Abstract
Within the complex aggregation process of amyloidogenic peptides into fibrils, early stages of aggregation play a central role and reveal fundamental properties of the underlying mechanism of aggregation. In particular, low-molecular-weight aggregates of the Alzheimer amyloid-β peptide (Aβ) have attracted increasing interest because of their role in cytotoxicity and neuronal apoptosis, typical of aggregation-related diseases. One of the main techniques used to characterize oligomeric stages is fluorescence spectroscopy. To this end, Aβ peptide chains are functionalized with fluorescent tags, often covalently bound to the disordered N-terminus region of the peptide, with the assumption that functionalization and presence of the fluorophore will not modify the process of self-assembly nor the final fibrillar structure. In this investigation, we systematically study the effects of four of the most commonly used fluorophores on the aggregation of Aβ (1–40). Time-resolved and single-molecule fluorescence spectroscopy have been chosen to monitor the oligomer populations at different fibrillation times, and transmission electron microscopy, atomic force microscopy and x-ray diffraction to investigate the structure of mature fibrils. Although the structures of the fibrils were only slightly affected by the fluorescent tags, the sizes of the detected oligomeric species varied significantly depending on the chosen fluorophore. In particular, we relate the presence of high-molecular-weight oligomers of Aβ (1–40) (as found for the fluorophores HiLyte 647 and Atto 655) to net-attractive, hydrophobic fluorophore-peptide interactions, which are weak in the case of HiLyte 488 and Atto 488. The latter leads for Aβ (1–40) to low-molecular-weight oligomers only, which is in contrast to Aβ (1–42). The disease-relevant peptide Aβ (1–42) displays high-molecular-weight oligomers even in the absence of significant attractive fluorophore-peptide interactions. Hence, our findings reveal the potentially high impact of the properties of fluorophores on transient aggregates, which needs to be included in the interpretation of experimental data of oligomers of fluorescently labeled peptides.
Introduction
Amyloid-forming proteins have attracted a great deal of attention in the last decades because of their association with a numerous group of degenerative conditions. Alzheimer’s, Huntington’s, Parkinson’s, Creutzfeldt-Jacob, and prion diseases are some examples of the most well-known neurodegenerative disorders, whereas the most well-known systemic ones are amyotrophic lateral sclerosis and type II diabetes (1). The etiology of all the aforementioned diseases seems to be found in the “defective” folding or “misfolding” of normally soluble, functional peptides and proteins and their subsequent conversion into intractable aggregates, also known as amyloid fibrils (2). The latter, however, have been found to also be a well-defined structural form for many proteins unrelated to diseases (3). This observation suggests, in fact, that the structural motif common to these aggregates is broadly accessible by various polypeptide chains and can also be functional (4, 5). Either way, amyloidogenic proteins seem to follow a common dynamical pathway toward fibril formation, characterized by an intermediate step of aggregation into small heterogeneous oligomers. The latter are also deemed to be the primary cause of cyto- and neurotoxicity (6). In Alzheimer’s disease, for instance, recent studies have demonstrated that cytotoxicity is indeed associated with the smaller, still-soluble species of Aβ oligomers, which lead to cell death by binding to the neuronal membrane and possibly by membrane permeabilization (7, 8, 9, 10, 11). The two most abundant amyloid allomorphs in the extracellular aggregates signature of advanced Alzheimer’s disease are Aβ (1–40) and Aβ (1–42) (12, 13), which will be named Aβ40 and Aβ42 further on. Generated by β-secretase cleavage of the intramembrane amyloid precursor protein (14), the latter peptides have an intrinsically disordered structure that can be subject to conformational changes toward secondary and tertiary structures. Numerous in vitro studies of short and medium-length amyloidogenic peptides have provided some clues to amyloid formation with an α-helix to β-sheet folding transition. However, the mechanisms triggering such conformational changes as well as aggregation are still unclear both in vivo and in vitro (1). Efforts to develop a more mechanistic understanding of how Aβ assembles into toxic species have been limited by significant experimental challenges. To start with, Aβ normally circulates in plasma and cerebrospinal fluid as a soluble peptide in nanomolar to picomolar concentrations (2, 15, 16), and its aggregates are metastable and highly heterogeneous (17, 18, 19). Additionally, the self-assembly dynamics are highly dependent on the environmental conditions like pH, temperature, ionic strength, or presence of metal ions (20) as well as concentration (21). Each of those factors can have enhancing or impeding effects in respect to aggregation, and thus protocols on how to handle such peptides should be selected with great care, trying to keep conditions as close as possible to the physiological ones. Given the especially low concentrations at which Aβ peptides are normally found, many of the most popular techniques, such as polyacrylamide gel electrophoresis or size exclusion chromatography, are not suitable for these studies. Thus, investigation methods that can work at much lower concentrations, such as optical techniques based on absorption and/or fluorescence detection, have been most widely employed to monitor in real time the kinetics of the early aggregation processes in solution (20). In the last two decades, techniques such as fluorescence absorbance (22), photobleaching (19, 23), self-fluorescent-quenching (20), fluorescence correlation spectroscopy (FCS) (24, 25, 26, 27), fluorescence cross-correlation spectroscopy (28, 29), Förster resonance energy transfer combined with FCS (28), and confocal two-color coincidence detection (30) but also fluorescence imaging of labeled peptides (19, 31, 32, 33) or binding of Thioflavine T (ThT) (34) or Congo Red (35) have been widely applied. Despite the great sensitiveness of such optical techniques, one drawback of using fluorescence is to be found in the inherent, yet necessary, modification of the original peptide system because of the attached fluorophore. The latter modification happens either through binding to aggregate-selective fluorophores, like Congo Red or ThT, or through N-terminus covalent binding (see Fig. 1 A). The N-terminus is considered to be a loose end of the primary peptide structure, which is thus supposed not to strongly contribute to its aggregation mechanisms nor structure formation (19, 30, 32). Nonetheless, other experimental studies have suggested quite the opposite. Modifications of the N-terminus (36, 37) or labeling with various fluorophores (38) can, in fact, interfere and modify the Aβ peptide self-assembly dynamics and, as a result, its structure formation. In support of these findings, more recent theoretical and numerical studies have also pointed out that chemical modifications of the protein surface, such as the addition of a fluorophore, do affect the physical properties of protein solutions. In particular, the chemical modification is found to be equivalent to the addition of a large hydrophobic patch with a large attractive potential energy well, significant even at low labeling fractions (39). To experimentally address the effect of N-terminus-bound fluorophores, we studied the early stages of aggregation of fluorescently labeled Aβ40 with four different fluorophores by time-resolved and single-molecule fluorescence experiments. More in detail, we have investigated the following fluorophores, Atto 488 and Atto 655 and HiLyte 488 and HiLyte 647, by a concerted approach of different techniques. With the help of time-resolved and single-molecule fluorescence spectroscopy, we could carefully monitor and characterize the early stages of aggregation of all the above-listed samples at a single-molecule level. X-ray diffraction (XRD) measurements have been used to test the local structure of the final fibrils, whereas imaging techniques such as atomic force microscopy (AFM) and transmission electron microscopy (TEM) have been used to directly visualize eventual effects translated up to the mesoscopic scale, such as changes in length or thickness, undetectable otherwise. As a result, from the single-molecule studies, we found different characteristic distributions of oligomer sizes that we relate to specific properties of the fluorophores. Fluorophores displaying net-attractive interactions with the peptide lead to stable high-molecular-weight (HMW) oligomers, which are not further participating in the fibril formation. On the other hand, fluorophores displaying less hydrophobic interactions tend to form low-molecular-weight (LMW) oligomers that are consumed with ongoing fibrillation. From a structural point of view, TEM and AFM images revealed a diverse scenario of mature fibrillar structures ranging from very straight and thick to thin and curvy, whereas the local structure of the β-sheets investigated via XRD showed no notable differences between differently labeled Aβ40 peptides. All the observations collected thus far encouraged the conclusion that nucleation and growth are restricted to small (labeled) oligomers without structural deviations from the wild-type oligomer: 1) XRD measurements show no substantial variation in the structure of the nucleation units, nor 2) is the fibrillation kinetics changed; 3) the detected stable HMW aggregates do not contribute to fibril formation because they stay detectable by single-molecule fluorescence and visible in AFM. Structural differences are only detected on the macroscopic scale, as supported by TEM images. Finally, oligomers formed by Aβ40 and by the disease-relevant peptide Aβ42 were compared using the fluorophore with the least interactions, Atto 488. A detailed comparison of the oligomer size distributions revealed that Aβ42 tends to form more of the larger oligomers than Aβ40. In contrast to the HMW oligomers of Aβ40, which were, if present, identified as fluorophore-induced off-pathway oligomers, Aβ42 forms on-pathway HMW oligomers, suggesting an alternative fibrillation and/or nucleation process.
