Abstract
FlowNMR has the aim of continuously monitoring processes that occur in conditions that are not compatible with being carried out within a closed tube. However, it is sample intensive and not suitable for samples, such as proteins or living cells, that are often available in limited volumes and possibly low concentrations. We here propose a dialysis-based modification of a commercial flowNMR setup that allows for recycling the medium while confining the sample (proteins and cells) within the active volume of the tube. This approach is demonstrated in the specific cases of in-cell NMR and protein-based ligand studies.
Introduction
NMR is the method of choice for characterizing proteins and their interactions (1, 2, 3, 4, 5, 6) and to study them in their functional environment (i.e., within living cells) (7, 8, 9). NMR is mostly conceived as a batch technique in which each individual sample is closed in a tube and has no interactions with the outer environment besides a limited exchange of volatile components. However, several experimental situations dramatically benefit from flow operation, like the characterization of catalytic events (10, 11, 12, 13).
NMR applications to living cells requires that the cells remain vital and metabolically active during the whole duration of the experiment to avoid the fact that alterations of metabolism caused by starvation impact the significance of the data; for this reason, NMR in living cells has been mostly limited to experiments of a few hours (14, 15). An NMR flow system can provide nutrients, remove byproducts, and ensure cell viability for extended periods of time. Most existing NMR bioreactor setups are focused on the metabolic flux analysis of living cells (16). These setups are usually designed for low-field NMR using 10-mm or wider-bore magnets and/or optimized for low cell densities in the NMR-active volume (17, 18, 19, 20). Bioreactors for in-cell NMR studies of proteins have been reported, consisting of custom-made flow systems adapted to 5-mm NMR tubes (21, 22, 23). These setups require the cells to be supported by a gel matrix, imposing a limit to the high cell densities necessary for in-cell NMR experiments.
Another application that would dramatically benefit from a flowNMR strategy is drug screening. Actually, the strategy of screening ligand libraries flowing them sequentially in the same immobilized target sample (target immobilized NMR screening (TINS)) is well established (24). In TINS, the target protein is bound to a solid support, and protein-ligand interactions are observed because differences arise in the ligand signal intensity with respect to a reference tube that does not contain the target. Although this is efficient for screening, the protein-level information detail that is important for further drug development (25) cannot be obtained.
Here, we describe the development of a flow apparatus and test its application and potential for 1) in-cell NMR and 2) structure-activity relationships in drug discovery.
Materials and Methods
Bioreactor setup
The optimal flow rate to maximize the exchange of chemicals was determined by flowing a 20 mmol dm−3 aqueous solution of ethanol in the outer NMR tube (of the InsightMR) filled with a 20 mmol dm−3 methanol solution. A flow of 50 μL/min provided the highest rate of exchange with a 1 MDa membrane. The flow of 50 μL min−1 gives a net velocity within the membrane of ∼0.4 cm s−1. The effect of the net flow can impact the acquisition of gradient-based experiments and is reminiscent of the convection effects, which are well known in diffusion-ordered spectroscopy (26, 27). Considering a simple experiment with two gradients with the same length (τ ∼ 1 ms), same intensity (G0 ∼ 5 mT cm−1), and opposite sign, separated by a given time (t1), the phase shift induced on a coherence of order −1 by a flow of speed v corresponds to Δφ = γG0vτt1. For a one-dimensional (1D) experiment with excitation sculpting, Δφ is expected to be ∼4°. We here note that this effect could become important in the case of an 1H-15N-heteronuclear single quantum coherence (HSQC) experiment aimed at observing a small molecule (which can be thus located within the inner part of the membrane). The Δφ can easily exceed 360°; however, the small ratio of the internal volume over the total (3.6%) reduces the amount of signal that can actually be affected. The effect of the flow on the larger molecules (which cannot access the inner part of the membrane) can be estimated on the basis of the velocity that would be attained in the absence of the membrane (i.e., 0.008 cm s−1), which gives a negligible phase shift in any experiment of practical interest. A closed glass capillary filled with 98% D2O was inserted to provide the lock signal.
Cell culture and transfection
HEK293T cells (CRL-3216; American Type Culture Collection, Manassas, VA) were maintained in Dulbecco’s modified Eagle’s medium high-glucose medium (Life Technologies, Carlsbad, CA) supplemented with 2 mmol dm−3 L-glutamine (Life Technologies), 100 U/mL penicillin and streptomycin (Life Technologies), and 10% fetal bovine serum (FBS; Gibco-Thermo Fisher Scientific, Waltham, MA, USA) in uncoated 75 cm2 plastic flasks and incubated at 37°C, 5% CO2 in a humidified atmosphere. The cells were transiently transfected with the pHLsec plasmid (28) containing the complementary DNA of human superoxide dismutase 1 (SOD1) (UniProtKB: P00441) using branched polyethylenimine ((PEI) average Mw 25 kDa; Sigma-Aldrich, St. Louis, MO), as previously described (14). A 1:2 DNA:PEI weight ratio (25 μg SOD1 DNA, 50 μg PEI per 75 cm2 flask) was used. Transient expression of SOD1 was carried out for 48 h in [U-15N]-BioExpress6000 medium (Cambridge Isotope Laboratories, Tewksbury, MA) supplemented with 2% FBS, 100 U/mL penicillin/streptomycin, and 10 μM ZnSO4.
Sample preparation for in-cell NMR
To preserve sample homogeneity and allow efficient diffusion of the nutrients through the sample, the cells were kept in suspension in fresh Dulbecco’s modified Eagle’s medium (2% FBS) in the presence of 30% Percoll. The amount of Percoll was tuned empirically to match the buoyant density of the cells at 37°C. A suspension of HEK293T cells from 2 × 75 cm2 flasks (500 μL containing ∼6 × 107 cells) was placed in the outer NMR tube. The bottom of the tube had been previously filled with 0.8% agarose gel to further confine the cells into the active volume of the 1H NMR coil.
