Abstract
Recent studies have shown that the small GTPase KRAS adopts multiple orientations with respect to the plane of anionic model membranes, whereby either the three C-terminal helices or the three N-terminal β-strands of the catalytic domain face the membrane. This has functional implications because, in the latter, the membrane occludes the effector-interacting surface. However, it remained unclear how membrane reorientation occurs and, critically, whether it occurs in the cell in which KRAS operates as a molecular switch in signaling pathways. Herein, using data from a 20 μs-long atomistic molecular dynamics simulation of the oncogenic G12V-KRAS mutant in a phosphatidylcholine/phosphatidylserine bilayer, we first show that internal conformational fluctuations of flexible regions in KRAS result in three distinct membrane orientations. We then show, using single-molecule fluorescence resonance energy transfer measurements in native lipid nanodiscs derived from baby hamster kidney cells, that G12V-KRAS samples three conformational states that correspond to the predicted orientations. The combined results suggest that relatively small energy barriers separate orientation states and that signaling-competent conformations dominate the overall population.
Introduction
C-terminally lipidated small GTPases of the RAS superfamily are peripheral membrane proteins that mediate a variety of crucial cellular processes, including cell growth, motility, and trafficking (1, 2, 3). They act as molecular switches by cycling between active GTP- and inactive GDP-bound conformational states (4, 5, 6). GDP/GTP exchange and GTP hydrolysis are catalyzed by nucleotide exchange factors and GTPase-activating proteins, respectively. Biochemical events that break this cycle, such as activating somatic mutations in RAS (7, 8, 9, 10), can lead to a variety of cancers and developmental disorders (11, 12). The absence of any effective drugs on the market to treat these diseases calls for alternative approaches to inhibiting abnormal signaling mediated by the RAS superfamily GTPases. The concept of membrane reorientation, the focus of the current work, offers a potential alternative to traditional approaches that primarily focus on the soluble catalytic domain of this class of proteins (13).
The RAS family of proteins, which in mammals includes the three major isoforms NRAS, HRAS, and KRAS, is tethered to the inner leaflet of the plasma membrane. Upon GTP binding, RAS proteins interact with downstream effectors to transduce extracellular signal to the nucleus. KRAS, the focus of this study, is arguably the most sought-after anticancer drug target because mutations, such as G12V and G12D, impair its GTPase activities and lead to a wide variety of cancers (8, 14). KRAS is made up of a bilobed catalytic domain (lobe 1: residues 1–86, and lobe 2: residues 87–166) and a flexible hypervariable region (HVR: residues 167–185) that contains a C-terminal polybasic and farnesylated lipid anchor. It has been shown that in addition to the lipid anchor, the catalytic domain of KRAS dynamically interacts with anionic model membranes via 1) α-helices 3–5 on lobe 2 (termed orientation state 1 (OS1)), 2) β-strands 1–3 on lobe 1 (OS2), or 3) an intermediate orientation (OS0) in which the helices are roughly perpendicular to the membrane plane (15, 16). OS1 and OS2 differ in the accessibility of functionally critical switch loops to partner proteins, suggesting that membrane reorientation in the cell may modulate KRAS signaling. However, our current understanding of this phenomenon is limited to inferences from structural models from molecular dynamics (MD) simulations or indirect spectroscopic techniques in simple model membranes (15, 16, 17, 18). While MD simulations typically access only relatively short length and timescales, the timescale of RAS reorientation in the complex cell membranes is likely too fast to resolve by experimental techniques alone.
In the current work, we combine atomistic MD simulation with single-molecule fluorescence resonance energy transfer (FRET) (smFRET) in native nanodiscs (NDs) to show that the catalytic domain of G12V-KRAS makes dynamic interactions with membrane lipids in three distinct orientations that are separated by relatively small energy barriers. Our results show that membrane orientation dynamics of KRAS is governed by internal fluctuations of the protein, and demonstrate that KRAS membrane reorientation is observable under near-native conditions.