Figure 1.
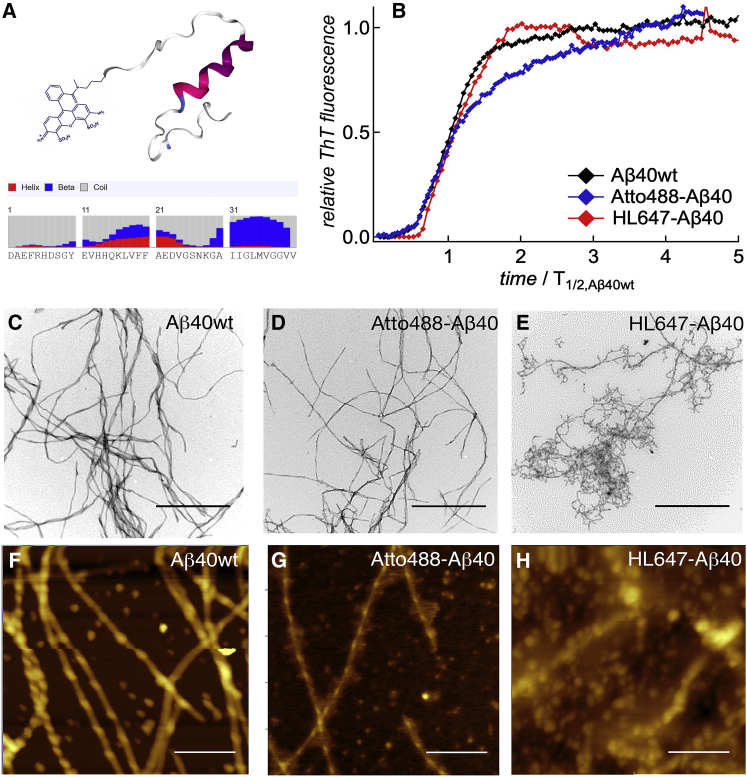
The effect of N-terminus-bound fluorophores on the fibrillation kinetics and fibril morphology of Aβ40. (A) Fluorescence labeling of the unstructured N-terminus of Aβ40 is expected to have the least effect on structure formation (presentation is based upon Protein Data Bank (PDB) structure 2LFM (55) and plotted by the NGL viewer (56)). Inset: amino acid sequence of Aβ40 with secondary structure prediction using RaptorX (57). (B) The time-dependent ThT fluorescence of Aβ40wt without (black) and with 5% of Atto488-Aβ40 (blue) or HL647-Aβ40 (red). The time axis is normalized to the half time of Aβ40wt. (C–H) Representative TEM (C–E) and AFM (F–H) images of the fibrils after 24 h of incubation. The scale bar represents 500 and 200 nm for TEM and AFM, respectively. To see this figure in color, go online.
Materials and Methods
Samples
Wild-type Aβ (1–40) and Aβ (1–42) peptides (DAEFRHDSGY EVHHQKLVFF AEDVGSNKGA IIGLMVGGVV(IA), Aβ40wt and Aβ42wt) and an N-terminus cysteine mutant, C-Aβ40, as well as Atto488-Aβ40 and Atto488-Aβ42, the Aβ peptides labeled with Atto 488 (ATTO-TEC, Siegen, Germany), were synthesized and purified by the Core Unit of Peptide Technologies of the University Leipzig (purity >98%). C-Aβ40 has been further used for labeling with Atto 655 (ATTO-TEC) in our laboratories (see below). The labeled peptides with HiLyte 488 and HiLyte 647 (HL488-Aβ40, HL647-Aβ40) have been purchased from Anaspec, Fremont, CA (purity >95%). To obtain different degrees of labeling (DOL), labeled and unlabeled Aβ peptides have been mixed with specific ratios. For example, for measurements with a DOL of 5%, the labeled peptide samples were mixed with the wild type to reach a final ratio of 1:20. Fluorescence labeling with Atto655: 1 mg/mL of C-Aβ40 was dissolved in phosphate buffer at pH 9.2 in the presence of 1 mM tris(2-carboxyethyl)phosphine (Sigma Aldrich, Schnelldorf, Germany) and dialyzed in vacuum against the same buffer but at pH 7.5 and tris(2-carboxyethyl)phosphine free. Atto 655 maleimide (ATTO-TEC) was added at a 1.3:1 molar ratio, and the solution was stored for 2 h in the dark at room temperature. Labeled Aβ40 was separated from the unbound free dye using Sephardex G-25 gel filtration (PD Minitrap G-25 column; GE Healthcare, Waukesha, WI). The final yield of the labeled protein was 5.1%.
Oligomerization and fibrillation
Following an established protocol (37), lyophilized peptides were first dissolved in 25 mM sodium phosphate buffer (pH 9.2) containing 150 mM NaCl. Under these conditions, FCS analysis of all samples displayed a single monomer decay without the contribution of large aggregates (see Fig. S1). The Aβ-samples were then either dialyzed against the same buffer with a pH of 7.5 (4 h, MWCO 1000) or diluted by the same buffer but at a lower pH to reach pH 7.5. The final protein concentrations were determined by absorption measurements and were adjusted to 40 μM if not otherwise stated. For fibrillation, Aβ solutions were incubated at 37°C and shaken with 400–450 rpm for 24 h with different sampling times. The appropriate incubation times to monitor oligomers or fibrils were chosen by TEM imaging, which, e.g., for Aβ40wt, revealed small aggregates after 6 h of incubation and mature fibrils after 24 h (see Fig. S2).
ThT assays
A 2.5 mM ThT (Sigma Aldrich) stock solution was prepared in 25 mM sodium phosphate buffer (pH 7.5) containing 150 mM NaCl and stored at 4°C in the dark. The solution was filtered through 0.2 μm filters and diluted 1:10 in the same buffer before use. Aβ solutions were mixed with the ThT solution to a final ThT concentration of 20 μM. The samples were incubated in a well plate reader (FLUOstar Omega; BMG Labtech, Ortenberg, Germany) at 37°C and 400 rpm. Measurements were taken every 300 s using 450 and 480 nm for excitation and detection, respectively.
TEM
5 μL of Aβ40 solution was dropped on Formvar/Cu grids with mesh 200 (Ted Pella, Redding, CA). After 3 min of waiting time, the grids were first cleaned in water for 60 s and then negatively stained with 1% (w/v) uranyl acetate for a further 60 s. TEM images were taken with an electron microscope (EM 900; Zeiss, Jena, Germany) at 80 kV acceleration voltage.
AFM
10 μL of fibrillated Aβ solution was pipetted on top of freshly cleaved mica (V1-grade, 10 mm diameter; Ted Pella). After a 1 min waiting time to allow sedimentation of the heavier fibrillar structures, it was gently rinsed with clean water and dried in air. AFM measurements were taken with a Multimode 8 AFM (Bruker, Billerica, MA) with NSG30 cantilevers (NT-MDT Spectrum Instruments, Limerick, Ireland) in net-repulsive tapping mode, choosing a target amplitude of 500 mV and a 5% peak offset.
X-ray scattering
The fibrillated dispersions of the Aβ peptides were placed in ultracentrifugation at 60,000 rpm for 1 h for fibril sedimentation. The thus-obtained pellets were then either transferred into a ring-shaped aluminum holder (2 mm thick and with a central hole of 1.5 mm diameter) or scraped out with standard capillaries from Hilgenberg (Malsfeld, Germany) of borosilicate glass with 3.8 or 1 mm outer diameter and 0.005 or 0.001 mm thickness and left to dry overnight. Small and wide angle x-ray scattering experiments (SAXS: qSAXS = 0.01–1.0 Å−1, WAXS: qWAXS = 0.4–3.2 Å−1) were performed in transmission mode using a SAXSLAB laboratory setup (Retro-F) equipped with an AXO microfocus x-ray source and an AXO multilayer x-ray optic (AXO Dresden GmbH, Dresden, Gemany) used as monochromator for Cu-Kα radiation (λ = 0.154 nm). A two-dimensional (2D) detector (PILATUS3 R 300K; DECTRIS, Baden, Switzerland) was used to record the 2D scattering patterns. The measurements were performed at room temperature in vacuum. The measured scattering intensities were corrected for background and absorption.
