Abstract
BACKGROUND: Tumor infiltrating lymphocytes (TILs) are widely considered a key sign of the immune interaction between host and tumor, and potentially prognostic biomarkers of good or bad outcome in many cancers, included invasive breast cancer (BC). However, results about the association between TIL typology, location and BC prognosis, are controversial. The aim of the study was to evaluated the prognostic significance of TIL subtypes (CD4+, CD8+, FOXP3+ T cells) and their location (stromal “s” and intratumoral “i” CD4+ and CD8+) in BC patients, focusing on the association between these markers and immunocheckpoint molecules such as cytotoxic T lymphocyte antigen 4 (CTLA-4), programmed cell death ligand 1 (PD-L1) and its receptor (PD-1). METHODS: CD4+, CD8+, FOXP3+, CTLA4+, PD-L1+ and PD-1+ expression was examined by immunohistochemistry on tissue microarrays (TMAs) from 180 BC patients. Univariate and Kaplan–Meier analyses of disease free survival (DFS) were performed to evaluate the prognostic significance of marker expression. RESULTS: Total CD8+ T cells were not significantly associated with DFS. Differently, patients with iCD8+ and sCD8+ overexpression showed a trend toward respectively a worse (P = .050) and a better 5-years DFS (P = .064). Interestingly, TIL expression of both PD-1+ and PD-L1+, was significantly associated with iCD8+ (P = .0004; P < .0001 respectively), while only TIL expression of PD-1 was associated with sCD8+ (P = .001). CONCLUSION: Our data show that iCD8+ T cells, but no sCD8+ T cells identify a subgroup of patients with poor DFS and this could be due to the overexpression of PD-L1/PD-1 pathway.
Introduction
Breast cancer (BC) is the leading cause of mortality in women worldwide [1]. Despite advances in the diagnosis and treatment, new biomarkers of prognosis are needed to develop target therapies and improve patient survival. Recently, several studies demonstrated the importance of the microenvironment in cancer progression [2], focusing on the function and interaction between immune cells and cancer cells, in a variety of solid tumors, including BC [3]. Tumor infiltrating lymphocytes (TILs) are considered a selected population of T-cells with a higher specific immunological reactivity against tumor cells [4]. TILs play dual role in cancer, by either suppressing or promoting tumor growth [4]. The anti- and pro-tumoral function of TILs in BC could be TIL subtype dependent. Indeed cytotoxic T cells (CD8+ T cells) have been reported to be associated with improved clinical outcome in different phenotypes of BC [5], [6], [7], [8]. Conversely, the prognostic role of regulatory T cells (Treg), defined as forkhead box protein 3 (FOXP3+) T cells, remains to define. Infact, BC with FOXP3+ TILs have been reported having both a worse [9], [10] and a better prognosis [11], [12]. Moreover, other authors reported that the expression of FOXP3+ has no a dominant role in BC prognosis [13].
If the clinical significance of TILs in BC is still arguable, their location (stromal TILs, sTILs; intratumoral TILs, iTILs) is even less studied, and the conclusions remain debatable. Chen et al. demonstrated that the presence of intratumoral CD8+ cytotoxic lymphocytes was a favorable prognostic marker in node negative BC [14], while Wang et al. recently reported that no CD4+ neither CD8+ T cells, at either location (tumor-host interface and within intratumoral stroma) were significantly associated with relapse free survival or overall survival in triple negative BC (TNBC) [15]. Thus, the issue of TIL role and prognostic significance in BC is still an open field of research, which should be investigated in light of the new molecular mechanisms that enable tumor cells to escape from the host immune system. BC cells express the immunocheckpoint molecules to evade antitumor immune responses, to grow progressively and to metastasize. One of these, is the programmed cell death ligand 1 (PD-L1) and its receptor, the programmed cell death 1 (PD-1). Both are expressed in TILs and in different cancer types including melanoma, ovarian, lung and BC. [16]. The interaction PD-L1/PD1 inhibits T-cell activation, induces T lymphocyte apoptosis and blocks immune responses against cancer [17]. Data concerning the correlation between PD-L1 and BC prognosis are numerous but conflicting [18], [19].
Another immune checkpoint molecule is the cytotoxic T lymphocyte antigen 4 (CTLA-4). CTLA4 is a CD28 homologous, normally expressed at low levels on the surface of naïve effector T cells and Treg cells [20]. After stimulation of T naïve cells, CD8+, CD4+ and FOXP3+ T cells upregulate CTLA4, which reduces T-cell activation [20]. In BC, CTLA4 expression has been little studied and, similarly to PD-L1, the relation with the prognosis remains elusive.
Here we present a study on 180 BC patients. The aim was to evaluate whether the different subtypes of TILs (CD4+, CD8+, FOXP3+ T cells) and their location (sCD4 and iCD4; sCD8 and iCD8), in association with PD-L1/PD-1 and CTLA4 expressed by TILs and tumor cells, could identify an immunophenotype with a prognostic significance. These evaluations may shed new light on the possible clinical use of these immune markers as new therapeutic targets.
Methods
Patients and Clinicopathological Characteristics
This study involved a retrospective, not consecutive, series of 180 female patients diagnosed with invasive breast cancer at the IRCCS Istituto Tumori “Giovanni Paolo II” of Bari between 1994 and 2012. Clinicopathological characteristics of the study populations are included in Supplementary Table 1. Histological type, tumor size, lymph node status, histological grade, estrogen receptor (ER) status, progesterone receptor (PgR) status, proliferative activity (MIB1), Human epidermal growth factor receptor 2 (HER2/neu) status, were obtained from the Pathology Department of our Institute.
Tumors with ER or PgR expression were scored as positive when nuclear immunoreactivity was present in >10% of tumor cells. For the proliferative activity (Ki67 index), assessed by MIB1 nuclear staining, the cut-off value of 20% positive cells was adopted, and tumors with proliferative activity >20% were considered highly proliferating. This cut-off represents the median value of the scores relative to all breast tumor samples analyzed in these years within our Institute. The HER2/neu was scored as 0, 1+, 2+ or 3+ using a monoclonal antibody (MoAb clone CB11, Novocastra Laboratories Ltd., Newcastle, UK), in accordance with the Herceptest scoring system (Food and Drug Administration accepted): 0 = no membranous immunoreactivity or <10% of cells reactive; 1+ = incomplete membranous reactivity in >10% of cells; 2+ = ≥10% of cells with weak to moderate complete membranous reactivity; and 3+ = strong and complete membranous reactivity in >10% of cells. Cytoplasmic immunoreactivity was ignored. Cases scoring 0 and 1+ were classified as negative. HER2/neu was considered to be positive if immunostaining was 3+ or if a 2+ result showed gene amplification by fluorescence in situ hybridization (FISH). In FISH analyses, each copy number of HER2 gene and its centromere 17 (CEP17) reference was counted. The interpretation followed the criteria of the ASCO/CAP 2007 guidelines for HER2 testing in breast cancer; the cases were considered positive if the HER2/CEP17 ratio was higher than 2.2 [21]. “The 16.2% (26/160) of patients received chemotherapy (CT), the 40% (64/160) chemotherapy + hormone therapy (CT+HT) and the 43.8% of patients (70/160) received only hormone therapy (HT). For the 11.1% of patients (20/180) the treatment data were not available”.
