Abstract
Despite advances in early diagnosis and treatment, cancer remains the major reason for mortality worldwide. The Runt-related transcription factor (RUNX) family has been reported to participate in diverse human diseases. However, little is known about their expression and prognostic values in human leukemia. Herein, we conducted a detailed cancer versus normal analysis. The mRNA expression levels of the RUNX family in various kinds of cancers, including leukemia, were analyzed via the ONCOMINE and GEPIA (Gene Expression Profiling Interactive Analysis) databases. We observed that the mRNA expression levels of RUNX1, RUNX2, and RUNX3 were all increased in most cancers compared with normal tissues, especially in leukemia. Moreover, the expression levels of RUNX1, RUNX2, and RUNX3 are also highly expressed in almost all cancer cell lines, particularly in acute myeloid leukemia (AML) cell lines, analyzed by Cancer Cell Line Encyclopedia (CCLE) and European Bioinformatics Institute (EMBL-EBI) databases. Further, the LinkedOmics and GEPIA databases were used to evaluate the prognostic values. In survival analyses based on LinkedOmics, higher expression of RUNX1 and RUNX2 indicated a better overall survival (OS), but with no significance, whereas increased RUNX3 revealed a poor OS in leukemia. In addition, the GEPIA dataset was also used to perform survival analyses, and results manifested that the expression of RUNX1 and RUNX2 had no remarkable correction with OS in leukemia, but it showed highly expressed RUNX3 was significantly related with poor OS in leukemia. In conclusion, the RUNX family showed significant expression differences between cancer and normal tissues, especially leukemia, and RUNX3 could be a promising prognostic biomarker for leukemia.
Keywords: Runt-related transcription factor, leukemia, ONCOMINE, Cancer Cell Line Encyclopedia
Introduction
The Runt-related transcription factors (RUNXs), known as the polyomavirus enhancer-binding protein 2 (PEBP2) or core-binding factor (CBF), belong to an ancient family of metazoan genes involved in developmental processes.1 They are heterodimers of α and β subunits. In humans, the α subunit comprises three proteins, RUNX1 (AML1/CBFA2/PEBP2aB), RUNX2 (AML3/CBFA1/PEBP2aA), and RUNX3 (AML2/CBFA3/PEBP2aC), that contain an evolutionarily conserved 128-amino acid Runt domain responsible for DNA binding and heterodimerization with the β subunit. The β subunit includes a single protein, RUNX β (also known as PEBP2β or CBFβ), that does not contain a DNA-binding domain but allosterically enhances the DNA-binding activity of the α subunit and regulates its turnover by protecting it from ubiquitin-proteasome-mediated degradation.2, 3 Through multiple protein-interacting partners, RUNX proteins have been involved in diverse signaling pathways and cellular processes. RUNX transcription factors contribute to hematopoiesis and are frequently implicated in hematologic malignancies. All three RUNX isoforms are expressed at the earliest stages of hematopoiesis.
RUNX1–3 are key transcriptional regulators involved in several major developmental pathways including hematopoiesis, neurogenesis, and skeletogenesis.1 RUNX1, known as acute myeloid leukemia 1 because of the discovery of its gene sequence from a human patient with acute myeloid leukemia,4 is among the most frequently mutated genes in human leukemias.5, 6, 7 Over the past 20 years, studies have elucidated many important functions of RUNX1 in hematopoietic development, hematopoietic stem cell homeostasis, and various blood malignancies. Goyama et al.8 reported that RUNX1 overexpression inhibited the growth of CB cells by inducing myeloid differentiation, whereas a certain level of RUNX1 activity was required for the growth of acute myeloid leukemia (AML) cells. They also found that human AML cells are more sensitive to RUNX1 inhibition probably due to their reduced RUNX activities.8 In mice, Runx1 is crucial for the generation and maintenance of hematopoietic stem cells (HSCs).9 Hence RUNX1 is considered a tumor suppressor in myeloid neoplasms, and inactivating RUNX1 mutations have frequently been found in patients with myelodysplastic syndrome (MDS) and cytogenetically normal AML.10 Haploinsufficiency of RUNX2 is associated with cleidocranial dysplasia (CCD), an autosomal dominant skeletal disorder.11 Moreover, RUNX2 is overexpressed in breast cancer and promotes breast cancer bone metastasis by increasing integrin α5-mediated colonization.12 Kuo et al.13 also reported that Runx2 induced AML in cooperation with Cbfbeta-SMMHC in mice. Therefore, RUNX2 is generally regarding as an oncogene in AML. RUNX3 is expressed in a wide range of tissues and has diverse biological functions. It plays a crucial role in regulation of epithelial homeostasis in the gastrointestinal tract.14, 15 In humans, RUNX3 is a key regulator of gene expression in major developmental pathways, and it has been implicated in a multitude of cancers where it can function as a tumor suppressor or oncogene.16, 17 RUNX3 is reported to be a tumor suppressor in a variety of cancers including gastric, colon, and breast cancer.16, 18 Nevertheless, it has also been revealed that RUNX3 overexpression is frequently observed and is well correlated with malignant behaviors in head and neck cancer.17 Furthermore, RUNX3 is frequently expressed in the nuclei of ovarian cancer cell lines and plays an oncogenic role in ovarian cancer.16 In addition, Selvarajan et al.19 identify RUNX3 overexpression in natural killer/T cell lymphoma (NKTL) with functional oncogenic properties. Hence the specific role of RUNX3 in different cancers has been disputed. Although to date, little is known about the expression pattern, functional role, and prognostic role of RUNX3 in leukemia.