Sample preparation for drug screening
The protein hCAII was expressed, purified, and prepared for NMR analysis as previously reported (29). Experiments were performed on samples of the 15N isotopically enriched carbonic anhydrase II at the protein concentration of 0.3 mmol dm−3 in buffered solution (10 mmol dm−3 HEPES). The interaction of the zinc(II)-bound enzyme with the benzoate and furosemide ligands was performed at pH 7.2. The buffer used for the interaction with benzoate contained the ligand at the concentration of 20 mmol dm−3; the buffer used for the interaction with furosemide instead contained the latter ligand at 2 mmol dm−3 concentration. The buffer used for demetallation of Zn(II)-hCAII is 10 mmol dm−3 sodium phosphate (pH 6) and 50 mmol dm−3 2,6-pyridinedicarboxylic acid (dipicolinic acid (DPA)) (30). The cobalt(II) was added at the concentration of 1 mmol dm−3, exchanging the buffer to 10 mmol dm−3 HEPES (pH 6.8).
In-cell NMR spectra
1D 1H excitation sculpting and two-dimensional (2D) 1H-15N band-selective optimized-flip-angle short-transient heteronuclear multiple quantum correlation (31) in-cell NMR spectra were recorded at 310 K on a Bruker (Billerica, MA) AVANCE DRX 700 MHz spectrometer equipped with a triple resonance cryoprobe. 1D 31P in-cell NMR spectra were recorded at 310 K on a Bruker AVANCE III HD 400 MHz spectrometer equipped with a broadband observe probe. The spectra were processed with the Bruker TOPSPIN software and analyzed with Bruker Dynamic Center suite.
hCAII NMR spectra
The 1D excitation-sculpting 1H spectra and the 2D 1H-15N-HSQC (32) spectra of hCAII for screening were recorded on a Bruker AVANCE DRX 700 spectrometer equipped with a triple resonance cryoprobe with the temperature set to 298 K, processed with the Bruker TOPSPIN software, and analyzed with the program Computer Aided Resonance Assignment (ETH Zurich, Zürich, Switzerland). A total of 185 and 172 pseudocontact shift could be collected for the spectra of the paramagnetic protein in the presence of benzoate and furosemide, respectively. The tensor parameters have been calculated using the FANTEN user interface (33).
The flow used during the acquisition of spectra was of 0.05 mL/min, and around 30 min were needed for complete buffer exchange.
Results and Discussion
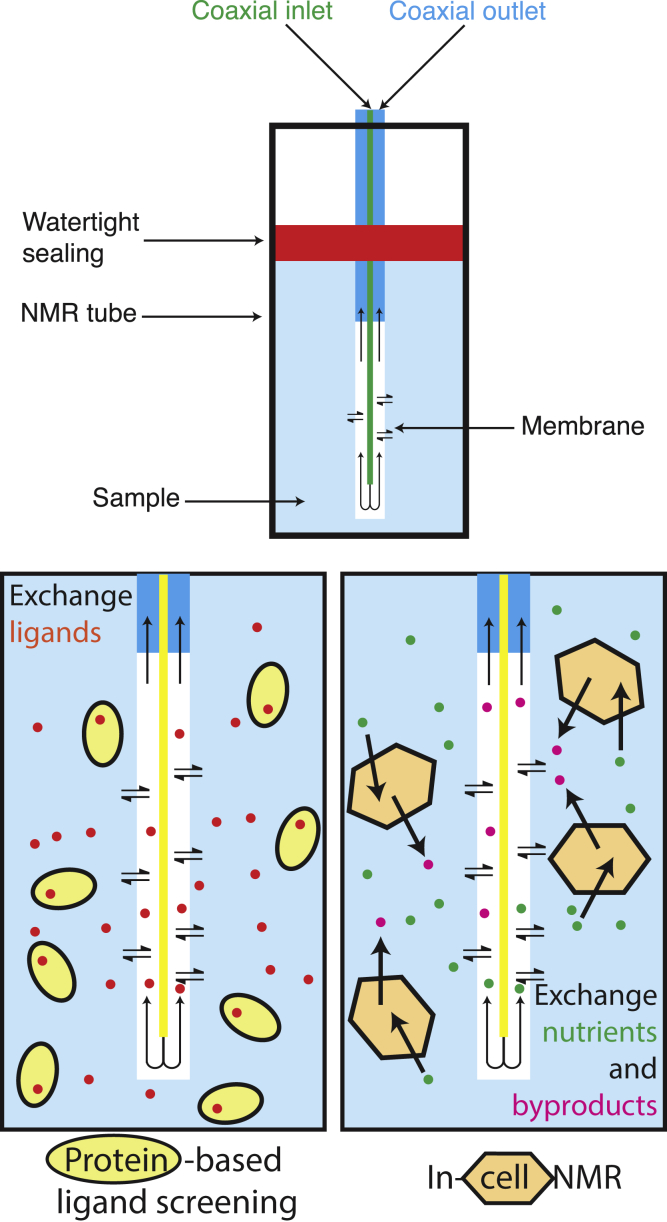
The bioreactor design that we here propose is made of the following elements: the NMR tube, a watertight sealing to confine the sample within the active volume of the tube, and a microdialysis membrane through which small molecules can be freely exchanged. This setup makes it possible to monitor events triggered by changes in the chemical composition of the medium/buffer as they are occurring. In this experimental setup (Fig. 1), the watertight sealing is provided by the commercial InsightMR device (11), and the flow occurs within a cylindrical, custom membrane (CMA20; CMA Microdialysis, a division of Harvard Bioscience, Solna, Sweden; see Table S1) in which inlet and outlet are coaxial. The outer NMR tube has 5-mm outer diameter and 4.2-mm inner diameter. The watertight sealing sets the total height of the sample to 38 mm; thus, the total volume is 526 μL. The microdialysis membrane has an outer diameter of 0.8 mm and reaches the bottom of the tube with a total internal volume of 19 μL.
