Abstract
Human serum albumin (HSA) has been identified as an important regulator of amyloid-β (Aβ) fibrillization both in blood plasma and in cerebrospinal fluid. Fatty acids bind to HSA, and high serum levels of fatty acids increase the risk of Alzheimer’s disease. In vitro, fatty-acid-loaded HSA (FA·HSA) loses the protective effect against Aβ fibrillization, but the mechanism underlying the interference of fatty acids on Aβ-HSA interactions has been unclear. Here, we used molecular dynamics simulations to gain atomic-level insight on the weak binding of monomeric Aβ40 and Aβ42 peptides with apo and FA·HSA. Consistent with recent NMR data, C-terminal residues of the Aβ peptides have the highest propensities for interacting with apo HSA. Interestingly, the Aβ binding residues of apo and FA·HSA exhibit distinct patterns, which qualitatively correlate with backbone flexibility. In FA·HSA, both flexibilities and Aβ binding propensities are relatively even among the three domains. In contrast, in apo HSA, domain III shows the highest flexibility and is the primary target for Aβ binding. Specifically, deformation of apo HSA creates strong binding sites within subdomain IIIb, around the interface between subdomains IIIa and IIIb, and at the cleft between domains III and I. Therefore, much like disordered proteins, HSA can take advantage of flexibility in forming promiscuous interactions with partners, until the flexibility is quenched by fatty-acid binding. Our work explains the effect of fatty acids on Aβ-HSA binding and contributes to the understanding of HSA regulation of Aβ aggregation.
Introduction
Amyloid plaques caused by the aggregation of amyloid-β (Aβ) peptides are a pathological hallmark of Alzheimer’s disease (1). Plasma proteins sequester Aβ and are potent inhibitors of Aβ amyloidosis (2, 3). Human serum albumin (HSA), the most abundant protein in blood plasma, binds Aβ and inhibits Aβ fibrillization in vitro (3, 4, 5, 6, 7, 8, 9, 10, 11, 12) and in an animal models (13) and has been explored as a therapy for Alzheimer’s disease (14). HSA is responsible for the transport of many endogenous hydrophobic molecules, including fatty acids, and hydrophobic therapeutic drugs. Fatty acids negate the inhibitory effect of HSA on Aβ fibrillization (8, 12). This observation is particularly intriguing and biologically relevant, given that high dietary levels of fatty acids increase the risk of developing Alzheimer’s disease (15, 16, 17). The mechanism by which fatty acids interfere with HSA’s protective inhibition of Aβ fibrillization has been unclear. The aim of this study was to elucidate this mechanism through comparative molecular dynamics (MD) simulations of the weak binding of Aβ peptides with apo and fatty-acid-loaded HSA (FA·HSA) (Fig. 1).
Figure 1.
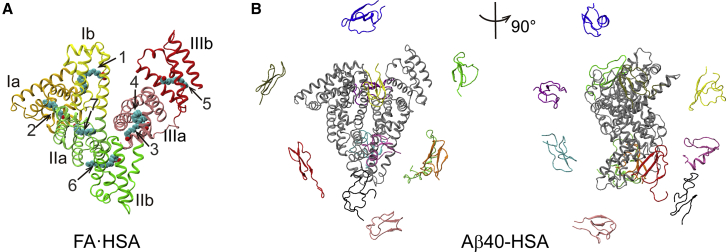
Starting structures for MD simulations. (A) The crystal structure of HSA bound with seven palmitic acids (PDB: 1E7H) used for simulations of isolated and Aβ40-bound FA·HSA. Subdomains Ia, Ib, IIa, IIb, IIIa, and IIIb are shown in gold, yellow, light green, dark green, pink, and red, respectively; palmitic acids are shown as spheres. (B) Two views of the 12 starting positions of Aβ40 (in color) around HSA (in gray). To see this figure in color, go online.
Aβ peptides are derived from the proteolytic cleavage of the transmembrane amyloid precursor protein by β- and γ-secretases (18). They occur in sequence lengths ranging from 36 to 43 amino acids. The amyloid plaques in the Alzheimer’s brain mainly contain the 40- and 42-residue species. Aβ40 is the most abundantly produced, and Aβ42 is less abundant but more prone to aggregation (1). Although Aβ peptides are found in blood plasma and cerebrospinal fluid (CSF) at similar concentrations (0.1–0.5 nM) (19, 20), Aβ fibrillization occurs exclusively in the central nervous system. Endogenous plasma proteins, most notably HSA, are capable of binding Aβ and suppressing amyloid formation (2, 3). HSA can bind ∼90% of Aβ40 and Aβ42 in blood plasma (4). In the CSF, HSA is still the most abundant protein, but its concentration markedly decreases from ∼640 μM in the plasma (21) to a typical 3 μM (22). Aβ sequestration by plasma HSA may also help with the clearance of Aβ from the brain to the blood (23). It is worth noting that serum levels of Aβ-HSA complexes were significantly lower in Alzheimer’s patients than in controls, even though HSA levels were similar in the two groups (24).
Aβ fibrillization (e.g., as measured by thioflavin T fluorescence) starts with a lag phase for nucleation, is then followed by a rapid growth phase, and ends with a plateau with mature fibrils. HSA lengthened the lag phase and reduced the total amount of fibrils formed (7). These effects grew with increasing HSA concentration and became statistically significant at 5 μM, just above the physiological concentration in CSF. These observations implicate interactions with Aβ monomers or prenucleus oligomers in HSA’s inhibitory effect. Although this and other earlier studies disagreed on whether HSA binds Aβ monomers (5, 7, 25, 26) or oligomers (6, 27, 28), recent experiments have consistently shown that HSA targets both species as well as protofibrils and fibrils at different phases of fibrillization (10, 11, 29). Although the binding affinity of HSA for Aβ monomers is lower than for protofibrils (submillimolar versus micromolar), it is still physiologically relevant given the submillimolar HSA plasma concentration (29).