Materials and methods
MD simulation
A 20 μs-long MD simulation was conducted on Anton2 and was started from the final snapshot from a previous simulation of G12D-KRAS bound to a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine /1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine bilayer (5:1 molar ratio) (16) after replacing aspartate to valine at position 12. The protein-bilayer system was solvated by TIP3P waters plus 192 Na+ and 96 Cl− ions, resulting in a 150,000-atoms system. We used the CHARMM36m force field for KRAS (19), an in-house parameter for GTP, and CHARMM36 for lipids (20). A 2.5 fs time step and default parameters, including the U-series cutoffs, were used for the production simulations. Coordinates were saved every 100 ps for analysis. The trajectory was analyzed primarily based on the z component of the distance between Cα atoms of residues 132 and 183 (ζ) and the angle between a vector from residue 5 to residue 9 Cα atoms and the membrane normal (Θ) (see Supporting Materials and Methods for details).
Extraction of G12V-KRAS in native NDs
To measure FRET distance between fluorophore-labeled residues 132 and 183 (ζ∗), we generated a hemagglutinin (HA)-His-tagged G12V-KRAS with D132C/T183C/C118S mutations (G12V-KRAS∗). In this construct, D132C and T183C provide surface-exposed cysteines for site-specific fluorescent labeling, whereas C118S eliminates the only surface cysteine to avoid nonspecific labeling. The C118S mutation does not affect structure or function (21). Similarly, we do not expect major structural or functional effects from mutations at the surface residues D132 or T183. The His tag was added to aid surface immobilization (see Supporting Materials and Methods), and HA to enhance expression of the Cys mutant KRAS. The construct was expressed in baby hamster kidney (BHK), cells grown to confluency, and styrene-maleic acid (SMA) extracted in NDs, as follows. Intact cells suspended in phosphate-buffered saline (PBS) containing 20 μg/mL deoxyribonuclease and 0.5 mM phenylmethylsulfonyl fluoride were treated with 2% SMA for 1 h at room temperature under rotation. After ultracentrifugation at 100,000 × g for 1 h at 4°C, the supernatant containing ND-bound G12V-KRAS∗ was collected and quantified using Western blotting (see Fig. 3 B). On the day of analysis, the supernatant containing ND-G12V-KRAS∗ was incubated with a 1:4 molar ratio of Alexa Fluor 555 (donor) and Alexa Fluor 647 (acceptor) maleimide (Invitrogen, Carlsbad, CA) for 1 h at room temperature under rotation. Excess dye was removed using a PD-10 desalting column (GE Healthcare Life Sciences, Waukesha, WI).
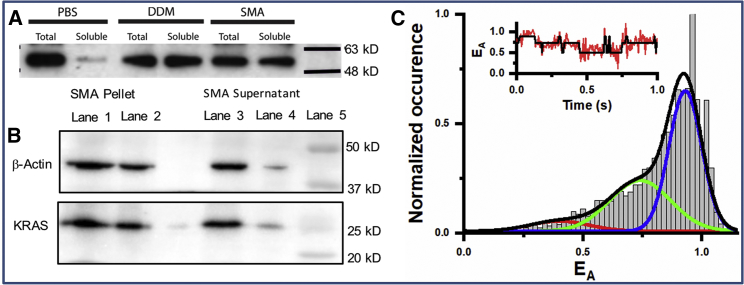
Figure 3.
(A) Western blots of GFP-tagged G12V-KRAS from cell extracts without a solubilizing agent (PBS) and after treatment with DDM or SMA (see text). (B) Western analysis, using an anti-HA antibody, of different volumes of SMA-solubilized cell extract pellet (lanes 1–2) or supernatant (lanes 3–4) is shown, with β-actin used as loading control. (C) FRET efficiency (EA) histogram fitted to three Gaussians, indicating three distinct conformational states, with high FRET states in blue, mid-FRET states in green, and low FRET states in red. A representative EA trajectory indicating fast transitions between states is shown as inset, with the actual signal in red and state transitions highlighted in black. To see this figure in color, go online.
