Abstract
Capillary electrophoresis-mass spectrometry (CE-MS) using a sheathless porous tip interface emerged as an attractive tool in metabolomics thanks to its numerous advantages. One of the main advantages compared to the classical co-axial sheath liquid interface is the increased sensitivity, while maintaining the inherent properties of CE, such as a high separation efficiency and low sample consumption. Specially, the ability to perform nanoliter-based injections from only a few microliters of material in the sample vial makes sheathless CE-MS a well-suited and unique approach for highly sensitive metabolic profiling of limited sample amounts. Therefore, in this work, we demonstrate the utility of sheathless CE-MS for metabolic profiling of biomass-restricted samples, namely for 20 µm-thick tissue sections of kidney from a mouse model of polycystic kidney disease (PKD). The extraction method was designed in such a way to keep a minimum sample-volume in the injection vial, thereby still allowing multiple nanoliter injections for repeatability studies. The developed strategy enabled to differentiate between different stages of PKD and as well changes in a variety of different metabolites could be annotated over experimental groups. These metabolites include carnitine, glutamine, creatine, betaine and creatinine. Overall, this study shows the utility of sheathless CE-MS for biomass-limited metabolomics studies.
Introduction
Polycystic kidney disease (PKD) is a complex clinical entity unifying a group of diseases that results in renal cyst development1. The animal models and the rodents in particular are essential for the understanding of the biochemical mechanisms of the disease, providing information which would be impossible to get from the observational human studies only. The trend towards a more responsible use of the laboratory animals stimulates the changes in analytical methodologies demanding analytical workflows for the volume restricted biological samples. Metabolomics is one of the areas where such methods are highly needed. For instance, the onset and progression of autosomal dominant polycystic kidney disease (ADPKD) is commonly explained as an effect of the dysregulated expression of the PKD1 or PKD2 genes. The encoded proteins are associated with remodeling of the tubular architecture and as such, directly or indirectly, control multiple signaling cascades in renal epithelial cell. This, in turn, leads to a profound metabolic remodeling of the affected cells. Consequently, it raises a question whether such remodeling contributes to the PKD pathophysiology2,3. Here, we would like to address this question using kidney-specific PKD1-deletion mice, of which we analysed single 20 μm tissue sections.
There are different technical solutions available, but here we are presenting a report on using capillary electrophoresis – mass spectrometry (CE-MS) for metabolic profiling of such material. CE-MS offers many advantages that makes it a useful tool in metabolomics studies. One can mention minimal sample requirement (usually only a few nanoliters are injected) or the high separation efficiency due to the flat flow profile of the electro-osmotic flow where only longitudinal diffusion is contributing to band broadening in CE4. Moreover, the method has a proven track record for the analysis of polar and charged compounds5. However, as every analytical technique, CE-MS has its weaknesses. Limitations of CE include low migration time repeatability and relatively poor detection sensitivity due to the low injection volume. Moreover, in the case of employing a sheath-liquid interface an extra dilution occurs thereby compromising the sensitivity even further4. In the past years, significant developments addressed these limitations and resulted in improved interfacing techniques for CE-MS among them the sheathless porous tip interface6. The porous tip interface (offered by SCIEX as CESI sprayer) can substantially improve the detection sensitivity (i.e., up to 100-fold) as compared to using a conventional co-axial sheath-liquid interface7,8. Another advantage of the porous tip sprayer interface is reduced ion suppression, which is inherent to the use of flow rates below 20 nL/min in ESI-MS9. For a variety of reasons, the potential of CESI-MS for metabolomics has hardly been explored. Only a limited number of studies were reported using urine7,10, plasma11,12, CSF11, and cells13–16. Thus, with this report we demonstrate the utility of CESI-MS for metabolic profiling of biomass-restricted tissue material and, at the same time, we provide a descriptive study of the metabolic changes associated with PKD progression1.
Results and Discussion
For decades, laboratory animals and rodents in particular are being used as the main experimental tool of the modern biology. They offer a unique possibility for modeling the human diseases in vivo, getting insight into the systemic and local effects of a given pathology. Miniaturization of the analytical workflows is one of the most obvious demands. With regard to metabolomics the diversity of the mammalian metabolome implies availability of several micro-scale strategies which could be adopted to a particular class of the metabolites or the sample types. CE-MS has a proven track record as a micro-scale method, known for its possibility to provide complex biological profiles from only a limited amount of material. Here, we demonstrate the feasibility of this approach for an animal model for PKD using a specific type of biomass-restricted samples, namely 20 µm-thick tissue sections of mouse kidneys. The set of the samples consisted of four groups. The two wild type groups (Wt) at the beginning of the experiment and 14 weeks into the experiment (Wt0 and Wt14). The other two groups represent a mild (MCK) and advanced (ESCK) kidney damage conditions.
