Abstract
Super-resolution (SR) microscopy has been used to observe structural details beyond the diffraction limit of ∼250 nm in a variety of biological and materials systems. By combining this imaging technique with both computer-vision algorithms and topological methods, we reveal and quantify the nanoscale morphology of the primary cilium, a tiny tubular cellular structure (∼2–6 μm long and 200–300 nm in diameter). The cilium in mammalian cells protrudes out of the plasma membrane and is important in many signaling processes related to cellular differentiation and disease. After tagging individual ciliary transmembrane proteins, specifically Smoothened, with single fluorescent labels in fixed cells, we use three-dimensional (3D) single-molecule SR microscopy to determine their positions with a precision of 10–25 nm. We gain a dense, pointillistic reconstruction of the surfaces of many cilia, revealing large heterogeneity in membrane shape. A Poisson surface reconstruction algorithm generates a fine surface mesh, allowing us to characterize the presence of deformations by quantifying the surface curvature. Upon impairment of intracellular cargo transport machinery by genetic knockout or small-molecule treatment of cells, our quantitative curvature analysis shows significant morphological differences not visible by conventional fluorescence microscopy techniques. Furthermore, using a complementary SR technique, two-color, two-dimensional stimulated emission depletion microscopy, we find that the cytoskeleton in the cilium, the axoneme, also exhibits abnormal morphology in the mutant cells, similar to our 3D results on the Smoothened-measured ciliary surface. Our work combines 3D SR microscopy and computational tools to quantitatively characterize morphological changes of the primary cilium under different treatments and uses stimulated emission depletion to discover correlated changes in the underlying structure. This approach can be useful for studying other biological or nanoscale structures of interest.
Introduction
Revealing the three-dimensional (3D) nanoscale membrane structure of biological cells in both a noninvasive and precise manner remains a challenging problem. Structural components that define the cell outer morphology have been examined using various conventional fluorescence microscopy methods (1, 2), but the resolution is ultimately limited by diffraction (∼250 nm). Super-resolution (SR) fluorescence microscopy circumvents the diffraction limit by either imaging and localizing a sparse set of single molecules (SMs) separated in time (3, 4, 5) or by shrinking the effective excitation point-spread function (PSF) via stimulated emission depletion (STED) microscopy (6, 7, 8). The relatively noninvasive nature of optical microscopy can then be utilized for nanoscale structural analysis.
The first technique, SM SR microscopy, requires the structure of interest to be densely labeled with a fluorophore that has at least two states with distinct emissive (on-off) properties, and this mechanism is crucial to force sparsity in each imaging frame (9). Active control of the labels can be achieved either optically (e.g., illumination with a near-UV light source for photoactivation) (10, 11, 12, 13) or nonoptically (e.g., label reacting with a nearby ligand in a reversible fashion to induce blinking) (14, 15, 16). Each molecule is then fitted with a mathematical function that determines its position with a precision that scales as ∼, where N is the number of photons detected, and the locations are eventually merged together to create a reconstructed image with an enhanced resolution of typically ∼20–40 nm for fluorescent protein labels, with improved precisions for small-molecule emitters providing more photons. However, determining 3D position information is difficult because of the standard PSF being symmetric about the focal plane and only observable over a relatively small axial range (∼500 nm) compared to typical structures of interest (17). To address these issues, recent developments in PSF engineering have made it possible to encode depth information within the PSF shape of the SM emitter, including astigmatism (18), multiplane (19), 4Pi (20), and Fourier domain manipulation (21, 22, 23, 24, 25, 26). In particular, the double-helix (DH) PSF allows for 3D SM localization because individual emitters appear as two spots on the camera, which revolve around one another in different z-positions, and thus the angle between the two lobes encodes z information over a relatively large axial depth of field (∼2 μm) (27). The DH-PSF is generated by moving the camera away from the intermediate image plane by four focal lengths f and then placing an optical phase mask in the collection path equidistant between two lenses of focal length f, referred to as a 4f system (28). This technique has been used to study nanohole arrays across large fields of view (29), protein localization patterns within bacteria (30, 31, 32), and organelles inside mammalian cells (33).
An alternate SR microscopy method, STED microscopy (6, 7, 8), requires two laser beams that work in concert to shrink the effective PSF of the excitation spot in a confocal scanning configuration. The excitation beam is focused to a diffraction-limited spot onto a sample and pumps the molecules into the first electronic excited state. A second overlapping depletion beam at longer wavelengths is shaped optically into a donut with a dark center, selectively forcing emitters that lie at the edge of the excitation spot to produce far-red stimulated emission light that is filtered out. In this way, only the fluorophores at the very center of the donut undergo normal fluorescence emission, which yields an imaging resolution better than the diffraction limit. Images are generated by scanning the coaligned laser beams over the region of interest (34, 35). This technique has been used to examine nitrogen-vacancy centers (36), neurons (37), centriole proteins (35, 38, 39), the ciliary transition zone (40), and many other structures and proteins at a high resolution (41, 42).
In this work, we employed both imaging techniques to study the primary cilium in cells, which is a tubular structure ∼2–6 μm long and ∼200–300 nm in diameter. The overall structure is based on the axoneme (43), a column of ninefold symmetric microtubule doublets running through the center of the cilium. The cilium is covered by the ciliary membrane, an extension of the plasma membrane, which has a distinct protein-lipid composition compared to the rest of the cell (44). This nonmotile, antenna-like sensory organelle is essential for facilitating proper signal transduction, cell-to-cell communication, and proper regulation of cell division in mammalian cells (45, 46). Impaired ciliary function leads to a collection of human diseases termed “ciliopathies” with a broad range of pathologies of various severities including polydactyly (multiple digits), cystic kidney disease, obesity, and retinal degeneration (47). One reason these debilitating diseases arise is due to impaired expression of intraflagellar transport (IFT) complexes, which is a family of proteins that move components required for constructing the cilium up and down along the structure (48). When IFT is functioning incorrectly, cilia are often found completely missing or misshapen when observed using fluorescence or electron microscopy (EM) (49, 50, 51, 52). Although recent studies using SR microscopy have elucidated the distribution of a plethora of proteins residing within the primary cilium (35, 53, 54, 55), there remains a need for quantitative approaches to characterize the ciliary membrane shape.