HSA is a transport protein of fatty acids. It is composed of three homologous domains (I, II, and III); each can be further divided into two subdomains (a and b) comprising six and four helices, respectively (Fig. 1 A). Together, the three domains bind up to seven fatty acids and in doing so experience relatively minor internal conformational changes (backbone root mean-square deviation (RMSD) at 1.3, 0.8, and 1.4 Å, respectively) but do undergo more significant rearrangements in their relative orientations in crystal structures (30, 31). The fatty acids are almost completely buried inside HSA. Recently, Algamal et al. (8) combined saturation-transfer difference NMR and mutagenesis to investigate the competition of myristic acid and Aβ oligomers for binding with HSA domain III. Several mutations that disrupted myristic-acid binding did not impede Aβ binding, suggesting that their interactions with HSA are distinct. Additional data indicated a separate Aβ binding site on each of the two subdomains of domain III and moreover identified the 494–515 segment within subdomain IIIb as important for binding Aβ oligomers. Mass spectrometry and small-angle X-ray scattering by Choi et al. (11) revealed a single Aβ monomer bound at the cleft between domains I and III and found no gross change in HSA structure upon Aβ binding. NMR relaxation and saturation-transfer difference NMR data of Algamal et al. (29) have also provided residue-level information for Aβ monomers interacting with HSA. For Aβ40, the C-terminal residues 31–40 formed the primary interaction region, with the central 12–24 segment contributing additional interacting residues. The Aβ42 affinity for HSA was approximately twofold weaker, with the C-terminus again showing the strongest evidence for interacting with HSA. Together, these studies have presented valuable details on Aβ-HSA binding, but none has yielded any direct information for how fatty acids interfere with this binding.
To uncover the mechanism underlying the interference of Aβ-HSA binding by fatty acids, here, we carried out MD simulations of monomeric Aβ40 and Aβ42 binding to apo and FA·HSA. Consistent with the NMR data of Algamal et al. (29), C-terminal residues of the Aβ peptides have the highest propensities for interacting with apo HSA. Whereas FA·HSA is relatively rigid and shows no strong site preference for Aβ binding, domain III of apo HSA is highly flexible and the primary target for Aβ binding. Specifically, deformation of apo HSA creates strong binding sites within subdomain IIIb, around the interface between subdomains IIIa and IIIb, and at the cleft between domains III and I. Therefore, much like disordered proteins, HSA can take advantage of flexibility in forming promiscuous interactions with partners, until the flexibility is quenched by fatty-acid binding.
Methods
System preparation
To select representative conformations of monomeric Aβ40 and Aβ42, 1000-ns MD simulations were carried out for each, starting from their NMR structures (Protein Data Bank (PDB): 2LFM (32) and 1Z0Q (33)) in aqueous solution (or with 30% organic cosolvent in the latter case). The secondary structure contents in the MD simulations are consistent with those measured by circular dichroism (CD) spectroscopy (34). For Aβ40, the β-strand and α-helix contents are 27 and 1%, respectively, by MD, compared to 25 and 5% by CD. For Aβ42, corresponding percentages are 33 and 2% by MD and 27 and 6% by CD. After dihedral principal component analysis of 60,000 sampled conformations from each simulation, the free-energy surface converted from the histogram in the first two dihedral principal components was used to select 10 representative conformations (Fig. S1, A and C). The secondary structures in these conformations were mainly β-strands and turns, in line with their approximate 30% populations each in the overall simulation (Fig. S2). This selection was complemented by a mostly random coil conformation from the simulation and the partially α-helical NMR structure (Fig. S1, B and D); these additions correct for the slight overestimation of β-strand and slight underestimation of α-helix in the 1000-ns simulation. The resulting 12 Aβ40 or Αβ42 conformations were each used to start a simulation of complex formation with HSA.
Three Aβ-HSA systems at 1:1 stoichiometry were simulated: Aβ40 with apo HSA (Aβ40-HSA), Aβ40 with palmitic-acid-bound HSA (Aβ40-FA·HSA), and Aβ42 with apo HSA (Aβ42-HSA). Starting structures of Aβ40-HSA were generated by placing the 12 Aβ40 conformations randomly at ∼10 Å away from apo HSA (PDB: 1AO6 (30)). By replacing the apo HSA with palmitic-acid-bound structure (PDB: 1E7H (31)) or replacing the Aβ40 conformations with Aβ42 conformations, starting structures for the second or third system were then obtained.
MD simulations
All simulations were performed with GROMACS 4.6.7 software package (35). For the protein molecules, the GROMOS 53a6 force field (36) was used. For palmitic acid, force field parameters were taken from a previous study (37). For each system, the solute molecules were energy minimized in vacuum and then solvated by SPC (simple point charge) water in an octahedron box with a minimal distance of 12 Å from the box boundaries. Sodium and chloride ions were added to achieve charge neutralization and a 150-mM salt concentration. The whole system was energy minimized, first with protein heavy atoms constrained and then without any constraint. Subsequently, the system was equilibrated at constant NPT (particle number-pressure-temperature) for 100 ps with protein heavy atoms constrained. In the final production run, these constraints were removed. The temperature was maintained at 300 K by the velocity-rescale coupling method (38) (with a coupling constant of 0.1 ps), and the pressure was maintained at one bar by the Parrinello-Rahman pressure coupling method (39, 40) (with a coupling constant of 2 ps and a compressibility of 4.5 × 10−5 bar−1). Protein bond lengths and water geometries were constrained using LINCS (41) and SETTLE (42), respectively. The simulation time step was 2 fs. Long-range electrostatic interactions were treated by the particle mesh Ewald method (43) with a real space cutoff of 10 Å; van der Waals interactions were calculated using a cutoff of 10 Å.
Lengths of the production runs of the different systems are listed in Table S1. Snapshots were saved every 10 ps. The 1000-ns MD simulations for Aβ40 and Aβ42 were already mentioned; the last 600 ns were used for analysis. As references, simulations for apo and FA·HSA were carried out in duplicates, each lasting 200 ns and with the last 150 ns for analysis. For the three Aβ-HSA systems, duplicate runs of 200 ns each were carried out for each of the 12 Aβ starting conformations; the last 10 ns were saved for analysis.
Analysis
Built-in tools in GROMACS (35) and VMD (44) scripts were used for analysis. Dihedral principal components were calculated using g_angle, g_covar, and g_anaeig in GROMACS. Backbone root mean-square fluctuation (RMSF) and RMSD were calculated using g_rmsf and g_rms, respectively, in GROMACS. Secondary structures of Aβ were determined by the DSSP (45) program.
Binding propensity was measured by the percentage of snapshots in which a particular residue of one protein was in contact with any residue of the partner protein. Contact was defined using a 5.4-Å cutoff distance between two heavy atoms. For each snapshot, the position of Aβ relative to HSA was represented by the center-of-mass coordinates of Aβ after superimposing HSA to the starting structure using backbone atoms. For each complex system, all poses sampled in the last 10 ns of the 24 trajectories were partitioned into clusters by the k-means algorithm. All structure figures were prepared in VMD or PyMOL (46).
Data availability
The data reported here, including the MD simulation trajectories, are available upon request.