FRET data acquisition and analysis
The labeled protein was diluted 5× and immobilized on a glass surface for smFRET measurements, as detailed in the Supporting Materials and Methods. A custom-built PicoQuant MicroTime 200 Fluorescence Lifetime Microscope was used for smFRET measurements at 80 MHz using pulsed interleaved excitation. The 532 nm (LDH-D-TA-530; PicoQuant, Berlin, Germany) and 637 nm (LDH-D-C-640; PicoQuant) lasers were used to characterize the efficiency of energy transfer between molecules potentially showing FRET. The sample was excited and observed through a 100× oil immersed lens (100× 1.4 NA; Olympus, Tokyo, Japan) while immobilized on a scanning x-y-z piezo stage (P-733.2CD; Physik Instrumente, Auburn, MA). The photons emitted from the sample postexcitation were separated through a dual-band dichroic beam splitter (Zt532/640rpc-UF3; adaptive high frequency [AHF]/Chroma, Los Angeles, CA) and sent to two single-photon avalanche photodiodes (SPCM CD3516H; Excelitas Technologies, Fremont, CA). The 550 nm (FF01-582/64; AHF/Semrock, Rochester, NY) and 650 nm (2XH690/70; AHF) emission filters were used for the donor and acceptor channels, respectively. ROXS was used during all data acquisitions. The photon counts were acquired at 1-ms resolution, binned to 10 ms, denoised with wavelet decomposition, and plotted as separate histograms showing the occurrence of photons FRETing at their observed efficiencies.
Step transition and state identification (STaSI) analysis (22) was then used to determine the number of states that best describes the distribution of FRET efficiencies found in the obtained FRET data. The free energy (in terms of kBT) associated with each STaSI-identified state (ΔG) was calculated by setting the most populated state to 0 kBT and using the frequency of state occurrences (OState) relative to the most populated state (ORef) as . The energy barriers between states were assumed to be of first-order kinetics and calculated using the Arrhenius equation: given . By substitution and rearrangement, Ea = ln((A∗tbin)/(Strans∗tres)). Ea represents the energy of activation in kBT, k is the rate constant of the transition between states in transitions per millisecond, A is the preexponential factor set to (10 ms)−1, Strans represents the number of transitions between states, tbin represents the total number of time bins, and tres is the duration of each time bin in milliseconds. The concentration of the starting state was taken as the STaSI-derived fractional occupancy of that state. Forward and reverse energies of activation were averaged. Data were analyzed using Origin (OriginLab, Northampton, MA), MATLAB (The MathWorks, Natick, MA), and Excel (Microsoft, Tulsa, OK). For smFRET experiments, after filtering out the molecules that failed the anticorrelation check, the number of particles was n = 13.
Results and discussion
G12V-KRAS adopts three distinct orientations with respect to the membrane surface
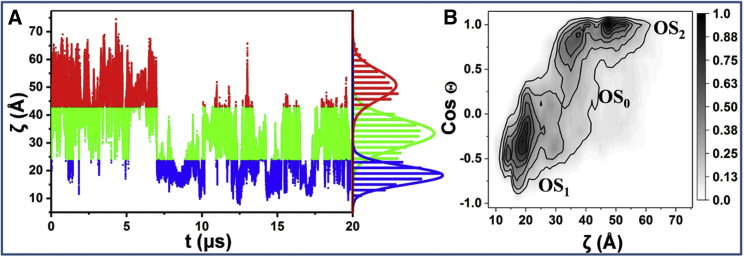
We conducted a 20 μs MD simulation of G12V-KRAS bound to a phosphatidylcholine/phosphatidylserine (POPC/POPS) bilayer and defined two reaction coordinates to analyze the trajectory: 1) the distance between Cα atoms of D132 on α4 and T183 on the lipid anchor (ζ), and 2) the angle between a vector along β1 (residues 5–9) and the membrane normal (Θ). The time evolution of ζ shows major conformational fluctuations (Fig. 1 A). For example, helix α4 remained away from the membrane-interacting residue T183 (and hence away from the membrane surface) until ∼7 μs (ζ > 40 Å), with occasional excursions to the bilayer surface (ζ < 25 Å). After that, the protein predominantly fluctuates between two distinct states with average ζ = 33.3 ± 0.2 and ζ = 18.6 ± 0.1 Å, rarely visiting larger ζ values. The cosine of Θ, which measures the orientation of the catalytic domain with respect to the bilayer normal, exhibits similar fluctuations and primarily samples three dominant orientations characterized by Θ ≈ 90, Θ > 90°, and Θ < 90° (Fig. S1). The resulting two-dimensional histogram, (ζ, CosΘ), which yielded three distributions centered at (18.6, −0.5), (33.3, 0.9), and (49.7, 1) (Fig. 1 B), demonstrates that G12V-KRAS samples the three orientation states OS1, OS2, and OS0 observed in G12D-KRAS (16). Unlike the previous study, however, the longer timescale of the current simulation enabled us to observe transitions from OS2, in which the effector-binding loop is occluded by the membrane, to OS1, in which the loop is solvent accessible. Although there is no direct transition between OS1 and OS2, transitions between OS1 and OS0 or OS2 and OS0 are frequent and occur at a sub-μs timescale. However, despite the comparatively long 20 μs simulation that we have conducted, the limited number of transition events prevented us from investigating the relative likelihood of the different states. A much longer run or multiple runs of tens of microseconds each would be required to quantitatively describe the thermodynamic stability and transition kinetics of the different orientation states. Therefore, we used the smFRET data described later to roughly estimate the population of the states.