Here, we describe a method for revealing and quantifying the membrane structure of the primary cilium using a combination of SR microscopy techniques and computer-vision algorithms. In mouse embryonic fibroblast (MEF) cells, we genetically express and label a crucial transmembrane ciliary protein smoothened (SMO) with a bright fluorophore, add a blinking buffer for stochastic optical reconstruction microscopy (STORM) (56), and perform 3D SM SR using the DH-PSF with a precision of 10–25 nm. Our processed 3D molecular positions of SMO are then fed into the Poisson surface reconstruction (PSR) algorithm, which produces 3D triangulated surface meshes representing the shape of each primary cilium membrane. Computing the mean (H) and Gaussian (K) curvature provides a quantitative picture of the surface shape at a high resolution. Upon impairment of the retrograde transport machinery through genetic modification or treatment, the narrowing and bulging near the tip of the cilium is more severe. Surface information allows us to calculate the Willmore energy (WE) (57) as an appropriate metric for measuring its overall characteristic shape and shows a significant increase in our mutant cells. Furthermore, when imaging primary cilia in IFT25 mutant cells using two-color two-dimensional (2D) STED microscopy, we observe fluorescently labeled α-tubulin spanning the entire primary cilium and, in some cases, occupying the bulging ciliary membrane at the tip, features that we did not find in wild-type (WT) MEF cells. By combining SR fluorescence microscopy and quantitative surface meshing, this method of shape analysis may be used to study a broad range of nanoscale tubular structures in a more quantitative and precise way.
Materials and Methods
Cell culture
MEF cells were generated, which stably express SMO proteins with an extracellular SNAP tag (SNAP-SMO) and Pericentrin-YFP (PACT-YFP) as a basal marker. Cell lines used were 1) WT: SMO−/−, SNAP-SMO, PACT-YFP and 2) IFT25: IFT25−/−, SNAP-SMO, PACT-YFP, as described in (58).
Cellular conditions
The following cellular conditions were used: 1) WTSAG, WT cell line treated with 100 nM SMO agonist (SAG) for 4 h; 2) , WT cell line treated with 100 nM SAG for 4 h, then treated with 10 μM ciliobrevin D (cb) for 1 h; 3) IFT25NoAg, IFT25 knockout cell line not treated with any agonist at all; 4) IFT25SAG, IFT25 knockout cell line treated with 100 nM SAG for 4 h.
Sample preparation for 3D SR microscopy
MEF cells are first labeled with Benzylguanine-Alexa647 (S9136S; NEB, Ipswich, MA), then fixed with 4% paraformaldehyde, and finally treated with a quenching solution of 10 mM NH4Cl. The samples are washed 3× with 1× phosphate-buffered saline (PBS) (pH 7.4), then either imaged immediately or stored at 4°C up to 1 week before being discarded.
3D SM microscopy imaging
Experiments are performed on a customized inverted microscope (IX71; Olympus, Tokyo, Japan), where our sample is mounted on a piezoelectric stage (PI-Nano, Irvine, CA) and is in contact with an oil-immersion objective (100×, 1.4 numerical aperture, UPLanSAPO; Olympus). New imaging buffer is added for each primary cilium imaged (1–2 h), which consists of glucose oxidase (G2133; Sigma-Aldrich, St. Louis, MO), catalase from bovine serum (C100; Sigma-Aldrich), 100 mM Tris-HCl (pH 8.0) (15568025; Thermo Fisher Scientific, Waltham, MA), 10% (w/v) glucose solution (49139; Sigma-Aldrich), 140 mM β-mercaptoethanol (M6250; Sigma-Aldrich), and H2O (Nanopure, Waltham, MA) (56). We locate one primary cilium and image SNAP-SMO-Alexa647 and PACT-YFP using the 641 nm (Coherent Cube, 100 mW; Coherent, Santa Clara, CA) and 514 nm (Coherent Sapphire, 100 mW) lasers, respectively. When we place the cilium at the center of the field of view, the DH-phase mask is carefully placed at the Fourier plane in our 4f system (Fig. S1). The DH-phase mask has the effect of optically splitting the standard PSF into two lobes in which the midpoint determines its x, y position, whereas the angular orientation provides z information, which is separately calibrated (see Supporting Materials and Methods). To begin imaging, the intensity of the 641 nm laser (1–5 kW/cm2) is increased, the collection of labels is allowed to bleach down until SM concentrations are achieved, and imaging frames are recorded in the presence of blinking. Over the next hour, irradiation from a secondary 405 nm (Obis, 100 mW) laser is used to maintain photoblinking labels at suitable densities. Red fluorescent fiducial beads are also imaged simultaneously several microns away from the cilium to correct for sample drift. Detected fluorescence is recorded using a silicon electron-multiplying charge coupled device camera (DU-897U-CS0-#BV; Andor Xion, Belfast, UK) at a speed of 20 frames/s (50 ms/frame) with an electron-multiplying gain of 200 and further analyzed using the easyDHPSF program, which is freely available online (59).
Meshing
Using MeshLab (ISTI-CNR, Pisa, Italy), we apply the PSR algorithm to our 3D data to create a triangulated surface as described in the Supporting Materials and Methods. The results are exported in a “∗.ply” file format, which is used to plot our 3D mesh in MATLAB (The MathWorks, Natick, MA) and for our curvature analysis.
Curvature analysis
We then fit a surface to a “patch” of points, which consists of the first and second nearest neighbors to each vertex of the mesh (Fig. S4), to the following form:
where its coefficients are used to calculate the mean curvature (H) and Gaussian Curvature (K) as follows (60):
We can then calculate the Willmore energy (57) by the following expression:
where Hi, Ki, and ai are the mean curvature, Gaussian curvature, and surface area of the ith triangle of the mesh, respectively. The Willmore energy density is defined as WE,D = WE/A, where .
Sample preparation for 2D STED
Cell samples are fixed with 4% paraformaldehyde for 15 min at 25°C, washed with 1× PBS, then immersed in a blocking solution consisting of 0.1% TritonX-100, 1% normal donkey serum (017-000-121; Jackson ImmunoResearch, West Grove, PA), and 1× PBS for 30 min at 25°C. We stain our samples with primary and secondary antibodies, targeting SNAP-SMO with Atto647N and α-tubulin with Star520SXP.
Two-color 2D STED microscopy
STED images were collected on a bespoke two-color fast-scanning STED microscope (Fig. S6). Briefly, laser pulses of 750 nm for the depletion beam and 530 and 635 nm for the excitation beams are scanned along the fast axis using a 7.5 kHz resonant mirror and along the slow axis using a piezo stage. Fluorescence is detected through a 0.7 and 0.8 Airy units pinhole (in the red and green channels, respectively) sequentially on a Si avalanche photodiode between 550 and 615 nm for the green channel and between 660 and 705 nm for the red channel. The images are acquired using a custom LabVIEW algorithm running on an field programmable gate array. See Supporting Materials and Methods for details.