Tissue Microarray (TMA) and Immunohistochemistry
TILs (CD4, CD8, FOXP3) and imunocheckpoint marker (CTLA4, PD-L1, PD-1) expressions were examined by immunohistochemistry on tissue microarrays (TMAs) containing 540 tumor tissue cores from 180 breast cancer patients. For the construction of TMA, we followed the methods of Mangia et al. 2017 [22]. In detail, TMAs were generated using all available formalin-fixed and paraffin-embedded (FFPE) breast tumor tissue blocks. Three different regions of tumors were identified and marked on hematoxylin and eosin stained sections. The selection of tumor areas was made by choosing the regions with the highest quantity of lymphocyte infiltrate. Sections were matched to their corresponding paraffin blocks (donor blocks), and three tumor cores with a diameter of 1 mm were punched from these tumor regions of each donor block and precisely arrayed into a new recipient paraffin block (TMA block) using the Galileo Tissue MicroArrayer CK 4500 (Transgenomic). Each sample was arrayed in triplicate cores to minimize tissue loss and to overcome tumor heterogeneity. The three cores were representative of the whole specimen. Four μm-thick slices were cut from the TMA blocks and transferred to slides which were processed and stained by the indirect immunoperoxidase method using the BenchMark XT automated staining instrument (Ventana Medical Systems, Tucson, AZ, USA). All solutions were from Ventana Medical Systems unless otherwise specified. Slides underwent deparaffinization with the EZ PREP solution, and antigen retrieval with Cell Conditioning solution 1 (56 min, 95 °C). The following step was the incubation with the specific primary antibodies. The dilution of the primary antibodies was based on preliminary dilution experiments. The different analyzed biomarkers, dilution, source/clone, the staining localization of antibody and the cut off [median values of immunoreactive lymphocyte numbers, and cancer cell percentages or Immunohistochemical score (HIS)] used to classify positive versus negative cases, are shown in Supplementary Table 2. The UltraView Universal diaminobenzidina tetrachloride (DAB) detection kit was used to detect the protein expression. Slides were counterstained with Hematoxylin and Bluing Reagent respectively for 12 min and 4 min.
Tonsil tissue was used as positive control for each biomarker, whereas the negative control was prepared by replacing the primary antibody with a nonimmune immunoglobulin of the same isotype. Positive and negative controls were included in each staining run.
Quantification of TILs (CD4+, CD8+, FOXP3+) and Immunocheckpoint Markers (CTLA4, PD1, and PD-L1)
Quantification of the CD4+, CD8+, FOXP3+ and immunocheckpoint proteins (CTLA4, PD-1 and PD-L1) was performed manually by two observers blind to patient outcome and clinicopathological characteristics. Any discrepancies between the two observers were resolved by re-examination and consensus. The expression of CD4, CD8 and FOXP3 was evaluated in TILs and expressed as the number of positive cells counted in each TMA core at ×400 magnification (×40 objective). The mean of three readings relative to the three TMA cores, for each tumor sample, was calculated and represented the number of positive TIL subtypes of each tumor. If one TMA core was uninformative, or either lost or contained no tumor tissue or infiltrate, the overall score applied was that of the remaining cores of the triplicate. Furthermore, the cases in which all three TMA cores were uninformative, were considered non-assessable and excluded from the analyses. Lymphocytes in contact or within the tumor epithelium were scored as intratumoral (i), whereas lymphocytes in the interstitial space or in the stromal areas were defined stromal (s). The median value of immunoreactive cells was used as cut off for: CD4 (≥36), sCD4 (≥26), iCD4 (0), CD8 (≥64), sCD8 (≥36), iCD8 (≥3), FOXP3 (≥1), (Supplementary Table 2). The expression of immunocheckpoint proteins (CTLA4, PD-1 and PD-L1) was evaluated both in total TILs (expressed as the number of positive cells) and in tumor cells (expressed as the percentage of positive cells). The median value of immunoreactive cells was used as cut off for: CTLA4 (>0), PD-1 (>0), PD-L1 (≥1). The cut off for PD-L1 expression in tumor cells (≥1%) was in accordance with international guidelines [23] (Supplementary Table 2).
The number and the percentage of positive and negative samples for immune marker expression, in TILs and tumor cells, categorized according to the median values, are shown in Table 1 (a, b).
Table 1.
Immune Marker Expression in TILs and Tumor Cells
| a | ||||||||
|---|---|---|---|---|---|---|---|---|
| TILs | CD4 |
CD8 |
FOXP3 |
|||||
| Negative | Positive | Negative | Positive | Negative | Positive | |||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||
| 80 (49) | 83 (51) | 85 (49.7) | 86 (50.3) | 75 (44.4) | 94 (55.6) | |||
| TIL location | sCD4 |
iCD4 |
sCD8 |
iCD8 |
||||
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| 80 (49.1) | 83 (50.9) | 86 (52.8) | 77 (47.2) | 84 (49.4) | 86 (50.6) | 82 (48) | 89 (52) | |
| s = stromal | ||||||||
| i = intratumoral | ||||||||
| b | ||||||
|---|---|---|---|---|---|---|
| TILs | CTLA4 |
PD-1 |
PD-L1 |
|||
| Negative | Positive | Negative | Positive | Negative | Positive | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| 100 (57.8) | 73 (42.2) | 114 (73) | 42 (27) | 131 (79) | 35 (21) | |
| Tumor cells | CTLA4 |
PD-1 |
PD-L1 |
|||
| Negative | Positive | Negative | Positive | Negative | Positive | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| 180 (100) | 0 (0) | 180 (100) | 0 (0) | 160 (96) | 7 (4) | |
Statistical Analyses
In order to analyze the association between immune marker expression and patient clinical characteristics, and among markers themselves, we used the chi-square and Fisher's exact tests for the dichotomized variables. The results from the immunohistochemical analyses of immune markers, were assessed in relation to disease-free survival (DFS) and overall survival (OS). DFS (in months) was defined as the time from diagnosis to the date of loco-regional or distant recurrence, second invasive breast carcinoma, second primary cancer and/or death without evidence of breast cancer or to the date of last contact. OS (in months) was defined as the time from diagnosis to the date of last contact or of death from any cause. Univariate Cox regression analysis of DFS was to estimate the hazard ratio (HR) and 95% confidence intervals (95% CIs) in order to evaluate the prognostic relevance of the marker expression (for OS it could not be completed due to the low number of deaths). Survival curves were calculated according to the Kaplan–Meier analysis and compared by the log-rank test. All statistical differences were considered significant at a level of P < .05. Statistical analyses were performed using SPSS software version 15 (SPSS Inc., Chicago, IL, USA).
Results
Clinicopathological Characteristics
The tumor characteristics of the 180 patients with invasive BC are listed in Supplementary Table 1. The median age at diagnosis was 57 years (range 31–80). The 52.8% of patients were younger than 57 years. The majority had an invasive ductal carcinoma (IDC) (83.9%) and a moderate histological grade (G2) (58%). Tumors with tumor size ≤2 cm were the 64.6% and the axillary lymph node status was negative in the 62.4% of patients. The majority of patients had ER (82%) and PgR (64%) positive status, low MIB1 (67.8%) and HER2/neu negative (84.7%); 8.3% were triple negative breast cancers (TNBC). Complete follow up was available only for 160/180 (88.9%) patients with a median value of 63 months (range 3–203).
Associations Between Immune Marker Expression and Clinicopathological Characteristics
Negative and positive expression of immune markers, of the whole cohort of tumor samples, is reported in Table 1 (a, b). The mean values for sCD4+ (72 cells), iCD4+ (16 cells), sCD8+ (76 cells) and iCD8+ (36 cells) numerical counts, show that stromal lymphocytes are predominant compared to intratumoral lymphocytes. T-Student test shows that there are no significant differences between sCD4+ and sCD8+ T cells (P = .76), while iCD8+ are significantly more abundant than iCD4+ (P < .05).
A summary of significant associations between TILs (CD4+, CD8+, FOXP3+), TIL location (sCD4+, iCD4+; sCD8+, iCD8+) and clinicopathological characteristics of patients, are respectively reported in Table 2a, Table 2b.
Table 2a.
