day (white bars) and night (gray bars). For detailed values see supplementary table S4, Supplementary Material online. Enzymes present in all strains are labeled with filled boxes. Descriptions of the abbreviations for the enzymes and the corresponding gene locus for each strain are listed in supplementary table S5, Supplementary Material online.
The abundance of TCA cycle intermediates was similar at day and night for Chlorogloeopsis and Synechocystis, whereas all other strains showed a reduced abundance during the night (fig. 9). Anabaena showed the highest citrate level, but the following intermediates of the TCA cycle occurred in lower concentrations compared with the other strains. Additionally, we could detect isocitrate in Calothrix and Synechocystis. This observation was independent from the used medium.
The lack of any 2-oxoglutarate dehydrogenase in the genome annotations corresponds to the well-known fact that cyanobacteria do not possess a conventional TCA cycle. This step is bypassed by a 2-oxoglutarate decarboxylase (2OGDC, EC 4.1.1.71) and a succinate-semialdehyde dehydrogenase (SSDH, EC 1.2.1.79; Zhang and Bryant 2011), two enzymes that could be annotated in all strains (supplementary table S5, Supplementary Material online). However, while we detected almost all metabolites of the TCA cycle, we could not detect succinate semialdehyde, which might reflect a complex formation of both enzymes resulting in a fast processing of the intermediate (fig. 9). Metabolite levels in Calothrix indicated a higher activity of the pathway from 2-oxoglutarate into glutamine instead of succinate.
Furthermore, 2-oxoglutarate has previously been reported to act as a sensor for the nitrogen status of the cell (Muro-Pastor et al. 2005). However, the observed glutamate concentrations do not indicate a nitrogen limitation in our experimental set-up. Another possible bypass of the TCA cycle is the GABA shunt (Zhang et al. 2016). The necessary glutamate decarboxylase (GAD, EC 4.1.1.15) has been exclusively annotated in Synechocystis (sll1641) and this cyanobacterium showed higher amounts of GABA during the night (fig. 9).
The left part of the TCA cycle also showed some differences between the isolates (fig. 9): A malate dehydrogenase (MDH, EC 1.1.1.37) was annotated in all strains but Chroococcidiopsis, where this enzyme is replaced by the irreversible malate quinone oxidoreductase (MQO, EC 1.1.5.4; supplementary table S5, Supplementary Material online). The malate, fumarate, and succinate levels were lowest in Chroococcidiopsis and Calothrix. Chlorogloeopsis showed lower amounts of fumarate and succinate compared with Synechocystis, but malate was more abundant.
Intermediates of the TCA cycle can be replenished by two well characterized reactions: Via the PEP carboxylase (PEPC, EC 4.1.1.31) and the malic enzyme (ME, EC 1.1.1.39), which have been annotated in all strains (fig. 9). The genes for the glyoxylate shunt (EC 2.3.3.9 and EC 4.1.3.1) and the phosphoenolpyruvate carboxykinase (PEPCK, EC 4.1.1.49) are only present in Chlorogloeopsis (supplementary table S5, Supplementary Material online) and were acquired by HGT as described above. The presence of citramalate, 2-methylmaleate and 2-methylcitrate especially in Nostoc may point towards an alternative pathway of anaplerosis.
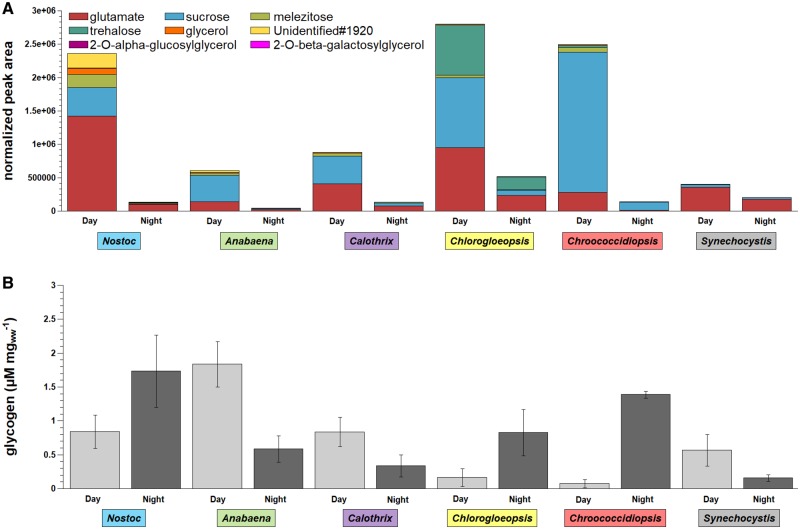
Preferences of Compatible Solutes and Small Storage Compounds
All six investigated strains showed high intracellular levels of glutamate and sucrose, but in different amounts and ratios (fig. 10A). Sucrose had the lowest abundance in Synechocystis and the highest abundance in Chroococcidiopsis. Sucrose and trehalose were already described as compatible solutes in cyanobacteria (Klähn and Hagemann 2011; Meissner et al. 2015). We mainly found trehalose in Chlorogloeopsis and in minor amounts in Chroococcidiopsis. 2-O-β-galactosylglycerol could be detected in all strains, but 2-O-α-glucosylglycerol was specific for Synechocystis. However, both compounds were only detected in minor amounts compared with the dominant compounds glutamate and sucrose. The trisaccharide melezitose was found in all strains except for Synechocystis. Additionally, glycerol and one so far unidentified compound (Unidentified#1920), which showed mass spectrometrical features of a modified monosaccharide, were found in considerable concentrations in Nostoc (fig. 10A).