Results and Discussion
3D SR microscopy reveals heterogeneity in membrane shape
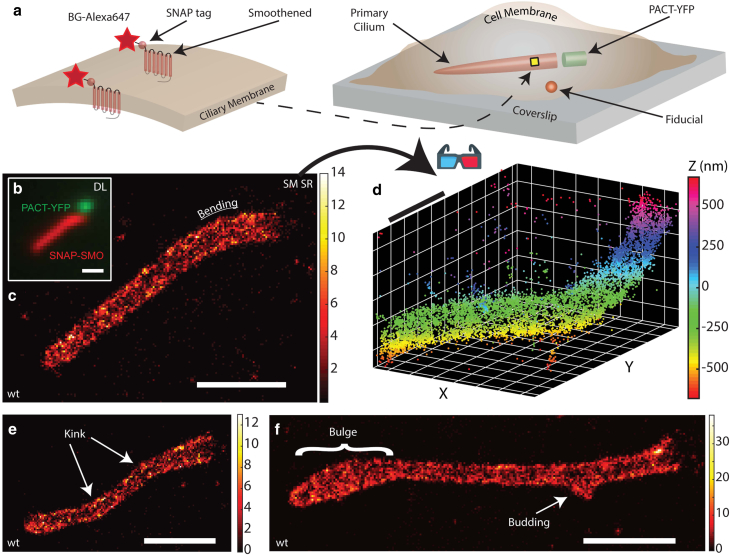
To study the ciliary membrane, we chose the transmembrane protein SMO as the labeling target because these proteins are known to move in a largely diffusive manner and be relatively dense during Hedgehog pathway activation (58, 61, 62). Our baseline MEF cells, denoted as WT, were treated with SAG (63), which activates the Hedgehog signaling pathway and triggers SMO to localize specifically to the primary cilium (Fig. 1 a). SMO is conveniently labeled extracellularly using cells expressing a genetic fusion to the SNAP tag (SNAP), which reacts with a benzylguanine-derivatized fluorophore, here Alexa647. (It would also have been possible to label other ciliary membrane proteins such as the somatostatin receptor 3, but we chose SMO because of the availability of a cell line with permanent incorporation of the SNAP label.) After SNAP-SMO is covalently labeled with the Alexa647 fluorophore and chemically fixed, we first imaged our samples in the presence of a blinking buffer, using a 641 nm laser to detect a diffraction-limited image of SNAP-SMO, then used a 514 nm laser to detect PACT-YFP, which is a fluorescent marker of the primary cilium base (Fig. 1 b). To gain as much information of the ciliary surface, a longer-than-normal time (∼60 min) is spent imaging blinking labels for one sample, in which each detected molecule appears as two spots that both roughly take on the shape of a Gaussian on our camera. We capture molecules within a 2-μm-thick axial range with a high spatial precision and a relatively large xy area (40 × 40 μm), which allows us to simultaneously image SNAP-SMO and a nearby bright fiducial marker used to correct for sample drift (Fig. 1 c). The single emitters produced an average signal of ∼8700 photons and a mean background level of ∼216 photons/pixel, yielding a ∼10–25 nm spatial resolution with several thousands of localizations for each cilium (Fig. S2). Our complete 3D reconstructions, or “point clouds,” reveal localizations densely packed within the ciliary membrane in a near-homogenous fashion (Fig. 1 d). By looking at cilia in 3D on many WT MEF cells, we observe a variety of distinct morphological classes, such as bending, kinking, bulging, and the occasional budding with a diameter of ∼100 nm (Fig. 1 c, e, and f). In addition, we also detect a few SNAP-SMO molecules outside of the primary cilium, likely on the plasma membrane and not included in our analysis. Despite the fact that these cells were grown under identical conditions, the wide range of heterogeneity in shape from one cilium to another is clearly apparent. Furthermore, our results demonstrate that the underlying structure of the primary cilium is not a simple cylinder-hemispheric cap as it is frequently modeled.
Figure 1.
Labeling and 3D SR imaging of the ciliary membrane. (a) SNAP-SMO proteins, where the SNAP protein is on the extracellular side, are covalently labeled with benzylguanine-Alexa647 along the ciliary membrane, which is usually found near the coverslip surface. PACT-YFP indicates the base of the cilium, and a nearby bright fiducial is used to correct for spatial drift. (b) Overlaid diffraction-limited images of the SNAP-SMO and PACT-YFP in chemically fixed control MEF cells that were treated with SAG. 3D SR microscopy using the double-helix point spread function (DH-PSF) was performed to obtain a localization map of SNAP-SMO molecules along one primary cilium, reconstructed as a (c) 2D histogram, where the bin size is 20 nm and the color bar represents the number of localizations per bin, and (d) 3D scatterplot. For SNAP-SMO distributions of other primary cilia, there is evidence of (e) kinking, (f) bulging, and (f) budding within the same control MEF cells. Scale bars, 1 μm. To see this figure in color, go online.
2D surface fitting captures the ciliary membrane surface and enables curvature quantification
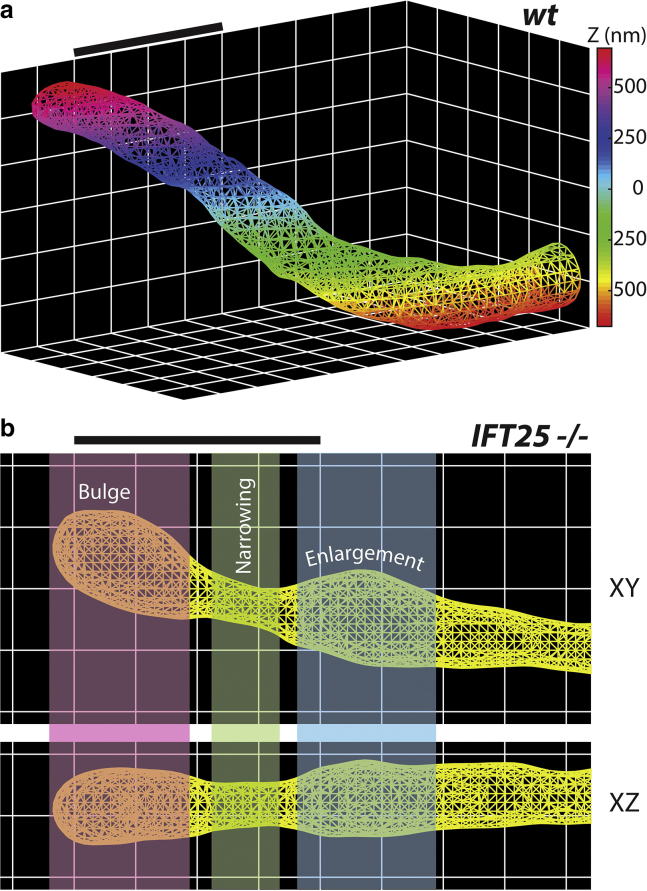
To extract the complex surface of the ciliary membrane, we took advantage of the very dense surface-point sampling (typically ∼1000 points per μm2) and applied the PSR algorithm in the freely available package MeshLab (64). The advantages of this algorithm are that it is robust to noisy, nonuniform point cloud data and outperforms other commonly used surface-fitting methods (65). We prepared our data set by first selecting only the points that reside on the primary cilium, but if the localization density was sparse, the cilium was not analyzed. Because the algorithm works best when the underlying surface is topologically closed, we selected several points near the base of the cilium to determine a plane of best fit. The point cloud was then copied and rotated 180° about the plane axis, producing a rotationally symmetric point cloud made up of two identical shapes. Using the proper fitting parameters, we then generated 2D triangular meshes of each primary cilium, providing us with a clearer picture of the surface (Fig. 2 a). Rather than having to navigate through a point cloud, we are able to observe the contours of the surface with greater clarity from many different perspectives (Videos S1 and S2). Similar to our SR-reconstructed images, we can better identify regions of bulging, narrowing, and enlargement along the ciliary membrane, which are prominent features in IFT25 mutant cells (Fig. 2 b). In addition, the vertices of the mesh allowed us to obtain the ciliary axis, a centroid line that spans from the base to the tip of the primary cilium (Fig. 3). If necessary, surface meshes for WT and mutant cells can also be 3D printed using standard plastic filament material, a true 3D representation of the meshes shown in Fig. 2 (Fig. S3).