Figure 1.
(A) Time evolution of ζ during a 20 μs MD simulation of GTP-bound G12V-KRAS; the normalized density distribution of the distance is shown on the right-hand side. (B) Normalized number density distribution of conformations was projected onto a plane defined by the reaction coordinates ζ and CosΘ. Color scale indicates low (white) through high (black) density. To see this figure in color, go online.
Intrinsic conformational dynamics underlies the membrane reorientation of G12V-KRAS
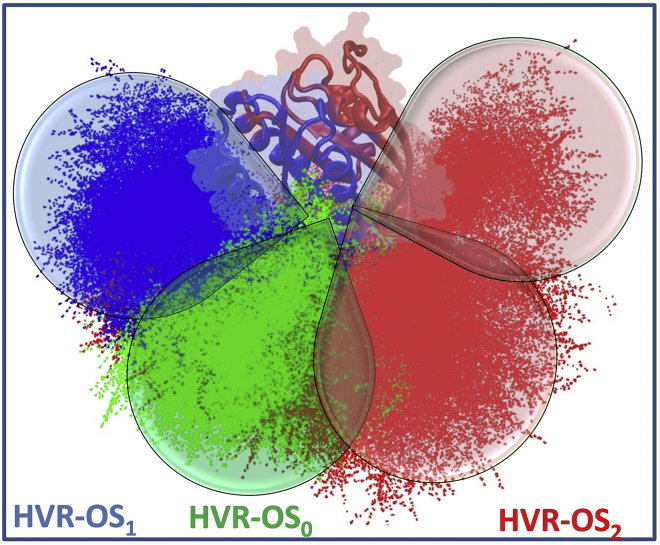
Fig. 2 shows that the transition between orientation states is underpinned by internal conformational fluctuations of the protein. In particular, an interdomain motion via a hinge region at the C-terminus of helix 5 alters the disposition of the HVR relative to the catalytic domain. As a result, the HVR faces lobe 2 in OS1 (blue in Fig. 2), is toward lobe 1 in OS2 (red), and lies somewhere in between in OSo (green). Despite significant fluctuations within each group, especially in OS2 in which two subpopulations are apparent, the three ensembles are clearly separate with little overlap (highlighted by cones in Fig. 2). This result directly links internal conformational fluctuation to membrane reorientation. We would like to emphasize that, although the different orientations are sampled spontaneously, they may be stabilized by protein-lipid electrostatic interactions as suggested by several previous reports (16, 18, 23). That intrinsic flexibility underlies the membrane orientation dynamics of G12V-KRAS is further confirmed by performing another 20 μs MD simulation using the CHARMM36 force field (24). In this trajectory, a very stable salt bridge between side chains of Lys176 on the HVR and Asp47 on the β2-β3 loop “locked” the protein in the OS2 state for the duration of the simulation (Fig. S2). Together, these results demonstrate that the disposition of the HVR relative to the two lobes of the catalytic domain is an important determinant of G12V-KRAS membrane reorientation.
Figure 2.