Single-molecule fluorescence spectroscopy
Sample preparation
All single-molecule experiments were performed in 25 mM sodium phosphate buffer with 150 mM NaCl at pH 7.5 unless otherwise stated. For single-molecule fluorescence spectroscopy, 5 μL of the Aβ solutions was diluted to peptide concentrations of approximately 0.8 nM (DOL 5%) or 80 pM (DOL 100%), depending on the degree of labeling. After dilution, the samples were placed on top of a coverslip for measurements. In case of an initial intensity loss due to adsorption of aggregates to the glass surface, the droplet was repeatedly renewed until no intensity change was observed within the first 5 min. The typical measurement time was 1 h.
Measurement setup
Fluorescence measurements were conducted on a confocal microscope capable to perform polarization and time-dependent fluorescence measurements. The home-built optical microscope equipped with a tunable fiber laser (1 ps pulse width, 80 MHz pulse frequency; TVIS, Toptica, Germany) and a diode laser (640 nm, LDH Series, 90 ps pulse width; PicoQuant, Berlin, Germany), a single-mode fiber (SuperK FD7; NKPhotonics, Cologne, Germany), and a 60X/1.20 W PlanApo objective (Zeiss) is described elsewhere (40). The sampled fluorescence was collected and split into a parallel and a perpendicular polarization component by a broadband polarization-dependent beam-splitter (Melles Griot, Rochester, NY) and focused onto two single-photon avalanche diodes (SPCM-AQR-14; Excelitas, Vaudreuil-Dorion, Canada). Photon arrival times were recorded by a single-photon-counting board with 25 ps time resolution (TimeHarp 260 nano; PicoQuant).
Single-molecule data analysis
Intensity time traces were analyzed by selecting photons of either detection channel by a threshold criterium of 0.2 photons/ms and combining them into single bursts. Only bursts containing more than twice the mean number of photons as detected for monomeric solutions were used to characterize oligomer solutions. To adjust for quenching effects, a fluorescence-lifetime-corrected photon number for each burst, Nphot, was determined using the equation Nphot = (N − Nbg)(τF,mon/τF,burst), where N is the number of detected burst photons, Nbg the fluorescent background of the buffer, and τF,mon/burst the average fluorescence lifetime of the monomer or burst, as used, e.g., by (31). The applied selection criteria were optimized to reliably select not only bright molecules but also less intense molecules with long dwell times in the focus volume. The oligomers were characterized by a relative photon number which was used to determine the apparent concentrations, c, by the equation , where d is the dilution, β the experimentally determined encounter rates, τd the average dwell time of all oligomers belonging to one oligomer species, ω0 the width of the focus volume, and NA the Avogadro number (25). For more details, we refer to Supporting Materials and Methods.
Time-dependent fluorescence decay and anisotropy
Time-dependent fluorescence anisotropy experiments are used to measure the time-dependent depolarization of the fluorescence light due to rotation of the emission dipole moment of the dye. The characteristic time constants of the anisotropy decay are capable to reveal hindered rotation of the attached fluorophore and hence provide information with respect to attractive fluorophore-peptide interactions, which are often referred to as stickiness. The time window of the experiment is the picosecond to nanosecond range, limited at short times by the pulse widths of the laser and the time response of the detectors (approximately 300 ps) and at long times by the fluorescence lifetimes. In case of unhindered rotation, the fluorescence anisotropy decays by a single exponential. As investigated in this study, the characteristic time constants for freely diffusive fluorophores, , were between 450 ps and 700 ps (well below 1 ns). The variations were related to the different sizes and shapes of the fluorophores. Fluorophores attached to peptides display a slower and potentially more complex decay. The rotation of the dye is either slowed down (by fast binding and unbinding to the peptide) or the dye molecules adsorbs to the peptide, which leads to a second decay component, , that can be related to the hydrodynamic radius, Rh, of the peptide-fluorophore complex following the Einstein-Smoluchowski equation of the rotational diffusion with , where Dr is the rotational diffusion coefficient, η the solvent viscosity, kb the Boltzmann constant, and T the temperature. To identify both processes, the normalized anisotropy was fitted using a two-component exponential tail-fit of the following form: . The ratio of the amplitudes refers to a second-rank order parameter, S2, with S2 = Af + p/(Af + Af + p), which can be further used to determine a rate of relative reorientation of the fluorophore in respect to the peptide, , with (41). The offset, y0, which reflects the presence of few large aggregates that rotate on timescales longer than the fluorescence lifetime, was subtracted if present.
Results and Discussion
In the following, the experiments that were performed to reveal the effects of N-terminus functionalization of the Aβ40 peptide with various fluorescent tags will be described and discussed in terms of 1) fibrillation kinetics, 2) fibril morphology, 3) LMW- and HMW-oligomeric species, and 4) fibrillar structure. Finally, the different oligomer sizes as found for Aβ40 and Aβ42 will be discussed.
Fibrillation kinetics of Aβ40
Two fundamental criteria to verify successful amyloid aggregation in time are the increase of β-sheet-rich structures and the final formation of bundles of extended β-sheets called fibrils. A common way to verify the β-sheet content of a sample and to follow the kinetics of β-sheet formation is to monitor the increase of the fluorescence of ThT. ThT is a small dye that binds to β-sheet-rich structures and shows enhanced fluorescence upon binding. Fig. 1 B displays the time-dependent increase of the ThT fluorescence in the presence of Aβ40wt without and with 5% of the fluorescently labeled peptides Atto488-Aβ40 or HL647-Aβ40. We found—within the accuracy of the measurement—no major differences for the lag times and the half times, T1/2, which are the characteristic times of the onset time of the rise of the fluorescence and the time at which 50% of the maximum of the ThT fluorescence is reached. This result is in agreement with other literature findings, e.g., (30).
Aβ40 fibril morphologies
To investigate the final morphologies of the mature fibrils, we used TEM and AFM imaging. The Fig.1, C–H shows representative images for Aβ40wt and two of the labeled peptides (more images can be found in Fig. S4). The TEM image of Aβ40wt displays elongated and twisted fibrils. Detailed analysis of the AFM image (Fig. 1 F) revealed a ribbon-like structure of ∼20 nm width and 5 nm height, twisted every 120 nm. All these structural properties are in agreement with the common features of amyloid fibrils found in literature (42, 43). Fibrils of Aβ40wt formed in the presence of 5% of labeled peptides show slightly different morphologies, ranging from long and straight—which is very similar to the wild-type form and was observed, e.g., for Atto488A-β40 (Fig. 1 D)—to thin and highly curved for HL647-Aβ40 (Fig. 1 E). Interestingly, the DOL did not affect the final morphology. Even with a DOL of 100%, Atto488-Aβ40 still formed elongated fibrils (see Fig. S4). Notably, the AFM image of HL647N-Aβ40 in Fig. 1 H displays, in addition to the fibrils, also unstructured and soft material, whereas for Atto488-Aβ40 and Aβ40wt, only a few spherical structures are seen (Fig. 1 G).
Oligomer size distributions of Aβ40
If the concept of nucleation and growth is applied to the process of fibrillation, the formation of nuclei is the rate-limiting step of aggregation (26, 27, 44). Oligomers below a critical size are either thermodynamically unstable (concept of one-step nucleation) or are stable but lacking in structure (concept of multistep nucleation). However, if the size and/or structure of the critical nucleus is reached, larger aggregates will form rapidly. With increasing time, all amyloid oligomers should participate in the fibrillar growth and finally be consumed. In terms of oligomer size distributions, this signifies that at short incubation times, at least three populations should be found: 1) a fraction of monomeric peptides, 2) a smaller fraction of (un)stable oligomers, and 3) one of rapidly aggregating larger oligomers. Toward longer incubation times, one would expect a larger fraction of prefibrillar to fibrillar aggregates and a small fraction of monomers that did not aggregate. Moreover, aggregates that did not participate in the fibrillar growth because of lacking structure might also continue to grow with time and form large but unstructured oligomers. This will be fraction number 4.