Figure 2.
2D surfaces of the ciliary membrane. MeshLab is used to create a triangular mesh using the input point cloud for (a) control MEF primary cilium and (b) IFT25 mutant cell, in which different colored regions indicate—from left to right—bulging at the tip of the cilium, narrowing, and then enlargement. Scale bars, 1 μm. To see this figure in color, go online.
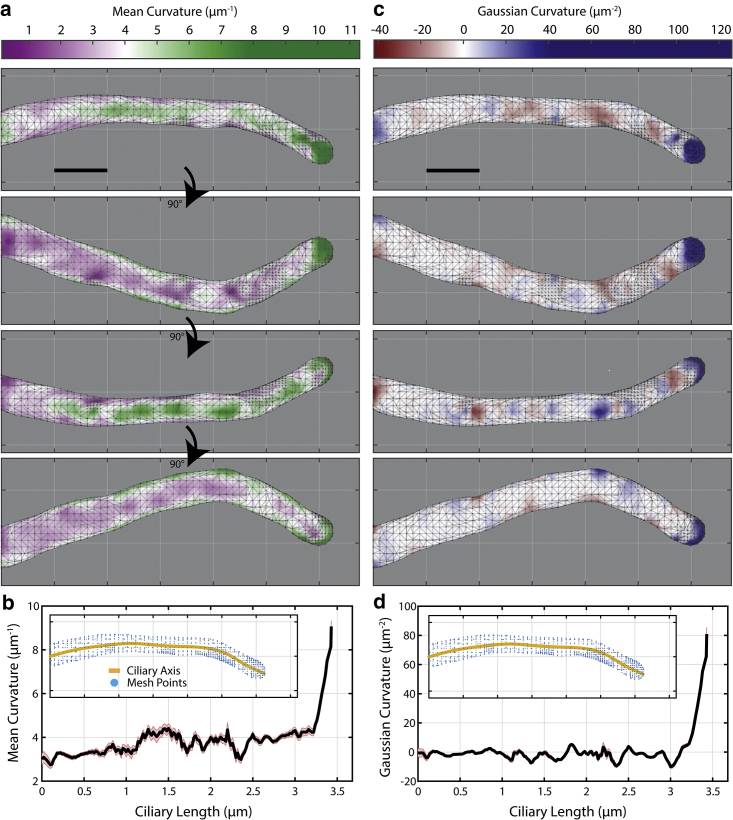
Figure 3.
Calculating mean and Gaussian curvature along mesh. (a) Mean curvature (H) and (c) Gaussian curvature (K) are calculated for each vertex along the mesh for a control MEF cell and displayed here as heat maps. Each panel is a 90° rotation about the horizontal axis. The average (b) H and (d) K is calculated over a 100-nm-wide window that slides along the ciliary axis in 20 nm steps. (Inset figures) Yellow lines indicate the measured ciliary axis; blue points are the vertices of the mesh. Reporting is mean ± SEM. Scale bars, 0.5 μm. To see this figure in color, go online.
Camera flythrough, which starts above the mesh, then follows along the ciliary axis, and finally pans away. This is the same mesh as shown in Fig. 2 a. White bounding box dimensions, 2750 nm x 1500 nm x 1500 nm; video speed, 30 frames/s. The HSV color scaling represents z in the image over a range from −175.8 to 1194.5 nm.
Camera flythrough, which starts above the mesh, then follows along the ciliary axis, and finally pans away. This is the same mesh as shown in Fig. 2 b. White bounding box dimensions, 3700 nm x 900 nm x 600 nm; video speed, 30 frames/s. The HSV color scaling represents z in the image over a range from −196.4 to 227.1 nm.
To quantify the local shape of the primary cilium, we decided to use curvature as an appropriate metric. Curvature (for a 2D curve) is inversely proportional to the radius of an osculating circle at a point along the curve. In our 3D problem, 2D cross sections through a point on the surface oriented at different angles will result in a large range of curvature values. The most important values are the two principal curvatures, denoted as k1 and k2, which are the minimal and maximal curvature values, respectively. These two metrics are more meaningful when they are combined to calculate the mean curvature H, which is expressed as (k1 + k2)/2, and the Gaussian curvature K, which is expressed as k1 × k2 (66). These two metrics can be evaluated along the surface mesh of the primary cilium, which provides us with a better visualization for how it is shaped (Fig. 3). A K surface heat map highlights regions of curvature arising from one of the following shapes: 1) sphere, 2) cylinder, or 3) saddle point. When comparing one cilium to another with these metrics, it is easy to recognize morphological differences and locate where regions of unusual or interesting behavior occur. Further, by measuring the surface area, A, and the length of the cilium, l, we can then compute an approximate diameter along the shaft of the primary cilium (Table 1; see Supporting Materials and Methods). When plotting the values of A and l cell by cell in a 2D scatterplot, overall, they follow a linear relationship, as expected (Fig. S5). None of the diameters are significantly different across all four conditions, so we concentrate in the following on the curvature measures.
Table 1.
Measuring the Length, Surface Area, and Diameter of the Primary Cilium for Different Conditions from 3D SR Imaging
| WTSAG | IFT25NoAg | IFT25SAG | ||
|---|---|---|---|---|
| Number of cells | 17 | 14 | 16 | 12 |
| Ciliary length (μm) | 2.821 ± 0.658 | 2.564 ± 0.779 | 3.170 ± 0.640 | 3.083 ± 0.371 |
| Surface area (μm2) | 2.403 ± 0.631 | 1.960 ± 0.610 | 2.594 ± 0.728 | 2.475 ± 0.349 |
| Diameter (nm) | 272 ± 40 | 244 ± 29 | 258 ± 41 | 257 ± 31 |
Ciliary length is simply the complete length from the tip to the base of the cilium, summing over the areas of all the triangles is the surface area, and the diameter displayed is computed from a model assuming the cilium is a cylinder with a hemispheric cap (Fig. S5). Reporting mean ± SD.