Figure 1.
Experimental setup. The microdialysis membrane is represented in white, and the outlet is coaxial with the inlet. The sample is confined within the active volume of the NMR tube through a watertight sealing (red in the scheme). The solution flows inside the membrane through the inlet (green), exchanges with the solution outside of the membrane, and flows out through the outlet (cyan) because of the pressure created by a pump. To see this figure in color, go online.
The functioning of the flowNMR setup is thus the following: 1) the sample is confined between the NMR tube and the microdialysis membrane; 2) the fresh solution flows from the inlet to the outlet, which is coaxial to the inlet, and the diffusion of small molecules occurs between the sample volume and the inner part of the membrane; and 3) the slight overpressure pushes the solution toward the outlet.
For the in-cell NMR application, a 1 MDa microdialysis membrane was used to allow for the diffusion of the high molecular weight components of the serum contained in the growth medium while confining the cells. Our bioreactor design can keep suspended human cells metabolically active at high cell densities and allows for monitoring time-dependent intracellular processes over a span up to 42 h.
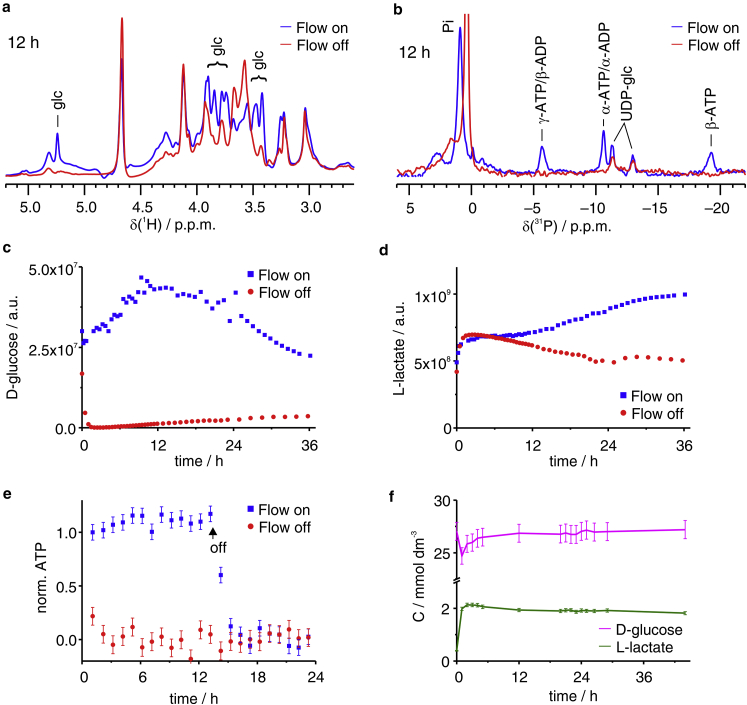
The metabolic activity of the cells as a function of time was evaluated through 1D 1H and 1D 31P in-cell NMR (Fig. 2, a and b; Fig. S1). In the 1H spectra, signals from glucose (Fig. 2 a) and lactate (Fig. S2) were identified. As expected, D-glucose was higher in the bioreactor than in a closed-tube control sample, in which it was depleted in 1–2 h (Fig. 2 c). The initial increase in D-glucose suggests that the cells recovered from an initial state of starvation (occurred during sample preparation), whereas the subsequent decrease indicates an increased metabolic activity due to cell growth. L-lactate, a product of anaerobic glycolysis, also appeared to slowly increase in the bioreactor after the initial recovery phase (Fig. 2 d), consistent with an increased glucose consumption over time. In another experiment, we have probed the intracellular ATP concentration, which remained constant during 13 h of 31P NMR measurement with a continuous flow of nutrients, decreasing quickly after the flow was stopped, whereas in the closed-tube sample it had already decreased by ∼80% after the first hour (Fig. 2 e, the time at which the flow was stopped is indicated by an arrow). Samples from the used medium were analyzed by 1H NMR to quantify the major metabolites (Fig. 2 f; Fig. S3) and show a continuous efflux of metabolic products.
Figure 2.
(a) D-glucose region of the 1H NMR spectra and (b) 31P spectra of cells in the bioreactor (blue) and in the control sample (red) collected after 12 h. (c and d) Integrals of D-glucose (anomeric signal, c) and L-lactate (methyl signal, d) as a function of time in the bioreactor (blue) and in the control sample (red). (e) Normalized integral of the 31P signal of ATP γP was measured as a function of time in the bioreactor (blue) and in the control sample (red)—in the bioreactor, the flow was halted after 13 h. (f) Concentration of D-glucose (magenta) and L-lactate (green) in the spent medium as a function of time. Error bars are calculated from the SD of the spectral noise. To see this figure in color, go online.