Fig. 10.
—Comparison of small storage compounds and glycogen. Shown are (A) normalized peak areas of the most abundant metabolites detected in the day and night samples and (B) glycogen concentrations in µM per mg wet weight (ww) for Nostoc sp. DSM 107007 (blue), Anabaena variabilis DSM 107003 (green), Calothrix desertica DSM 106972 (purple), Chlorogloeopsis sp. PCC 6912 (yellow), Chroococcidiopsis cubana DSM 107010 (red), and Synechocystis sp. PCC 6803 (gray). For detailed values see supplementary table S6, Supplementary Material online.
After the night cycle, sucrose was almost completely depleted (1–11% left) in all strains except for Synechocystis (supplementary table S6, Supplementary Material online). The sucrose level in Synechocystis was still at 59% at the end of the night cycle. The glutamate level decreased in Synechocystis by 50% and by 75–97% in all other strains. Cyanobacteria are known to produce glycogen as major carbon storage compound (Ball and Morell 2003). In our study, the highest glycogen level was observed in Anabaena after 16 h of illumination (fig. 10B). Anabaena and Synechocystis showed overall the lowest content of small storage compounds. Thus, both strains degraded ∼70% and Calothrix ∼60% of the stored glycogen during the night cycle. Noticeable is the increasing glycogen level in Nostoc, Chlorogloeopsis and Chroococcidiopsis during the night cycle (fig. 10B). These three strains also showed the highest di and trisaccharide levels during the days, which are drastically reduced during the night cycle (fig. 10A). In the case of Chroococcidiopsis Chl a increases during the night cycle, indicating a possible preference of disaccharide sugars over glycogen with respect to pigment synthesis.
Discussion
Phylogeny and Taxonomy of Cyanobacteria
The generic assignment of cyanobacteria was previously based on morphological criteria dividing the strains in five different sections showing stable patterns of structure and development (Rippka et al. 1979). However, this classification is bothered by the recurring occurrence of possibly analogous morphological traits in different lineages that is contradictory to state-of-the-art phylogenies (Shih et al. 2013; Gonzalez-Esquer et al. 2016). The actual dilemma in cyanobacterial taxonomy is exemplified by numerous incongruencies that were observed in our well resolved MLSA trees. The scientific description of new cyanobacteria was historically performed by phycologists in a subsidiary discipline of botany, and morphological characteristics were hence and according to the International Code of Nomenclature for algae, fungi and plants (“Botanical Code”) the main criteria for their classification. However, it is well known that some morphologically complex cyanobacteria lose their typical traits, which are diagnostic for field collected samples, if they are cultured under defined conditions in the laboratory. At least three reasons might explain the discrepancies observed in subclade B1 (fig. 3), which is exemplified by the wrong designation of one of the two “S. hofmannii” strains (PCC 7110, UTEX2349). Apart from 1) a simple initial misclassification and 2) a mix up of strains, which might be not retraceable due to an altered morphology in culture, the cyanobacterial classification might moreover be bothered by the 3) independent development of analogous structures in evolutionary distantly related taxa (convergent evolution). Accordingly, the 16S rRNA as Gold Standard of microbial taxonomy (“Prokaryotic Code”) was already used for the reclassification of cyanobacteria, which is exemplified by the scattered distribution of supposed isolates of the genus Chroococcidiopsis (Fewer et al. 2002) and the placement of several Microcoleus strains in the novel genus Coleofasciculus (Siegesmund et al. 2008). The paradigm shift in cyanobacterial taxonomy that started with the reclassification of “blue-green algae” as cyanobacteria by Roger Stanier (Stanier and Cohen 1977; Rippka and Cohen-Bazire 1983) should ultimately result in a natural classification, that is, a hierarchically clustering of monophyletic lineages.