To further quantify the cilium’s overall morphology and how much the surface deviates from a sphere, we calculated its Willmore energy (WE), which is expressed by the following:
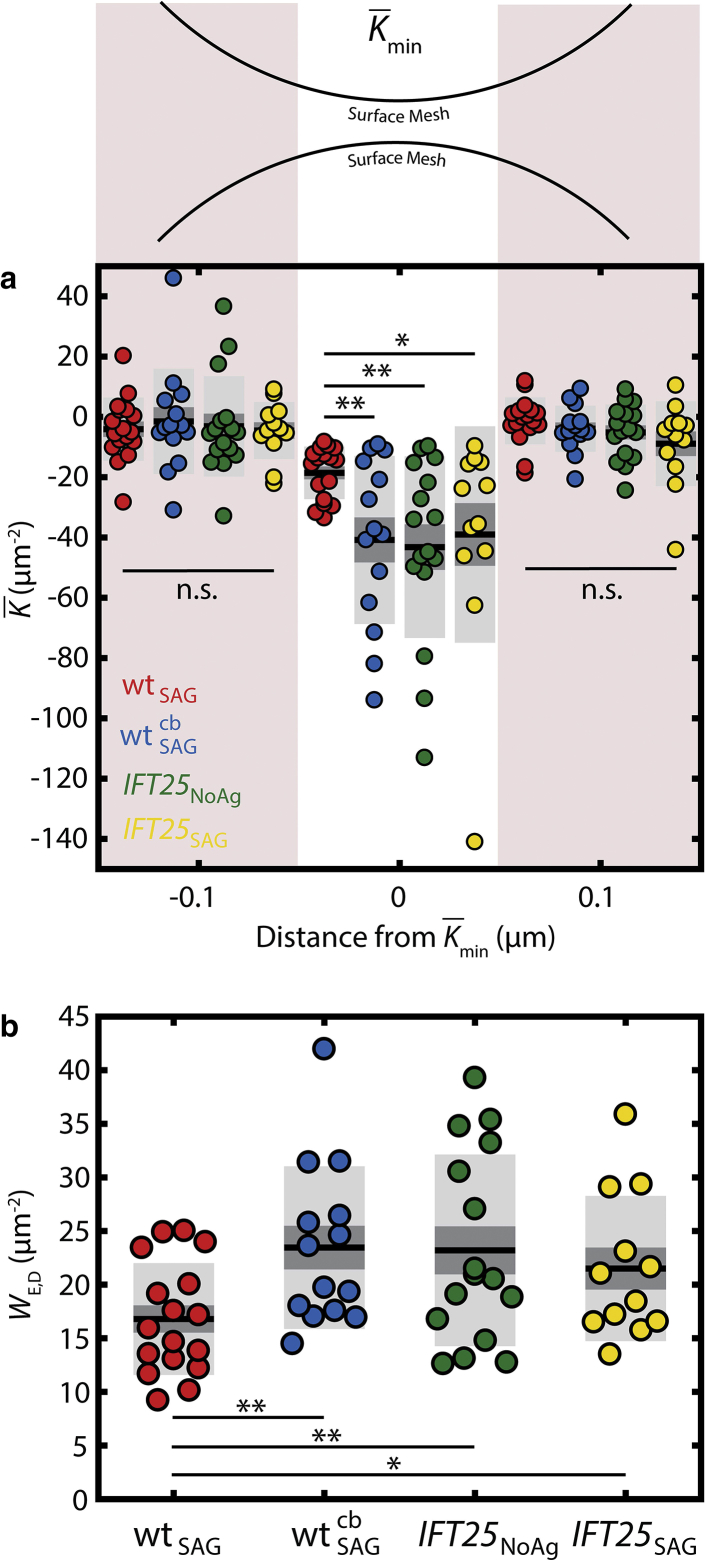
where Hi, Ki, and ai are the mean curvature, Gaussian curvature, and surface area of the ith triangle of the mesh, respectively. The intuition behind WE is that it is a strictly positive quantity with a minimal value of 0 when the object is a sphere. For a surface that is characterized to be morphologically more complex, which includes being stretched in a certain direction or severely indented, WE will take on a larger value. However, a normal cilium that is unusually long with a large surface area can have a value of WE comparable to one that is relatively short but very narrow and bulbous. To offset the variability in surface area and to ensure a fairer comparison, we computed the total surface area of one mesh, , and define here the Willmore energy density WE,D = WE/A. By calculating this value for all of our primary cilia, we found it to be a useful metric for comparing overall shape phenotype.
Altered morphology is detected in cells with impaired retrograde transport
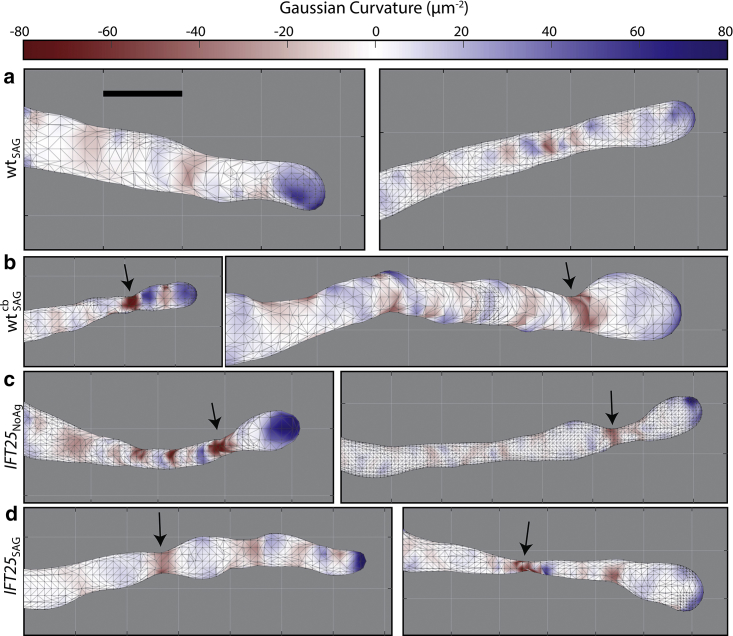
We characterized the surfaces of our meshes by calculating their respective H and K profiles, in which large negative values of K correspond to regions of severe narrowing of the mesh. WT MEF cells treated with SAG, WTSAG, exhibited a cylindrical structure with a hemispheric cap at the tip (Fig. 4 a). To see if we could observe a different phenotype in our WT MEF cells when transport is modified, we treated them with cb and performed 3D SM microscopy. This small-molecule inhibitor of the motor cytoplasmic dynein perturbs retrograde protein trafficking within the primary cilium (67). It has been previously shown that prolonged incubation of cells with cb leads to complete cilia loss, but within a 4-h incubation period, cilia are still present, although with disrupted retrograde transport (67). As expected (Fig. 4 b), in cells treated with both SAG and cb, , increased bulging and narrowing were observed.
Figure 4.
Comparing Gaussian Curvature (K) heat maps for different representative primary cilia across all conditions. (a) WTSAG cells exhibit a cylindrical structure with a hemispheric cap. (b) cells are shown to have a severe narrowing region near the tip of the cilium, denoted by black arrows that highlight the red, negative K regions. (c) IFT25NoAg cells also show narrowing near the tip, whereas interestingly, (d) IFT25SAG cells had narrowing closer to the base of the primary cilium in a few cells. Heat maps are all on the same scale. Scale bars, 0.5 μm. To see this figure in color, go online.