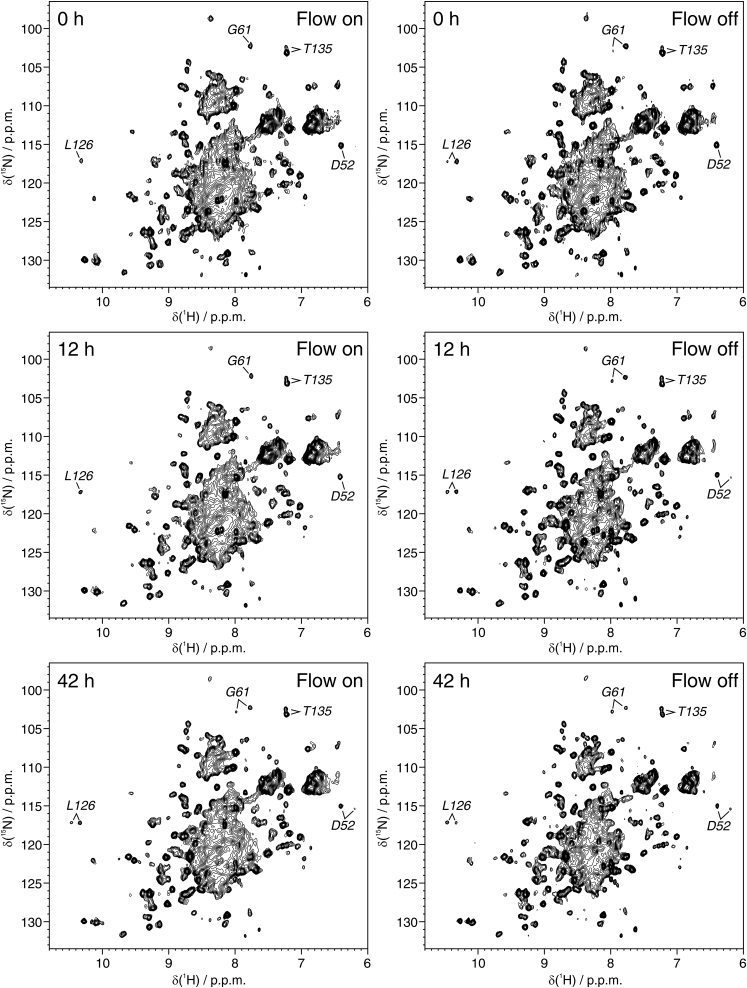
These data indicate that the bioreactor can sustain the metabolic activity of mammalian cells at high density for a prolonged time. We thus applied this setup to monitor the spectral changes of intracellular [U-15N]-human SOD1 over time. 2D 1H-15N spectra were continuously recorded over 40 h on both the bioreactor and a closed-tube control sample. Zinc-bound, dimeric SOD1 (E,Zn-SOD1) has been previously shown to be exceptionally stable, even in a stressed cellular environment (34). As a result, the signals from [U-15N]-SOD1 were still detected in both samples after 42 h (Fig. 3).
Figure 3.
2D 1H-15N short-transient heteronuclear multiple quantum correlation spectra recorded at different times on HEK293T cells expressing [U-15N]-SOD1 in the presence and in the absence of nutrient flow. Top row: 0 h; middle row: 12 h; bottom row: 42 h; left column: flow on; right column: flow off. Signals diagnostic of the formation of non-native Zn,Zn-SOD1 metalation state are labeled.
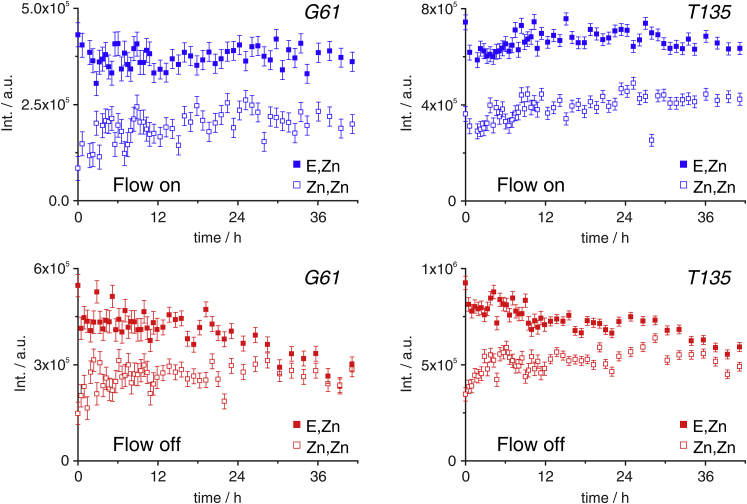
Interestingly, a non-native metalation state of SOD1 with two zinc ions per monomer (Zn,Zn-SOD1) (35) slowly built up in the closed-tube sample, whereas the native E,Zn-SOD1 decreased (Fig. 2, h and i). Instead, in the bioreactor, both species remained constant (with lower Zn,Zn-SOD1 than E,Zn-SOD1, at the level that was attained in the sample preparation stage in which nutrient deprivation occurred, Fig. 4). This effect is likely to be a consequence of dysregulated zinc homeostasis. We note that the supernatant of the bioreactor sample after 42 h of continuous flow contained some protein, suggesting that leakage had occurred to some extent (data not shown). However, several factors likely contributed to this outcome, including the initial nutrient deprivation and the final handling of the cell sample. Overall, the 31P NMR data show that cell metabolic activity (and therefore integrity) is totally preserved for at least 13 h, although the 1H-15N NMR spectra of SOD1 suggest that even longer experimental times can be reached.
Figure 4.
Integrals of G61 (left) and T135 (right) signals in different metalation states as a function of time in the bioreactor (top) and in the control sample (bottom). Error bars are calculated from the SD of the spectral noise. To see this figure in color, go online.