Potential of Sheathless CE-MS for Metabolic Profiling of Biomass-Restricted Samples
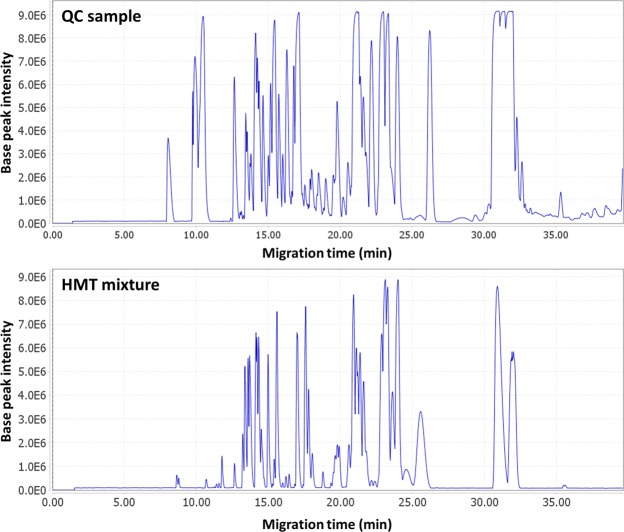
There are several technical solutions for integration of CE and MS6,15,17–19. Here, we use a CESI-MS platform, a sheathless CE-MS configuration, where CE separation and ESI ionization work as a single process8. Moreover, following the recent work of Kammeijer et al.20 in which a dopant-enriched nitrogen gas (DEN-gas) was employed to further increase the detection sensitivity of CESI-MS, we also used the DEN-gas attenuated version of CESI. To test the general performance of this approach as a tool for metabolic profiling, standard metabolite mixtures consisting of sixty compounds in an equimolar concentration (HMT mixture) have been analyzed. This mixture includes the major metabolite classes such as amino acids, betaines, lactams, purines and nucleosides. Figure 1 shows a base peak electropherogram (BPE) obtained by CESI-MS for the standard metabolite mixture. It is important to note that it was possible to obtain low nanomolar detection limits for most metabolites in this standard mixture, this being in agreement with previous CESI-MS metabolomics studies7. The second Figure in the panel (Fig. 1) shows the BPE of a quality control (QC) sample, i.e. a pooled sample containing equally volumes of all samples used in this study. Not surprisingly, the complexity of the QC sample exceeds the one of the standard metabolite mixture. Moreover, despite a general pattern similarity, there is a clear difference between the metabolite mixture and QC sample in the responses observed for individual metabolites. Supplementary Fig. S-1 clearly illustrates this point by showing extracted ion electropherograms for various selected metabolites from the different chemical families.
Figure 1.
Comparison of base peak electropherograms of standard metabolite mixture consisting of 60 metabolites at 50 µM (A) and a QC sample of tissue sections (B) obtained with the sheathless CE-MS platform. Experimental conditions: BGE, 10% (v/v) acetic acid (pH 2.3); voltage, +20 kV; sample injection, 1 psi × 60 s (1.4% total volume of the capillary, i.e. roughly 9 nL); 91 cm × 30 µm i.d. × 150 µm o.d. fused-silica capillary.
A detailed description of the analytical sequence is given in section 4.5. Apart from using a DEN-gas enhanced version of the CESI-MS the sequence has been organized as a standard profiling experiment using a block acquisition approach, where every block includes a random selection of the measured samples, QC samples and the analytical standards. CE-based metabolomics methods are usually criticized for low migration-time reproducibility21. Anticipating the problem, we took a special care on the alignment of the data using an in-house built tool, msalign2, which takes advantage of the superior mass accuracy of modern TOF instruments for migration-time correction21. Supplementary Fig. S-2 illustrates the effect of the alignment for measured QC samples (n = 11), where it can be seen that the variability of migration times was strongly reduced. We have also introduced a rigorous filtering step including in the analysis only the features which had a relative standard deviation (RSD) below 30% in the QC samples.
Exploratory analysis of polycystic kidney disease tissue samples
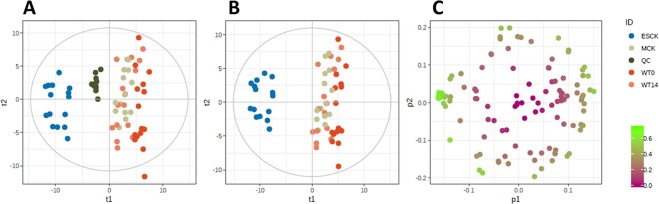
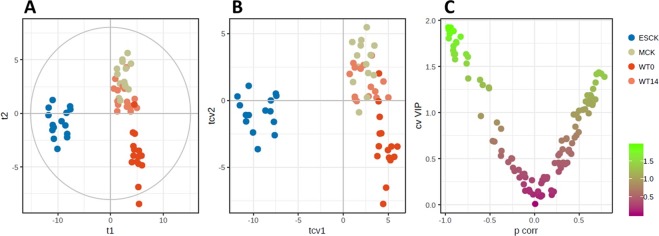
The final data matrix after application of the 30% RSD filtering includes only 112 metabolic features. Even though the number of identified metabolic features seems rather low compared to other metabolomic studies13, it should be taken into account that the starting material was limited. In addition, the lower number in identification could be considered as an advantage, since data can be easily overfitted during multivariate modelling when limited samples are available (N = 60 mice). The number of detected metabolic features might have been enlarged if preconcentration approaches, e.g. dynamic pH junction22, were used. The essential first analysis step of the metabolic profiling experiment is evaluation of the analytical consistency and description of the main sources of the variance. The initial principal component analysis (PCA) modeling revealed a strong outlier - a MCK sample. Despite using probabilistic quotation