The current study showed that the 16S-rRNA gene is still a useful phylogenetic marker for an initial taxonomic assessment of new cyanobacteria (fig. 4). The RpoB is due to an improved resolution the marker of choice to quickly determine a strain’s positioning in the (cyanobacterial) tree of life (Case et al. 2007; Petersen and Wagner-Döbler 2017) and the MLSA analysis of 43 universal proteins resulted in a statistically well-supported tree that is in agreement with recent reference studies (Shih et al. 2013; Walter et al. 2017). Our phylogenetic analysis of 208 cyanobacterial genomes recovered all previously described clades (fig. 3B, supplementary fig. S1, Supplementary Material online), but the comparably low statistical support of the tree’s backbone reflects the limits of the MLSA approach. We suggest to choose phylogenomic trees, which are based on thoroughly curated complete sets of orthologous proteins (Bartling et al. 2018), as a starting point for the future (re)classification of cyanobacteria. In contrast to the recently proposed “standardized bacterial taxonomy based on genome phylogeny” that allows to classify uncultured microbes (Parks et al. 2018), we recommend to reconcile the phylogenomic approach with the biological background in order to solve the current problems in cyanobacterial nomenclature (Oren and Ventura 2017).
Strain Specific Metabolism and Day and Night Regulation of Pigments and the Central Metabolism
We did not observe major visible changes by the comparison of day and night sampled cyanobacteria on the level of autofluorescence and cell form or size, but striking differences on the molecular level. Overall, the metabolic activity was lower in the night cycle, corresponding to the reduced pigment content of the cells, with the exception of Chroococcidiopsis. Interestingly, this effect is less pronounced in Synechocystis and Chlorogloeopsis. Despite the small difference in the used medium and the different cultivation temperatures, both showed similar metabolic levels in the TCA cycle after the day and the night cycle and only the upper part of the glycolysis, the CBB cycle and the OPP pathway appeared less active during the night. As we additionally see major differences in the distribution of storage compounds, the regulation of the central carbon metabolism is most likely linked to their availability during the night cycle.
Glutamate was not yet described as compatible solute in cyanobacteria (Klähn and Hagemann 2011) and might act as small storage compound because of the decreased glutamate level at night in all strains. A higher glutamate decarboxylase activity and GABA accumulation was observed for Synechocystis with the addition of glutamate to the medium (Kanwal et al. 2015). Thus, the higher GABA level and the decreasing glutamate level indicate a degradation of glutamate via GABA during the night cycle in Synechocystis. Interestingly, while Synechocystis still possessed 50% of its day cycle glutamate concentration, Nostoc reduced its glutamate concentration by 93%. It is possible that glutamate was converted to phycobiliproteins in the night cycle as Nostoc was the only cyanobacterium gaining in phycobiliprotein concentration and in contrast to Synechocystis did not convert glutamate to GABA. As phycobilisome pigments are involved in the channeling of solar energy a higher phycobiliprotein concentration after the light not the dark cycle was expected. In A. platensis, for example, PC pigment was found to be more abundant during the end of the day compared with the end of the dark phase (Matallana-Surget et al. 2014). However, in carbon-limited and nitrogen-sufficient batch cultures of a red alga which were grown in darkness, the PC concentration increased (Sloth et al. 2006). Therefore, the phycobiliprotein concentration might have been influenced by different nitrogen availability.
The main known compatible solutes in cyanobacteria are sucrose and trehalose (freshwater environment), glucosylglycerol and glucosylglycerate (marine environment) and glycine betaine (hypersaline environment; Klähn and Hagemann 2011). We could identify glucosylglycerol exclusively in Synechocystis, but our cultivation conditions represented low salt stress conditions. Thus, contrary to the described salt-stress induced accumulation of sucrose (Blumwald and Tel-Or 1982; Blumwald et al. 1983; Erdmann 1983; Reed et al. 1984), our study rather indicates sucrose as a small storage compound that was used at night. It was almost completely depleted by all strains except for Synechocystis that had still 59% left and showed overall the lowest sucrose content. A low sucrose level in Synechocystis was already observed in comparison to Microcystis (Meissner et al. 2015). Trehalose is another compatible solute found in Microcystis and Synechocystis (Meissner et al. 2015), but we could not detect trehalose in Anabaena and Synechocystis. Trehalose was only highly abundant in Chroococcidiopsis and especially Chlorogloeopsis. Again, the trehalose level drastically decreased during the night and could not be assigned to salt-stress.
We could detect glycogen in all strains but in different amounts. The glycogen levels during the day cycle were decreasing with increasing small storage compounds. Remarkable is the increase of glycogen during the night cycle in Nostoc, Chlorogloeopsis, and especially Chroococcidiopsis. The highly abundant small storage compounds accumulated during the day could be the carbon source to build up glycogen during the night in these three strains. Thus, the assumption that glycogen is the major storage compound and used during the night, as it is described for Synechocystis (Gründel et al. 2012), is not necessarily applicable for all cyanobacteria.