Results
We simulated the binding of single Aβ40 to Aβ42 monomers to apo and palmitic-acid-bound HSA. These systems are denoted as Aβ40-HSA, Aβ40-FA·HSA, and Aβ42-HSA. At the start, each monomer took 1 out of 12 conformations and was positioned randomly at a distance of 10 Å away from an HSA molecule (Figs. 1 B and S3). 1 of the 12 conformations was an NMR structure (PDB: 2LFM for Aβ40 (32) or 1Z0Q for Aβ42 (33)); the other 11 conformations were sampled from a 1000-ns simulation (see Methods; Fig. S1). The starting structures of apo and FA·HSA were from PDB: 1AO6 and 1E7H, respectively. For each of the 12 starting poses, two replicate runs of 200 ns were performed. As references, two replicate runs were also performed for apo and FA·HSA without Aβ.
C-terminal region of Aβ is the primary binding site for HSA
In the simulations, each Aβ molecule diffuses from the solvent to the HSA surface and settles into a binding site until the end. The Aβ-HSA complexes become stabilized after 150 ns, as indicated by, for example, steady RMSDs of the HSA molecules (Fig. S4, A and B). We extended 4 of the 24 Aβ40-HSA runs to up to 500 ns and 2 of the Aβ40-FA·HSA runs to 400 ns. The snapshots at 150 ns, 200 ns, and the end of these runs look very similar (Fig. S4, C and D). Snapshots in a 10-ns window, from 190 to 200 ns of each trajectory, were analyzed, and the data are reported below.
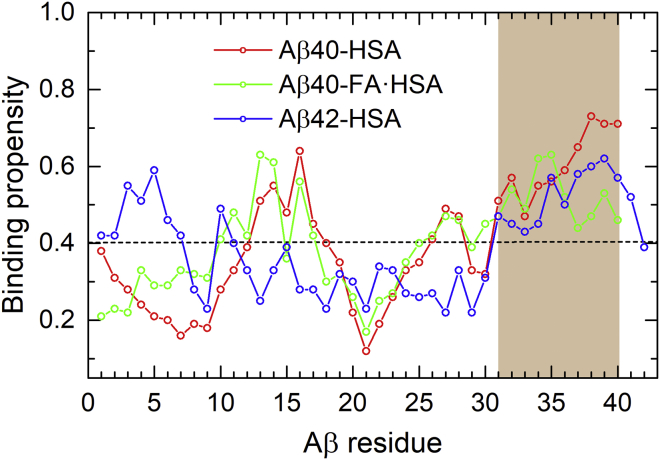
We calculated the HSA binding propensity of each Aβ residue, defined as the percentage of saved snapshots in which a particular Aβ residue is in contact with HSA. The results are shown in Fig. 2, where the average propensity over all the Aβ residues in the three complex systems is indicated by a horizontal dash to serve as a baseline. The average propensity is 40%, which corresponds to an average of 16 Aβ residues in contact with HSA.
Figure 2.
HSA binding propensities of Aβ residues in three complexes: Aβ40-HSA (red), Aβ40-FA·HSA (green), and Aβ42-HSA (blue). The average binding propensity of the three systems is shown as a horizontal dashed line. The major HSA binding site identified by the MD simulations, consistent with NMR data (29), is highlighted by brown shading. To see this figure in color, go online.
For Aβ40-HSA, two Aβ40 segments show relatively high propensities for binding HSA. The stronger of the two is composed of the C-terminal residues 31–40 (Fig. 2, brown shading); the central residues 12–18 also have above-average propensities. The binding pattern is preserved to some extent when HSA is loaded with fatty acids but with a notable drop in binding propensity for the very C-terminal four residues. For Aβ42 binding to apo HSA, the C-terminal residues 31–41 again exhibit relatively high binding propensities, but now the central region no longer shows any sign of preferential binding. These residue-specific results for the Aβ peptides binding to apo HSA are largely in agreement with the recent NMR data of Algamal et al. (29), who identified the Aβ C-terminal region as the primary site for binding HSA; Aβ40 central residues also contributed to binding, whereas the HSA affinity of Aβ42 was weaker.
We did not detect any significant shift in Aβ conformation upon binding to HSA or FA·HSA. For example, residue-specific populations of Aβ40 for β-strand and random coil are similar, whether in the 1000-ns simulation (Fig. S2) or in the 200-ns simulations for binding to HSA or FA·HSA.
Aβ binding propensities of HSA residues correlate with backbone flexibilities and are suppressed by fatty-acid binding
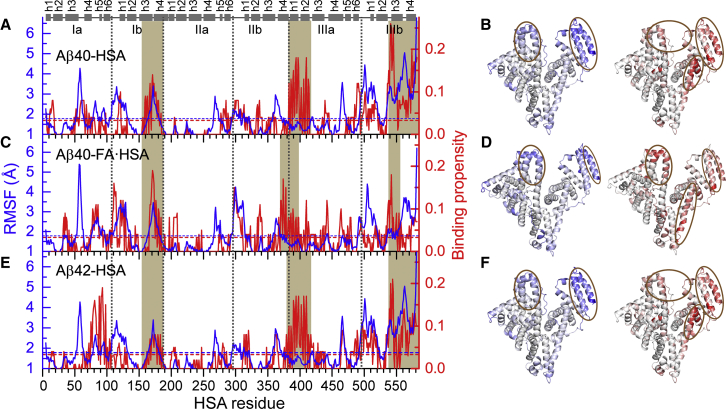
To confirm that fatty acid presence affects Aβ-HSA interactions in our MD simulations, we calculated Aβ binding propensities of HSA residues. Apo and FA·HSA indeed show different Aβ binding patterns, which apparently correlate with backbone RMSFs of apo and FA·HSA in the absence of Aβ (Fig. 3). In particular, residues having high Aβ binding propensities (Fig. 3, brown shading) generally also have high RMSFs, except for residues 384–415 (helices h1 and h2 in subdomain IIIa) in Aβ40-HSA and Aβ42-HSA where high binding propensities are not matched by high RMSFs. Moreover, the relative magnitudes of binding propensities among the three domains parallel those of RMSFs.
Figure 3.
An apparent correlation between backbone RMSFs of isolated apo or FA·HSA and Aβ binding propensities of HSA residues in three complex systems. (A), (C), and (E) display RMSFs (blue curves) and Aβ binding propensities (red curves) for Aβ40-HSA, Aβ40-FA·HSA, and Aβ42-HSA, respectively. Average values for each system are indicated by blue and red horizontal dashed lines; segments with high RMSFs or high binding propensities are highlighted by brown shading. Subdomain boundaries are indicated by vertical dashed lines; at the top, helices are represented by cylinders. (B), (D), and (F) display the RMSFs (left) and binding propensities (right) as mapped onto the crystal structures of apo or FA·HSA. Regions with high RMSFs and binding propensities are represented by intense blue and red colors, respectively, and highlighted by brown ovals. To see this figure in color, go online.