normalization (PQN), abnormally low signal intensities were observed in this sample, indicating an analytical failure as a possible cause and consequently the sample was removed from further analysis. The score plot of the PCA model built after reprocessing the data without the outlier is presented in Fig. 2A. The model requires just three principal components to cover the first 50% of the variance, while the first two principal components covered 43% of the variance. The QC samples clustered together which indicates a proper analytical consistency and analytical variance within the limits expected for the CESI-MS method. It is worth mentioning that metabolic profiles for QC samples were not analyzed from multiple injections using the same injection vial, but were obtained from independent aliquots which had to be reconstituted in 2.5 µL of water every day before the start of the acquisition sequence. Figure 2B shows the score plot of the PCA model built without QC samples, similar characteristics were observed when compared to Fig. 2A, with only three components 50% of the variance was covered, whereas the first two components covered 44% of the variance. The score plot clearly shows that the end-stage cystic kidney (ESCK) group, which forms a distinct cluster, strongly influences the model. Figure 2C shows the loadings plot of the model presented in Fig. 2B. The plot is colored according to the modelling power emphasizing the cluster of the variables relevant for the model. As a next step we built a multi-class PLS-DA model using the experimental groups as the class ID. Figure 3A,B show the score plot and cross-validated score plot of the resulting model, respectively. The model parameters (R2X 0.75, R2Y 0.94, Q2 0.5) and CV-ANOVA values (F = 13,1, p-value = 2.6 × 10−30) indicate a strong model. However, as well as in the PCA, samples from the ESCK group strongly influence the model, which complicates the interpretation of the VIP values (Fig. 3C). Apparently, the strongest ones will mainly explain the differences between the ESCK group and the remaining samples.
Figure 2.
(A) PCA of log-transformed data. (B) PCA of data in (A), excluding QC samples. (C) PCA loading plot of 112 variables in (B), colored according to the modeling power of (B).
Figure 3.
(A) PLS-DA score plot for the four groups of samples analyzed by the sheathless CE-MS platform. (B) Cross-validated score plot of the PLS-DA plot. (C) VIP vs p(corr) plot, colored according to the VIP values from (B).
Context-specific multivariate modelling
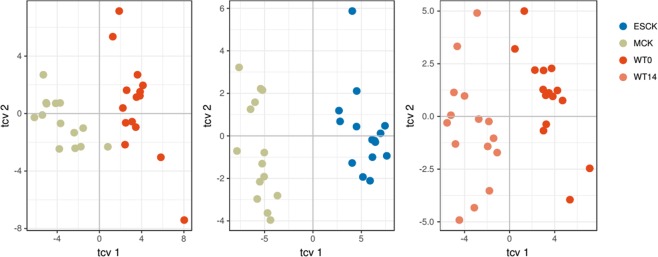
To get a more specific description of the metabolic changes between the groups, several two-class PLS-DA models were built, namely Wt0 (wildtype at timepoint 0) vs MCK (to explain the differences between non-PKD and mild PKD) and MCK vs ESCK (to explain a transition from mild to advanced stage of PKD). Finally, to get insight into metabolic changes associated with the aging of the animals, an additional model Wt0 vs Wt14 (Wt + 14 weeks) was built. Figure 4 shows the cross-validated score plots for all three models and Table 1 summarizes the model’s parameters. Despite the differences, all three models are valid. In agreement with the first two models (PCA and PLS-DA) the strongest model explained the differences between MCK and ESCK groups (highest values of R2X, R2Y, Q2 and F and the lowest p-value for CV-ANOVA from all models). From each model a subset of the most influential variables was selected by using the model specific variable importance on projection (VIP) values with an arbitrary cut-off of 1.35. Supplementary Fig. S-3 shows a Venn diagram summarizing all selected variables for the three models. Table 2 summarizes the features annotated according to the guidelines outlined in section 4.7.
Figure 4.
PLS-DA models for the three different pairwise comparisons performed in this work.
Table 1.
Number of latent variables, quality parameters (R2X, R2Y, Q2 and CV-ANOVA F and p-values) for the pairwise PLS-DA.
| PLS-DA model | Number of latent variables | R2X | R2Y | Q2 | CV-ANOVA | |
|---|---|---|---|---|---|---|
| F | p-value | |||||
| Wt0 vs MCK | 3 | 0.440 | 0.987 | 0.923 | 35.0 | 3.6 × 10−10 |
| MCK vs ESCK | 3 | 0.569 | 0.988 | 0.967 | 95.0 | 1.4 × 10−14 |
| Wt0 vs Wt14 | 2 | 0.311 | 0.969 | 0.857 | 37.8 | 2.9 × 10−14 |
Table 2.