The presence of citramalate and 2-methylcitrate pointed towards alternative pathways of replenishing TCA cycle intermediates and providing precursors for biomass productions. Citramalate biosynthesis and its subsequent conversion to 2-oxobutanoate as an isoleucine precursor has been characterized in Cyanothece (Wu et al. 2010). We could detect the intermediate 2-methylmaleate (citraconate) in four strains. The downstream metabolite 2-oxobutanoate was only present in traces in our samples and not quantifiable. Beside its role as an isoleucine precursor, it may be converted further as a commonly characterized side-activity of the pyruvate synthase (PYS, EC 1.2.7.1) to propanoyl-CoA, which is a precursor of 2-methylcitrate but not yet described in cyanobacteria (Textor et al. 1997). The 2-methylcitrate synthase (2MCS, EC 2.3.3.5) could be annotated in all six analyzed strains (supplementary table S5, Supplementary Material online). Enzymes with a citrate and a 2-methylcitrate synthase activity are frequently annotated as a second citrate synthase (Gerike et al. 1998; Horswill and Escalante-Semerena 1999). In the following steps of the cycle, 2-methylcitrate may be converted in a similar manner as citrate, and the methylisocitrate lyase (MICL, EC 4.1.3.30) catalyzes the formation of succinate and pyruvate. This enzyme has not been annotated in cyanobacteria, but we detected homologs in four of the six analyzed strains (supplementary table S5, Supplementary Material online). According to our data, this proposed pathway was more important in Nostoc and Chlorogloeopsis compared with the well-studied Synechocystis strain.
Evolution of the Primary Metabolism
The very limited distribution of the glyoxylate shunt and the PEPCK in <5% of the investigated strains exemplified the impact of HGT on the evolution and actual composition of the primary metabolism in cyanobacteria. Metabolically relevant genes and pathways were once horizontally recruited from heterotrophic bacteria and subsequently transferred between the distinct cyanobacterial (sub)clades. The prerequisite for the detection of HGTs was the establishment of a solid phylogenetic backbone of cyanobacterial evolution in form of the comprehensive MLSA tree, which also allowed to pinpoint secondary losses of the respective genes. The glyoxylate shunt plays an important role in oxidative stress response of bacteria (Ahn et al. 2016), but this role is poorly explored despite more than sixty years of metabolic investigation in the model organism E. coli (Dolan and Welch 2018). Its actual metabolic function for Chlorogloepsis and seven other cyanobacteria remains to be investigated. One starting point is the surprising co-occurrence of the MLS-ICL gene cluster with the PEPCK in four cyanobacteria of subclade B1 that suggests from an evolutionary perspective rather a specific functional correlation than a simple coincidence.
Synechocystis sp. PCC 6803 as a Model Strain
Carotenoid composition has been used to classify cyanobacteria in the past, already highlighting the possibility of different metabolic pathways. Synechocystis sp. PCC 6803 showed the highest change in its carotenoid composition during the day–night cycle, thereby showing a different response than the other cyanobacteria of this study. Synechocystis is also considered as a common model strain for the central carbon metabolism in cyanobacteria (Eisenhut, Huege, et al. 2008; Eisenhut, Ruth, et al. 2008; Knoop et al. 2013; Osanai et al. 2014; Schwarz et al. 2014; Meissner et al. 2015; Jablonsky et al. 2016). While the overall assumptions of the general metabolism are well reflected by Synechocystis, cyanobacteria harbor an enormous metabolic versatility. Cyanobacteria of clade A were reported to have severe alterations in glycolysis and TCA cycle based on sequence analysis (Beck et al. 2012). But even among more closely related strains, we detected differences in the abundance of intermediates and the anaplerosis of the TCA cycle: Chlorogloeopsis and Synechocystis were the only analyzed strains that showed a similar abundance of TCA cycle intermediates during the day and the night. Only Nostoc, and to a lower extent Anabaena and Chlorogloeopsis, showed increased amounts of 2-methylcitrate as a putative intermediate of succinate replenishment. The CBB cycle is the widely studied and commonly used CO2 fixation pathway in cyanobacteria (Schwarz et al. 2013). Consequently, we detected intermediates of the CBB cycle in all analyzed strains. However, under light exposure Anabaena showed low abundant ribulose-1, 5-bisphosphate and in contrast activity of the OPP pathway based on the abundance of 6-phosphogluconate.
At the level of di and trisaccharides, Synechocystis differed from the other analyzed cyanobacteria. The preferred disaccharide was sucrose for all strains, which was less abundant in Synechocystis. Glutamate was found as one of the most abundant metabolites in all strains. However, the storage compounds differed in their ratio between all strains. Further specific compounds were detected in Nostoc and Chlorogloeopsis indicating variable capabilities for small storage compounds. Glycogen could be detected in all strains at day and night, but we observed major differences especially for Nostoc, Chlorogloeopsis, and Chroococcidiopsis that showed higher levels during the night.
Conclusion
Our comparison of different phylogenetic markers showed that the molecular differentiation on genus level is not trivial and exceeds the resolving power of the 16S rRNA gene. Genome sequencing and state-of-the-art MLSA analyses allowed a reliable localization of new taxa in the tree of life and phylogenomic analyses should pave the way for the challenges in cyanobacterial taxonomy. Comparative genome analyses of >200 cyanobacteria showed that the program CheckM (Parks et al. 2015), which is mainly used to assess the quality of single-cell and metagenomes, is also a very useful in silico tool for the proof of cyanobacterial axenicity.