This setup also allows for confining a protein of interest within the active volume of the NMR tube and then flowing ligands to characterize their interaction with the protein, as it is done in TINS. In this case, the protein can reorient freely, allowing for monitoring the effect of ligands through the protein resonances, although the ligands can exchange. This approach is proven on the pharmaceutical target human carbonic anhydrase II (hCAII 29 kDa; see Supporting Material for the details of sample preparation) (36) but can be applied to any protein-ligand system, including larger objects that are not routine samples for drug discovery by NMR (37, 38) and is particularly useful for samples with limited availability, especially if combined with 3-mm tubes. This strategy can be adapted to highly automated operations, which may contribute to leverage the time and cost burden of NMR-based library screening. In this setup, the microdialysis membrane had a cutoff of 20 kDa to confine the protein in the NMR tube.
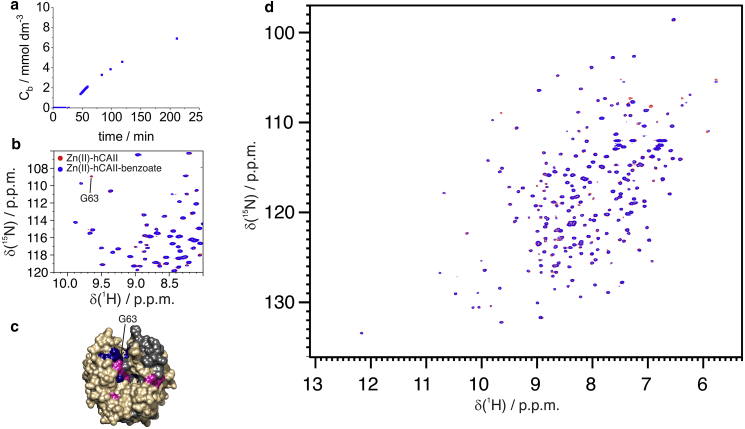
We have tested the interaction of the zinc(II)-loaded hCAII with one low-affinity ligand, benzoate (Kd in the millimolar range (39)), and one high-affinity ligand, furosemide (Kd in the nanomolar range). A buffer containing 20 mmol dm−3 benzoate was flown through the system. 1D 1H spectra were acquired to detect the increase of the concentration of the ligand in the tube (Fig. 5 a), and 2D 1H-15N HSQC (Fig. 5, b and c) were acquired to detect changes in the protein spectrum. Under these conditions (i.e., a flow rate of 50 μL min−1 inside a membrane that takes 3.6% of the total volume), the effect on signal intensity of the net flow inside the membrane when acquiring gradient-based experiments is negligible (see Materials and Methods). Benzoate binding was in fast exchange; therefore, chemical shift changed continuously upon increase of the ligand concentration, and the intensity of some peaks was reduced (Fig. 5, b and c; Fig. S4), in particular close to the active site (Fig. 5 d).
Figure 5.
(a) Concentration of benzoate within the NMR tube as a function of time while flowing a benzoate solution at 20 mmol dm−3. (d) Overlay of the spectra of free-Zn-hCAII (red) and benzoate-Zn-hCAII (blue) and (b) detail of the spectra where the drop in intensity of G63 is observed. (c) Mapping of chemical shift perturbations (pink) and intensity changes (blue) on hCAII upon interaction with benzoate. To see this figure in color, go online.
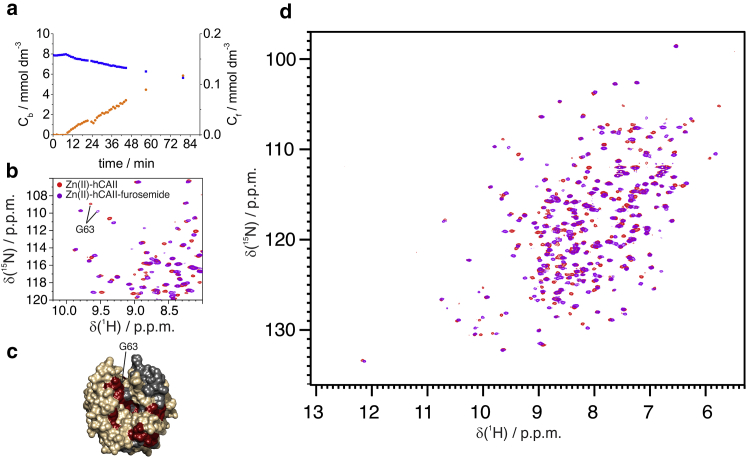
When two ligands have different affinities, this setup allows for probing the displacement of the first ligand; thus, still in the presence of benzoate, we started the flow of a buffer containing furosemide at 2 mmol dm−3 concentration (Fig. 6 a). Furosemide binding was in slow-exchange regime, and its effects on the spectrum can be readily observed, even when the concentration of benzoate is still high (Fig. 6, b and c); the signals of the furosemide-bound protein appeared in the spectrum with the simultaneous disappearance of those of the benzoate-bound protein. The chemical shift perturbations are obviously larger than in the case of benzoate, and the residues experiencing the largest effects were in a wider region on the protein surface (Fig. 6 d; Fig. S5). In both cases, the extent of the perturbation was comparable with that observed in the standard non-flow-based protein-ligand interaction experiments (Fig. S6). The outflow of the ligands had a half-time of ∼3.2 h (Fig. S7).
Figure 6.
(a) Concentration of benzoate and furosemide within the NMR tube as a function of time while flowing a furosemide solution at 2 mmol dm−3. (d) Overlay of the spectra of free-Zn-hCAII (red) and furosemide-Zn-hCAII (magenta) and (b) detail of the spectra in the same spectral region as Fig. 5c. (c) Mapping of chemical shift perturbations (red) upon interaction with furosemide. To see this figure in color, go online.