Annotation of variables with VIP values higher than 1.35 (highlighted in bold) in at least a pairwise PLS-DA model from the CE-MS metabolomics analysis of PKD samples.
| Level of assignment | Annotation | Abbreviation | Migration time (min) | Detected specie | Experimental m/z | m/z error | VIP values of PLS-DA | ||
|---|---|---|---|---|---|---|---|---|---|
| Wt0 vs Wt14 | Wt0 vs MCK | MCK vs ESCK | |||||||
| Confirmed with standard | Creatinine | Cre | 13.4 | [M+H]+ | 114.0671 115.0697 | 0.9 mDa | 2.03 1.75 | 1.56 1.29 | 1.30 1.36 |
| Carnitine | Carn | 15.4 | [M+H]+ | 163.1159 (isotopic profile of 162.1139) 164.1173 | 1.4 mDa | 0.37 0.31 | 0.91 0.95 | 1.55 1.58 | |
| Creatine | Cr | 17.0 | [M+H]+ | 133.0796 (isotopic profile of 132.0782) | 1.4 mDa | 1.49 | 1.42 | 0.20 | |
| Glutamine | Gln | 21.0 | [M-NH3+H]+ | 130.0503 | 0.5 mDa | 1.27 | 1.42 | 0.59 | |
| Betaine | Btn | 22.9 | [M+H]+ | 119.0899 (isotopic profile of 118.0884) | 2.1 mDa | 0.12 | 0.44 | 1.39 | |
| Tentative annotation | 1-methylhistidine | mHis | 14.6 | [M+H]+ | 170.0925 171.0955 | 0.1 mDa | 2.31 2.29 | 2.17 2.20 | 0.25 0.26 |
| Acetylcholine | Ach | 14.7 | [M+H]+ | 146.1178 | 0.2 mDa | 0.91 | 1.47 | 0.72 | |
| 4-Guanidinobutanoic acid | Gba | 15.0 | [M+H]+ | 146.0927 | 0.3 mDa | 1.04 | 1.44 | 1.65 | |
| 2-Aminooctanoic acid | AoA | 15.2 | [M+H]+ | 160.1334 161.1367 | 0.2 mDa | 1.53 1.54 | 1.81 1.86 | 0.94 0.96 | |
| Methylguanine (different isomers are possible) | mGu | 15.2 | [M+H]+ | 166.0725 | 0.2 mDa | 0.46 | 0.61 | 1.55 | |
| γ-L-Glutamylputrescine or N5-(1-imino-3-butenyl)-L-ornithine | Gput | 15.4 | [M-H2O+H]+ | 200.1393 | 0.6 mDa | 1.55 | 1.71 | 1.01 | |
| Formylisoglutamine | fGln | 15.4 | [M+H]+ | 175.0715 | 0.2 mDa | 0.14 | 1.04 | 1.60 | |
| Imidazolelactic acid | ILA | 16.3 | [M+H]+ | 157.0609 | 0.1 mDa | 1.66 | 1.66 | 0.73 | |
| Asn-Hydroxyproline or Hydroxyprolyl-Asn | P1 | 16.5 | [M+H]+ | 246.1083 | 0.1 mDa | 1.41 | 1.29 | 1.57 | |
| Argininic acid | ArgA | 16.5 | [M+H]+ | 176.1030 | 0.0 mDa | 0.20 | 0.19 | 1.65 | |
| Propionyl-L-carnitine | Pcarn | 16.6 | [M+H]+ | 218.1386 219.1417 | 0.1 mDa | 1.36 1.37 | 0.92 0.90 | 1.28 1.27 | |
| Homocitrulline | Hcit | 16.8 | [M+H]+ | 190.1185 | 0.1 mDa | 1.09 | 0.30 | 1.64 | |
| Glycyl-lysine or lysyl-glycine | P2 | 17.4 | [M+H]+ | 204.1342 | 0.1 mDa | 0.68 | 0.28 | 1.63 | |
| N-α-Acetyl-L-arginine | Aarg | 17.7 | [M+H]+ | 217.1286 | 0.9 mDa | 1.75 | 0.82 | 1.54 | |
| 3-Hydroxyisovalerylcarnitine | HIVC | 17.7 | [M+H]+ | 262.1647 | 0.2 mDa | 2.07 | 0.43 | 1.47 | |
| (1) Phe-Thr or Thr-Phe (2) Hydroxyproline-Leu(Ile), or Leu(Ile)-Hydroxyproline | P3 | 18.5 | [M+H]+ (1) [M+Na]+ (2) | 267.1339 268.1370 | 0.9 mDa | 1.70 1.68 | 1.57 1.86 | 1.61 1.65 | |
| 2-[3-carboxy-3-(methylammonio)propyl]-L-histidine | CMHis | 19.0 | [M+H]+ | 271.1400 | 0.1 mDa | 0.52 | 1.27 | 1.64 | |
| Proline betaine | ProB | 23.1 | [M+H]+ | 144.1023 | 0.4 mDa | 1.86 | 2.18 | 1.39 | |
| γ-carboxyglutamic acid | Gla | 32.1 | [M+H]+ | 192.0501 | 0.2 mDa | 1.26 | 0.83 | 1.35 | |
| 5-Hydroxyindoleacetylglycine | hINaG | 32.1 | [M-H2O+H]+ | 231.0764 | 0.6 mDa | 1.88 | 1.67 | 1.56 | |
| Pantothenic acid | PA | 32.2 | [M-H2O+H]+ | 202.1074 | 0.5 mDa | 0.06 | 0.15 | 1.45 | |
Biological Relevance of the Annotated Metabolites
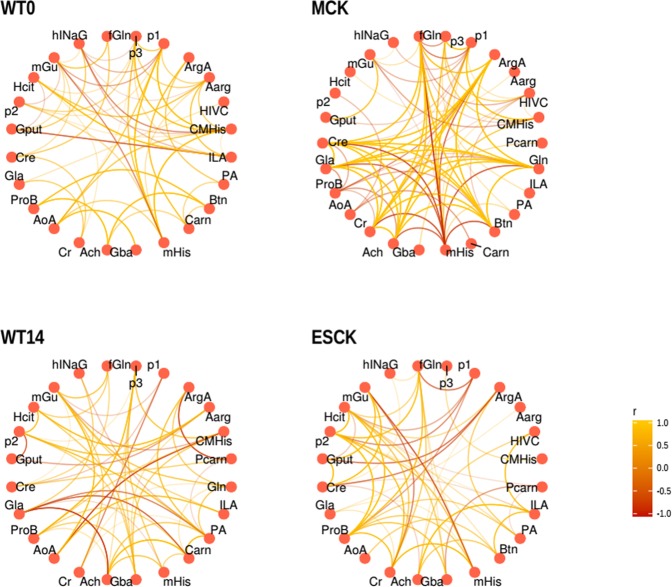
Table 2 summarizes all annotated features, however, the degree of confidence for the given annotations is varying. Figures S-4 and S-5 from supporting information display the box-plots for metabolites annotated unequivocally and tentatively, respectively. From this point the most logical question is to which degree the metabolites present in this table describe biology associated with the applied experimental design. There are several metabolites like e.g. creatinine, creatine, carnitine, and glutamine which have a proven track record of being associated with a renal physiology. The very fact that these metabolites are on the list illustrates that the selected analytical workflow is capable of targeting the relevant structures. Yet, verifying the results of a metabolomics experiment with the published data only provides hardly any novel information. Something that a post-genomics technology such as metabolomics is meant to do. Mapping the annotated structures to the biochemical pathways provides even less