To this end, time-dependent intensity traces of Aβ40 peptides with different fluorophores attached were acquired at 0 h and after 4 h and 24 h of incubation time. TEM imaging revealed for Aβ40wt after 6 h spherical and prefibrillar oligomers only (see Fig. S2), which is in agreement with an estimated lag time of ∼10 h, as determined by ThT assays under identical conditions (37). In the time window of 2–6 h, single-molecule experiments revealed oligomer distributions that did only slightly change, indicative of a quasi-equilibrium status between monomers and oligomers. We chose in the following a constant incubation time of 4 h to investigate and compare the effect of fluorescence markers on oligomer size distributions.
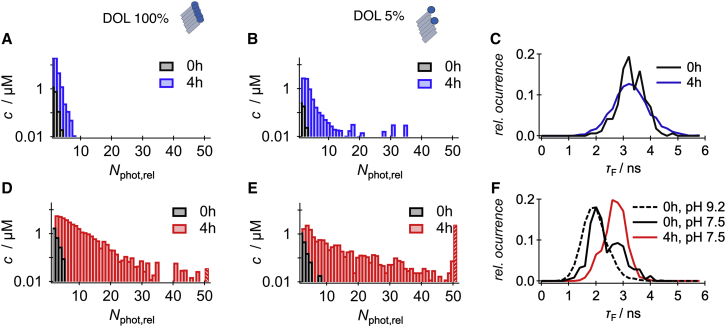
Fig. 2, A and D display the oligomer distributions as their concentration in function of monomer-normalized number of photons, , for Atto488-Aβ40 and HL647-Aβ40 peptides, respectively. The initial monomer peptide concentration was 40 μM. The distributions of the two peptides deviated strongly at any of the investigated sampling times: although Atto488-Aβ40 peptides did not display HMW oligomers, which are oligomers with values of Nphot,rel larger than 10, 20% of the oligomers of HL647-Aβ40 peptides were referred to be HMW oligomers. A reduction of the DOL to 5% allowed us to measure at higher peptide concentrations, which enhanced the probability to detect HMW aggregates (Fig. 2, B and E). Under this condition, the dye-dependent variations appeared even stronger: while only 5% of the oligomers of Atto488-Aβ40 were HMW oligomers, 59% of the oligomers of HL647-Aβ40 belonged to this category. Moreover, the detection probability of very large aggregates with Nphot,rel > 50 increased for HL647, which is in clear contrast to Atto488-Aβ40 peptides. Notably, the fluorescence properties of HL647 appeared to be more sensitive to aggregation: although monomers and oligomers of Atto488-Aβ40 had similar fluorescence lifetimes of 3.1–3.2 ns (Fig. 2 C), the fluorescence lifetimes of HL647-Aβ40 peptides significantly increased if oligomers were formed. In Fig. 2 F, the normalized distributions of the fluorescence lifetimes of HL647-Aβ40 are shown for two different conditions, before aggregation and after 4 h of incubation. The monomeric peptides were first dissolved in buffer at pH 9.2 and the fluorescence lifetime distribution displayed a single distribution with a peak maximum at 1.9 ns. After changing the pH to 7.5, the majority of molecules displayed a small shift of the fluorescence lifetime to 2.0 ns, which can be explained by a weak pH dependence of the emission properties of the fluorophore itself. In addition, aggregation was initiated, and some oligomers were instantaneously formed that displayed an average fluorescence lifetime of 2.8 ns. After 4 h of incubation, all fluorophores displayed a fluorescence lifetime of 2.8 ns. The shift arises from the solvatochromic property of the dye, which leads to longer fluorescence lifetimes in less polar environments. Hence, the increase in the fluorescence lifetime indicates an increased solvent shielding of the fluorophore, e.g., due to hydrophobic burial. Fibrils, however, showed an overall quenched fluorescence with a fluorescence lifetime of 1.3 and 1.0 ns for Atto488 and HL647, respectively. The fluorescence quenching is based on the Förster mechanism of close fluorophores and was earlier found to scale with fibril growth (20).
Figure 2.
The effect of the DOL of N-terminus-bound fluorophores on oligomer size distributions of Aβ40. (A, B, D, and E) Oligomer size distributions in terms of relative photon number, , using 100% (A and D) and 5% (B and E) of labeled Aβ40 peptides is shown. The oligomer distributions are shown before (black) and after (colored) 4 h of incubation for Atto488-Aβ40 (blue) and HL647-Aβ40 (red). The last bar in the histograms sums up all oligomers above Nphot,rel > 50. (C and F) The distributions of the fluorescence lifetimes, τF, before (black) and after (colored) 4 h of incubation for Atto488 (blue) and HL647 (red). The dashed line in (F) displays HL647-Aβ40 dissolved at pH 9.2; all other measurements were conducted at pH 7.5. The burst selection criteria were chosen such that the probability to include monomers in the histograms is very low. To see this figure in color, go online.
Finally, the question should be addressed how the relative photon numbers of the discussed oligomer distributions can be related to size. After correcting for different fluorescence lifetimes, the obtained photon number, Nphot,rel, is expected to be proportional to the aggregation number and to depend on the individual dwell times in the focus volume. Hence, any increase in the number of fluorophores as well as the reduction of the translational diffusion coefficient will lead to enhanced photon counts. However, the unexpectedly narrow distribution of the molecular brightness of the oligomers led to an apparent linear relationship between the average diffusion time, τd, and Nphot,rel, which justifies relating Nphot,rel directly to size: Rh ∝ τd ∝ Nphot,rel (see Fig. S3 for more details).
HMW oligomers of Aβ40
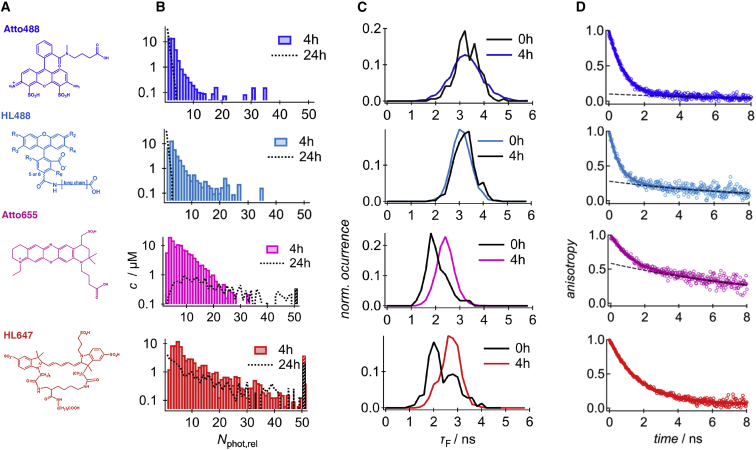
In the following, the effects of four different fluorophores were investigated. To enhance the detection of HMW oligomers, which we call oligomers with Nphot,rel larger than 10, a low DOL of 5% was used. Fig. 3 displays the result and links characteristic features in terms of oligomer size distributions, the shift of fluorescence lifetimes upon aggregation, and the time-resolved fluorescence anisotropy of monomeric peptides. Starting with the oligomer size distributions, we found for Atto488-Aβ40 peptides a narrow distribution that can be characterized by the mean value (first moment, μ1) of = 2.7 and a width (root of the second moment, ) of 4.7 (see Table 1). Just very rarely, some large aggregates with Nphot, rel > 50 were detected. After 24 h of incubation, the detected oligomer sizes were again strongly reduced and comparable to the distribution before fibrillation (see also Fig. 2 B). Oligomers of HL488-Aβ40 peptides had a slightly increased mean value of 4.1 . The width of the distribution was 6.3 , but like for Atto488, most of the oligomers were consumed by the end of the fibrillation process. In comparison, Atto655-Aβ40 formed larger oligomers with a mean of 7.4 and a distribution width of 8.8 . HL647-Aβ40 displayed a clear shift to even larger aggregates. The mean was 10.6 and the width 14.9 . Very large aggregates of above 50 were frequently detected but not included in the calculation of the moments of the distribution. It can be assumed that these large aggregates are no oligomers in the narrow sense. Notably, the larger aggregates maintained in the solution, even after 24 h of incubation, whereas the fraction of smaller aggregates seemed to be reduced. Fig. 3 C compares the fluorescence lifetimes of the detected bursts before and after 4 h of incubation for all investigated fluorophores. It appeared that dyes as Atto655 and HL647 that induced HMW aggregates also displayed a clear shift to higher fluorescence lifetimes during aggregation. We refer this shift to hydrophobic interactions of the dyes and to solvent shielding upon aggregation. In contrast, the fluorescence lifetime of Atto488 and HL488 did not show any difference during aggregation, which reveals a more hydrophilic nature and indicates that Atto488 and HL488 stayed exposed to the solvent during aggregation.