In apo HSA, domains I and II are less flexible than domain III, evidenced by average RMSFs at 1.8, 1.4, and 2.2 Å, respectively (Fig. 3, A and E, blue curves). Similarly, average Aβ40 binding propensities of domains I (2.4%) and II (1.8%) are lower than that of domain III (5.6%) (Fig. 3 A, red curve). The latter correspond to averages of 5, 3, and 11 residues in contact with Aβ. The same trend holds true for Aβ42 binding (Fig. 3 C, red curve), with average propensities of the three domains at 2.8, 1.4, and 5.4%, respectively. These similar results between Aβ40-HSA and Aβ42-HSA attest to the statistical reproducibility of the disparate Aβ binding patterns among the three domains, especially because the two Aβ isoforms started with different secondary structure distributions in the MD simulations (Figs. S1 and S2).
Overall, Aβ binding residues of apo HSA are concentrated in domain III (Fig. 3, B and F). The C-terminal residues 540–582 (helices h3 and h4 in subdomain IIIb) display the highest RMSFs and also the highest binding propensities for both Aβ40 and Aβ42. Another cluster of residues with high binding propensities comes from helices h1, h2, and h6 in subdomain IIIa and the linker between subdomains IIIa and IIIb, though among these elements, only the linker has high RMSFs. Lastly, subdomain IIIb, in particular, helices h1 and h2, contributes to a third cluster that also involves subdomain Ib, including helices h1 and h4. In comparison, most residues in domain II have below-average RMSFs and binding propensities.
In contrast to the disparate patterns across domains, the three domains of FA·HSA display nearly even backbone flexibilities (Fig. 3 C, blue curve), with averages at 1.9, 1.6, and 1.8 Å, respectively. Correspondingly, domain-specific average Aβ binding propensities are also very similar (Fig. 3 C, red curve), with values at 3.3, 2.7, and 3.6%, respectively. At the C-terminus, the binding propensities are all but lost in subdomain helix h4 and diminish in h3 to a level comparable to the highest values in domains I and II. The result is a much more dispersed distribution of Aβ binding residues among the three domains (Fig. 3 D).
In short, the Aβ binding residues on apo and FA·HSA exhibit distinct patterns, which qualitatively correlate with backbone flexibility. In FA·HSA, both flexibilities and Aβ binding propensities are relatively even among the three domains. In contrast, in apo HSA, domain III shows the highest flexibility and is the primary target for Aβ binding.
Number of domain-specific contacts hints at distinct Aβ-HSA interaction strengths in the absence and presence of fatty acids
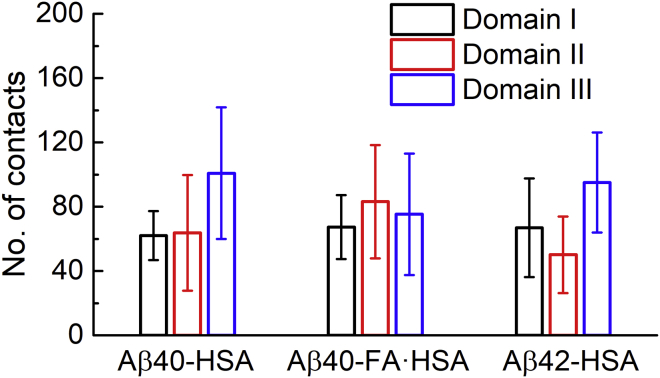
The concentration of Aβ binding residues in domain III of apo HSA suggests that this domain presents strong binding sites for Aβ. As a first approximation, the more HSA atoms form contacts with the bound Aβ, the stronger the binding. We therefore calculated the domain-specific number of contacts (i.e., the number of heavy atoms in a given HSA domain that simultaneously formed contacts with Aβ) for each snapshot in the last 10 ns of simulations. The average was then calculated for each trajectory and, to focus on close Aβ encounters with a particular HSA domain, those trajectories with averages less than 10 contacts for the said domain were filtered; a final average was taken over the average contacts of the remaining trajectories. These domain-specific average contacts for the three systems are shown in Fig. 4. In apo HSA, domain III, on average, forms 100 contacts with Aβ40, significantly larger than the averages for domain I (62 contacts) and domain II (64 contacts). The same trend is followed for Aβ42 binding to apo HSA, with domain III forming 95 contacts but domains I and II forming only 66 and 55 contacts, respectively. In the presence of fatty acids, average contacts formed by the three domains are close to each (67, 83, 70, respectively), with the difference between each pair within standard error. From these data, we conclude that apo HSA harbors strong Aβ binding sites in domain III, but FA·HSA presents relatively weak binding sites dispersed throughout the three domains.
Figure 4.
The average numbers of contacts formed by each HSA domain with Aβ in the three systems. To see this figure in color, go online.
Clustering of binding poses identifies three major Aβ40 binding sites within or around domain III in apo HSA
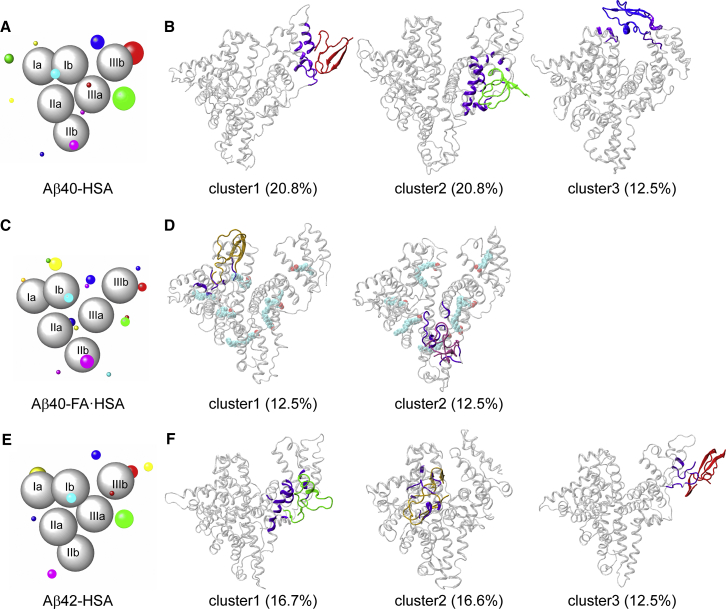
To define Aβ binding sites, we clustered Aβ positions around HSA in the last 10 ns of simulations. In Fig. 5, A, C, and E, we display the clustering results for the three complex systems. HSA subdomains are represented by gray spheres; Aβ clusters are shown as colored spheres, centered at cluster centers and with radii proportional to cluster sizes. Each cluster is formed from the binding of multiple Aβ starting conformations, and no preference for particular Aβ conformations, as judged by residue-specific populations of β-strand and random coil, is discernible. Representative conformations of major clusters are shown in Fig. 5, B, D, and F. As can now be expected, Aβ40-HSA and Aβ42-HSA show similar results and differ from Aβ40-FA·HSA.