practical information. The flexibility of the mammalian biochemistry is such that a causal relation between the metabolites cannot be reconstructed from the profiling data without an experiment exploring a product-substrate relationship. To this end, we have explored an unbiased way of the data interpretation which uses the information available within the dataset: experimental design and the relative abundances of the metabolites. However, instead of a comparison between the experimental groups we have visualized the data in a form of the correlational relationships between metabolites within each experimental group. Figure 5 shows the results of such visualization. Notably, the most complex correlation structure at the given threshold of the inclusion (0.6) was observed within the MCK group. One of the “hubs” in this network is methylhistidine showing strong negative correlations to a number of the metabolites including creatine, creatinine, glutamine and a downstream product of the histidine metabolism, formylisoglutamine. It is interesting to note that the correlational structure of the MCK samples includes a number of the expected associations like e.g. creatine, creatinine, creatine-glutamine and some which till recent were not mentioned in the literature in the context of the renal physiology. It is important to keep in mind that the relationships visualized in Fig. 5 are not actual biochemical interactions but rather guidelines which can be used for follow-up research. To our opinion, creating the sets of the ideas for further validation in a more focused experimental design/setting is exactly the purpose of global metabolic profiling. Yet, one cannot ignore the fact that methylhistidine offers a few possibilities for speculation about its possible physiological role. For instance, the imidazole ring of methylhistidine was for long time considered as a prime source of the anti-oxidative activity of the histidine derivatives. Moreover, it has been shown that naturally occurring histidine containing the di-peptides carnosine and anserine have ability to chelate metals.
Figure 5.
Correlational relationships between the annotated metabolites within each of the four experimental groups. For abbreviations see Table 2.
Conclusions
Here, we present a first application of sheathless CE-MS (CESI-MS) for metabolic profiling of biomass-restricted tissue samples, namely individual sections of mouse kidney. After application of a strict data processing routine, consistent metabolic profiles were obtained covering more than 100 metabolic features, although minute amounts of sample were used (histological samples). Note that the number of metabolic features might have been increased if preconcentration strategies, such as dynamic pH junction, were used. The recorded metabolic profiles clearly differentiated the experimental groups included in this pilot study. Using a combination of non-supervised and supervised multivariate analysis we have dissected a subset of the metabolites underlying the differences between the experimental groups (non-PKD young mice, adult mice with PKD in mild state, adult mice with PKD in end-stage, and non-PKD adult mice). Several annotated compounds such as glutamine, carnitine, creatinine and creatine are well known for their roles in renal physiology. Using a data visualization procedure based on the correlational relationships between the metabolites, we could provide leads for further research. Overall, this study demonstrates the feasibility of sheathless CE-MS as a platform for metabolic profiling of biomass-restricted tissue samples, and it may open new possibilities for a better understanding of biological processes in sample-limited cases.
Methods
Chemicals and reagents
Sodium hydroxide (NaOH) was obtained from Merck (Darmstadt, Germany). Glacial acetic acid, water, hydrochloric acid (HCl), glutamine, carnitine, creatine, and creatinine were purchased from Sigma-Aldrich (Steinheim, Germany) and were of analytical grade or higher. Acetonitrile (MeCN) of LC–MS grade originated from Biosolve (Valkenswaard, The Netherlands). A standard metabolite mixture comprised of roughly 60 metabolites, with a concentration of 50 µM in water, was obtained from Human Metabolome Technologies (Tokyo, Japan), the so-called “HMT” mixture.