Relationship Between Immune Cell Markers and Clinicopathological Characteristics
| Characteristics | CD4 |
CD8 |
FOXP3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||||
| Age | |||||||||
| ≤57 | 36 (45) | 52 (62.7) | .0238 | 39 (45.9) | 50 (58.1) | .1087 | 42 (56) | 46 (49) | .3611 |
| >57 | 44 (55) | 31 (37.3) | 46 (51.1) | 36 (41.9) | 33 (44) | 48 (51) | |||
| Histological type | |||||||||
| IDC | 70 (87.5) | 69 (83.1) | .3163 | 71 (83.5) | 72 (83.7) | .9273 | 63 (82.9) | 78 (83.9) | .6803 |
| ILC | 7 (8.75) | 6 (7.2) | 9 (10.6) | 8 (9.3) | 9 (11.8) | 8 (8.6) | |||
| Other | 3 (3.75) | 8 (9.6) | 5 (5.9) | 6 (7) | 4 (5.3) | 7 (7.5) | |||
| Tumor size (cm) | |||||||||
| ≤2 cm | 44 (55.7) | 56 (68.3) | .0995 | 56 (65.9) | 53 (62.4) | .6314 | 52 (69.3) | 55 (59.1) | .172 |
| >2 cm | 35 (44.3) | 26 (31.7) | 29 (34.1) | 32 (37.6) | 23 (30.7) | 38 (40.9) | |||
| Lymph node status | |||||||||
| Negative | 52 (65.8) | 47 (57.3) | .2675 | 56 (65.9) | 48 (57.1) | .243 | 42 (56.8) | 60 (64.5) | .307 |
| Positive | 27 (34.2) | 35 (42.7) | 29 (34.1) | 36 (42.9) | 32 (43.2) | 33 (35.5) | |||
| Histological grade | |||||||||
| G1 | 5 (6.3) | 6 (7.5) | .1212 | 5 (6) | 9 (10.7) | .033 | 8 (10.7) | 6 (6.7) | .6756 |
| G2 | 52 (65.8) | 40 (50) | 57 (68.7) | 41 (48.8) | 44 (58.7) | 51 (56.7) | |||
| G3 | 22 (27.9) | 34 (42.5) | 21 (25.3) | 34 (40.5) | 23 (30.6) | 33 (36.6) | |||
| Receptor status | |||||||||
| ER-negative (≤10%) | 10 (12.5) | 20 (24.7) | .047 | 13 (15.3) | 17 (20.2) | .4003 | 12 (16.2) | 18 (19.4) | .5997 |
| ER-positive (>10%) | 70 (87.5) | 61 (75.3) | 72 (84.7) | 67 (79.8) | 62 (83.8) | 75 (80.6) | |||
| PgR-negative (≤ 10%) | 23 (28.8) | 33 (40.7) | .1102 | 28 (32.9) | 34 (40.5) | .3095 | 21 (28.4) | 41 (44.1) | .0369 |
| PgR-positive (> 10%) | 57 (71.2) | 48 (59.3) | 57 (67.1) | 50 (59.5) | 53 (71.6) | 52 (55.9) | |||
| MIB1 | |||||||||
| Negative (≤20%) | 62 (77.5) | 43 (53.1) | .0011 | 64 (75.3) | 51 (60.7) | .0421 | 54 (73) | 61 (65.6) | .3062 |
| Positive (>20%) | 18 (22.5) | 38 (46.9) | 21 (24.7) | 33 (39.3) | 20 (27) | 32 (34.4) | |||
| HER2/neu | |||||||||
| Negative (0,1+) | 73 (91.2) | 61 (76.3) | .0101 | 75 (88.2) | 67 (80.7) | .1783 | 64 (86.5) | 75 (81.5) | .3889 |
| Positive (3+) | 7 (8.8) | 19 (23.7) | 10 (11.8) | 16 (19.3) | 10 (13.5) | 17 (18.5) | |||
Table 2b.
Relationship Between Stromal (s) and Intratumoral (i) CD4 and CD8 Lymphocytes and Clinicopathological Characteristics
| Characteristics | sCD4 |
iCD4 |
sCD8 |
iCD8 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||||
| Age | ||||||||||||
| ≤ 57 | 35 (43.8) | 53 (63.9) | .1569 | 49 (57) | 38 (49.4) | .9174 | 35 (41.7) | 53 (61.6) | .1479 | 43 (52.4) | 46 (51.7) | 1.0000 |
| > 57 | 45 (56.2) | 30 (36.1) | 37 (43) | 39 (50.6) | 49 (58.3) | 33 (38.4) | 39 (47.6) | 43 (48.3) | ||||
| Histological type | ||||||||||||
| IDC | 70 (87.5) | 69 (83.1) | .0781 | 71 (82.5) | 67 (87) | .6520 | 68 (81) | 74 (86.1) | .6536 | 70 (85.4) | 73 (82) | .5117 |
| ILC | 8 (10) | 5 (6) | 9 (10.5) | 5 (6.5) | 10 (11.9) | 7 (8.1) | 6 (7.3) | 11 (12.4) | ||||
| Other | 2 (2.5) | 9 (10.9) | 6 (7) | 5 (6.5) | 6 (7.1) | 5 (5.8) | 6 (7.3) | 5 (5.6) | ||||
| Tumor size (cm) | ||||||||||||
| ≤ 2 cm | 42 (53.2) | 58 (70.7) | .0216 | 57 (67.1) | 43 (56.6) | .1712 | 53 (63.1) | 56 (65.9) | .7050 | 62 (75.6) | 47 (53.4) | .0026 |
| > 2 cm | 37 (46.8) | 24 (29.3) | 28 (32.9) | 33 (43.4) | 31 (36.9) | 29 (34.1) | 20 (24.4) | 41 (46.6) | ||||
| Lymph node status | ||||||||||||
| Negative | 53 (67.1) | 46 (56.1) | .1519 | 54 (62.8) | 45 (60) | .7166 | 52 (61.9) | 52 (61.9) | 1.0000 | 54 (65.9) | 50 (53.2) | .0883 |
| Positive | 26 (32.9) | 36 (43.9) | 32 (37.2) | 30 (40) | 32 (38.1) | 32 (38.1) | 28 (34.1) | 44 (46.8) | ||||
| Histological grade | ||||||||||||
| G1 | 4 (5.1) | 7 (8.6) | .1624 | 8 (9.5) | 4 (4.7) | .4442 | 6 (7.3) | 8 (9.5) | .6145 | 9 (11) | 5 (5.9) | .0039 |
| G2 | 51 (65.4) | 41 (50.6) | 50 (59.5) | 51 (60) | 51 (62.2) | 46 (54.8) | 56 (68.3) | 42 (49.4) | ||||
| G3 | 23 (29.5) | 33 (40.8) | 26 (30.9) | 30 (35.3) | 25 (30.5) | 30 (35.7) | 17 (20.7) | 38 (44.7) | ||||
| Receptor status | ||||||||||||
| ER-negative(≤10%) | 8 (10) | 22 (27.2) | .0052 | 21 (24.7) | 10 (13.2) | .0636 | 9 (10.7) | 21 (25) | .0156 | 17 (21) | 13 (14.8) | .2908 |
| ER-positive (>10%) | 72 (90) | 59 (72.8) | 64 (75.3) | 66 (86.8) | 75 (89.3) | 63 (75) | 64 (79) | 75 (85.2) | ||||
| PgR-negative (≤10%) | 22 (27.5) | 34 (42) | .0538 | 35 (41.2) | 21 (27.6) | .0716 | 27 (32.1) | 35 (41.7) | .2009 | 34 (42) | 28 (31.8) | .1711 |
| PgR-positive (> 10%) | 58 (72.5) | 47 (58) | 50 (58.2) | 55 (72.4) | 57 (67.9) | 49 (58.3) | 47 (58) | 60 (68.2) | ||||
| MIB1 | ||||||||||||
| Negative (≤20%) | 59 (73.8) | 46 (56.8) | .0239 | 59 (69.4) | 46 (60.5) | .2373 | 61 (72.6) | 53 (63.1) | .1863 | 62 (76.5) | 53 (60.2) | .0231 |
| Positive (> 20%) | 21 (26.2) | 35 (43.2) | 26 (30.6) | 30 (39.5) | 23 (27.4) | 31 (36.9) | 19 (23.5) | 35 (39.8) | ||||
| HER2/neu | ||||||||||||
| Negative (0.1+) | 72 (90) | 62 (77.5) | .0321 | 73 (86.9) | 61 (80.3) | .2555 | 73 (86.9) | 68 (81.9) | .3751 | 71 (87.7) | 71 (81.6) | .2790 |
| Positive (3+) | 8 (10) | 18 (22.5) | 11 (13.1) | 15 (19.7) | 11 (13.1) | 15 (18.1) | 10 (12.3) | 16 (18.4) | ||||
Positive CD4 expression was observed in patients ≤57 years. (P = .0238). Negative CD4 expression was associated with positive ER (P = .047) and both negative MIB1 (P = .0011) and HER2/neu (P = .0101). Stromal CD4 expression showed the same associations of CD4 (P = .0052, P = .0239, P = .0321 respectively). Moreover, positive sCD4 expression was associated with tumor size ≤2 cm (P = .0216) and negative sCD4 expression was observed in patients with positive PgR with a trend of significance (P = .0538). Negative CD8 expression was associated with the histological grade G2 (P = .033) and negative MIB1 (P = .0421). sCD8 expression was associated with positive ER (P = .0156), while negative iCD8 expression showed the same associations of CD8 (P = .0039, P = .0231) and with a tumor size ≤2 cm (P = .0026). For what concern FOXP3+, we only found an association between negative FOXP3 and positive PgR status (P = .0369). Finally, we investigated the relationships between immunocheckpoint molecule expression (CTLA4, PD-1 and PD-L1) and clinicopathological characteristics (Table 3). Negative CTLA4 expression was associated with the histological grade G2 (P = .0103). Significant associations were found between the expression of PD-1 and PD-L1 in TILs and the lymph node status (P < .0001, P < .0001 respectively). Negative PD-L1 expression in TILs was associated with both negative MIB1 and HER2/neu status (P = .0028, P = .0054 respectively). Negative PD-L1 expression in tumor cells was significantly associated with the histological types considered (P = .0287).