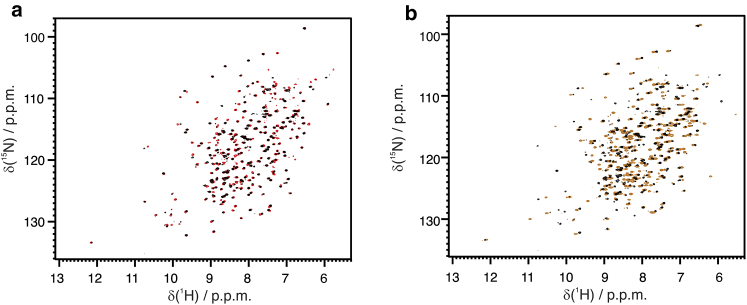
This setup is convenient for monitoring metal substitution as well. Zinc(II) was removed from hCAII by flowing a buffer containing 50 mmol dm−3 DPA through the system. The formation of apo-hCAII (Fig. 7 a) was completed within 3 h when DPA concentration reached its equilibrium value. The excess of DPA and its zinc complex were washed from the NMR tube by flowing a new buffer containing 1 mmol dm−3 cobalt(II) (Fig. 7 b). The spectra obtained in this way for the cobalt(II)-hCAII also show signals of the zinc(II)-hCAII (∼35% of the total) because of the 25,000-fold higher affinity of hCAII for zinc(II) than for cobalt(II) and because of the inertia of demetallation (40) that causes accumulation of the zinc-bound form even in the presence of traces of zinc(II). The presence of the diamagnetic species is, however, not a drawback for measuring the paramagnetic shifts.
Figure 7.
Overlay of the 2D 1H-15N HSQC of (a) free, zinc(II)-loaded hCAII (red); free, apo-hCAII (black); (b) apo-hCAII (black); and free, cobalt(II)-loaded hCAII (orange). To see this figure in color, go online.
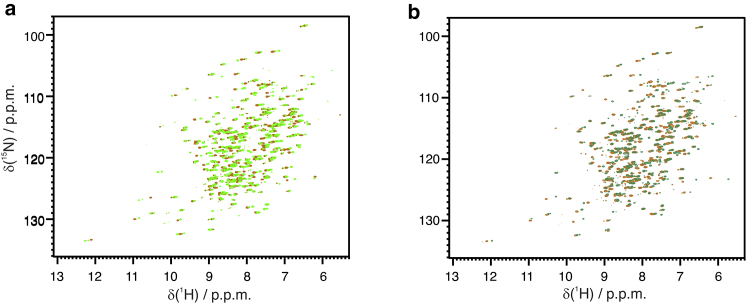
The interaction with benzoate and furosemide was then repeated for the cobalt(II)-hCAII. The paramagnetic effects such as relaxation enhancement and hyperfine shifts make the experiments more sensitive to protein-ligand interactions (41, 42). The interaction of benzoate with the protein was apparent both in the spectrum of the ligand, which was in fast exchange between the free and the bound (broadened and shifted) form (Fig. S8) and in that of the protein (Fig. 8 a). The benzoate ligand imposed a different coordination on the metal site (possibly pentacoordinate, with one water molecule and a benzoate ion acting as monodentate ligand (39)), imparting a different orientation of the magnetic susceptibility anisotropy tensor (Fig. 9 a) with respect to the free enzyme (Fig. S9; Table S2) (29).
Figure 8.
(a) Overlay of the spectra of free-Co-hCAII (orange) and benzoate-Co-hCAII (light green) and (b) of free-Co-hCAII (orange) and furosemide-Co-hCAII (dark green). To see this figure in color, go online.
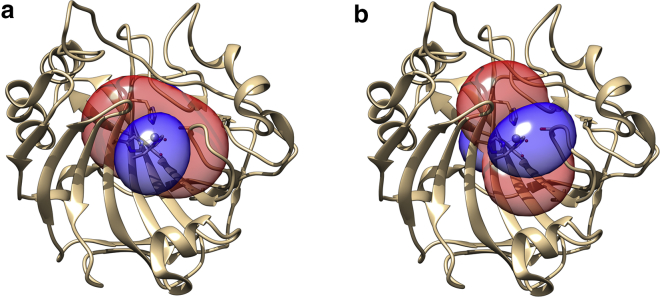
Figure 9.
Ribbon representation of the enzyme showing the metal in the active site as a sphere and the iso-pseudocontact shirt surfaces (blue: +1 ppm and red −1 ppm), obtained upon binding of benzoate (a) or furosemide (b). An example of fit is given in Fig. S11. To see this figure in color, go online.
Furosemide binding was in slow exchange also for the cobalt(II)-hCAII, and the variations of the chemical shifts were also clearly visible in this case (Fig. 8 b). Upon binding of furosemide, the geometry of the metal site was maintained (Figs. 9 b and S9; Table S2).
Ligands diffused slowly through the tube, so the strong binder furosemide molecules bound as soon as they encounter a protein molecule. Therefore, a diffusion front of furosemide was formed. The consequences of this behavior are apparent only in the paramagnetic sample; the benzoate molecules that are behind the furosemide front and those that are beyond are in slow exchange with one another, so that those that are behind show the spectra typical of the free molecule, whereas those beyond still experience shift and broadening because they remain in fast exchange with the paramagnetic enzyme molecules that are not yet reached by furosemide molecules (Fig. S10).