Figure 5.
Clustering of Aβ positions around HSA and representative binding poses in major clusters. (A, C, and E) Binding clusters represented by colored spheres with radii proportional to cluster sizes. HSA subdomains are shown as gray spheres. (B, D, and F) Representative snapshots of major clusters (i.e., those with cluster sizes exceeding 10% of total snapshots). HSA residues in contact with Aβ are colored in violet. To see this figure in color, go online.
For Aβ40-HSA, 10 clusters are obtained, and the first three accounts for more than 50% of total snapshots. Domain III is involved in all the three major clusters. Cluster 1 defines an Aβ40 binding site within the lower part of subdomain IIIb, encompassing a portion of the linker from subdomain IIIa, the region from the C-terminus of helix h2 to the middle of h3, and most of h4. In cluster 2, the binding site is around the IIIa-IIIb interface, comprising the exposed surface of subdomain IIIa (lined by helices h1, h2, and h6 and the very N-terminal portion of the IIIa-IIIb linker), and the N-terminal half of IIIb h3 facing IIIa. The binding site of cluster 3 is at the cleft between subdomains IIIb and Ib, including helices h1 and h2 on the IIIb side and h1 on the Ib side. We note that the NMR data of Algamal et al. (8) indicated a separate Aβ binding site on each of the two subdomains of domain III and, more specifically, the 494–515 segment as part of one such site. Our MD simulations also identify one site in each subdomain; the 494–515 segment overlaps with our cluster 1 site and borders our cluster 2 site. The mass spectrometry data of Choi et al. (11) revealed a binding site at the cleft between domains I and III; this is similar to our cluster 3 site.
Aβ42-HSA displays a largely similar clustering pattern. Among 11 clusters, the first three account for 45% of total snapshots. Two of these three major clusters define binding sites nearly identical to those in Aβ40-HSA, one around the interface between subdomains IIIa and IIIb (cluster 2 in Aβ40-HSA), and the other within IIIb (cluster1 in Aβ40-HSA). A new binding site emerges on the backside of subdomain Ia, centered around helix h5.
The presence of fatty acids changes the cluster distribution. In Aβ40-FA·HSA, Aβ40 positions all over the three HSA domains are partitioned into 15 clusters. Only two of these have a cluster size greater than 10% of total snapshots; the rest are all small clusters. Of the two larger clusters, one has the binding site within subdomain Ia, lined by the h2-h3 and h4-h5 loops and the linker to subdomain Ib; the other has the binding site within subdomain IIb, lined by the linker from IIa to helices h2 and h4.
Apparently, the presence of fatty acids breaks the strong Aβ40 binding sites in and around domain III; instead, multiple weak binding sites appear throughout the three domains. To begin to understand why Aβ and fatty acid are competitive in binding to HSA, we next examine how Aβ40 is accommodated in the three major binding sites in apo HSA, with an eye toward potential conflict with fatty-acid binding.
Intrinsic flexibility allows deformation of apo HSA to create binding sites for Aβ
The three major Aβ40 binding sites on apo HSA are already illustrated in Fig. 5 B. Now we contrast the conformational space sampled by Aβ40-bound HSA against that sampled by isolated FA·HSA. We also make a corresponding comparison between isolated apo HSA and isolated FA·HSA to dissect the contrast between Aβ40-bound HSA and FA·HSA into a contribution due to the release of fatty acids and a contribution further induced by Aβ40 binding.
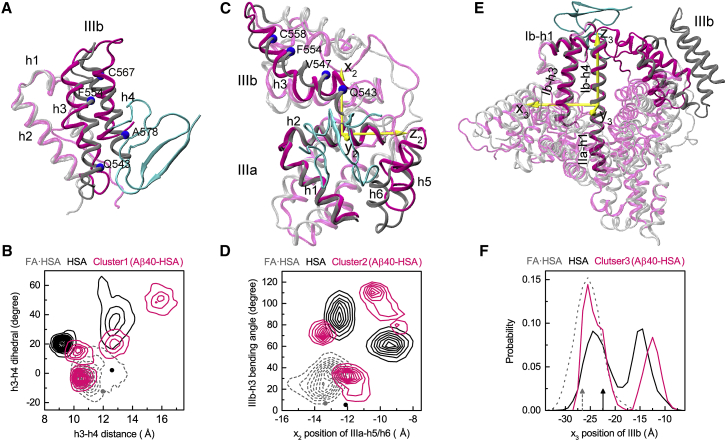
The first binding site is within subdomain IIIb where Aβ40 mainly interacts with the lower portions of helices h3 and h4. Compared to the crystal structure of FA·HSA, the h3-h4 separation and relative tilt angle in Aβ40-bound HSA are significantly different. We measure these two parameters by the distance between h3 and h4 and the dihedral angle formed by the Cα atoms of Q543, F554, C567, and A578 (Fig. 6 A). In the crystal structures, the h3-h4 distances are similar for FA·HSA and apo HSA, at 12.0 and 12.6 Å, respectively. The h4 C-terminus has a slight outward tilt relative to h3 for FA·HSA, whereas the two helices are nearly parallel for apo HSA, with the interhelical dihedral angle defined above at −12.2 and 2.3°, respectively. These values are indicated by a gray dot and dark dot in Fig. 6 B, where the distribution of the two parameters in the snapshots making up binding cluster 1 of the Aβ40-HSA simulations is displayed as magenta contours. The corresponding two-dimensional distributions in the isolated FA·HSA and apo HSA simulations are also presented as gray dashed and dark contours, respectively.
Figure 6.
Conformational changes of apo HSA accompanying Aβ40 binding to the three major sites. (A), (C), and (E) show the superposition of HSA in the representative poses (magenta) with the FA·HSA crystal structure (gray); Aβ40 is in cyan. (B), (D), and (F) show quantitative measurements of structural differences among Aβ40-bound, apo, and FA·HSA. In (B), the distance and relative tilt angle between the subdomain IIIb h3 and h4 helices are measured. The latter is defined as the dihedral formed by the Cα atoms of Q543, F554, C567, and A578. Dark and gray dots indicate values in the crystal structures. In (D), the movement of IIIa-h5/h6 relative to IIIa-h2 and the bending angle of IIIb-h3 are measured. The former is according to the position of IIIa-h5/h6 along the x2 axis, which is perpendicular to the helical axis of IIIa-h2 and points toward IIIb. The latter is defined as the angle between two vectors, one connecting Q543–V547 and the other connecting F554–C558. Dark and gray dots indicate values in the crystal structures. In (F), the movement of IIIb relative to the long helix connecting Ib-h4 and IIa-h1 is measured, as measured by the IIIb center of mass along the x3 axis. The latter is perpendicular to the helical axis of the long helix and points toward IIa-h3. Black and gray rrowsa indicate values in the crystal structures. To see this figure in color, go online.