Table 3.
Relationship Between Immunocheckpoint Markers and Clinicopathological Characteristics
| Characteristics | CTLA4 in TILs |
PD-1 in TILs |
PD-L1 in TILs |
PD-L1 in Tumor Cells |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||||
| Age | ||||||||||||
| ≤ 57 | 53 (53) | 38 (52.1) | .9021 | 55 (48.2) | 25 (59.5) | .2113 | 68 (51.9) | 19 (54.3) | .8025 | 83 (51.9) | 5 (71.4) | .3105 |
| > 57 | 47 (47) | 35 (47.9) | 59 (51.8) | 17 (40.5) | 63 (48.1) | 16 (45.7) | 77 (48.1) | 2 (28.6) | ||||
| Histological type | ||||||||||||
| IDC | 83 (83) | 62 (84.9) | .9142 | 98 (86) | 35 (83.3) | .4111 | 108 (82.5) | 31 (88.6) | .2273 | 135 (84.4) | 5 (71.4) | .0287 |
| ILC | 10 (10) | 7 (9.6) | 7 (6.1) | 5 (11.9) | 16 (12.2) | 1 (2.8) | 17 (10.6) | 0 (0) | ||||
| Other | 7 (7) | 4 (5.5) | 9 (7.9) | 2 (4.8) | 7 (5.3) | 3 (8.6) | 8 (5) | 2 (28.6) | ||||
| Tumor size (cm) | ||||||||||||
| ≤ 2 cm | 63 (64.3) | 48 (65.8) | .8423 | 72 (63.7) | 23 (54.8) | .309 | 86 (65.6) | 18 (52.9) | .1714 | 99 (62.3) | 6 (85.7) | .2079 |
| > 2 cm | 35 (35.7) | 25 (34.2) | 41 (36.3) | 19 (45.2) | 45 (34.4) | 16 (47.1) | 60 (37.7) | 1 (14.3) | ||||
| Lymph node status | ||||||||||||
| Negative | 57 (58.2) | 48 (65.8) | .3132 | 70 (61.9) | 26 (28) | <.0001 | 74 (100) | 25 (73.5) | <.0001 | 95 (60.1) | 5 (71.4) | .5493 |
| Positive | 41 (41.8) | 25 (34.2) | 43 (38.1) | 67 (72) | 0 (0) | 9 (26.5) | 63 (39.9) | 2 (28.6) | ||||
| Histological grade | ||||||||||||
| G1 | 11 (11.2) | 4 (5.6) | .0103 | 8 (7.1) | 3 (7.5) | .164 | 13 (10.1) | 1 (7.1) | .1903 | 13 (8.3) | 1 (14.3) | .2979 |
| G2 | 64 (65.3) | 35 (49.3) | 69 (61.6) | 18 (45) | 80 (62) | 12 (85.8) | 91 (58.4) | 2 (28.6) | ||||
| G3 | 23 (23.5) | 32 (45.1) | 35 (31.3) | 19 (47.5) | 36 (27.9) | 1 (7.1) | 52 (33.3) | 4 (57.1) | ||||
| Receptor status | ||||||||||||
| ER-negative (≤10%) | 18 (18.2) | 10 (13.9) | .4539 | 18 (16.1) | 9 (21.4) | .4362 | 21 (16.2) | 7 (20.6) | .5407 | 27 (17.1) | 2 (28.6) | .4348 |
| ER-positive (> 10%) | 81 (81.8) | 62 (86.1) | 94 (83.9) | 33 (78.6) | 109 (83.8) | 27 (79.4) | 131 (82.9) | 5 (71.4) | ||||
| PgR-negative (≤ 10%) | 32 (32.3) | 28 (38.9) | .3744 | 41 (36.6) | 16 (38.1) | .8647 | 43 (33.1) | 16 (47.1) | .1304 | 55 (34.8) | 4 (57.1) | .2277 |
| PgR-positive (> 10%) | 67 (67.7) | 44 (61.1) | 71 (63.4) | 26 (61.9) | 87 (66.9) | 18 (52.9) | 103 (65.2) | 3 (42.9) | ||||
| MIB1 | ||||||||||||
| Negative (≤20%) | 70 (77.7) | 71 (74.7) | .5289 | 78 (69.6) | 24 (57.1) | .1441 | 96 (73.8) | 16 (47.1) | .0028 | 110 (69.6) | 3 (42,9) | .1358 |
| Positive (> 20%) | 29 (29.3) | 24 (25.3) | 34 (30.4) | 18 (42.9) | 34 (26.2) | 18 (52.9) | 48 (30.4) | 4 (57.1) | ||||
| HER2/neu | ||||||||||||
| Negative (0,1+) | 85 (85.9) | 59 (83.1) | .622 | 94 (83.9) | 34 (82.9) | .882 | 113 (87.6) | 23 (67.6) | .0054 | 133 (84.7) | 4 (57.1) | .0543 |
| Positive (3+) | 14 (14.1) | 12 (16.9) | 18 (16.1) | 7 (17.1) | 16 (12.4) | 11 (32.4) | 24 (15.3) | 3 (42.9) | ||||
Associations Between TILs and Immunocheckpoint Marker Expression
We first analyzed the association between total immune cell markers (CD4, CD8 and FOXP3) and immunocheckpoint markers (CTLA4, PD-1, PD-L1) (Table 4a). Considering the dichotomized variables we observed that both CD4 and CD8 expressions were significantly associated with CTLA4 (P = .005, P = .0002, respectively), PD-1 (.0004, .0005, respectively) and PD-L1 (P < .0001, P < .0001, respectively) expressions in total TILs; while only CD4 expression was significantly associated with PD-L1 expression in tumor cells (P = .014). Moreover, FOXP3 expression was associated with CTLA4 and PD-L1 expressions in TILs (P < .0001, P < .0001).
Table 4.