Figure 3.
The effect of N-terminus-bound fluorophores on oligomer size distributions of Aβ40 at low DOL. (A) Chemical structures as provided by the manufacturer (Atto dyes) or by (38) and (20) for HL488 and HL647, (B) oligomer size distributions in terms of relative photon numbers of labeled Aβ40 peptides (DOL 5%) after 4 and 24 h, (C) fluorescence lifetime distributions before and after 4 h of fibrillation, and (D) time-dependent anisotropy of the fluorescent labels before fibrillation. To see this figure in color, go online.
Table 1.
Characteristics of Labeled Aβ40 Peptides and Their Oligomeric Aggregates
| Peptide | Oligomer Size Distribution (DOL 5%) |
Fluorescence Lifetime |
Anisotropy Decay Characteristics |
||||
|---|---|---|---|---|---|---|---|
| μ1 | /ns | /ns | S2 | /ns | /ns−1 | ||
| Atto488-Aβ40 | 2.7 ± 0.2 | 4.7 ± 0.4 | 3.2 ± 0.1 | 3.1 ± 0.1 | 0.12 ± 0.01 | 0.51 ± 0.02 | 0.288 ± 0.005 |
| HL488-Aβ40 | 4.1 ± 0.2 | 6.3 ± 0.4 | 3.2 ± 0.1 | 3.1 ± 0.1 | 0.28 ± 0.01 | 0.68 ± 0.03 | 0.168 ± 0.013 |
| Atto655-Aβ40 | 7.4 ± 0.2 | 8.8 ± 0.4 | 1.9 ± 0.1 | 2.4 ± 0.1 | 0.68 ± 0.03 | 0.54 ± 0.09 | 0.081 ± 0.023 |
| HL647-Aβ40 | 10.6 ± 0.2 | 14.9 ± 0.4 | 2.0 ± 0.1 | 2.8 ± 0.1 | – | 1.77 ± 0.02a | 0.094 ± 0.003a |
Shown are the first moments, , and the roots of the second moments, , of the oligomer size distributions, the average fluorescence lifetimes, and parameters describing the anisotropy decays. The initial monomer concentration was 40 μM.
For HL647-Aβ40, the parameters refer to a single exponential decay.
The normalized time-dependent anisotropy measurements of Fig. 3 D display the time-dependent depolarization of the fluorescence emission of the dyes, which can be due either to fast orientation fluctuations of the dipolar moment of the dye around its own axis (fast component) or to the rotation of the dye-peptide complex (slow component). For Atto488-Aβ40 peptides, the majority displayed a fast decay, with τr = 510 ± 20 ps. However, there was also a small fraction of 12% with a longer decay time of 7.00 ± 0.18 ns, which refers to a hydrodynamic radius, Rh, of ∼1.9 nm. This value is slightly larger than the hydrodynamic radius of 1.6–1.7 nm found for unlabeled Aβ40 peptides (45, 46). HL488-Aβ40 peptides showed a similar behavior: in addition to a fast anisotropy decay, 28% of the fluorophores displayed a decay time on the order of the fluorophore-peptide complex. Comparing the corresponding relative rates of reorientation, Atto488 dyes displayed with 0.29 ns−1 the higher rate (see Table 1). In the same manner, Atto655-Aβ40 showed two different decay times, but the fraction of fluorophore-peptide complexes was much larger as compared to Atto488 and HL488. 68% of Atto655-Aβ40 displayed a slow decay with 6.08 ± 0.05 ns. This finding correlates with strongly reduced rates of reorientation of 0.08 ns−1. We assume that the enhanced interaction is mediated by the increased hydrophobicity of the dye. In contrast to all other dyes, HL647-Aβ40 peptides apparently displayed a single decay only, which might be related to their different chemical structure. However, the time constant of the decay was 1.77 ± 0.02 ns, which is larger than the decay of 0.89 ± 0.05 ns as observed for peptides at elevated pH (pH 9.2). Hence, at pH 7.5, all fluorophores strongly interact with the Aβ40 peptides, and the apparent rate of reorientation is 0.09 ns−1 only.
Fibrillar structure of Aβ40
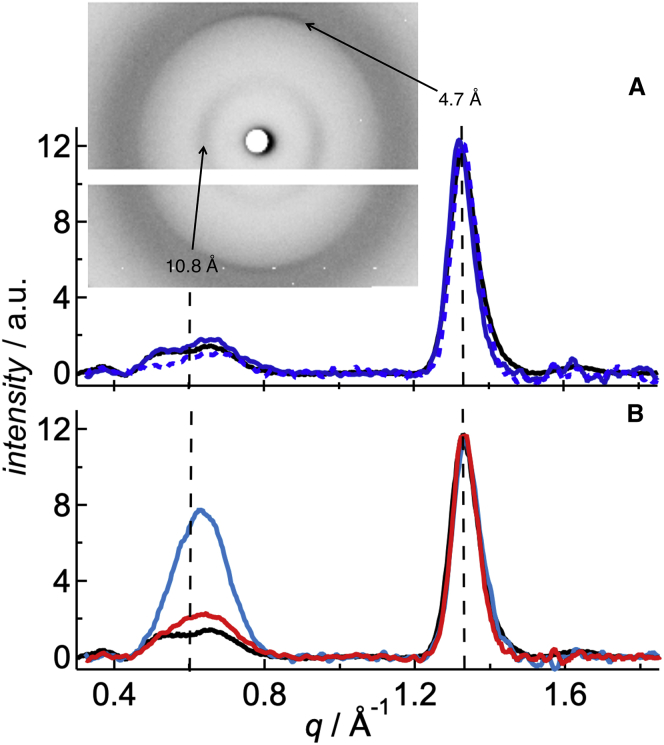
To investigate whether the enhanced interaction of fluorophores with the peptide has any impact to the fibrillar structure, x-ray scattering experiments were conducted. XRD can be used for determining the atomic or molecular structure of crystals or crystal-like samples. Relative distances and absolute positions of the first neighboring atoms can be deduced from the scattering intensity and scattering directions of the x-rays deviated by the atomic electronic densities. The first XRD measurements of Aβ amyloid fibrils trace back to the late 1960s (47, 48) and revealed a typical cross-β structure, with β-sheets arranged in parallel to the fibril axis and their constituent β-strands perpendicular to the fibril axis. The latter resulted in 4.7–4.8 Å meridional reflections and a 10 Å equatorial reflection, respectively, which are considered the hallmarks of a cross-β structure (49). All amyloid proteins or peptides showed indeed very similar XRD patterns (42). In Fig. 4, just the meaningful WAXS region between q = 0.4 and 1.8 Å−1 of the curves is reported after amorphous background subtraction and normalization with respect to the areas below the second peak, which indicate the crystalline content because of β-sheet formation. The full accessible q-range is shown in Fig. S5. The vertical lines in Fig. 4 indicate the peak positions of Aβ40wt, which are related to the β-strand distances along the fibril axis and the β-sheet distances perpendicular to it. The first refers to the peak at q = 1.32 Å−1, indicating a distance of 4.75 Å between β-strands, and the latter to the peak at 0.58 Å−1, which is related to a distance of 10.8 Å between β-sheets. The equatorial reflections appear broader and weaker than their meridional equivalents, implying a much lower crystalline order in directions perpendicular to the fiber axis than parallel to it. The scattering intensity of the labeled peptides displayed the same two peaks with q-values ranging from 1.32 to 1.34 Å−1 for the meridional reflections and from 0.63 to 0.65 Å−1 for the equatorial ones, corresponding to distances ranging between 4.70 and 4.75 Å and 9.6–10.8 Å, respectively. More precisely, all the HiLyte-labeled samples (HL488-Aβ40 and HL647-Aβ40) showed a peak maximum that refers to a distance of ∼10.0 Å, which is significantly lower compared to Aβ40wt. In contrast, Atto488-Aβ40 peptides showed at a low DOL of 5% no difference from Aβ40wt. At an increased DOL of 100%, however, a shift to even higher q-values with an average inter-β-sheet distance of 9.6 Å was found. The common reflection for the distances of the β-strands (4.75 Å) indicates very similar, if not identical, nuclei. In other words, the fluorophores did not affect the basic unit cell of the nuclei because this structure would propagate during growth. However, the lateral order of the fibrillar bundles was modified, which is depicted by the dye-dependent differences of the other peak.