Animals, Experimental Design and Sample Collection
Oral administration of tamoxifen was conducted at Post Natal (PN) days 40–42 to inducible kidney-specific PKD1-deletion mice (tam-KspCad-CreERT2;PKD1lox2–11;lox2–11 mice) as previously reported23. At 8–12 weeks after gene disruption a subgroup of mice was sacrificed (MCK group: mild cystic kidneys). These mice displayed tubular dilations and small cysts accompanied by increased 2KW/BW% (1.9–3.3%). Another group of mice was sacrificed at renal failure (ESCK group: end stage cystic kidneys) having large 2KW/BW% (11–17%). A blood urea concentration of ≥20 mmol/L was used as an indicative of renal failure. In addition, there were two control group of mice having normal kidneys and not receiving tamoxifen, mice of age PN40 (Wt0) and at age of PN40 + 14 weeks (Wt14). Therefore, the study was composed of a total of four groups (Wt0, Wt14, MCK, and ESCK) having five mice each, being twenty the total number of subjects.
Snap-frozen kidneys were attached to a cryotome table using the embedding medium for frozen tissue KP-CryoCompound (Klinipath BV, The Netherlands). Three 20 µm-thick sections of the same kidney were obtained per mouse. Sections were collected in 1.5 mL microtubes and they were kept at −80 °C until sample preparation was conducted.
All the animal experiments were approved by the Animal Experiment Ethics Committee of Leiden University Medical Center and the Commission Biotechnology in Animals of the Dutch Ministry of Agriculture and all the methods were performed in accordance with their guidelines and regulations.
Sample preparation
Sample preparation was a continuation of the work by Sánchez-López et al. in which the samples were analyzed on liquid chromatography-MS platforms (both RPLC, and HILIC)24. Upon evaporation and reconstitution of the samples in 80:20 (MeCN:water, v/v), 7.5 µL were evaporated for 15 min at room temperature in a SpeedVac instrument (Eppendorf, model 5301) and reconstituted in 2.5 µL of water for sheathless porous tip CE-MS analysis. QC pool for the CE-MS platform was prepared by taking 1 µL of each sample (N = 20) and mixing thoroughly.
Sheathless CE-MS conditions
CE analyses were performed on a Sciex/Beckman Coulter CESI 8000 system (Sciex, Framingham, MA) equipped with a temperature-controlled sample tray, kept at 10 °C, and a power supply able to deliver up to 30 kV. Separation was performed using a bare fused silica capillary (91 cm × 30 μm i.d. × 150 μm o.d.) and carried out by applying a voltage of 20 kV without any pressure applied. Prior to each sample injection, the capillary was rinsed with 0.1 M NaOH for 2.5 min, water for 4 min, 0.1 M HCl for 2.5 min, water for 4 min, and background electrolyte (BGE) for 4 min. The BGE was a solution of 10% (v/v) acetic acid (pH 2.3). All samples were hydrodynamically injected applying a pressure of 1 psi for 60 s (which corresponds to 1.4% of the capillary volume, injecting 9 nL). A plug of BGE was injected after the sample injection by applying a pressure of 0.5 psi for 25 s (corresponding to 0.3% of the capillary volume). CE vials were built “in-house” by manually cutting the tip of a PCR microtube and introducing it in a Sciex/Beckman Coulter 1.5 mL vial by means of a stainless spring (see Fig. S-6). This setup enabled working with vial volumes as low as 2.5 µL per sample.
The CE apparatus was coupled to a UHR-QqTOF maXis Impact HD mass spectrometer from Bruker Daltonics (Bremen, Germany) via a sheathless porous tip CE-MS interface based on a custom-made platform from Sciex/Beckman Coulter (Brea, CA) allowing for an optimal position of the capillary porous tip in front of the MS nanospray shield (Bruker Daltonics). Dopant enriched nitrogen (DEN)-gas at 0.2 bar was used along the sequence. To this end, an in-house made polymer cone was slid onto the housing of the porous tip allowing for a coaxial sheath flow of the DEN-gas around the ESI emitter. The concentration of acetonitrile in the DEN-gas corresponds to ∼4% (mole percentage). All CE-MS experiments were carried out in ESI positive mode with a capillary voltage of 1,000 V. Drying gas flow (N2) rate was 1.2 L/min at 150 °C. MS data was acquired between m/z 50 and 1,300 with a spectral acquisition rate of 1 Hz. For MS/MS experiments, all parameters were similar to the ones for MS, except for the spectra acquisition that was registered from m/z 30 to 1,200. Collision energy was set to 20 V. MS and MS/MS data were acquired with DataAnalysis 4.2 (Build 387, Bruker Daltonics).
CE-MS metabolomics sequence
At the beginning of the metabolic profiling sequence, blanks, QC pools and standard metabolite mixtures from Human Metabolome Technologies (HMT) were injected in the CE-MS platform. Samples of PKD study were randomized for injection, and a QC pool was injected after seven samples were injected. The HMT mixture was injected at least once per day to assess the metabolite annotation (the metabolomic sequence lasted for 5 days).