Conclusions
The use of a microdialysis membrane represents an ideal solution to provide flowNMR capabilities to all those applications that require confinement of the sample within the active volume. We have demonstrated that this approach can be successfully applied to maintain cell viability, significantly extending the timeframe over which in-cell NMR can be reliably performed and enabling, in principle, real-time NMR studies of intracellular processes; likewise, we have demonstrated that it is possible to monitor the interaction of a target protein with different ligands as they are flowing through the system. Speed of the screening, cost reduction, flexibility, possibility of automation, and quality of the information that can be retrieved are the key requirements for the success of ligand-library-screening strategies. The approaches potentially available up to now fail in at least one of these points, whereas the development of the flowNMR strategy here reported has the potential of fulfilling all these requirements by the confinement of the protein target.
Author Contributions
L. Banci, M.F., E.L., and E.R. conceived the research. E.L. and E.R. designed the experiments. A.C., M.F., E.L., E.R., and M.P. conceived the experimental setup. L.C., S.G., E.L., and E.R. performed the NMR experiments. L. Barbieri grew and transfected the human cells. S.G. expressed and purified hCAII. L. Barbieri, L. Banci, L.C., M.F., E.L., and E.R. analyzed the data. All authors wrote the manuscript. M.P. and A.C. are employees of Bruker UK, manufacturer and supplier of NMR hard- and software solutions that have been used in this research.
Acknowledgments
The authors dedicate this work to the memory of Prof. Stefano Caldarelli.
This work has been supported by Fondazione Cassa di Risparmio di Firenze, Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) PRIN 2012SK7ASN and MEDINTECH (CTN01_001177_962865), the EC contracts iNEXT (653706), West-Life (675858), Instruct-ULTRA (731005), and the PhosAgro/UNESCO/IUPAC Green Chemistry for Life (4500319942). The authors acknowledge the support and the use of resources of Instruct-ERIC and the support of the University of Florence CERM-TT, Recombinant Proteins JOYNLAB.
Editor: Timothy Cross.
Footnotes
Eleven figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)34454-0.
Contributor Information
Enrico Luchinat, Email: eluchinat@cerm.unifi.it.
Enrico Ravera, Email: ravera@cerm.unifi.it.
Supporting Material
References
- 1.Sánchez-Pedregal V.M., Reese M., Carlomagno T. The INPHARMA method: protein-mediated interligand NOEs for pharmacophore mapping. Angew. Chem. Int.Engl. 2005;44:4172–4175. doi: 10.1002/anie.200500503. [DOI] [PubMed] [Google Scholar]
- 2.Pellecchia M., Bertini I., Siegal G. Perspectives on NMR in drug discovery: a technique comes of age. Nat. Rev. Drug Discov. 2008;7:738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalvit C. NMR methods in fragment screening: theory and a comparison with other biophysical techniques. Drug Discov. Today. 2009;14:1051–1057. doi: 10.1016/j.drudis.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Saxena K., Schwalbe H. eMagRes. American Cancer Society; 2015. The role of NMR in target identification and validation for pharmaceutical R&D; pp. 305–314. [Google Scholar]
- 5.Erlanson D.A., Fesik S.W., Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 6.Nitsche C., Otting G. NMR studies of ligand binding. Curr. Opin. Struct. Biol. 2018;48:16–22. doi: 10.1016/j.sbi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Plitzko J.M., Schuler B., Selenko P. Structural Biology outside the box-inside the cell. Curr. Opin. Struct. Biol. 2017;46:110–121. doi: 10.1016/j.sbi.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Luchinat E., Banci L. New structural and functional insights from in-cell NMR. Emerg. Top. Life Sci. 2018 doi: 10.1042/ETLS20170136. Published online February 6, 2018. [DOI] [PubMed] [Google Scholar]
- 9.Li C., Zhao J., Liu M. Magnetic resonance spectroscopy as a tool for assessing macromolecular structure and function in living cells. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 2017;10:157–182. doi: 10.1146/annurev-anchem-061516-045237. [DOI] [PubMed] [Google Scholar]
- 10.Khajeh M., Bernstein M.A., Morris G.A. A simple flowcell for reaction monitoring by NMR. Magn. Reson. Chem. 2010;48:516–522. doi: 10.1002/mrc.2610. [DOI] [PubMed] [Google Scholar]
- 11.Foley D.A., Bez E., Marquez B.L. NMR flow tube for online NMR reaction monitoring. Anal. Chem. 2014;86:12008–12013. doi: 10.1021/ac502300q. [DOI] [PubMed] [Google Scholar]
- 12.Silva Elipe M.V., Milburn R.R. Monitoring chemical reactions by low-field benchtop NMR at 45 MHz: pros and cons. Magn. Reson. Chem. 2016;54:437–443. doi: 10.1002/mrc.4189. [DOI] [PubMed] [Google Scholar]
- 13.Hall A.M.R., Broomfield-Tagg R., Hintermair U. Online monitoring of a photocatalytic reaction by real-time high resolution FlowNMR spectroscopy. Chem. Commun. (Camb.) 2017;54:30–33. doi: 10.1039/c7cc07059d. [DOI] [PubMed] [Google Scholar]
- 14.Barbieri L., Luchinat E., Banci L. Characterization of proteins by in-cell NMR spectroscopy in cultured mammalian cells. Nat. Protoc. 2016;11:1101–1111. doi: 10.1038/nprot.2016.061. [DOI] [PubMed] [Google Scholar]