Association Between TILs and Immunocheckpoint Markers
| a | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| CD4 |
CD8 |
FOXP3 |
|||||||
| Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||||
| CTLA4 in TILs | |||||||||
| Negative | 54 (70.1) | 38 (48.1) | .005 | 59 (71.9) | 36 (43.4) | .0002 | 59 (80.8) | 35 (37.6) | <.0001 |
| Positive | 23 (29.9) | 41 (51.9) | 23 (28.1) | 47 (56.6) | 14 (19.2) | 58 (62.4) | |||
| PD-1 in TILs | |||||||||
| Negative | 60 (85.7) | 45 (59.2) | .0004 | 63 (85.1) | 46 (59.7) | .0005 | 47 (78.3) | 60 (67.4) | .148 |
| Positive | 10 (14.3) | 31 (40.8) | 11 (14.9) | 31 (40.3) | 13 (21.7) | 29 (32.6) | |||
| PD-L1 in TILs | |||||||||
| Negative | 72 (96) | 45 (59.2) | <.0001 | 77 (96.2) | 49 (60.5) | <.0001 | 68 (93.1) | 59 (66.3) | <.0001 |
| Positive | 3 (4) | 31 (40.8) | 3 (3.8) | 32 (39.5) | 5 (6.9) | 30 (33.7) | |||
| PD-L1 in tumor cells | |||||||||
| Negative | 75 (100) | 70 (90.9) | .014 | 79 (98.7) | 76 (92.7) | .117 | 71 (95.9) | 85 (95.5) | .890 |
| Positive | 0 (0) | 7 (9.1) | 1 (1.3) | 6 (7.3) | 3 (4.1) | 4 (4.5) | |||
| b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sCD4 |
iCD4 |
sCD8 |
iCD8 |
|||||||||
| Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | Negative |
Positive |
P | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||||
| CTLA4 in TILs | ||||||||||||
| Negative | 51 (66.2) | 41 (51.9) | .070 | 53 (66.2) | 40 (52.6) | .084 | 53 (64.6) | 42 (50.6) | .069 | 49 (62.8) | 46 (52.9) | .198 |
| Positive | 26 (33.8) | 38 (48.1) | 27 (33.8) | 36 (47.4) | 29 (35.4) | 41 (49.4) | 29 (37.2) | 41 (47.1) | ||||
| PD-1 in TILs | ||||||||||||
| Negative | 60 (84.5) | 45 (60) | .001 | 60 (76.9) | 45 (67.2) | .191 | 63 (84) | 46 (60.5) | .001 | 61 (85.9) | 48 (60) | .0004 |
| Positive | 11 (15.5) | 30 (40) | 18 (23.1) | 22 (32.8) | 12 (16) | 30 (39.5) | 10 (14.1) | 32 (40) | ||||
| PD-L1 in TILs | ||||||||||||
| Negative | 62 (84.9) | 55 (70.5) | .035 | 71 (91) | 46 (63) | <.0001 | 67 (83.7) | 59 (72.8) | .094 | 70 (92.1) | 56 (65.9) | <.0001 |
| Positive | 11 (15.1) | 23 (29.5) | 7 (9) | 27 (37) | 13 (16.3) | 22 (27.2) | 6 (7.9) | 29 (34.1) | ||||
| PD-L1 in tumor cells | ||||||||||||
| Negative | 72 (98.6) | 73 (92.4) | .118 | 79 (100) | 66 (90.4) | .005 | 77 (96.2) | 78 (95.1) | .725 | 75 (98.7) | 80 (93) | .122 |
| Positive | 1 (1.4) | 6 (7.6) | 0 (0) | 7 (9.6) | 3 (3.8) | 4 (4.9) | 1 (1.3) | 6 (7) | ||||
s = stromal.
i = intratumoral.
We secondly analyzed the association between immune cell marker subtypes respect to their location (sCD4, iCD4, sCD8, iCD8) and immunocheckpoint markers (CTLA4, PD-1, PD-L1) (Table 4b). The expression of sCD4 was associated with PD-1 and PD-L1 expression in TILs (P = .001, P = .035 respectively), and iCD4 expression was associated with PD-L1 expression in TILs and in tumor cells (P < .0001, P < .005, respectively). Stromal CD8 was associated with PD-1 expression in TILs (P = .001) and iCD8 expression was associated with both PD-1 and PD-L1 expressions in TILs (P = .0004, P < .0001 respectively).
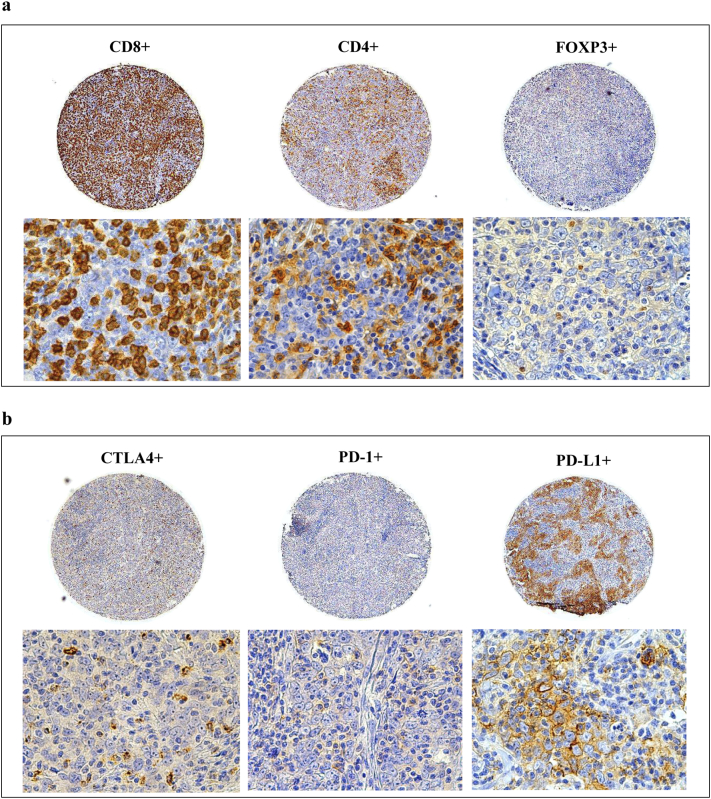
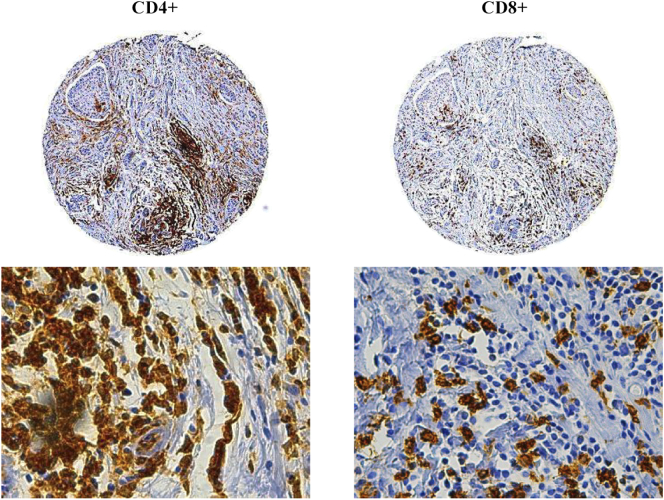
Figure 1 shows some examples of CD4, CD8, FOXP3, CTLA4, PD-1 and PD-L1 immunohistochemical staining patterns in TMA samples. Supplementary Figure 1 shows some examples of stromal CD4+ and CD8+ T cells.
Figure 1.
Immunoreactivity of TIL subtypes and immunocheckpoint molecules on breast cancer (BC) tissue microarrays (TMAs).
Immunohistochemical expression of CD4+, CD8+, FOXP3+ (a) and PD-L1, PD-1, CTLA4 (b) on the same TMA sample. Panoramic views of the TMA cores at original magnification 50×, and detail views at original magnification 630×.
Supplementary Figure 1.
Immunoreactivity of stromal CD4+ and CD8+ T cells on breast cancer (BC) tissue microarrays (TMAs). Immunohistochemical expression of stromal CD4+ and CD8+ on the same TMA sample. Panoramic views of the TMA cores at original magnification 50×, and detail views at original magnification 630×.