During the MD simulations, the two h3-h4 parameters of FA·HSA stay close to their crystal values and exhibit small variations. In comparison, in the apo HSA simulations, the h4 C-terminus tilts inward relative to h3 (with the interhelical dihedral angle ranging approximately from 15 to 65°); the h3-h4 distance also diverges from the crystal value, with one population sampling around 9.5 Å and another population sampling around 13 Å. The significant conformational variations involved are consistent with the high RMSFs of h3 and h4 (Fig. 3 A). The cluster-1 snapshots of Aβ40-HSA fall into four subsets, one (40% of snapshots) overlapping with the FA·HSA set, two (20% each) near the apo subsets, and a new subset (20%) with a much more inwardly tilted h4 and much wider h3-h4 separation. The picture that emerges is then that HSA becomes much more flexible upon releasing fatty acids; taking advantage of the increased conformational flexibility, HSA and Aβ40 adapt to each other and form preferential interactions at the site within subdomain IIIb in this case and, as detailed next, at the other two major sites as well.
The second binding site is around the IIIa-IIIb interface where Aβ40 interacts with helices h1, h2, h6 of IIIa, and h3 of IIIb. In the Aβ40-HSA simulations, IIIa-h1/h2 are rigid (Fig. 3 A) and align well with the counterparts in the crystal structure of FA·HSA, but h6, along with h5, moves toward IIIb, and IIIb-h3 becomes bent (Fig. 6 C). To measure the h5 and h6 movement, we introduce a local coordinate system based on the principal axes of IIIa-h2, with the z2 axis along the helical axis and the x2 axis toward IIIb. We also define the bending angle of IIIb-h3 as the angle between two vectors, one connecting Q543–V547 and the other connecting P554–C558. The values of these two parameters in the crystal structures of FA·HSA and apo HSA are close to each other, at (−13.3 Å, 6.9°) and (−12.1 Å, 5.5°), respectively (Fig. 6 D, gray and dark dots). In the simulations, FA·HSA stays close to the crystal structure according to these parameters (Fig. 6 D, gray dashed contours). In contrast, apo HSA deviates from the crystal structure with either significant IIIb-h3 bending or both IIIb-h3 bending and IIIa-h5/h6 movement toward IIIb. The wide span in IIIb-h3 bending angle contributes to the high RMSFs of this helix (Fig. 3 A); although the large spread in x2 of IIIa-h5/h6 at first sight seems incompatible with the low RMSFs of these helices, we note that both the upstream h4-h5 loop and the downstream IIIa-IIIb linker are highly flexible. Aβ40-bound HSA shows even greater variability, presumably to attain robust fit between the molecules.
The third binding site is at the cleft between domains I and III where Aβ40 mainly interacts with Ib-h1 and IIIb-h1/h2. Relative to the crystal structure of FA·HSA, interdomain conformational changes accompany Aβ40 binding. In particular, using the rigid helix connecting Ib and IIa (Ib-h4 and IIa-h1) as reference, IIIb moves closer toward Ib in the direction of Ib-h3. We measure this movement by the x3 coordinate in a local coordinate system defined based on the principal axes of the just-mentioned long helix (Fig. 6 E). In the crystal structures, IIIb is closer to Ib for apo than for FA·HSA (Fig. 6 F, dark and gray arrows). In the simulations, x3 of FA·HSA stays near its crystal value (Fig. 6 F, gray dashed curve). For apo HSA, one population preserves the moderate shift in x3 as seen in the crystal structures, but a second population has IIIb moving much closer toward Ib (Fig. 6 F, dark curve). Underlying this movement is the intrinsic flexibility of domain IIIb (Fig. 3 A). Aβ40-bound HSA behaves similarly to apo HSA, except that in the second population, the movement of IIIb toward Ib is even greater. The movement reshapes the interdomain cleft to accommodate the Aβ40 molecule.
In short, Aβ40 binding is accompanied by HSA deformation within IIIb, around the IIIa-IIIb interface, and at the cleft between domains III and I. The deformation is directly associated with the intrinsic flexibility of apo HSA. The presence of fatty acids quenches the conformational flexibility and therefore prevents conformational changes needed for optimal Aβ binding.
Discussion
We have carried out comparative MD simulations of Aβ peptides binding to apo and fatty acid-loaded HSA so as to provide atomic insights into the interference of fatty acids with Aβ-HSA binding. Our simulations identify the C-terminal region of Aβ as the primary interaction site for apo HSA, consistent with previous NMR data (29). Distinct patterns of Aβ binding propensities on apo and FA·HSA demonstrate the effects of fatty acids on Aβ-HSA binding. The binding propensities qualitatively correlate with backbone flexibilities. Whereas FA·HSA has moderate flexibility and Aβ binding propensities that are even among the three domains, apo HSA concentrates the highest flexibility and binding propensities in domain III. It harbors three major Aβ binding sites within subdomain IIIb, around the interface between subdomains IIIa and IIIb, and at the cleft between domains III and I. Our first site overlaps with the 494–515 segment identified by Algamal et al. (8), and our third site is similar to the one found by Choi et al. (11).
Our MD simulations have further suggested conformational flexibility as the basis for the competition between Aβ and fatty acids in binding to HSA. The high flexibility of apo HSA allows it to readily deform to accommodate Aβ in forming optimal binding sites. The presence of fatty acids quenches the conformational flexibility of HSA and thereby interferes with preferential Aβ-HSA interactions. Note that this competitive mechanism operates with fatty acids and Aβ occupying different sites, the former buried inside HSA, whereas the latter is on the surface, in line with a previous observation that these two types of substances have distinct determinants for HSA interactions (8). In addition to fatty acids, cholesterol and drugs such as tolbutamide and warfarin have been reported to compete with Aβ in binding tο ΗSA binding (3, 12). It was speculated that these molecules might bind to the same sites as Aβ (3) or the hydrophobic tail of Aβ might partially insert into the buried pockets (12). The flexibility-based mechanism proposed here stands as an alternative.