Figure 4.
The effect of N-terminus-bound fluorophores on the fibrillar structure of Aβ40. Relative scattering intensity of the amyloid fibrils in the range of 0.4 and 1.8 Å−1 after azimuthal integration, background subtraction, and renormalization with respect to the crystalline β-sheet content. (A) Aβ40wt (black line) and Atto488-Aβ40 (blue solid line for DOL 5% and blue dashed line for DOL 100%). (B) Aβ40wt (black line) and HL488-Aβ40 (blue line) and HL647-Aβ40 (red line), both DOL 5%. Inset: 2D image of Atto488-Aβ40 (DOL 5%) with the typical meridional and equatorial peaks reflections at 4.75 and 10.8 Å, respectively. To see this figure in color, go online.
Comparison of Aβ40 with Aβ42
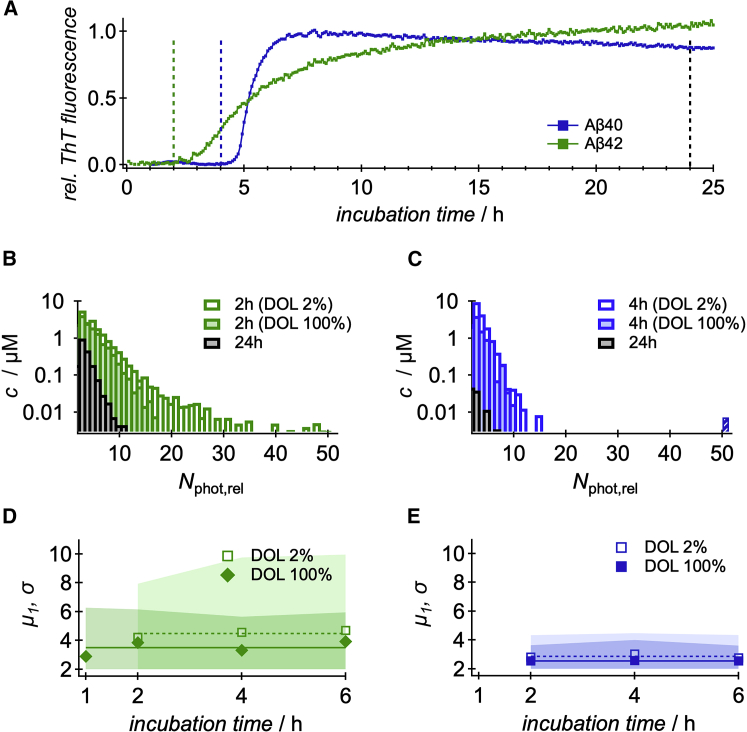
Because of the excellent labeling properties of Atto488, this fluorescence tag was further used to compare the oligomer distributions of Aβ40 and Aβ42. As for Aβ40, this fluorescence marker did not affect the kinetics of Aβ42 fibrillation nor display pronounced interactions with the Aβ42 peptide (see Fig. S6). The time-dependent anisotropy revealed for Atto488-Aβ42 a fast decay of 0.76 ± 0.3 ns and a slow component of 16%, with a decay time of 7.7 ± 0.9 ns. The slightly longer decay time of the slow component corresponds to an increased hydrodynamic radius of approximately 2.0 nm for the fluorophore-peptide complex. The resulting rate of reorientation for the bound fluorophore was 0.187 ± 0.013 ns−1, which is comparable to the properties of Atto488-Aβ40 or HL488-Aβ40 (see Table 1). The overall fibrillation of Aβ42 peptides appears to be much faster compared to Aβ40 (50). To slow down the fibrillation and to meaningfully compare the oligomeric states of the two Aβ peptides, the initial monomer concentrations were reduced to 10 μM. For this concentration, the lag times of Aβ42 and Aβ40 were in the range of 1.8 ± 0.9 and 5.8 ± 1.1 h, respectively (see Fig. 5 A). It should be noted that micromolar concentrations of the fluorophore ThT accelerate the fibrillation of Aβ40 (37, 51), and the determined lag times are a lower limit for ThT-free aggregation. For this reason, the aggregation of Aβ40 and Aβ42 was compared for a wide range of incubation times. Interestingly, the obtained oligomer distributions for Aβ40 and Aβ42 with respect to Nphot,rel did not vary for the investigated time range, which is displayed by almost constant mean values and SDs (see Fig. 5, D and E). The plots in Fig. 5, B and C display three representative oligomer size distributions for each peptide: for low and high DOL before the growth phase and for high DOL after the growth phase. The use of a low DOL improves the probability to detect large oligomers, as discussed earlier. The comparison of the distributions reveals for Aβ42 peptides a much broader oligomer size distribution than for Aβ40: although the probability of Aβ40 to exceed 10 is ∼0.1%, a significant fraction (6%) of Aβ42 oligomers are larger. The time-averaged first and second moments of the distributions are summarized in Table 2 and show always larger values for Aβ42, e.g., the low DOL samples revealed oligomers with a mean size of 2.88 and 4.49 for Aβ40 and Aβ42, respectively. The widths of the distributions were 3.30 and 6.35 . It should be noted that the fluorescence lifetimes of the Aβ42 oligomers remained unaffected by the aggregation (τF = 3.2 ns), which is in contrast to the oligomers induced by HL647 or Atto655 for Aβ40 (see Fig. 3). After 24 h of fibrillation, TEM images displayed for both Aβ peptides elongated, mature fibrils without any effect of the DOL to the fibril morphologies (see Fig. S7). However, after 24 h, most of the HMW oligomers of Aβ42 were consumed, but a significant fraction of small oligomers with a mean of 2.78 and reduced width of 3.58 could still be found (Fig. 5 B). The latter is not the case for Aβ40, indicating a more efficient fibrillation.
Figure 5.
Comparison of Aβ40 and Aβ42. (A) Time dependence of the ThT fluorescence for Aβ40 (blue) and Aβ42 (green) at 10 μM monomer concentration. (B and C) Oligomer size distributions of Atto488-Aβ42 (B) and Atto488-Aβ40 (C) in terms of relative photon number, , for different DOLs and for two characteristic times: before the growth phase (colored bars) and after 24 h of incubation (black bars). (D and E) Mean values and SDs of the oligomer size distributions for Atto488-Aβ42 (D) and Atto488-Aβ40 (E) during early aggregation. The mean values are depicted as filled (DOL 100%) and open symbols (DOL 2%), whereas the SDs are represented by dark- (DOL 100%) and light- (DOL 2%) shaded areas, respectively. The values are listed in Table 2. To see this figure in color, go online.
Table 2.
Oligomer Size Distributions of Aβ40 and Aβ42 Peptides: Time Averages of the First Moments,, and of the Root of the Second Moments, and
| Peptide | Oligomer Size Distribution (DOL 100%) |
Oligomer Size Distribution (DOL 2%) |
||||
|---|---|---|---|---|---|---|
| μ1 | σ | μ1 | σ | |||
| Atto488-Aβ40 | 2.55 ± 0.03 | 1.20 ± 0.12 | 2.83 ± 0.06 | 2.88 ± 0.08 | 1.51 ± 0.04 | 3.30 ± 0.08 |
| Atto488-Aβ42 | 3.50 ± 0.24 | 2.50 ± 0.30 | 4.42 ± 0.11 | 4.49 ± 0.14 | 4.72 ± 0.50 | 6.53 ± 0.45 |
The initial monomer concentration was 10 μM.