We examined cells in which IFT25 was genetically knocked out, which allows SMO to densely accumulate to the primary cilium despite the absence of pathway activation. 3D SM microscopy on many different cilia in untreated IFT25 cells, IFT25NoAg, shows an altered morphology (Fig. 4 c), similar to the morphological features found in our cells. In particular, there was a larger proportion of cells that exhibited a bulbous tip, which was apparent upon generating the surface meshes. As a related condition, IFT25 cells treated with SAG, IFT25SAG, should have an added accumulation of SMO, and these cells showed a similar phenotype to the IFT25 cells without SAG (Fig. 4 d). This suggests that increased bulging effects in the primary cilium are largely due to the absence of transport complexes and the accumulation of non-SMO proteins rather than driven strictly by SMO protein accumulation.
To quantitatively distinguish distinct categories of morphology, we calculated the curvatures and Willmore energies adjacent to a constriction. The most significant constriction occurs at the position of minimal Gaussian curvature K (Kmin). By computing the average K, , at the constriction and on both sides of the position of the Kmin value, the average curvature is significantly smaller for each of the three mutant cases compared to the WT condition, indicating a more severe narrowing near this region (Fig. 5 a). Furthermore, when calculating WE,D for every cilium under the different mutant conditions, the value was on average higher among IFT25 cells compared to the WT case and was also higher when WT cells are treated with cb (Fig. 5 b).
Figure 5.
Quantitatively detecting changes in primary cilia morphology in MEF cells. When calculating the value and the average values in the two neighboring 100 nm-wide regions, (a) there is a significant decrease in found in mutant cells compared to the control WT MEF cells. (b) WE,D measures the overall shape of the object and is found to be significantly higher in mutant cells. Reporting is mean ± SEM (dark gray) and mean ± SD (light gray). (∗∗p < 0.01, ∗p < 0.05). To see this figure in color, go online.
Our 3D SR methods for quantifying the morphology of the primary cilium clearly reveal measurable morphological changes when the retrograde transport machinery is severely impaired in MEF cells, either by genetic knockout of IFT25 or inhibition of dynein-2 function. Our observations are similar to previous EM-based studies revealing a bulging tip for primary cilia in cells carrying mutations for ciliary proteins that are necessary for proper retrograde transport, including Dnchc2 and THM1 (50, 68). Changes in ciliary membrane morphology can be subtle under certain physiological conditions, but by combining 3D SM microscopy and existing quantitative methods from differential geometry, we were able to reveal and characterize features that would have been impossible to observe using conventional microscopy techniques. Although we show IFT25 to have at the very least an indirect effect on the membrane shape, the direct mechanism of these structural defects in the ciliary membrane is a subject of future study.
Characterizing the membrane and axoneme simultaneously using two-color 2D STED microscopy
Based on previous studies (69), we hypothesized the axoneme, a ninefold symmetric microtubule doublet structure providing the central core of the primary cilium, may also be altered in our IFT25 mutant cells. We observed both SMO and the α-tubulin component of the axonemal microtubules using primary and secondary antibodies staining in fixed MEF cells. To achieve a resolution beyond the diffraction limit, we utilized STED microscopy, which provided both a secondary verification of the morphological changes detailed above and also allowed us to correlate the relative distributions of SMO and the inner cytoskeleton of the same primary cilium. Although our axial z resolution remains diffraction limited, the projection of the additional fluorophores in z onto the xy plane provides enhanced brightness along the sides of the cylindrically shaped primary cilium, whereas the top and bottom surfaces appear dimmer. With an xy full-width half-maximal resolution of 50–100 nm, we were able to better resolve the spatial distribution of SMO along the ciliary membrane and obtained an enhanced resolution of the axoneme in our STED images compared to the corresponding confocal images (Fig. S7). In addition, we were able to measure the diameters of the membrane and the axoneme (Fig. S9), which are consistent with previous EM studies of the primary cilium (70). We find that, as expected, the ciliary membrane diameter is larger than the axonemal diameter by ∼25%, but these diameters are not significantly different when comparing WT to IFT25 cells.
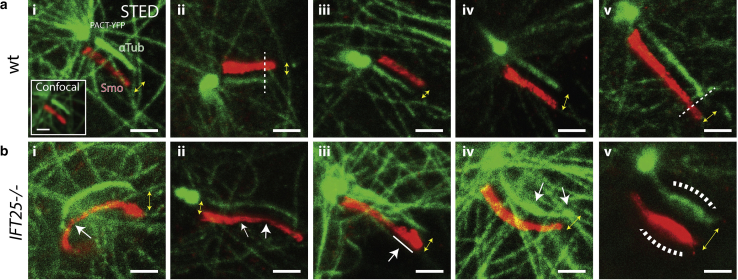
For the WT MEF cells, the ciliary membrane and the axoneme were both found to be cylindrical in shape, and the axoneme often stopped a few hundred nanometers before reaching the cilium tip (∼60%) (Fig. 6 a). Although a very small proportion of WT cilia had a bulged membrane near the tip of the cilium (∼20%), the axoneme still did not extend all the way to the tip (Document S1. Supporting Materials and Methods and Figs. S1–S9, Video S1. Control WT MEF Primary Cilium 3D Mesh, Video S2. IFT25 Mutant MEF Primary Cilium 3D Mesh, Document S2. Article plus Supporting Material). Despite this, the tip still maintained a semi-hemispheric structure in 2D, which is consistent with our 3D SR results. In contrast, most of the IFT25 mutant cells imaged exhibited clear bulging near the tip of the ciliary membrane, consistent with our 3D SR results. Furthermore, a larger proportion of these cells had antibody-labeled α-tubulin proteins spanning the entire primary cilium length and occupying the bulging ciliary tip (∼90%) (Fig. 6 b). Although the images of the axoneme in some IFT25 mutant cells were found to lose the normal cylindrical structure near the tip resembling a large bulge (∼20%), it is possible that this region is occupied by monomeric α-tubulin proteins (Fig. S8 b). An unlikely but alternative explanation is the depletion of IFT25 could also result in lower molecular crowding at the tip, which makes α-tubulin more accessible to antibodies. Based on these observations, one may speculate that when IFT25 is absent, a larger number of ciliary proteins accumulate at the tip, leading to a bulge in the ciliary membrane. Therefore, IFT25 clearly plays some role in ensuring that the primary cilium maintains a normal shape, even though previous studies show this protein is not required for ciliogenesis.
Figure 6.