Immune Marker Expression and Patient Outcome
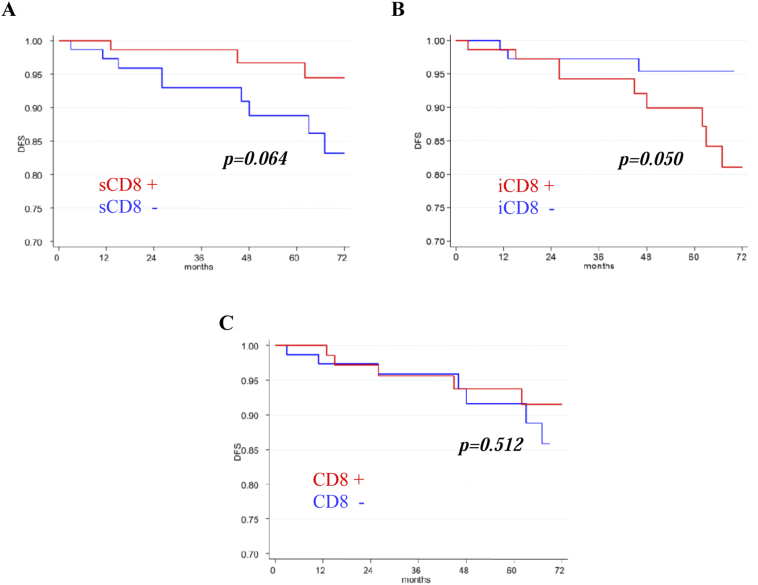
Univariate survival analysis was carried out including the expressions of immune markers with respect to DFS (Table 5). We found that the phenotypes with negative sCD8 and positive iCD8 showed a trend toward a shorter 5-year DFS (P = .064 and P = .050 respectively). No significant association was found between DFS and the other markers. Kaplan–Meier curves confirmed the results of the univariate analysis. Patients with negative sCD8 (Figure 2A) and patients with positive iCD8 (Figure 2B) tended toward a poorer DFS (P = .064 and P = .050 respectively). No difference, respect to DFS, was found between the subgroups of patients with positive and negative CD8 expression (P = .512) (Figure 2C). The analyses respect to OS could not be performed due to the low number of events (deaths).
Table 5.
Univariate Analysis with Respect to DFS
| N. Pts | N. Events | % 5-Year DFS (95% CI) |
P | HR (95% CI) |
|
|---|---|---|---|---|---|
| Overall | 160 | 13 | 92 (88–97) | - | - |
| CD4 | |||||
| Negative | 73 | 7 | 92 (85–99) | 1.00 | |
| Positive | 70 | 6 | 90 (82–99) | .906 | 1.07 (0.36–3.19) |
| CD8 | |||||
| Negative | 77 | 7 | 92 (84–99) | 1.00 | |
| Positive | 74 | 5 | 94 (88–100) | .512 | 0.68 (0.22–2.15) |
| FOXP3 | |||||
| Negative | 65 | 5 | 95 (89–100) | 1.00 | |
| Positive | 84 | 7 | 91 (83–98) | .793 | 1.17 (0.37–3.68) |
| CTLA4 | |||||
| Negative | 87 | 9 | 90 (83–97) | 1.00 | |
| Positive | 65 | 3 | 96 (91–100) | .193 | 0.43 (0.12–1.59) |
| PD-1 in TILs | |||||
| Negative | 103 | 10 | 91 (85–97) | 1.00 | |
| Positive | 36 | 3 | 91 (82–100) | .895 | 0.92 (0.25–3.33) |
| PD-L1 in TILs | |||||
| Negative | 115 | 8 | 93 (88–99) | 1.00 | |
| Positive | 31 | 4 | 88 (74–100) | .228 | 2.06 (0.62–6.85) |
| PD-L1 in tumor cells | |||||
| Negative | 141 | 11 | 92 (87–97) | 1.00 | |
| Positive | 6 | 1 | 100 | .534 | 1.89 (0.24–14.69) |
| sCD4 | |||||
| Negative | 75 | 7 | 92 (86–99) | 1.00 | |
| Positive | 68 | 6 | 90 (82–99) | .832 | 1.13 (0.38–3.35) |
| iCD4 | |||||
| Negative | 77 | 6 | 92 (86–99) | 1.00 | |
| Positive | 66 | 7 | 90 (82–99) | .474 | 1.49 (0.50–4.42) |
| sCD8 | |||||
| Negative | 76 | 9 | 89 (81–97) | 1.00 | |
| Positive | 75 | 3 | 97 (92–100) | .064 | 0.31 (0.08–1.15) |
| iCD8 | |||||
| Negative | 75 | 3 | 95 (90–100) | 1.00 | |
| Positive | 76 | 9 | 90 (82–98) | .050 | 3.42 (0.93–12.67) |
s = stromal.
i = intratumoral.
Figure 2.
Kaplan–Meier survival curves.
Disease free survival (DFS) according to sCD8 (A), iCD8 (B) and total CD8 (C) expressions in 160 breast cancer patients.
Discussion
The prognostic role of TILs has been demonstrated in BC [23]. It is generally thought that high TIL infiltration correlates with complete response rate and improve survival [24]. However, it remains yet to be elucidated which subset of TILs has a role in BC progression and prognosis. In the adaptive immune response against cancer, CD8+ cytotoxic T cells play one of the most important role in killing cancer cells [25]. CD8+ T cells have been shown having a protective prognostic effect in many cancer types, such as colorectal [26], lung [27], esophageal [28], ovarian [29], renal [30], pancreatic [31], liver [32] and breast [5], [6], [7], [8] cancers.
In the present study, we found that total CD8+ T cells were not significantly associated with DFS. However, we observed that the impact of CD8+ T cells on BC DFS depended on their location. Indeed, univariate analysis reported that patients with iCD8+ T cell overexpression showed a trend toward a worse 5-years DFS, while those with sCD8+ T cell overexpression showed a trend toward a better 5-years DFS. These data were also confirmed by the Kaplan Meier curves.
Our results are opposite to those found by Chen et al. who found a strong infiltrate of iCD8+ T cells associated with improved BC OS and DFS. [14]. However, the study of Chen was conducted on the subgroup of node negative BC, while in our study, the 36% of all invasive BC samples presented a positive lymph node status. In addition, our data show that iCD8+ but no sCD8+ expression was associated with more aggressive clinicopathological characteristic tumors, such as size >2 cm, high histological grade and high proliferative activity and this is consistent with the negative prognostic role referred to iCD8+ T cells. Considering the different median values for iCD8+ and sCD8+, we also observed that there was no difference in the percentage of tumor samples with iCD8+ and sCD8+ T cell overexpression (52% versus 50.6% respectively). Moreover, analyzing the numerical counts of TILs we observed that sCD8+ were more than iCD8+, thus we hypothesized that the negative impact of the latter on survival did not depend on the number of TILs. A biological explanation for the different effect of sCD8+ and iCD8+ T cells on BC prognosis could be related to the activation of molecular mechanisms that block their cytotoxic function against cancer cells. Stromal CD8+ T cells expression, still protects the host against cancer development, because TIL function has not yet been impaired. Conversely, iCD8+ T cell function, is negatively affected and compromised by the presence of the tumor mass. One of the mechanisms which could inhibit CD8+ T cell function is the expression of immunocheckpoint molecules such PD-1 and PD-L1. The inhibition of PD-L1 emerges as a reliable therapeutic option in several tumor types including BC. It is known that PD-L1 binding to PD-1 blocks T cell activation and the immune response against tumor [17], [33]. The characterization of PD-L1 expression as a negative prognostic factor of poor clinical outcome in BC and other tumors has been reported [34], [35]. A recent meta-analysis showed an association between shorter OS and PD-L1 expression in all types of BC [18]. Other authors found an association between TIL PD-L1 expression and DFS in TNBC, but no effect on OS or with regard to PD-L1 tumoral expression [36].