Data treatment and data analysis
MS data files obtained by the CE-MS platforms were recalibrated based on sodium acetate clusters and were exported in the.mzXML format. Alignment was performed by an in-house tool, msalign221. Peak picking was carried out on the aligned data using XCMS R-package (The Scripps Research Institute, La Jolla, CA) based on the centWave algorithm using the following settings: maximum tolerated m/z deviation in consecutive scans, 15 ppm; chromatographic peak width, 5–25 s; scan range, 60–2325 s; scan range to process, m/z 50–1,300; noise, 20,000; prefilter step, at least 3 peaks with intensity > 50,000; m/z center of the feature, wMean (intensity weighted mean of the feature m/z values); signal-to-noise ratio threshold, 50; minimum difference in m/z for peaks with overlapping migration time, 0.01 min. After peak picking, peak grouping was performed with the following parameters: bandwidth, 2; and m/z width of 0.01. After peak picking and grouping, variables having RSD values above 30% in the QC were excluded.
Before multivariate analysis, the data was normalized using the PQN algorithm. PQN is based on the determination of the most probable dilution factor by considering the distribution of the quotients of the signals of a certain sample by those of the QC samples25. Multivariate statistical analysis was carried out by means of SIMCA 14 (Umetrics, Umeå, Sweden).
Base peak electropherogram and extracted ion electropherogram Figures were plotted using MZmine 226. The data visualization was performed using R version 3.4 and packages, “Rcpm”, “ggplot”, “igraph”, “ggraph”, “corrr”. For Fig. 5 the data set was divided according to the conditions MCK, ESCK, Wt0, Wt14, and a correlation matrix was built on every subset separately and converted into a long format; this output was filtered to retain only the correlations with absolute value above 0.6 and passed to the “ggraph” function using “circular” outline for visualization.
Metabolite annotation
Highly influencing variables in the PLS-DA models were tentatively identified by looking up their m/z values in different databases (METLIN (http://metlin.scripps.edu), HMDB (http://hmdb.ca), and KEGG (http://genome.jp/kegg)), establishing 20 ppm as the error width. The identity of carnitine, glutamine, creatinine, betaine and creatine could be confirmed by matching the migration time and MS/MS spectra pattern obtained from standards. Tentative identities were annotated based on databases.
Supplementary information
Acknowledgements
This research was supported by the Astellas Pharma B.V. and SCIEX. We thank Kimberley Veraar (LUMC) for technical assistance. The Developing Interventions to halt Progression of ADPKD (DIPAK) consortium is an inter-university collaboration in The Netherlands that is established to study Autosomal Dominant Polycystic Kidney Disease and to develop treatments for this disease. Partners are the department of Human Genetics at the University Medical Center Leiden (D.J.M. Peters), the departments of Nephrology at the University Medical Centers Groningen (R.T Gansevoort), Leiden (J.W. de Fijter), Nijmegen (J.F. Wetzels), and Rotterdam (R. Zietse), the department of Hepatology at the University Medical Center Nijmegen (J.P. Drenth). The DIPAK consortium is sponsored by the Dutch Kidney Foundation (grant number CP 10.12). We acknowledge the members of the consortium for support and the Dutch Kidney Foundation for funding. E.S.L., A.L.C., and M.L.M. thank the Ministry of Economy and Competitiveness (Spain) for research project CTQ2013-48740-P. E.S.L. and M.L.M. thank the same Ministry for project CTQ2016-76368-P. E.S.L. thanks the University of Alcalá for her pre-doctoral contract and for the research stay mobility grant in the Center for Proteomics and Metabolomics from the Leiden University Medical Center.
Author Contributions
E.S.L., D.J.M.P. and O.A.M. designed the research. E.S.L., G.S.M.K., O.A.M., performed the experiments. E.S.L., G.S.M.K. and O.A.M. analyzed the data. E.S.L., G.S.M.K., R.R. and O.A.M. wrote the paper. A.L.C. and M.L.M. co-supervised the work performed by E.S.L. in this article as a part of her PhD Thesis carried out in the University of Alcalá with international label. All authors reviewed final manuscript.
Data Availability
The dataset generated and analyzed during this study is included in the Supplementary Information file of this article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37512-8.