Summary
In this study, the impact of N-terminus labeling of Aβ40 peptides was investigated using common fluorophores with different spectral properties and hydrophobicities. In the following section, the experimental findings for each dye are summarized and briefly discussed.
Atto488-Aβ40 and Atto655-Aβ40
Fluorescence anisotropy measurements revealed that most of the bound Atto488 fluorophores display unhindered rotational motion, and only a low percentage of 12% of the dye molecules show cooperative rotation with the peptide. This finding correlates with the successful formation of elongated, mature fibrils, which were very similar to the morphology of Aβ40wt fibrils as found by TEM and AFM imaging. XRD revealed interstrand and inter-β-sheet distances identical to Aβ40wt. Atto488-Aβ40 formed only small oligomers—as found by single-molecule fluorescence experiments—which were consumed during fibrillation. Hence, Atto488-Aβ40 is an excellent candidate to study oligomerization and fibrillation. Atto655 resembles a hydrophilic dye in the red spectral range. Surprisingly, the fluorophore tended to stick to the peptide at the buffer conditions used, and the peptide formed shorter and thinner fibrils compared to Aβ40wt or Atto488-Aβ40. In addition, Atto655-Aβ40 supported the aggregation into HMW oligomers and hence should not be used for peptide labeling.
HL488-Aβ40 and HL647-Aβ40
HL488-Aβ40 and HL647-Aβ40 are two peptides that are commercially available and therefore often used in literature, e.g., (19, 30, 31, 32, 33, 38). A clear drawback of the use of these dyes is their pH dependence in terms of net-attractive interactions with the Aβ40 peptide itself, which was discussed for HL647, but could been observed somewhat more weakly for HL488 as well. At a pH of 7.5, 28% of the HL488 dyes and basically all of HL647 displayed slower rotational motion compared to pH 9.2. Moreover, HL647-Aβ40 formed large oligomers and promoted the formation of very large aggregates with Nphot,rel > 50, whereas for HL488-Aβ40, mainly small oligomers with a mean value of 4.1 were detected. The latter is in agreement with the small oligomers of up to six monomers, which were found, e.g., by photobleaching experiments of surface-attached HL488-Aβ40 peptides (19). The tendency of HL647-Aβ40 to form small fractions of large aggregates at the early stage of fibrillation is mentioned in literature as a side note (32). Interestingly, the differences in terms of interaction and oligomer sizes could be also linked to a different morphology of the fibrils after 24 h of fibrillation. Although HL488-Aβ40 formed straight and extended fibrils similar to Atto488-Aβ40, the fibrils of HL647-Aβ40 seemed to be thin and strongly bended. Delayed fibrillation kinetics could not be the reason for the different morphologies because standard ThT assays used to investigate the kinetics of β-sheet formation displayed no delay for HL647-Aβ40 compared to Aβ40wt. And since the local structure of the fibril as investigated by WAXS measurements did not show any deviation from the wild type, we concluded that for HL647-Aβ40, not the nucleation but the fibril growth is modified, e.g., by termination of the growth ends and hindered lateral stacking. Hence, care must be taken if mixtures of fluorophores are used, e.g., for two-color detection or cross-seeding experiments (28, 30, 52), or other highly hydrophobic dyes are attached, e.g., Atto647N (29, 53, 54).
Atto488-Aβ42
Aβ42 functionalized with Atto488 shows almost the same low level of fluorophore-peptide interactions as found for Aβ40 peptides. Hence, the HMW oligomers formed by Atto488-Aβ42 are unlikely to be induced by the fluorophore and indicate alternative pathways of fibrillation as proposed earlier, e.g., in (18). This finding is further supported by the missing shift in fluorescence lifetime of the oligomers and the consumption of the HMW oligomers during fibrillation. These properties are in clear contrast to fluorophore-induced HMW oligomers, e.g., of HL647-Aβ40 and Atto655-Aβ40.
Conclusions
Combining the results discussed above, we conclude that the formation of amyloid oligomers is very sensitive to the chosen fluorophore, whereas the fibrillation kinetics and the structures of the final fibrils are only marginally affected. In particular, the occurrence of HMW oligomers of N-terminally labeled Aβ40 peptides can be linked to hydrophobic properties of the attached dye, which is indirectly pictured by attractive fluorophore-peptide interactions and by reduced fluorescence quenching upon aggregation. Because HMW oligomers of Aβ40 tend to maintain in the solution after fibrillation, we further conclude that they do not contribute to the fibrillation. Consequently, LMW oligomers form the nucleus and control fibrillar growth, which explains the similar fibrillation kinetics of labeled peptides as well as the same internal structures along the axis of fibrillar growth (4.75 Å peak). Differences were found on larger scales, ranging from the distance between β-sheets (10 Å peak) to the overall morphology of the fibrils on the 100 nm scale (TEM images). Consequently, the ideal fluorophore that should be used to investigate oligomeric states of Aβ should not lead to HMW oligomers for Aβ40, which would superimpose with those responsible for fibrillation. From our studies, we found Atto488 and also HL488 to be promising candidates for such kinds of labeling functionalization. We demonstrated exemplarily for Atto488 that the low fluorophore-peptide interactions as detected for Aβ40 also hold for Aβ42. Hence, the dye was well suited to detect on-pathway HMW oligomers formed by Aβ42, and a comparison between Aβ40 and Aβ42 concerning oligomer distributions during fibrillation became feasible. We expect that Atto488 and HL488 may also applicable for other amyloid peptides.
Author Contributions
M.O. designed the research. J.W., S.D.S., and B.V. performed the research. J.W. and S.D.S. contributed equally. J.W., S.D.S., and M.O. analyzed the data. M.O., S.D.S., J.W., and J.B. wrote the manuscript.
Acknowledgments
The authors acknowledge Dr. Sven Rothemund (University Leipzig) for peptide synthesis, Monika Baumann (Martin-Luther-University Halle-Wittenberg) for initial TEM images, and Prof. Thomas Thurn-Albrecht (Martin-Luther-University Halle-Wittenberg) for providing access to the SAXS equipment and fruitful discussion.
The study was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB TRR 102, projects B12 and A06).
Editor: Jochen Mueller.
Footnotes
Supporting Materials and Methods and seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)34505-3.