On the metabolic level, Synechocystis sp. PCC 6803 as model strain is still the best understood cyanobacterium. However, in a more detailed view, other cyanobacteria show metabolic alterations and different metabolic potentials even in the central carbon metabolism that might be taken into account. For biotechnological approaches, broader screening for suitable strains may be the key to find strains that already accumulate required metabolic precursors or provide shortcuts, anaplerotic reactions or inactivated pathways without the necessity of a genetic modification.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Maike Lorenz (SAG culture collection, Göttingen, Germany) and Lucas Stal for strain provision and Franziska Klann, Gesa Martens, and Maja Marheine for excellent technical assistance. Finally, we would like to thank three anonymous reviewers for their very constructive criticism that led to a significant improvement of our manuscript.
Author Contributions
M.N.-S. and J.P. conceived of the study. S.E.W., P.H., C.B., M.J., M.R., and S.S. performed the experiments. S.E.W., P.H., C.B., S.H., H.B., J.O., M.N.-S., and J.P. analyzed the data. S.E.W., P.H., C.B., T.F., M.S., H.B., M.N.-S., and J.P. wrote the manuscript with input of all authors.
Footnotes
Data deposition: The five newly established cyanobacterial draft genomes have been deposited under the following accession numbers: 1) Nostoc sp. DSM 107007 = RSCN00000000, 2) Anabaena variabilis DSM 107003 = RSCM00000000, 3) Calothrix desertica DSM 106972 = RSCL00000000, 4) Chlorogloeopsis sp. PCC 6912 = RSCJ00000000, and 5) Chroococcidiopsis cubana DSM 107010 = RSCK00000000.
Literature Cited
- Ahn S, et al. 2016. Role of glyoxylate shunt in oxidative stress response. J Biol Chem. 29122:11928–11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. 2011. ea-utils: Command-line tools for processing biological sequencing data. Available from: https://github.com/ExpressionAnalysis/ea-utils.
- Ball SG, Morell MK.. 2003. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol. 541:207–233. [DOI] [PubMed] [Google Scholar]
- Baran R, et al. 2013. Functional genomics of novel secondary metabolites from diverse cyanobacteria using untargeted metabolomics. Mar Drugs. 1110:3617–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling P, Vollmers J, Petersen J.. 2018. The first world swimming championships of roseobacters – phylogenomic insights into an exceptional motility phenotype. Syst Appl Microbiol. Syst Appl Microbiol. 41(6):544–554. [DOI] [PubMed] [Google Scholar]
- Beck C, Knoop H, Axmann IM, Steuer R.. 2012. The diversity of cyanobacterial metabolism: genome analysis of multiple phototrophic microorganisms. BMC Genomics 131:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A, Bogorad L. 1973. Complementary chromatic adaption in a filamentous blue-green alga. J. Cell Biol. 58(2):419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Mehlhorn RJ, Packer L.. 1983. Studies of osmoregulation in salt adaptation of cyanobacteria with ESR spin-probe techniques. Proc Natl Acad Sci U S A. 809:2599–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Tel-Or E.. 1982. Osmoregulation and cell composition in salt-adaptation of Nostoc muscorum. Arch Microbiol. 1322:168–172. [Google Scholar]
- Boedeker C, et al. 2017. Determining the bacterial cell biology of Planctomycetes. Nat Commun. 8:14853.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers RM, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 358:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks JJ, Logan GA, Buick R, Summons RE.. 1999. Archean molecular fossils and the early rise of eukaryotes. Science 2855430:1033–1036. [DOI] [PubMed] [Google Scholar]
- Büdel B. 2011. Cyanobacteria: habitats and Species In: Lüttge U, Beck E, Bartels D, editors. Plant desiccation tolerance. Ecological studies. Vol. 215. Berlin, Heidelberg (Germany: ): Springer Berlin Heidelberg; pp. 11–21. doi:10.1007/978-3-642-19106-0_2 M4-Citavi. [Google Scholar]
- Case RJ, et al. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 731:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. 2016. The Entner–Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc Natl Acad Sci U S A. 11319:5441–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet L, Meunier L, et al. 2018. Consensus assessment of the contamination level of publicly available cyanobacterial genomes. PLoS ONE 137:e0200323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet L, Bertrand AR, et al. 2018. Metagenomic assembly of new (sub)arctic cyanobacteria and their associated microbiome from non-axenic cultures. Microb Genomics. 4(9):1–15. doi:10.1101/287730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Archibald JM.. 2017. Endosymbiosis: did plastids evolve from a freshwater cyanobacterium? Curr Biol. 273:R103–R105. [DOI] [PubMed] [Google Scholar]