References
- 1.Wilson PD. Mechanisms of disease: Polycystic kidney disease. New Engl. J. Med. 2004;350:151–164. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- 2.Harris PC, Torres VE. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J. Clin. Invest. 2014;124:2315–2324. doi: 10.1172/JCI72272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Happé H, Peters DJM. Translational research in ADPKD: lessons from animal models. Nat. Rev. Nephrol. 2014;10:587–601. doi: 10.1038/nrneph.2014.137. [DOI] [PubMed] [Google Scholar]
- 4.Barbas C, Moraes EP, Villaseñor A. Capillary electrophoresis as a metabolomics tool for non-targeted fingerprinting of biological samples. J. Pharm. Biomed. Anal. 2011;55:823–831. doi: 10.1016/j.jpba.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Hirayama A, Wakayama M, Soga T. Metabolome analysis based on capillary electrophoresis-mass spectrometry. TrAC-Trend. Anal. Chem. 2014;61:215–222. doi: 10.1016/j.trac.2014.05.005. [DOI] [Google Scholar]
- 6.Moini M. Simplifying CE–MS Operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal. Chem. 2007;79:4241–4246. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 7.Ramautar R, Busnel J-M, Deelder AM, Mayboroda OA. Enhancing the coverage of the urinary metabolome by sheathless capillary electrophoresis-mass spectrometry. Anal. Chem. 2012;84:885–892. doi: 10.1021/ac202407v. [DOI] [PubMed] [Google Scholar]
- 8.Haselberg R, Ratnayake CK, de Jong GJ, Somsen GW. Performance of a sheathless porous tip sprayer for capillary electrophoresis–electrospray ionization-mass spectrometry of intact proteins. J. Chromatogr. A. 2010;1217:7605–7611. doi: 10.1016/j.chroma.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Hankemeier T, Ramautar R. Next-generation capillary electrophoresis-mass spectrometry approaches in metabolomics. Curr. Opin. Biotechnol. 2017;43:1–7. doi: 10.1016/j.copbio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Hirayama A, Tomita M, Soga T. Sheathless capillary electrophoresis-mass spectrometry with a high-sensitivity porous sprayer for cationic metabolome analysis. Analyst. 2012;137:5026–5033. doi: 10.1039/c2an35492f. [DOI] [PubMed] [Google Scholar]
- 11.Ramautar R, et al. Metabolic profiling of mouse cerebrospinal fluid by sheathless CE-MS. Anal. Bioanal. Chem. 2012;404:2895–2900. doi: 10.1007/s00216-012-6431-7. [DOI] [PubMed] [Google Scholar]
- 12.Shyti R, et al. Plasma metabolic profiling after cortical spreading depression in a transgenic mouse model of hemiplegic migraine by capillary electrophoresis – mass spectrometry. Mol. BioSyst. 2015;11:1462–1471. doi: 10.1039/C5MB00049A. [DOI] [PubMed] [Google Scholar]
- 13.Canuto GAB, et al. CE-ESI-MS metabolic fingerprinting of Leishmania resistance to antimony treatment. Electrophoresis. 2012;33:1901–1910. doi: 10.1002/elps.201200007. [DOI] [PubMed] [Google Scholar]
- 14.Gulersonmez MC, Lock S, Hankemeier T, Ramautar R. Sheathless capillary electrophoresis-mass spectrometry for anionic metabolic profiling. Electrophoresis. 2016;37:1007–1014. doi: 10.1002/elps.201500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onjiko RM, Portero EP, Moody SA, Nemes P. In situ microprobe single-cell capillary electrophoresis mass spectrometry: metabolic reorganization in single differentiating cells in the live vertebrate (Xenopus laevis) embryo. Anal. Chem. 2017;89:7069–7076. doi: 10.1021/acs.analchem.7b00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirayama A, et al. Development of a sheathless CE-ESI-MS interface. Electrophoresis. 2018;39:1382–1389. doi: 10.1002/elps.201800017. [DOI] [PubMed] [Google Scholar]
- 17.Lapainis T, Rubakhin SS, Sweedler JV. Capillary Electrophoresis with Electrospray Ionization Mass Spectrometric Detection for Single-Cell Metabolomics. Anal. Chem. 2009;81:5858–5864. doi: 10.1021/ac900936g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxwell EJ, Zhong X, Zhang H, van Zeijl N, Chen DD. Decoupling CE and ESI for a more robust interface with MS. Electrophoresis. 2010;31:1130–1137. doi: 10.1002/elps.200900517. [DOI] [PubMed] [Google Scholar]
- 19.Sun L, Zhu G, Zhang Z, Mou S, Dovichi NJ. Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J. Proteome Res. 2015;14:2312–2321. doi: 10.1021/acs.jproteome.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kammeijer GSM, et al. Dopant enriched nitrogen gas combined with sheathless capillary electrophoresis-electrospray ionization-mass spectrometry for improved sensitivity and repeatability in glycopeptide analysis. Anal. Chem. 2016;88:5849–5856. doi: 10.1021/acs.analchem.6b00479. [DOI] [PubMed] [Google Scholar]
- 21.Nevedomskaya E, Derks R, Deelder AM, Mayboroda OA, Palmblad M. Alignment of capillary electrophoresis-mass spectrometry datasets using accurate mass information. Anal. Bioanal. Chem. 2009;395:2527–2533. doi: 10.1007/s00216-009-3166-1. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, MacDonald D, Huang X, Chen DD. Capture efficiency of dynamic pH junction focusing in capillary electrophoresis. Electrophoresis. 2016;37:1143–1150. doi: 10.1002/elps.201600008. [DOI] [PubMed] [Google Scholar]
- 23.Leonhard WN, et al. Curcumin inhibits cystogenesis by simultaneous interference of multiple signaling pathways: in vivo evidence from a Pkd1-deletion model. Am. J. Physiol-Renal. 2011;300:F1193–F1202. doi: 10.1152/ajprenal.00419.2010. [DOI] [PubMed] [Google Scholar]
- 24.Sánchez-López E, et al. A cross-platform metabolic workflow for volume-limited tissue samples. Application to the metabolomics analysis of polycystic kidney disease samples. Mol. BioSyst. 2017;13:1940–1945. doi: 10.1039/C7MB00245A. [DOI] [PubMed] [Google Scholar]
- 25.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in H-1 NMR metabonomics. Anal. Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 26.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset generated and analyzed during this study is included in the Supplementary Information file of this article.