Supporting Material
References
- 1.Harrison R.S., Sharpe P.C., Fairlie D.P. Amyloid peptides and proteins in review. Rev. Physiol. Biochem. Pharmacol. 2007;159:1–77. doi: 10.1007/112_2007_0701. [DOI] [PubMed] [Google Scholar]
- 2.Seubert P., Vigo-Pelfrey C., Swindlehurst C. Isolation and quantification of soluble Alzheimer’s beta-peptide from biological fluids. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 3.Knowles T., Vendruscolo M., Dobson C. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell. Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 4.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Lazo N.D., Samir K., Teolow B.T. Amyloid Proteins: The Beta Sheet Conformation and Disease. Chapter 17. Wiley-Blackwell; 2008. The amyloid β protein; pp. 384–491. [Google Scholar]
- 7.Dahlgren K.N., Manelli A.M., LaDu M.J. Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 8.Walsh D.M., Hartley D.M., Teplow D.B. Amyloid beta-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 9.Caughey B., Lansbury P.T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Biochem. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 10.Arispe N., Pollard H.B., Rojas E. Giant multilevel cation channels formed by Alzheimer disease amyloid beta-protein [A beta P-(1-40)] in bilayer membranes. Proc. Natl. Acad. Sci. USA. 1993;90:10573–10577. doi: 10.1073/pnas.90.22.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arispe N., Pollard H.B., Rojas E. Beta-amyloid Ca(2+)-channel hypothesis for neuronal death in Alzheimer disease. Mol. Cell. Biochem. 1994;140:119–125. doi: 10.1007/BF00926750. [DOI] [PubMed] [Google Scholar]
- 12.Glenner G.G., Wong C.W. Alzheimer’s disease and Down’s syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem. Biophys. Res. Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 13.Masters C.L., Simms G., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vassar R., Kovacs D.M., Wong P.C. The β-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lue L.F., Kuo Y.M., Rogers J. Soluble amyloid β peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am. J. Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLean C.A., Cherny R.A., Masters C.L. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann. Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 17.Bitan G., Lomakin A., Teplow D.B. Amyloid beta-protein oligomerization: prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J. Biol. Chem. 2001;276:35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- 18.Bitan G., Kirkitadze M.D., Teplow D.B. Amyloid beta -protein (Abeta) assembly: abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding H., Wong P.T., Steel D.G. Determination of the oligomer size of amyloidogenic protein β-amyloid(1-40) by single-molecule spectroscopy. Biophys. J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinn S.D., Dalgarno P.A., Penedo J.C. Real-time probing of β-amyloid self-assembly and inhibition using fluorescence self-quenching between neighbouring dyes. Mol. Biosyst. 2014;10:34–44. doi: 10.1039/c3mb70272c. [DOI] [PubMed] [Google Scholar]
- 21.Harper J.D., Lansbury P.T., Jr. Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 22.Hatai J., Motiei L., Margulies D. Analyzing amyloid beta aggregates with a combinatorial fluorescent molecular sensor. J. Am. Chem. Soc. 2017;139:2136–2139. doi: 10.1021/jacs.6b10809. [DOI] [PubMed] [Google Scholar]
- 23.Dukes K.D., Rodenberg C.F., Lammi R.K. Monitoring the earliest amyloid-β oligomers via quantized photobleaching of dye-labeled peptides. Anal. Biochem. 2008;382:29–34. doi: 10.1016/j.ab.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Tjernberg L.O., Pramanik A., Rigler R. Amyloid β-peptide polymerization studied using fluorescence correlation spectroscopy. Chem. Biol. 1999;6:53–62. doi: 10.1016/S1074-5521(99)80020-9. [DOI] [PubMed] [Google Scholar]
- 25.Orte A., Birkett N.R., Klenerman D. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garai K., Sahoo B., Maiti S. Quasihomogeneous nucleation of amyloid beta yields numerical bounds for the critical radius, the surface tension, and the free energy barrier for nucleus formation. J. Chem. Phys. 2008;128:045102. doi: 10.1063/1.2822322. [DOI] [PubMed] [Google Scholar]
- 27.Sahoo B., Nag S., Maiti S. On the stability of the soluble amyloid aggregates. Biophys. J. 2009;97:1454–1460. doi: 10.1016/j.bpj.2009.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wennmalm S., Chmyrov V., Tjernberg L. Highly sensitive FRET-FCS detects amyloid β-peptide oligomers in solution at physiological concentrations. Anal. Chem. 2015;87:11700–11705. doi: 10.1021/acs.analchem.5b02630. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y., Tian S., Hong T. Kinesin-1 inhibits the aggregation of amyloid-β peptide as detected by fluorescence cross-correlation spectroscopy. FEBS Lett. 2016;590:1028–1037. doi: 10.1002/1873-3468.12137. [DOI] [PubMed] [Google Scholar]
- 30.Narayan P., Orte A., Klenerman D. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-β(1-40) peptide. Nat. Struct. Mol. Biol. 2011;19:79–83. doi: 10.1038/nsmb.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schauerte J.A., Wong P.T., Gafni A. Simultaneous single-molecule fluorescence and conductivity studies reveal distinct classes of Abeta species on lipid bilayers. Biochem. 2010;49:3031–3039. doi: 10.1021/bi901444w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson R.D., Schauerte J.A., Steel D.G. Direct observation of single amyloid-β(1-40) oligomers on live cells: binding and growth at physiological concentrations. PLoS One. 2011;6:e23970. doi: 10.1371/journal.pone.0023970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esbjörner E.K., Chan F., Kaminski Schierle G.S. Direct observations of amyloid β self-assembly in live cells provide insights into differences in the kinetics of Aβ(1-40) and Aβ(1-42) aggregation. Chem. Biol. 2014;21:732–742. doi: 10.1016/j.chembiol.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie A.J., Brewer D.B. Optical properties of amyloid stained by Congo red: history and mechanisms. Micron. 2009;40:285–301. doi: 10.1016/j.micron.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Harry L. Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Prot. Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viet M.H., Nguyen P.H., Li M.S. Effect of the english familial disease mutation (H6R) on the monomers and dimers of Aβ40 and Aβ42. ACS Chem. Neurosci. 2014;5:646–657. doi: 10.1021/cn500007j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adler J., Scheidt H.A., Huster D. Local interactions influence the fibrillation kinetics, structure and dynamics of Aβ(1-40) but leave the general fibril structure unchanged. Phys. Chem. Chem. Phys. 2014;16:7461–7471. doi: 10.1039/c3cp54501f. [DOI] [PubMed] [Google Scholar]
- 38.Jungbauer L.M., Yu C., LaDu M.J. Preparation of fluorescently-labeled amyloid-beta peptide assemblies: the effect of fluorophore conjugation on structure and function. J. Mol. Recognit. 2009;22:403–413. doi: 10.1002/jmr.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinn M.K., Gnan N., McManus J.J. How fluorescent labelling alters the solution behaviour of proteins. Phys. Chem. Chem. Phys. 2015;17:31177–31187. doi: 10.1039/c5cp04463d. [DOI] [PubMed] [Google Scholar]
- 40.Roos M., Ott M., Saalwächter K. Coupling and decoupling of rotational and translational diffusion of proteins under crowding conditions. J. Am. Chem. Soc. 2016;138:10365–10372. doi: 10.1021/jacs.6b06615. [DOI] [PubMed] [Google Scholar]
- 41.Szabo A. Theory of fluorescence depolarization in macromolecules and membranes. J. Chem. Phys. 1984;81:150–167. [Google Scholar]
- 42.Sunde M., Serpell L.C., Blake C.C. Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 43.Fändrich M., Meinhardt J., Grigorieff N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion. 2009;3:89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crespo R., Rocha F.A., Martins P.M. A generic crystallization-like model that describes the kinetics of amyloid fibril formation. J. Biol. Chem. 2012;287:30585–30594. doi: 10.1074/jbc.M112.375345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danielsson J., Jarvet J., Gräslund A. Two-site binding of beta-cyclodextrin to the Alzheimer Abeta(1-40) peptide measured with combined PFG-NMR diffusion and induced chemical shifts. Biochem. 2004;43:6261–6269. doi: 10.1021/bi036254p. [DOI] [PubMed] [Google Scholar]
- 46.Tseng B.P., Esler W.P., Maggio J.E. Deposition of monomeric, not oligomeric, Abeta mediates growth of Alzheimer’s disease amyloid plaques in human brain preparations. Biochem. 1999;38:10424–10431. doi: 10.1021/bi990718v. [DOI] [PubMed] [Google Scholar]
- 47.Eanes E.D., Glenner G.G. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 48.Bonar L., Cohen A.S., Skinner M.M. Characterization of the amyloid fibril as a cross-beta protein. Proc. Natl. Acad. Sci. USA. 1969;131:1373–1375. doi: 10.3181/00379727-131-34110. [DOI] [PubMed] [Google Scholar]
- 49.Geddes A.J., Parker K.D., Beighton E. “Cross-beta” conformation in proteins. J. Mol. Biol. 1968;32:343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 50.Jarrett J.T., Berger E.P., Lansbury P.T., Jr. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 51.Xue C., Lin T.Y., Guo Z. Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R. Soc. Open Sci. 2017;4:160696. doi: 10.1098/rsos.160696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang C.-C., Althaus J.C., Gafni A. Synergistic interactions between Alzheimer’s Aβ40 and Aβ42 on the surface of primary neurons revealed by single molecule microscopy. PLoS One. 2013;8:e82139. doi: 10.1371/journal.pone.0082139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson R.D., Schauerte J.A., Gafni A. Single-molecule imaging reveals aβ42:aβ40 ratio-dependent oligomer growth on neuronal processes. Biophys. J. 2013;104:894–903. doi: 10.1016/j.bpj.2012.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altman R., Ly S., Voss J.C. Protective spin-labeled fluorenes maintain amyloid beta peptide in small oligomers and limit transitions in secondary structure. Biochim. Biophys. Acta. 2015;1854:1860–1870. doi: 10.1016/j.bbapap.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vivekanandan S., Brender J.R., Ramamoorthy A. A partially folded structure of amyloid-beta(1-40) in an aqueous environment. Biochem. Biophys. Res. Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rose A.S., Bradley A.R., Rose P.W. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 2018;34:3755–3758. doi: 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng J., Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins. 2011;79:161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.