- Dempo Y, Ohta E, Nakayama Y, Bamba T, Fukusaki E.. 2014. Molar-based targeted metabolic profiling of cyanobacterial strains with potential for biological production. Metabolites 42:499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez B, Ininbergs K.. 2014. Ecological importance of cyanobacteria In: Sharma NK, Rai AK, Stal LJ, editors. Cyanobacteria. Vol. 106. Chichester (UK): John Wiley & Sons, Ltd. pp. 41–63. doi:10.1002/9781118402238.ch3 M4-Citavi. [Google Scholar]
- Dittmann E, Fewer DP, Neilan BA.. 2013. Cyanobacterial toxins: biosynthetic routes and evolutionary roots. FEMS Microbiol Rev. 371:23–43. [DOI] [PubMed] [Google Scholar]
- Dolan SK, Welch M.. 2018. The glyoxylate shunt, 60 years on. Annu Rev Microbiol. 721:309–330. [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Huege J, et al. 2008. Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 1484:2109–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, et al. 2008. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci U S A. 10544:17199–17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann N. 1983. Organic osmoregulatory solutes in blue-green algae. Zeitschrift Für Pflanzenphysiologie. 1102:147–155. [Google Scholar]
- Fewer D, Friedl T, Büdel B.. 2002. Chroococcidiopsis and heterocyst-differentiating cyanobacteria are each other’s closest living relatives. Mol Phylogenet Evol. 231:82–90. [DOI] [PubMed] [Google Scholar]
- Gerike U, Hough DW, Russell NJ, Dyall-Smith ML, Danson MJ.. 1998. Citrate synthase and 2-methylcitrate synthase: structural, functional and evolutionary relationships. Microbiology 1444:929–935. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Esquer CR, et al. 2016. Cyanobacterial ultrastructure in light of genomic sequence data. Photosynth Res. 1292:147–157. [DOI] [PubMed] [Google Scholar]
- Gründel M, Scheunemann R, Lockau W, Zilliges Y.. 2012. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158:3032–3043. [DOI] [PubMed] [Google Scholar]
- Gueneli N, et al. 2018. 1.1-Billion-year-old porphyrins establish a marine ecosystem dominated by bacterial primary producers. Proc Natl Acad Sci U S A.115(30):E6978–E6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T, et al. 2013. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J Exp Bot. 6410:2943–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques M, Silva A, Rocha J.. 2007. Extraction and quantification of pigments from a marine microalga: a simple and reproducible method. Commun Curr Res Educ Top Trends Appl Microbiol. 586–593. [Google Scholar]
- Hoffmann P. 1976. Environmental diversity of middle precambrian stromatolites In: Walter MR, editor. Stromatolites. Amsterdam (Netherlands): Elsevier; pp. 599–611. [Google Scholar]
- Hofmann JD, et al. 2018. Metabolic reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front Microbiol. 9:1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horswill AR, Escalante-Semerena JC.. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol. 181:5615–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman J, et al. 2018. Cyanobacterial blooms. Nat Rev Microbiol. 168:471–483. [DOI] [PubMed] [Google Scholar]
- Jablonsky J, Papacek S, Hagemann M.. 2016. Different strategies of metabolic regulation in cyanobacteria: from transcriptional to biochemical control. Sci Rep. 6:33024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey SW, Wright SW, Zapata M.. 2011. Chlorophylls and carotenoids In: Roy S, Llewellyn CA, Egeland ES, Johnson G, editors. Phytoplankton pigments. Cambridge: Cambridge University Press; p. 845. [Google Scholar]
- Kaneko T, et al. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 33:109–136. [DOI] [PubMed] [Google Scholar]
- Kanwal S, Khetkorn W, Incharoensakdi A.. 2015. GABA accumulation in response to different nitrogenous compounds in unicellular cyanobacterium Synechocystis sp. PCC 6803. Curr Microbiol. 701:96–102. [DOI] [PubMed] [Google Scholar]
- Kashtan N, et al. 2014. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 3446182:416–420. [DOI] [PubMed] [Google Scholar]
- Kenyon CN, Rippka R, Stanier RY.. 1972. Fatty acid composition and physiological properties of some filamentous blue-green algae. Arch Microbiol. 833:216–236. [DOI] [PubMed] [Google Scholar]
- Klähn S, et al. 2015. Integrated transcriptomic and metabolomic characterization of the low-carbon response using an ndhR mutant of Synechocystis sp. PCC 6803. Plant Physiol. 1693:1540–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klähn S, Hagemann M.. 2011. Compatible solute biosynthesis in cyanobacteria. Environ Microbiol. 133:551–562. [DOI] [PubMed] [Google Scholar]
- Knoll AH, Summons RE, Waldbauer JR, Zumberge JE.. 2007. The geological succssion of primary producers in the oceans. The evolution of primary producers in the sea In: Falkowski P, Knoll AH, editors. Evolution of primary producers in the sea. Burlington (MA: ): Elesevier; pp. 133–163. doi:10.1016/B978-0-12-370518-1.50009-6. [Google Scholar]