Overlay of conformations showing three distinct orientations of the HVR with respect to the catalytic domain: HVR is oriented toward lobe 2 (blue) and lobe 1 (red) in conformations belonging to OS1 and OS2, respectively, with those in OS0 (green) being intermediate. The cones highlight large fluctuations of the HVR within each group and the presence of two subgroups in OS2. The alignment was done on the Cα atoms of the catalytic domain, excluding the flexible switches, using the equilibrated G12V-KRAS structure as a reference. To see this figure in color, go online.
smFRET confirms that G12V-KRAS bound to native nanodiscs samples three distinct conformational states that correspond to the predicted orientation states
A key question is whether the conformational and orientation dynamics we observe during simulations of simplified model systems apply to KRAS when it is bound to a more realistic model membrane. We addressed this question using native lipid NDs derived from cells using SMA copolymer and smFRET. To this end, we first tested whether KRAS can be isolated in NDs at high yield. BHK cells stably expressing green fluorescent protein (GFP)-tagged G12V-KRAS (MW = 48 kDa) were lysed by sonication and spun at 100,000 × g. The membrane fraction was resuspended and solubilized with n-dodecyl-β-D-maltopyranoside (DDM), SMA, or PBS as a negative control. Samples were spun again at 100,000 × g to pellet the insoluble protein, and the supernatant was immunoblotted. As shown in Fig. 3 A, without any solubilizing agent (PBS), little KRAS is found in the soluble fraction, whereas treatment with DDM to dislodge KRAS from membrane or SMA to isolate it in NDs showed efficient solubilization. Fig. 3 B further shows that non-GFP-labeled KRAS can be directly extracted from cells by SMA using the protocol described in Materials and Methods.
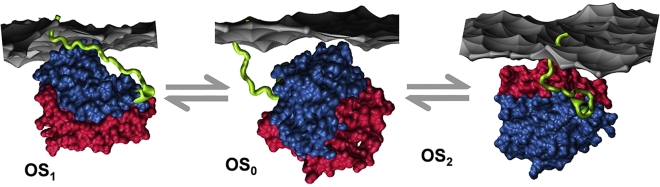
After extraction into NDs, the protein was labeled by Alexa dyes (see Materials and Methods) for smFRET measurements, with the His tag used to selectively immobilize ND-G12V-KRAS∗ particles on a glass surface. The normalized distribution of the FRET efficiency (EA) from 13 individual ND-G12V-KRAS∗ particles (Fig. 3 C) show that G12V-KRAS∗ exists in at least three distinct conformational states. STaSI analysis (22) yielded average EA values of 0.40, 0.74, and 0.94 for the three states. These values correspond to ζ∗ = 49, 39, and 29 Å, mirroring those of the MD-derived mean values of ζ (Fig. 1 A). The difference between some of the smFRET and MD distances is likely attributable to the labeled side chains being oriented in opposite directions. We therefore conclude that the three conformational states observed in the experiments correspond to OS2, OS0, and OS1, respectively (Fig. 4). Despite the possibility that the positively charged His tag might interact with the oppositely charged phosphatidylserine lipids and thereby potentially stabilize OS2, the population of the three states derived from smFRET is such that OS2 < OS0 < OS1. This suggests that the orientation with the effector-binding loop occluded by the membrane (OS2) is disfavored in this constitutively active KRAS mutant. The rates of transition between the states estimated from the smFRET data (see Materials and Methods) suggest low energy barriers, with the barrier between OS1 and OS0 (3.12 kBT) being somewhat smaller than that between OS2 and OS0 (3.73 kBT), consistent with the number of transition events observed in the MD simulation (Fig. 1 A). Transition between OS1 and OS2 appears less frequent. Combining this with the MD data and considering the relatively sparse sampling in the smFRET measurements, we propose that OS0 is an obligatory intermediate for the membrane reorientation of G12V-KRAS (Fig. 4).
Figure 4.
Summary of the three distinct membrane orientations of G12V-KRAS in dynamic equilibrium. Note the difference in the conformation and orientation with respect to lobe 1 (red) and lobe 2 (blue) of the HVR (green) among the three states. A portion of the membrane monolayer proximal to the protein is shown as gray surface. To see this figure in color, go online.