We found that PD-1 and PD-L1 expression levels are not associated with patient survival, but only with some adverse clinicopathological characteristics, such as a positive lymph node status or MIB1. Our data agree with those of Kitano A. et al. who reported that expression levels of PD-L1 and/or PD-1 are not sufficient prognostic factors in BC [37] and with those of Wang et al. who reported no effect on outcome for high expression of PD-L1, except for the basal like subtypes [19].
Studies aimed at clarifying the relationship between PD-1, PD-L1 and TILs in patients with BC are few [37], [38] and the relationship between total CD8+ T cells and PD-1/PD-L1 is even less studied. Chen et al. observed PD-L1 expression in patients with CD8+ low levels and reported an association between patients classified as PD-L1-high/CD8-low and a worse survival [39]. Moreover, to date, there are no studies on the relationship between PD-L1/PD-1 and CD8+ T cell location.
In our work, we analyzed the association between immunocheckpoint molecule (CTLA-4 and PD-L1/PD-1) expression and TIL subtypes with regard to their location. We did not find any association between sCD8+ or iCD8+ and CTLA4+. Interestingly, we showed that TIL expression of both PD-1 and PD-L1, was significantly associated with iCD8+ T cells increase, while only TIL expression of PD-1 was associated with positive sCD8, suggesting that the co-expression of PD-L1 and PD-1 is necessary to inhibit iCD8+ T cell function. The association of both these markers with the iCD8+ T cells could explain iCD8+ involvement in a worse prognosis. We also thought that the association of only PD-1 expression with sCD8+ T cells, was not sufficient to block sCD8+ function. This is consistent with the observed trend toward a better DFS of patients overexpressed sCD8+ T cells. Further survival analysis comparing patients overexpressing iCD8+ and PD-L1/PD-1 with those overexpressing iCD8+ and PD-1 are necessary to confirm our theory.
Our preliminary work presents some limits. One weakness is that there is no strong relationship between our markers and patient survival, probably due to the low number of deaths and relapses. Moreover, the major part of patients included in our retrospective case studies had favorable clinical pathological characteristics, chosen not on the basis of these parameters, but only on the availability of the follow up.
Another, is the use of TMAs which could eventually mislead in stromal TIL quantification. However, the construction of TMAs was made by selecting, for each core, tumor areas with significant amounts of infiltrate, so that the assessment of the stromal and intratumoral TILs could be performed for all samples studied. Each sample was also arrayed in triplicate cores to overcome tumor variability and heterogeneity. The indication of TIL evaluation and quantification, have been defined only for the whole TIL population, according to the International Working Group criteria [40], but not for their subtypes. However, all recent studies revealed significant differences when the evaluation of TILs was based on their location. In fact, in our work, the statistical significances we found with respect to patient survival, concerned the analysis of iCD8 and sCD8, but not total CD8 +.
Conclusions
In summary, our preliminary results show that iCD8+ T cells, but no sCD8+ T cells, have a negative impact on BC DFS and this effect could be due to the overexpression of PD-L1/PD-1 pathway. Indeed, although PD-L1 and PD-1 expression levels are not sufficient prognostic factors for survival, they are both significantly associated with the iCD8+ expression. In view of these data further studies, in a large cohort of patients are required to strengthen the statistical significance of our work. Moreover, the mechanisms inducing PD-L1/PD-1 activation in iCD8+ T cells are still to investigate.
Data Availability
All data generated or analyzed during this study were included in this published article.
The following are the supplementary data related to this article.
Tumor characteristics of 180 invasive breast cancer patients.
Concentration, source, staining of antibodies and cut off.
Ethical Guidelines
This study was approved by the Ethic Committee of Istituto Tumori “Giovanni Paolo II” with the reference number 234/CE 13–11-2017. Before undergoing routine surgery, all patients signed an informed consent form authorizing the Institute to utilize their removed biological tissue for research purposes according to ethical standards.
Authors' Contributions
IC and LS performed the experiments. AM and IC participated in the drafting of the manuscript. ES performed the statistical analyses. AS provided the tumor characteristics. NS helped to revise the manuscript. AM planned the study and is responsible for the critical and scientific supervision. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Francesco Fanelli for technical assistance.
Footnotes
Declaration of Interest: The authors declare that there is no conflict of interest regarding the publication of this paper.
Funding: This work was supported by funding from the Italian Ministry of Health “Ricerca Corrente 2018”.
Contributor Information
Ivana Catacchio, Email: ivana.84@libero.it.
Nicola Silvestris, Email: silvestrisnicola@libero.it.
Emanuela Scarpi, Email: emanuela.scarpi@irst.emr.it.
Laura Schirosi, Email: l.schirosi@oncologico.bari.it.
Anna Scattone, Email: a.scattoneanatopat@alice.it.
Anita Mangia, Email: a.mangia@oncologico.bari.it.
References
- 1.Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac J Cancer Prev. 2016;17(S3):43–46. doi: 10.7314/apjcp.2016.17.s3.43. [DOI] [PubMed] [Google Scholar]
- 2.Saponaro C, Vagheggini A, Scarpi E, Centonze M, Catacchio I, Popescu O, Pastena MI, Giotta F, Silvestris N, Mangia A. NHERF1 and tumor microenvironment: a new scene in invasive breast carcinoma. J Exp Clin Cancer Res. 2018;37(1):96. doi: 10.1186/s13046-018-0766-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 2011;25(24):2559–2572. doi: 10.1101/gad.169029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badalamenti G, Fanale D, Incorvaia L, Barraco N, Listì A, Maragliano R, Vincenzi B, Calò V, Iovanna JL, Bazan V. Role of tumor-infiltrating lymphocytes in patients with solid tumors: Can a drop dig a stone? Cell Immunol. 2018 doi: 10.1016/j.cellimm.2018.01.013. [pii:S0008-8749(18)30014-5] [DOI] [PubMed] [Google Scholar]
- 5.Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14(2):R48. doi: 10.1186/bcr3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, Earl HM, Poole CJ, Hiller L, Dunn JA. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 7.Miyashita M, Sasano H, Tamaki K, Hirakawa H, Takahashi Y, Nakagawa S, Watanabe G, Tada H, Suzuki A, Ohuchi N. Prognostic significance of tumor-infiltrating CD8+ and FOXP3+ lymphocytes in residual tumors and alterations in these parameters after neoadjuvant chemotherapy in triple-negative breast cancer: a retrospective multicenter study. Breast Cancer Res. 2015;17:124. doi: 10.1186/s13058-015-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao Y, Qu Q, Chen X, Huang O, Wu J, Shen K. The Prognostic Value of Tumor-Infiltrating Lymphocytes in Breast Cancer: A Systematic Review and Meta-Analysis. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0152500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, Lang R, Zhao J, Zhang X, Pringle GA, Fan Y, Yin D, Gu F, Yao Z, Fu L. CD8⁺ cytotoxic T cell and FOXP3⁺ regulatory T cell infiltration in relation to breast cancer survival and molecular subtypes. Breast Cancer Res Treat. 2011;130(2):645–655. doi: 10.1007/s10549-011-1647-3. [DOI] [PubMed] [Google Scholar]