- Knoop H, et al. 2013. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput Biol. 96:e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H, Zilliges Y, Lockau W, Steuer R.. 2010. The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiol. 1541:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, et al. 2017. Systems analysis of ethanol production in the genetically engineered cyanobacterium Synechococcus sp. PCC 7002. Biotechnol Biofuels. 10:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall L, Huege J, Catchpole G, Steinhauser D, Willmitzer L.. 2009. Assessment of sampling strategies for gas chromatography–mass spectrometry (GC–MS) based metabolomics of cyanobacteria. J Chromatogr B Analyt Technol Biomed Life Sci. 87727:2952–2960. [DOI] [PubMed] [Google Scholar]
- Kumar K, Mella-Herrera RA, Golden JW.. 2010. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 24:a000315–a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenz E, Fedewa EJ, Richardson TL.. 2011. Extraction protocols for the quantification of phycobilins in aqueous phytoplankton extracts. J Appl Phycol. 235:865–871. [Google Scholar]
- Matallana-Surget S, et al. 2014. Proteome-wide analysis and diel proteomic profiling of the cyanobacterium Arthrospira platensis PCC 8005. PLoS ONE 96:e99076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner S, Steinhauser D, Dittmann E.. 2015. Metabolomic analysis indicates a pivotal role of the hepatotoxin microcystin in high light adaptation of Microcystis. Environ Microbiol. 175:1497–1509. [DOI] [PubMed] [Google Scholar]
- Mimouni V, et al. 2012. The potential of microalgae for the production of bioactive molecules of pharmaceutical interest. Curr Pharm Biotechnol. 1315:2733–2750. [DOI] [PubMed] [Google Scholar]
- Mitra AK. 1950. Two new algae from Indian soils. Ann Bot New Ser. 144:457–464. [Google Scholar]
- Moreira D, et al. 2017. Description of Gloeomargarita lithophora gen. nov., sp. nov., a thylakoid-bearing, basal-branching cyanobacterium with intracellular carbonates, and proposal for Gloeomargaritales ord. nov. Int J Syst Evol Microbiol. 673:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ.. 2005. Ammonium assimilation in cyanobacteria. Photosynth Res. 832:135–150. [DOI] [PubMed] [Google Scholar]
- Nagy V, et al. 2018. Carotenoid glycoside isolated and identified from cyanobacterium Cylindrospermopsis raciborskii. J Food Compos Anal. 65:6–10. [Google Scholar]
- Narainsamy K, Cassier-Chauvat C, Junot C, Chauvat F.. 2013. High performance analysis of the cyanobacterial metabolism via liquid chromatography coupled to a LTQ-Orbitrap mass spectrometer: evidence that glucose reprograms the whole carbon metabolism and triggers oxidative stress. Metabolomics 91:21–32. [Google Scholar]
- Neumann-Schaal M, Hofmann JD, Will SE, Schomburg D.. 2015. Time-resolved amino acid uptake of Clostridium difficile 630Δerm and concomitant fermentation product and toxin formation. BMC Microbiol. 15:281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, et al. 2015. Elucidating butanol tolerance mediated by a response regulator Sll0039 in Synechocystis sp. PCC 6803 using a metabolomic approach. Appl Microbiol Biotechnol. 994:1845–1857. [DOI] [PubMed] [Google Scholar]
- Oren A, Ventura S.. 2017. The current status of cyanobacterial nomenclature under the “prokaryotic” and the “botanical” code. Antonie Van Leeuwenhoek. 11010:1257–1269. [DOI] [PubMed] [Google Scholar]
- Orf I, et al. 2016. Can cyanobacteria serve as a model of plant photorespiration? A comparative meta-analysis of metabolite profiles. J Exp Bot. 6710:2941–2952. [DOI] [PubMed] [Google Scholar]
- Osanai T, et al. 2014. Metabolomic analysis reveals rewiring of Synechocystis sp. PCC 6803 primary metabolism by ntcA overexpression. Environ Microbiol. 1610:3304–3317. [DOI] [PubMed] [Google Scholar]
- Parks DH, et al. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 3610:996–1004. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW.. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 257:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Wagner-Döbler I.. 2017. Plasmid transfer in the ocean – a case study from the Roseobacter group. Front Microbiol. 8:1350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. 1993. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 2122:5264–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Toledo RI, et al. 2017. An early-branching freshwater cyanobacterium at the origin of plastids. Curr Biol. 273:386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast P, et al. 2017. Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the Verrucomicrobial Subdivision 4. Front Microbiol. 8:202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RH, Richardson DL, Warr SRC, Stewart WDP.. 1984. Carbohydrate accumulation and osmotic stress in cyanobacteria. Microbiology 1301:1–4. [Google Scholar]
- Rippka R, Cohen-Bazire G.. 1983. The Cyanobacteriales: a legitimate order based on the type strain Cyanobacterium stanieri? Annu Microbiol. 134:21–36. [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY.. 1979. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. Microbiology 1111:1–61. [Google Scholar]