- 15.Koczula K.M., Ludwig C., Günther U.L. Metabolic plasticity in CLL: adaptation to the hypoxic niche. Leukemia. 2016;30:65–73. doi: 10.1038/leu.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris T., Eliyahu G., Degani H. Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells. Proc. Natl. Acad. Sci. USA. 2009;106:18131–18136. doi: 10.1073/pnas.0909049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macdonald J.M., Grillo M., James T.L. NMR spectroscopy and MRI investigation of a potential bioartificial liver. NMR Biomed. 1998;11:55–66. doi: 10.1002/(sici)1099-1492(199804)11:2<55::aid-nbm502>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 18.Keshari K.R., Kurhanewicz J., Macdonald J.M. Hyperpolarized (13)C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism. Magn. Reson. Med. 2010;63:322–329. doi: 10.1002/mrm.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shestov A.A., Mancuso A., Glickson J.D. Bonded cumomer analysis of human melanoma metabolism monitored by 13C NMR spectroscopy of perfused tumor cells. J. Biol. Chem. 2016;291:5157–5171. doi: 10.1074/jbc.M115.701862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong S., Eskandari R., Keshari K.R. Real-time quantitative analysis of metabolic flux in live cells using a hyperpolarized micromagnetic resonance spectrometer. Sci. Adv. 2017;3:e1700341. doi: 10.1126/sciadv.1700341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharaf N.G., Barnes C.O., Pielak G.J. A bioreactor for in-cell protein NMR. J. Magn. Reson. 2010;202:140–146. doi: 10.1016/j.jmr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubo S., Nishida N., Shimada I. A gel-encapsulated bioreactor system for NMR studies of protein-protein interactions in living mammalian cells. Angew. Chem. Int.Engl. 2013;52:1208–1211. doi: 10.1002/anie.201207243. [DOI] [PubMed] [Google Scholar]
- 23.Breindel L., DeMott C., Shekhtman A. Real-time in-cell nuclear magnetic resonance: ribosome-targeted antibiotics modulate quinary protein interactions. Biochemistry. 2018;57:540–546. doi: 10.1021/acs.biochem.7b00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegal G., Hollander J.G. Target immobilization and NMR screening of fragments in early drug discovery. Curr. Top. Med. Chem. 2009;9:1736–1745. doi: 10.2174/156802609790102400. [DOI] [PubMed] [Google Scholar]
- 25.Hajduk P.J., Meadows R.P., Fesik S.W. Discovering high-affinity ligands for proteins. Science. 1997;278 doi: 10.1126/science.278.5337.497. 497,499. [DOI] [PubMed] [Google Scholar]
- 26.Loening N.M., Keeler J. Measurement of convection and temperature profiles in liquid samples. J. Magn. Reson. 1999;139:334–341. doi: 10.1006/jmre.1999.1777. [DOI] [PubMed] [Google Scholar]
- 27.Swan I., Reid M., Morris G.A. Sample convection in liquid-state NMR: why it is always with us, and what we can do about it. J. Magn. Reson. 2015;252:120–129. doi: 10.1016/j.jmr.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 29.Cerofolini L., Staderini T., Luchinat C. Long-range paramagnetic NMR data can provide a closer look on metal coordination in metalloproteins. J. Biol. Inorg. Chem. 2018;23:71–80. doi: 10.1007/s00775-017-1511-y. [DOI] [PubMed] [Google Scholar]
- 30.Bertini I., Luchinat C., Vila A.J. The interaction of acetate and formate with cobalt carbonic anhydrase. An NMR study. Eur. J. Biochem. 1992;208:607–615. doi: 10.1111/j.1432-1033.1992.tb17225.x. [DOI] [PubMed] [Google Scholar]
- 31.Schanda P., Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 32.Bodenhausen G., Ruben D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–188. [Google Scholar]
- 33.Rinaldelli M., Carlon A., Luchinat C. FANTEN: a new web-based interface for the analysis of magnetic anisotropy-induced NMR data. J. Biomol. NMR. 2015;61:21–34. doi: 10.1007/s10858-014-9877-4. [DOI] [PubMed] [Google Scholar]
- 34.Banci L., Barbieri L., Aricescu A.R. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banci L., Barbieri L., Luchinat E. In-cell NMR in E. coli to monitor maturation steps of hSOD1. PLoS One. 2011;6:e23561. doi: 10.1371/journal.pone.0023561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Supuran C.T. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 37.Dias D.M., Ciulli A. NMR approaches in structure-based lead discovery: recent developments and new frontiers for targeting multi-protein complexes. Prog. Biophys. Mol. Biol. 2014;116:101–112. doi: 10.1016/j.pbiomolbio.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sivanandam V.N., Bernardo-Seisdedos G., Millet O. Biophysical Techniques in Drug Discovery. 2017. Chapter 3: receptor-based NMR techniques in drug discovery; pp. 44–66. [Google Scholar]
- 39.Bertini I., Luchinat C., Scozzafava A. Interaction of cobalt(II) bovine carbonic anhydrase with aniline, benzoate, and anthranilate. J. Am. Chem. Soc. 1977;99:581–584. doi: 10.1021/ja00444a043. [DOI] [PubMed] [Google Scholar]
- 40.Hunt J.A., Ahmed M., Fierke C.A. Metal binding specificity in carbonic anhydrase is influenced by conserved hydrophobic core residues. Biochemistry. 1999;38:9054–9062. doi: 10.1021/bi9900166. [DOI] [PubMed] [Google Scholar]
- 41.Bertini I., Fragai M., Terni B. Paramagnetic metal ions in ligand screening: the CoII matrix metalloproteinase 12. Angew. Chem. 2004;116:2304–2306. doi: 10.1002/anie.200353453. [DOI] [PubMed] [Google Scholar]
- 42.Guan J.Y., Keizers P.H., Siegal G. Small-molecule binding sites on proteins established by paramagnetic NMR spectroscopy. J. Am. Chem. Soc. 2013;135:5859–5868. doi: 10.1021/ja401323m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.