2D STED microscopy reveals structural changes in the axoneme for IFT25 knockout cells. SNAP-SMO (SMO) and α-tubulin (αTub) are stained with Atto647N and Star520SXP-labeled antibodies, respectively, in chemically fixed MEF cells and are subsequently imaged using a two-color confocal and two-color STED microscope. Yellow double arrows indicate both magnitude shift and direction between the two red and green images. (a) In control WT MEF cells, (i–v) the ciliary membrane exhibits a cylindrical shape, whereas the axoneme is frequently observed to not extend to the tip of the cilia. (ii) The dotted line indicates where the axoneme ends. (b) In IFT25 cells, results similar to the 3D SR data are observed, where (i) kinks and (ii) narrowing of both the ciliary membrane and axoneme are pointed out in white arrows. (iii) Bulging and budding often occurs at the tip, and the axoneme appears to extend all the way to the tip and (iv) sometimes has an uneven diameter throughout the cilium. (v) Severe swelling of the ciliary membrane and the axoneme is also observed, although this is relatively rare. Scale bars, 1 μm. Image contrast within each cell line condition is identical for both channels. To see this figure in color, go online.
Conclusions
We have demonstrated a quantitative approach for interrogating the primary cilium morphology in mammalian cells using SR fluorescence microscopy of a transmembrane protein of the ciliary surface. By combining 3D SR microscopy and a meshing algorithm, we obtain high-resolution images of the ciliary membrane surface, revealing a variety of nanoscale features. This high-resolution approach has revealed the existence of bulging and narrowing structural defects that were not seen in a previous report of diffraction-limited images of IFT25 mutants in MEF cells (70) yet are roughly consistent with swollen tips reported for IFT25 mutant mice sperm cells (69). Moreover, our analysis indicates significant changes in cilia morphology, manifested in our measurements of H, K, and WE,D. We are also able to demonstrate that WT cells treated with cb exhibit similar phenotypes to the IFT25 mutant cells. Therefore, impaired retrograde transport, either as a result of inhibiting retrograde motor dynein or due to a genetic deletion of IFT25, affects the overall shape of the ciliary membrane, primarily in form of bulging, which is indicative of an underlying mechanism related to the mobility of cargo in the cilium. We also resolved the axoneme structure within mammalian cells by implementing STED microscopy. Notably, the antibody-labeled α-tubulin proteins in the IFT25 mutant cells are observed to span the entire primary cilium. This suggests that IFT25 plays a role in properly maintaining the structure of the primary cilium, especially near the tip, even though it is not required for ciliogenesis. Our method for characterizing the biophysical and morphological properties of the ciliary membrane can be used to study other nanoscale structures, such as bacteria or the nuclear envelope, in a similarly quantitative manner, if the surfaces can be labeled with suitable dyes for SR microscopy.
Author Contributions
J.Y., L.E.W., and W.E.M. conceived the study. L.M. created the cell lines. J.Y. prepared biological samples for imaging, performed 3D SM microscopy experiments, and analyzed all data. C.J.C. performed two-color 2D STED experiments. J.Y., C.J.C., L.E.W., L.M., T.S., and W.E.M. wrote the manuscript.
Acknowledgments
We thank Dr. Gregory Pazour for providing us with the IFT25 mutant cell line, Dr. Rafe Mazzeo for helpful discussions, and Dr. Pete Dahlberg for the 3D printed models.
This work was supported in part by a National Science Foundation Graduate Fellowship (J.Y. and C.J.C.), a Stanford Bio-X Fellowship (L.E.W.), and the National Institute of General Medical Sciences through Grants No. R35-GM118067 (W.E.M.) and No. R01-GM121424 (T.S.).
Editor: Joseph Falke.
Footnotes
Supporting Materials and Methods, nine figures and two videos are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)34459-X.
Supporting Citations
Reference (59) appears in the Supporting Material.
Supporting Material
References
- 1.Stephens D.J., Allan V.J. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 2.So P.T., Dong C.Y., Berland K.M. Two-photon excitation fluorescence microscopy. Annu. Rev. Biomed. Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 3.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 4.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hell S.W., Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 7.Klar T.A., Jakobs S., Hell S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hell S.W. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 9.Sahl S.J., Moerner W.E. Super-resolution fluorescence imaging with single molecules. Curr. Opin. Struct. Biol. 2013;23:778–787. doi: 10.1016/j.sbi.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson R.M., Cubitt A.B., Moerner W.E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 11.Biteen J.S., Thompson M.A., Moerner W.E. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat. Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dertinger T., Colyer R., Enderlein J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proc. Natl. Acad. Sci. USA. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uno S.N., Kamiya M., Urano Y. A spontaneously blinking fluorophore based on intramolecular spirocyclization for live-cell super-resolution imaging. Nat. Chem. 2014;6:681–689. doi: 10.1038/nchem.2002. [DOI] [PubMed] [Google Scholar]
- 14.Dempsey G.T., Bates M., Zhuang X. Photoswitching mechanism of cyanine dyes. J. Am. Chem. Soc. 2009;131:18192–18193. doi: 10.1021/ja904588g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Linde S., Löschberger A., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 16.Dempsey G.T., Vaughan J.C., Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Diezmann A., Shechtman Y., Moerner W.E. Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem. Rev. 2017;117:7244–7275. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang B., Jones S.A., Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods. 2008;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalgarno P.A., Dalgarno H.I., Greenaway A.H. Multiplane imaging and three dimensional nanoscale particle tracking in biological microscopy. Opt. Express. 2010;18:877–884. doi: 10.1364/OE.18.000877. [DOI] [PubMed] [Google Scholar]
- 20.Schnitzbauer J., McGorty R., Huang B. 4Pi fluorescence detection and 3D particle localization with a single objective. Opt. Express. 2013;21:19701–19708. doi: 10.1364/OE.21.019701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavani S.R., Thompson M.A., Moerner W.E. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backer A.S., Backlund M.P., Moerner W.E. Single-molecule orientation measurements with a quadrated pupil. Opt. Lett. 2013;38:1521–1523. doi: 10.1364/OL.38.001521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shechtman Y., Sahl S.J., Moerner W.E. Optimal point spread function design for 3D imaging. Phys. Rev. Lett. 2014;113:133902. doi: 10.1103/PhysRevLett.113.133902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backer A.S., Backlund M.P., Moerner W.E. A bisected pupil for studying single-molecule orientational dynamics and its application to three-dimensional super-resolution microscopy. Appl. Phys. Lett. 2014;104:193701. doi: 10.1063/1.4876440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backer A.S., Moerner W.E. Extending single-molecule microscopy using optical Fourier processing. J. Phys. Chem. B. 2014;118:8313–8329. doi: 10.1021/jp501778z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shechtman Y., Weiss L.E., Moerner W.E. Precise three-dimensional scan-free multiple-particle tracking over large axial ranges with tetrapod point spread functions. Nano Lett. 2015;15:4194–4199. doi: 10.1021/acs.nanolett.5b01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson M.A., Lew M.D., Moerner W.E. Localizing and tracking single nanoscale emitters in three dimensions with high spatiotemporal resolution using a double-helix point spread function. Nano Lett. 2010;10:211–218. doi: 10.1021/nl903295p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gahlmann A., Ptacin J.L., Moerner W.E. Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano Lett. 2013;13:987–993. doi: 10.1021/nl304071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Diezmann A., Lee M.Y., Moerner W.E. Correcting field-dependent aberrations with nanoscale accuracy in three-dimensional single-molecule localization microscopy. Optica. 2015;2:985–993. doi: 10.1364/OPTICA.2.000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lew M.D., Lee S.F., Moerner W.E. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 2011;108:E1102–E1110. doi: 10.1073/pnas.1114444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M.K., Rai P., Moerner W.E. Small-molecule labeling of live cell surfaces for three-dimensional super-resolution microscopy. J. Am. Chem. Soc. 2014;136:14003–14006. doi: 10.1021/ja508028h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayas C.A., Wang J., Moerner W.E. Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 2018;115:E3712–E3721. doi: 10.1073/pnas.1721648115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustavsson A.K., Petrov P.N., Moerner W.E. 3D single-molecule super-resolution microscopy with a tilted light sheet. Nat. Commun. 2018;9:123. doi: 10.1038/s41467-017-02563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willig K.I., Harke B., Hell S.W. STED microscopy with continuous wave beams. Nat. Methods. 2007;4:915–918. doi: 10.1038/nmeth1108. [DOI] [PubMed] [Google Scholar]
- 35.Lau L., Lee Y.L., Moerner W.E. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 2012;102:2926–2935. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rittweger E., Han K.Y., Hell S.W. STED microscopy reveals crystal colour centres with nanometric resolution. Nat. Photonics. 2009;3:144–147. [Google Scholar]
- 37.Nägerl U.V., Willig K.I., Bonhoeffer T. Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl. Acad. Sci. USA. 2008;105:18982–18987. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y.L., Santé J., Stearns T. Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function. Mol. Biol. Cell. 2014;25:2919–2933. doi: 10.1091/mbc.E14-02-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang T.T., Chong W.M., Liao J.C. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 2018;9:2023. doi: 10.1038/s41467-018-04469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang T.T., Su J., Liao J.C. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci. Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T.T., Hampilos P.J., Liao J.C. Superresolution STED microscopy reveals differential localization in primary cilia. Cytoskeleton (Hoboken) 2013;70:54–65. doi: 10.1002/cm.21090. [DOI] [PubMed] [Google Scholar]
- 42.Weng R.R., Yang T.T., Liao J.C. Super-resolution imaging reveals TCTN2 depletion-induced IFT88 lumen leakage and ciliary weakening. Biophys. J. 2018;115:263–275. doi: 10.1016/j.bpj.2018.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirvis M., Stearns T., James Nelson W. Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem. J. 2018;475:2329–2353. doi: 10.1042/BCJ20170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohatgi R., Snell W.J. The ciliary membrane. Curr. Opin. Cell Biol. 2010;22:541–546. doi: 10.1016/j.ceb.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goetz S.C., Anderson K.V. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briscoe J., Thérond P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 47.Han Y.G., Alvarez-Buylla A. Role of primary cilia in brain development and cancer. Curr. Opin. Neurobiol. 2010;20:58–67. doi: 10.1016/j.conb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lechtreck K.F. IFT–cargo interactions and protein transport in cilia. Trends Biochem. Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huangfu D., Anderson K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran P.V., Haycraft C.J., Beier D.R. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacoby M., Cox J.J., Schurmans S. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009;41:1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- 52.Ko H.W., Norman R.X., Eggenschwiler J.T. Broad-minded links cell cycle-related kinase to cilia assembly and hedgehog signal transduction. Dev. Cell. 2010;18:237–247. doi: 10.1016/j.devcel.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang F., Sirinakis G., Bewersdorf J. Ultra-high resolution 3D imaging of whole cells. Cell. 2016;166:1028–1040. doi: 10.1016/j.cell.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi X., Garcia G., III, Reiter J.F. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat. Cell Biol. 2017;19:1178–1188. doi: 10.1038/ncb3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohli P., Höhne M., Schermer B. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep. 2017;18:1521–1535. doi: 10.15252/embr.201643846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halpern A.R., Howard M.D., Vaughan J.C. Point by point: an introductory guide to sample preparation for single-molecule, super-resolution fluorescence microscopy. Curr. Protoc. Chem. Biol. 2015;7:103–120. doi: 10.1002/9780470559277.ch140241. [DOI] [PubMed] [Google Scholar]
- 57.Bobenko A.I., Schröder P. Discrete Willmore flow. In: Desbrun M., Pottmann H., editors. Symp. Geometry Processing. Eurographics Assoc; 2005. pp. 101–110. [Google Scholar]
- 58.Milenkovic L., Weiss L.E., Moerner W.E. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc. Natl. Acad. Sci. USA. 2015;112:8320–8325. doi: 10.1073/pnas.1510094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lew M.D., von Diezmann A.R.S., Moerner W.E. Easy-DHPSF open-source software for three-dimensional localization of single molecules with precision beyond the optical diffraction limit. Protoc. Exch. 2013 [Google Scholar]
- 60.Garimella, R. V. and B. K. Swartz 2003. Curvature estimation for unstructured triangulations of surfaces. Los Alamos National Laboratory.
- 61.Milenkovic L., Scott M.P., Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J. Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corbit K.C., Aanstad P., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 63.Chen J.K., Taipale J., Beachy P.A. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cignoni P., Callieri M., Ranzuglia G. MeshLab: an open-source mesh processing tool. In: Scarano V., De Chiara R., Erra U., editors. Proceedings of the 2008 Eurographics Italian Chapter Conference. Eurographics Association; 2008. pp. 129–136. [Google Scholar]
- 65.Cao L., Verbeek F.J. Analytical evaluation of algorithms for point cloud surface reconstruction using shape features. J. Electron. Imaging. 2013;22:043008. [Google Scholar]
- 66.Raussen M. Aalborg University; Aalborg, Denmark: 2008. Elementary Differential Geometry: Curves and Surfaces. [Google Scholar]
- 67.Firestone A.J., Weinger J.S., Chen J.K. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.May S.R., Ashique A.M., Peterson A.S. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 69.Liu H., Li W., Zhang Z. IFT25, an intraflagellar transporter protein dispensable for ciliogenesis in somatic cells, is essential for sperm flagella formation. Biol. Reprod. 2017;96:993–1006. doi: 10.1093/biolre/iox029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keady B.T., Samtani R., Pazour G.J. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell. 2012;22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Camera flythrough, which starts above the mesh, then follows along the ciliary axis, and finally pans away. This is the same mesh as shown in Fig. 2 a. White bounding box dimensions, 2750 nm x 1500 nm x 1500 nm; video speed, 30 frames/s. The HSV color scaling represents z in the image over a range from −175.8 to 1194.5 nm.
Camera flythrough, which starts above the mesh, then follows along the ciliary axis, and finally pans away. This is the same mesh as shown in Fig. 2 b. White bounding box dimensions, 3700 nm x 900 nm x 600 nm; video speed, 30 frames/s. The HSV color scaling represents z in the image over a range from −196.4 to 227.1 nm.