The affinity of HSA for monomeric Aβ is relatively weak (dissociation constant in the submillimolar range (29)), and thus, Aβ-HSA binding can be qualified as nonspecific. In our MD simulations, nonspecificity is manifested by the multiple Aβ binding sites on the HSA surface and the multiple ways that Aβ interacts with HSA within each major binding site. It is possible that each HSA simultaneously binds multiple Aβ monomers. In this way, HSA would effectively take away monomeric Aβ from the pool competent for fibrillization. It is also possible that multiple sites on HSA are involved in binding several subunits of Aβ oligomers, protofibrils, or fibrils, and, conversely, larger Aβ assemblies simultaneously accommodate multiple HSA molecules. This scenario would explain the apparent micromolar affinity of HSA for Aβ oligomers or protofibrils, for which NMR data have indeed implicated binding sites on all the three domains of HSA (6). How HSA inhibits Aβ fibrillization is still a matter of debate (6, 7, 8, 10, 12). Our results are compatible with HSA binding Aβ monomers, oligomers, and larger assemblies and reinforces the notion that HSA inhibition occurs at multiple steps during the fibrillization process.
The MD simulations carried out here have addressed some pressing issues on Aβ-HSA interactions and their interference by fatty acids but leave much else to be desired. The lengths of the trajectories are still much too short, and the numbers of replicate simulations are still much too few. Hence, we were not able to observe possible Aβ conformational transitions upon binding HSA or the dissociation of Aβ from even the relatively weak binding sites on HSA (Fig. S4, C and D), let alone collect statistics on such unbinding events. It would also be interesting to carry out comparative simulations of Aβ oligomer binding to apo and FA·HSA, such as using discrete MD (47). This work has provided a foundation for such future studies.
The flexibility-based competition mechanism proposed here has many implications. First, this mechanism at its essence has much in common with a form of allosteric regulation that is mediated by stimulating or quenching of conformational flexibility prevalent at different timescales (48). So, the interference of fatty acids on Aβ-HSA binding can be viewed as a novel example of allosteric action where small molecules through quenching protein flexibility suppress the binding of partners at multiple surface sites. Second, flexibility has been thought as the key for intrinsically disordered proteins to form promiscuous interactions with partners. We now see that globular proteins like HSA can just as well use flexibility in achieving optimal binding with Aβ.
Third, we hypothesize that flexibility-based competition helps regulate the organization and traffic of proteins in cellular environments. In cellular environments, nonspecific interactions of proteins with their neighbors are ubiquitous and can facilitate subcellular localization and biological function (49). In the same way that fatty acids interfere with HSA-Aβ binding in in vitro studies (8, 12), hydrophobic molecules transported by HSA may coincidentally suppress the latter’s nonspecific interactions with other plasma proteins. The same mechanism may also explain competitive binding in other cellular contexts, including the possible competition between maltose and peptidoglycan for the maltose binding protein (50).
Author Contributions
C.G. designed research, performed research, analyzed data, and wrote the manuscript. H.-X.Z. designed research and wrote the manuscript.
Acknowledgments
This work was supported by National Institutes of Health grants GM118091 (to H.-X.Z.) and AG045703 (to Anant Paravastu).
Editor: Margaret Cheung.
Footnotes
Four figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)34455-2.
Contributor Information
Cong Guo, Email: congguo@shu.edu.cn.
Huan-Xiang Zhou, Email: hzhou43@uic.edu.
Supporting Material
References
- 1.Nasica-Labouze J., Nguyen P.H., Derreumaux P. Amyloid β protein and Alzheimer’s disease: when computer simulations complement experimental studies. Chem. Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo Y.M., Emmerling M.R., Roher A.E. High levels of circulating Abeta42 are sequestered by plasma proteins in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 1999;257:787–791. doi: 10.1006/bbrc.1999.0552. [DOI] [PubMed] [Google Scholar]
- 3.Bohrmann B., Tjernberg L., Nordstedt C. Endogenous proteins controlling amyloid beta-peptide polymerization. Possible implications for beta-amyloid formation in the central nervous system and in peripheral tissues. J. Biol. Chem. 1999;274:15990–15995. doi: 10.1074/jbc.274.23.15990. [DOI] [PubMed] [Google Scholar]
- 4.Biere A.L., Ostaszewski B., Selkoe D.J. Amyloid βετα-peptide is transported on lipoproteins and albumin in human plasma. J. Biol. Chem. 1996;271:32916–32922. doi: 10.1074/jbc.271.51.32916. [DOI] [PubMed] [Google Scholar]
- 5.Kuo Y.M., Kokjohn T.A., Roher A.E. Amyloid-βετα peptides interact with plasma proteins and erythrocytes: implications for their quantitation in plasma. Biochem. Biophys. Res. Commun. 2000;268:750–756. doi: 10.1006/bbrc.2000.2222. [DOI] [PubMed] [Google Scholar]
- 6.Milojevic J., Melacini G. Stoichiometry and affinity of the human serum albumin-Alzheimer’s Aβ peptide interactions. Biophys. J. 2011;100:183–192. doi: 10.1016/j.bpj.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanyon H.F., Viles J.H. Human serum albumin can regulate amyloid-β peptide fiber growth in the brain interstitium: implications for Alzheimer disease. J. Biol. Chem. 2012;287:28163–28168. doi: 10.1074/jbc.C112.360800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Algamal M., Milojevic J., Melacini G. Mapping the interactions between the Alzheimer’s Aβ-peptide and human serum albumin beyond domain resolution. Biophys. J. 2013;105:1700–1709. doi: 10.1016/j.bpj.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milojevic J., Costa M., Melacini G. In vitro amyloid-β binding and inhibition of amyloid-β self-association by therapeutic albumin. J. Alzheimers Dis. 2014;38:753–765. doi: 10.3233/JAD-131169. [DOI] [PubMed] [Google Scholar]
- 10.Wang C., Cheng F., Jia L. HSA targets multiple Aβ42 species and inhibits the seeding-mediated aggregation and cytotoxicity of Aβ42 aggregates. RSC Adv. 2016;6:71165–71175. [Google Scholar]
- 11.Choi T.S., Lee H.J., Kim H.I. Molecular insights into human serum albumin as a receptor of Amyloid-β in the extracellular region. J. Am. Chem. Soc. 2017;139:15437–15445. doi: 10.1021/jacs.7b08584. [DOI] [PubMed] [Google Scholar]
- 12.Bode D.C., Stanyon H.F., Viles J.H. Serum albumin’s protective inhibition of Amyloid-β fiber formation is suppressed by cholesterol, fatty acids and warfarin. J. Mol. Biol. 2018;430:919–934. doi: 10.1016/j.jmb.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Ezra A., Rabinovich-Nikitin I., Solomon B. Multifunctional effect of human serum albumin reduces Alzheimer’s disease related pathologies in the 3xTg mouse model. J. Alzheimers Dis. 2016;50:175–188. doi: 10.3233/JAD-150694. [DOI] [PubMed] [Google Scholar]