- Rivas-Ubach A, et al. 2018. Coping with iron limitation: a metabolomic study of Synechocystis sp. PCC 6803. Acta Physiol Plant. 40:1. [Google Scholar]
- Sandrini G, et al. 2016. Rapid adaptation of harmful cyanobacteria to rising CO2. Proc Natl Acad Sci U S A. 11333:9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartory DP, Grobbelaar JU.. 1984. Extraction of chlorophyll a from freshwater phytoplankton for spectrophotometric analysis. Hydrobiologia 1143:177–187. [Google Scholar]
- Saw JHW, et al. 2013. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a lava cave in Kīlauea Caldera, Hawai’i. PLoS ONE 810:e76376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe B. 1960. Zur autotrophen Vegetation in ariden Böden. Blaualgen und Lebensraum IV. Österreichische Bot Zeitschrift. 107(3–4):281–309. [Google Scholar]
- Schwarz D, Orf I, Kopka J, Hagemann M.. 2014. Effects of inorganic carbon limitation on the metabolome of the Synechocystis sp. PCC 6803 mutant defective in glnB encoding the central Regulator PII of cyanobacterial C/N acclimation. Metabolites 42:232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Orf I, Kopka J, Hagemann M.. 2013. Recent applications of metabolomics toward cyanobacteria. Metabolites 31:72–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schyns G, et al. 1997. Prochlorothrix hollandica PCC 9006: genomic properties of an axenic representative of the chlorophyll a/b-containing oxyphotobacteria. Res Microbiol. 1484:345–354. [DOI] [PubMed] [Google Scholar]
- Shih PM, et al. 2013. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A. 1103:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegesmund MA, Johansen JR, Karsten U, Friedl T.. 2008. Coleofasciculus gen. nov. (Cyanobacteria): morphological and molecular criteria for revision of the genus Microcoleus Gomont. J Phycol. 446:1572–1585. [DOI] [PubMed] [Google Scholar]
- Sloth JK, Wiebe MG, Eriksen NT.. 2006. Accumulation of phycocyanin in heterotrophic and mixotrophic cultures of the acidophilic red alga Galdieria sulphuraria. Enzyme Microb Technol. 38(1–2):168–175. [Google Scholar]
- Stadnichuk IN, Krasilnikov PM, Zlenko DV.. 2015. Cyanobacterial phycobilisomes and phycobiliproteins. Microbiology 842:101–111. [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY, Cohen BG.. 1977. Phototrophic prokaryotes: the Cyanobacteria. Annu Rev Microbiol. 311:225–274. [DOI] [PubMed] [Google Scholar]
- Steglich M, et al. 2018. Convergent loss of ABC transporter genes from Clostridioides difficile genomes is associated with impaired tyrosine uptake and p-cresol production. Front Microbiol. 9:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingley WD, et al. 2008. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci U S A. 105(6):2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllosi GJ, Boussau B, Abby SS, Tannier E, Daubin V.. 2012. Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations. Proc Natl Acad Sci U S A. 10943:17513–17518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaichi S, Maoka T, Masamoto K. 2001. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-alpha-L-fucoside), not rhamnoside. Plant Cell Physiol. 42(7):756–762. [DOI] [PubMed] [Google Scholar]
- Takaichi S, Mochimaru M, Maoka T, Katoh H. 2005. Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol. 46(3):497–504. [DOI] [PubMed] [Google Scholar]
- Takaichi S, Mochimaru M.. 2007. Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci. 64(19–20):2607–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 564:564–577. [DOI] [PubMed] [Google Scholar]
- Tanizawa Y, Fujisawa T, Nakamura Y.. 2018. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 346:1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 41:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, et al. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1685:428–436. [DOI] [PubMed] [Google Scholar]
- Walter JM, et al. 2017. Ecogenomics and taxonomy of Cyanobacteria phylum. Front Microbiol. 8:2132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, et al. 2010. Alternative isoleucine synthesis pathway in cyanobacterial species. Microbiology 1562:596–602. [DOI] [PubMed] [Google Scholar]
- Wu M, Eisen JA.. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 910:R151.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech H, et al. 2009. Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 914:3677–3697. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 185:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bryant DA.. 2011. The tricarboxylic acid cycle in cyanobacteria. Science 3346062:1551–1553. [DOI] [PubMed] [Google Scholar]
- Zhang S, Qian X, Chang S, Dismukes GC, Bryant DA.. 2016. Natural and synthetic variants of the tricarboxylic acid cycle in Cyanobacteria: introduction of the GABA shunt into Synechococcus sp. PCC 7002. Front Microbiol. 7:1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga M, Gueguen V, Pavon-Djavid G, Letourneur D.. 2017. Carotenoids from microalgae to block oxidative stress. BioImpacts 71:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.