- 10.Syed Khaja AS, Toor SM, El Salhat H, Faour I, Ul Haq N, Ali BR, Elkord E. Preferential accumulation of regulatory T cells with highly immunosuppressive characteristics in breast tumor microenvironment. Oncotarget. 2017;8(20):33159–33171. doi: 10.18632/oncotarget.16565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda N, Shimazu K, Naoi Y, Morimoto K, Shimomura A, Shimoda M, Kagara N, Maruyama N, Kim SJ, Noguchi S. Intratumoral regulatory T cells as an independent predictive factor for pathological complete response to neoadjuvant paclitaxel followed by 5-FU/epirubicin/cyclophosphamide in breast cancer patients. Breast Cancer Res Treat. 2012;136(1):107–116. doi: 10.1007/s10549-012-2245-8. [DOI] [PubMed] [Google Scholar]
- 12.West NR, Kost SE, Martin SD, Milne K, Deleeuw RJ, Nelson BH, Watson PH. Tumour-infiltrating FOXP3(+) lymphocytes are associated with cytotoxic immune responses and good clinical outcome in estrogen receptor-negative breast cancer. Br J Cancer. 2013;108(1):155–162. doi: 10.1038/bjc.2012.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Lee AH, Ellis IO, Green AR. An evaluation of the clinical significance of FOXP3+ infiltrating cells in human breast cancer. Breast Cancer Res Treat. 2011;127(1):99–108. doi: 10.1007/s10549-010-0987-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, Chen X, Zhou E, Chen G, Qian K, Wu X, Miao X, Tang Z. Intratumoral CD8⁺ cytotoxic lymphocyte is a favorable prognostic marker in node-negative breast cancer. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0095475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Shen T, Siegal GP, Wei S. The CD4/CD8 ratio of tumor-infiltrating lymphocytes at the tumor-host interface has prognostic value in triple-negative breast cancer. Hum Pathol. 2017;69:110–117. doi: 10.1016/j.humpath.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Gatalica Z, Snyder C, Maney T, Ghazalpour A, Holterman DA, Xiao N, Overberg P, Rose I, Basu GD, Vranic S. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomark Prev. 2014;23(12):2965–2970. doi: 10.1158/1055-9965.EPI-14-0654. [DOI] [PubMed] [Google Scholar]
- 17.Baptista MZ, Sarian LO, Derchain SF, Pinto GA, Vassallo J. Prognostic significance of PD-L1 and PD-L2 in breast cancer. Hum Pathol. 2016;47(1):78–84. doi: 10.1016/j.humpath.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Sun H, Zhao S, Wang Y, Pu H, Wang Y, Zhang Q. Expression of PD-L1 and prognosis in breast cancer: a meta-analysis. Oncotarget. 2017;8(19):31347–31354. doi: 10.18632/oncotarget.15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZQ, Milne K, Derocher H, Webb JR, Nelson BH, Watson PH. PD-L1 and intratumoral immune response in breast cancer. Oncotarget. 2017;8(31):51641–51651. doi: 10.18632/oncotarget.18305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Yang J, Jiao S, Li Y, Zhang W, Wang J. Cytotoxic T lymphocyte antigen 4 expression in human breast cancer: implications for prognosis. Cancer Immunol Immunother. 2015;64(7):853–860. doi: 10.1007/s00262-015-1696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2017;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 22.Mangia A, Scarpi E, Partipilo G, Schirosi L, Opinto G, Giotta F, Simone G. NHERF1 together with PARP1 and BRCA1 expression as a new potential biomarker to stratify breast cancer patients. Oncotarget. 2017;8:65730–65742. doi: 10.18632/oncotarget.19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin G, Fan X, Zhu W, Huang C, Zhuang W, Xu H, Lin X, Hu D, Huang Y, Jiang K. Prognostic significance of PD-L1 expression and tumor infiltrating lymphocyte in surgically resectable non-small cell lung cancer. Oncotarget. 2017;8(48):83986–83994. doi: 10.18632/oncotarget.20233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burugu S, Asleh-Aburaya K, Nielsen TO. Immune infiltrates in the breast cancer microenvironment: detection, characterization and clinical implication. Breast Cancer. 2017;24(1):3–15. doi: 10.1007/s12282-016-0698-z. [DOI] [PubMed] [Google Scholar]
- 25.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 26.Pagès F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27(35):5944–5951. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 27.Kawai O, Ishii G, Kubota K, Murata Y, Naito Y, Mizuno T, Aokage K, Saijo N, Nishiwaki Y, Gemma A. Predominant infiltration of macrophages and CD8(+) T Cells in cancer nests is a significant predictor of survival in stage IV non small cell lung cancer. Cancer. 2008;113(6):1387–1395. doi: 10.1002/cncr.23712. [DOI] [PubMed] [Google Scholar]
- 28.Cho Y, Miyamoto M, Kato K, Fukunaga A, Shichinohe T, Kawarada Y, Hida Y, Oshikiri T, Kurokawa T, Suzuoki M. CD4+ and CD8+ T cells cooperate to improve prognosis of patients with esophageal squamous cell carcinoma. Cancer Res. 2003;63(7):1555–1559. [doi:Published April 2003 PMID.12670904] [PubMed] [Google Scholar]
- 29.Hamanishi J, Mandai M, Abiko K, Matsumura N, Baba T, Yoshioka Y, Kosaka K, Konishi I. The comprehensive assessment of local immune status of ovarian cancer by the clustering of multiple immune factors. Clin Immunol. 2011;141(3):338–347. doi: 10.1016/j.clim.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M, Suzuki Y, Shintaku I, Nagura H, Ohtani H. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61(13):5132–5136. [doi: Published July 2001. PMID:11431351] [PubMed] [Google Scholar]
- 31.Fukunaga A, Miyamoto M, Cho Y, Murakami S, Kawarada Y, Oshikiri T, Kato K, Kurokawa T, Suzuoki M, Nakakubo Y. CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas. 2004;28(1):e26–e31. doi: 10.1097/00006676-200401000-00023. [PMID, 14707745] [DOI] [PubMed] [Google Scholar]
- 32.O'Beirne J, Farzaneh F, Harrison PM. Generation of functional CD8+ T cells by human dendritic cells expressing glypican-3 epitopes. J Exp Clin Cancer Res. 2010;29:48. doi: 10.1186/1756-9966-29-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He G, Zhang H, Zhou J, Wang B, Chen Y, Kong Y, Xie X, Wang X, Fei R, Wei L. Peritumoural neutrophils negatively regulate adaptive immunity via the PD-L1/PD-1 signalling pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34:141. doi: 10.1186/s13046-015-0256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muenst S, Soysal SD, Gao F, Obermann EC, Oertli D, Gillanders WE. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2013;139(3):667–676. doi: 10.1007/s10549-013-2581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muenst S, Schaerli AR, Gao F, Däster S, Trella E, Droeser RA. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2014;146(1):15–24. doi: 10.1007/s10549-014-2988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Wetherilt CS, Krishnamurti U, Yang J, Ma Y, Styblo TM, Meisel JL, Peng L, Siddiqui MT, Cohen C. Stromal PD-L1 Expression Is Associated With Better Disease-Free Survival in Triple-Negative Breast Cancer. Am J Clin Pathol. 2016;146(4):496–502. doi: 10.1093/ajcp/aqw134. [DOI] [PubMed] [Google Scholar]
- 37.Kitano A, Ono M, Yoshida M, Noguchi E, Shimomura A, Shimoi T, Kodaira M, Yunokawa M, Yonemori K, Shimizu C. Tumour-infiltrating lymphocytes are correlated with higher expression levels of PD-1 and PD-L1in early breast cancer. ESMO Open. 2017;2(2) doi: 10.1136/esmoopen-2016-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polónia A, Pinto R, Cameselle-Teijeiro JF, Schmitt FC, Paredes J. Prognostic value of stromal tumour infiltrating lymphocytes and programmed cell death-ligand 1 expression in breast cancer. J Clin Pathol. 2017;70(10):860–867. doi: 10.1136/jclinpath-2016-203990. [DOI] [PubMed] [Google Scholar]
- 39.Chen S, Wang RX, Liu Y, Yang WT, Shao ZM. PD-L1 expression of the residual tumor serves as a prognostic marker in local advanced brest cancer after neoadjuvant chemotherapy. Int J Cancer. 2017;140(6):1384–1395. doi: 10.1002/ijc.30552. [DOI] [PubMed] [Google Scholar]
- 40.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F. International TILs Working Group 2014. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tumor characteristics of 180 invasive breast cancer patients.
Concentration, source, staining of antibodies and cut off.
Data Availability Statement
All data generated or analyzed during this study were included in this published article.
The following are the supplementary data related to this article.
Tumor characteristics of 180 invasive breast cancer patients.
Concentration, source, staining of antibodies and cut off.