- 14.Boada M., Anaya F., Páez A. Efficacy and safety of plasma exchange with 5% albumin to modify cerebrospinal fluid and plasma Amyloid-β concentrations and cognition outcomes in Alzheimer’s disease patients: a multicenter, randomized, controlled clinical trial. J. Alzheimers Dis. 2017;56:129–143. doi: 10.3233/JAD-160565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmijn S., Launer L.J., Breteler M.M. Dietary fat intake and the risk of incident dementia in the Rotterdam Study. Ann. Neurol. 1997;42:776–782. doi: 10.1002/ana.410420514. [DOI] [PubMed] [Google Scholar]
- 16.Morris M.C., Evans D.A., Wilson R.S. Dietary fats and the risk of incident Alzheimer disease. Arch. Neurol. 2003;60:194–200. doi: 10.1001/archneur.60.2.194. [DOI] [PubMed] [Google Scholar]
- 17.Laitinen M.H., Ngandu T., Kivipelto M. Fat intake at midlife and risk of dementia and Alzheimer’s disease: a population-based study. Dement. Geriatr. Cogn. Disord. 2006;22:99–107. doi: 10.1159/000093478. [DOI] [PubMed] [Google Scholar]
- 18.Selkoe D.J. Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 19.Haass C., Selkoe D.J. Cellular processing of βετα-amyloid precursor protein and the genesis of amyloid β-peptide. Cell. 1993;75:1039–1042. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 20.Lame M.E., Chambers E.E., Blatnik M. Quantitation of amyloid beta peptides Aβ(1-38), Aβ(1-40), and Aβ(1-42) in human cerebrospinal fluid by ultra-performance liquid chromatography-tandem mass spectrometry. Anal. Biochem. 2011;419:133–139. doi: 10.1016/j.ab.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Doolittle D.F. W. B. Saunders Company; Philadelphia: 1987. The Molecular Basis of Blood Diseases. [Google Scholar]
- 22.Stevens R.W., Elmendorf D., Gaafar H.A. Application of fluoroimmunoassay to cerebrospinal fluid immunoglobulin G and albumin. J. Clin. Microbiol. 1979;10:346–350. doi: 10.1128/jcm.10.3.346-350.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boada M., Ortiz P., Páez A. Amyloid-targeted therapeutics in Alzheimer’s disease: use of human albumin in plasma exchange as a novel approach for Abeta mobilization. Drug News Perspect. 2009;22:325–339. doi: 10.1358/dnp.2009.22.6.1395256. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto K., Shimada H., Miki T. Serum levels of albumin-amyloid beta complexes are decreased in Alzheimer’s disease. Geriatr. Gerontol. Int. 2014;14:716–723. doi: 10.1111/ggi.12147. [DOI] [PubMed] [Google Scholar]
- 25.Rózga M., Kłoniecki M., Bal W. The binding constant for amyloid Abeta40 peptide interaction with human serum albumin. Biochem. Biophys. Res. Commun. 2007;364:714–718. doi: 10.1016/j.bbrc.2007.10.080. [DOI] [PubMed] [Google Scholar]
- 26.Costa M., Ortiz A.M., Jorquera J.I. Therapeutic albumin binding to remove amyloid-β. J. Alzheimers Dis. 2012;29:159–170. doi: 10.3233/JAD-2012-111139. [DOI] [PubMed] [Google Scholar]
- 27.Milojevic J., Raditsis A., Melacini G. Human serum albumin inhibits Abeta fibrillization through a “monomer-competitor” mechanism. Biophys. J. 2009;97:2585–2594. doi: 10.1016/j.bpj.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milojevic J., Esposito V., Melacini G. Understanding the molecular basis for the inhibition of the Alzheimer’s Abeta-peptide oligomerization by human serum albumin using saturation transfer difference and off-resonance relaxation NMR spectroscopy. J. Am. Chem. Soc. 2007;129:4282–4290. doi: 10.1021/ja067367+. [DOI] [PubMed] [Google Scholar]
- 29.Algamal M., Ahmed R., Melacini G. Atomic-resolution map of the interactions between an amyloid inhibitor protein and amyloid β (Aβ) peptides in the monomer and protofibril states. J. Biol. Chem. 2017;292:17158–17168. doi: 10.1074/jbc.M117.792853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugio S., Kashima A., Kobayashi K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharya A.A., Grüne T., Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000;303:721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- 32.Vivekanandan S., Brender J.R., Ramamoorthy A. A partially folded structure of amyloid-beta(1-40) in an aqueous environment. Biochem. Biophys. Res. Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomaselli S., Esposito V., Picone D. The alpha-to-beta conformational transition of Alzheimer’s Abeta-(1-42) peptide in aqueous media is reversible: a step by step conformational analysis suggests the location of beta conformation seeding. Chembiochem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 34.Fezoui Y., Teplow D.B. Kinetic studies of amyloid beta-protein fibril assembly. Differential effects of alpha-helix stabilization. J. Biol. Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 35.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 36.Gunsteren W.F.V., Billeter S.R., Tironi I.G. Verlag der Fachvereine Hochschulverlag AG an der ETH Zurich; Zurich, Switzerland: 1996. Biomolecular simulations: The GROMOS96 manual and user guide. [Google Scholar]
- 37.Tsfadia Y., Friedman R., Gutman M. Molecular dynamics simulations of palmitate entry into the hydrophobic pocket of the fatty acid binding protein. FEBS Lett. 2007;581:1243–1247. doi: 10.1016/j.febslet.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 38.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101–014107. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 39.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 40.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 41.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 42.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 43.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 46.Schrödinger, LLC . Version 1.8. Schrödinger, LLC; 2015. The PyMOL Molecular Graphics System. [Google Scholar]
- 47.Pilkington E.H., Xing Y., Ke P.C. Effects of protein corona on IAPP amyloid aggregation, fibril remodelling, and cytotoxicity. Sci. Rep. 2017;7:2455. doi: 10.1038/s41598-017-02597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo J., Zhou H.X. Protein allostery and conformational dynamics. Chem. Rev. 2016;116:6503–6515. doi: 10.1021/acs.chemrev.5b00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin S., Zhou H.X. Protein folding, binding, and droplet formation in cell-like conditions. Curr. Opin. Struct. Biol. 2017;43:28–37. doi: 10.1016/j.sbi.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miklos A.C., Sumpter M., Zhou H.X. Competitive interactions of ligands and macromolecular crowders with maltose binding protein. PLoS One. 2013;8:e74969. doi: 10.1371/journal.pone.0074969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported here, including the MD simulation trajectories, are available upon request.