Conclusions
In this work, we 1) used smFRET in native NDs to show for the first time, to our knowledge, that oncogenic mutant KRAS exists in three interconverting conformational states in a membrane of near-native lipid composition; and 2) demonstrated using MD that these conformational states represent distinct membrane orientations (Fig. 4). We also found that G12V-KRAS favors OS1, with an accessible effector-binding loop, over OS2, in which the effector-binding loop is occluded by the membrane. We propose that the population of these states can be altered by mutation or stabilized by specific interactions with lipids.
Author Contributions
A.A.G. designed and supervised the project. P.P. performed MD simulations and analyzed the data. D.L., D.D., and V.J. performed smFRET and analyzed the data. H.L., S.S.-B., D.D., Y.Z., and J.F.H. performed cell biology experiments. P.P. and D.L. wrote the first draft of the manuscript. A.A.G., V.J., and J.F.H. provided resources. All authors read and approved the final version of the manuscript.
Acknowledgments
This work was supported in part by the National Institutes of Health grants R01GM100078 and R35GM122528 (V.J.), Extreme Science and Engineering Discovery Environment grant MCB150054, Cancer Prevention and Research Institute of Texas grant DP150093, and the Houston Area Molecular Biophysics program no. T32GM008280-28 (D.L.). Anton 2 computer time was provided by the Pittsburgh Supercomputing Center through grant R01GM116961 from the National Institutes of Health. The Anton 2 machine at Pittsburgh Supercomputing Center was generously made available by D.E. Shaw Research.
Editor: Kalina Hristova.
Footnotes
Supporting Materials and Methods and two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)34506-5.
Supporting Material
References
- 1.Macara I.G., Lounsbury K.M., Bar-Sagi D. The RAS superfamily of GTPases. FASEB J. 1996;10:625–630. doi: 10.1096/fasebj.10.5.8621061. [DOI] [PubMed] [Google Scholar]
- 2.Wennerberg K., Rossman K.L., Der C.J. The RAS superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 3.Goitre L., Trapani E., Retta S.F. The RAS superfamily of small GTPases: the unlocked secrets. Methods Mol. Biol. 2014;1120:1–18. doi: 10.1007/978-1-62703-791-4_1. [DOI] [PubMed] [Google Scholar]
- 4.Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 5.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 6.Wittinghofer A., Vetter I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 7.Cox A.D., Der C.J. RAS history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubbert S., Shannon K., Bollag G. Hyperactive RAS in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 9.Bos J.L. RAS oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 10.Karnoub A.E., Weinberg R.A. RAS oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vega F.M., Ridley A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 12.Li G., Marlin M.C. Rab family of GTPases. Methods Mol. Biol. 2015;1298:1–15. doi: 10.1007/978-1-4939-2569-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash P., Gorfe A.A. Membrane orientation dynamics of lipid-modified small GTPases. Small GTPases. 2017;8:129–138. doi: 10.1080/21541248.2016.1211067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malumbres M., Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 15.Mazhab-Jafari M.T., Marshall C.B., Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash P., Zhou Y., Gorfe A.A. Oncogenic K-RAS binds to an anionic membrane in two distinct orientations: a molecular dynamics analysis. Biophys. J. 2016;110:1125–1138. doi: 10.1016/j.bpj.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorfe A.A., Hanzal-Bayer M., McCammon J.A. Structure and dynamics of the full-length lipid-modified H-RAS protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 18.Cao S., Chung S., Buck M. K-RAS G-domain binding with signaling lipid phosphoinositides: PIP2 association, orientation, function. bioRxiv. 2018 doi: 10.1074/jbc.RA118.004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mott H.R., Carpenter J.W., Campbell S.L. Structural and functional analysis of a mutant RAS protein that is insensitive to nitric oxide activation. Biochemistry. 1997;36:3640–3644. doi: 10.1021/bi962790o. [DOI] [PubMed] [Google Scholar]
- 22.Shuang B., Cooper D., Landes C.F. Fast step transition and state identification (STaSI) for discrete single-molecule data analysis. J. Phys. Chem. Lett. 2014;5:3157–3161. doi: 10.1021/jz501435p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory M.C., McLean M.A., Sligar S.G. Interaction of KRAS4b with anionic membranes: a special role for PIP2. Biochem. Biophys. Res. Commun. 2017;487:351–355. doi: 10.1016/j.bbrc.2017.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.