The dysregulated expression level of RUNX factors and their relationship with clinicopathological features and prognosis have been partly reported in human leukemia. To the best of our knowledge, bioinformatics analysis has yet been applied to explore the role of the RUNX family in leukemia. RNA and DNA research, an essential component of biological and biomedical studies, have been revolutionized with the development of microarray technology.20 On the basis of the analyses of thousands of gene expressions or variation in copy numbers published online, we analyzed in detail the expression and mutations of different RUNX factors in patients with leukemia to determine the expression patterns, potential functions, and distinct prognostic values of transcription factors (TFs) in leukemia.
Results
Transcriptional Levels of RUNXs in Patients with Lung Cancer
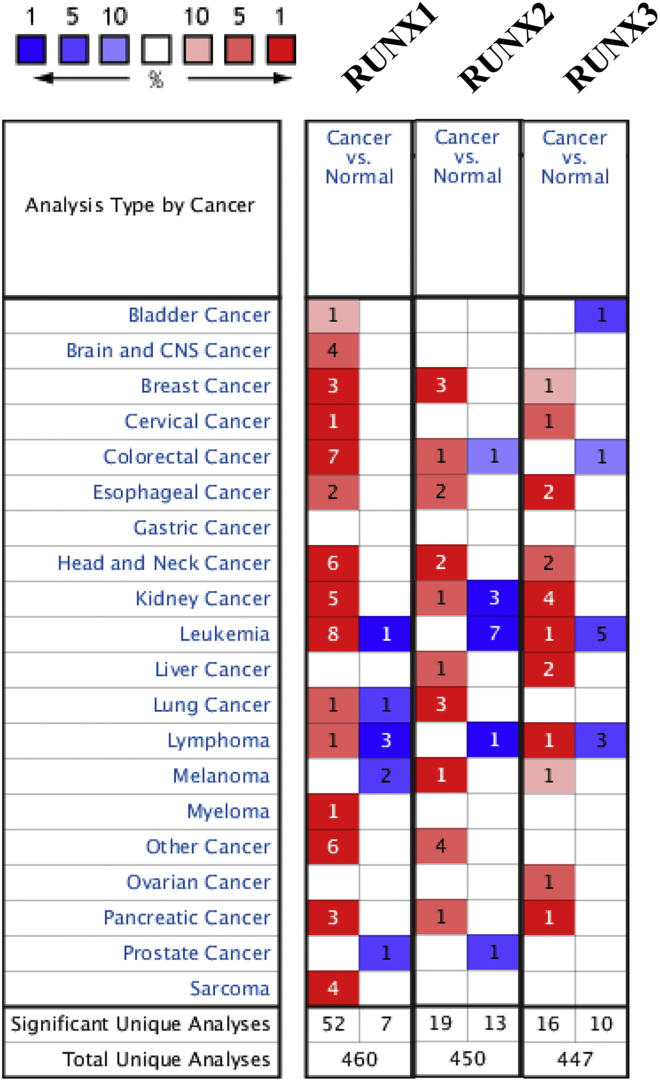
Three RUNX factors have been identified in mammalian cells. We compared the transcriptional levels of RUNXs in cancers with those in normal samples by using ONCOMINE databases (Figure 1). The mRNA expression levels of RUNX1 were significantly upregulated in patients with leukemia in six datasets. In Haferlach Leukemia 2’s dataset,21 RUNX1 is overexpressed compared with that in the normal samples in T cell acute lymphoblastic leukemia with a fold change of 2.554, and in pro-B acute lymphoblastic leukemia with a fold change of 2.559 (Table 1). In Stegmaier Leukemia’s dataset,22 RUNX1 is also overexpressed in AML with a fold change of 4.907. Haferlach Leukemia’s dataset21 showed RUNX1 expression factor with increased expression; that is, RUNX1 has a fold change of 2.464 in patients with T cell acute lymphoblastic leukemia, a fold change of 2.105 in patients with AML, a fold change of 2.342 in patients with B cell acute lymphoblastic leukemia, and a fold change of 2.161 in patients with pro-B acute lymphoblastic leukemia (Table 1). Haslinger Leukemia’s dataset23 indicated that RUNX1 overexpression is also found in chronic lymphocytic leukemia with a fold change of 3.066. In Valk Leukemia’s dataset,24 RUNX1 is overexpressed in AML with a fold change of 2.128. In Andersson Leukemia’s dataset,25 RUNX1 is overexpressed in T cell acute lymphoblastic leukemia with a fold change of 2.735. In Coustan-Smith Leukemia’s dataset,26 RUNX1 is overexpressed in B cell childhood acute lymphoblastic leukemia with a fold change of 3.126 and in T cell childhood acute lymphoblastic leukemia with a fold change of 5.366.
Figure 1.
The Transcription Levels of RUNX Factors in Different Types of Cancers (ONCOMINE)
Table 1.
The Significant Changes of RUNX Expression in Transcription Level between Different Types of Leukemia (ONCOMINE Database)
| Gene ID | Types of Leukemia versus Normal | Fold Change | p Value | t Test | References |
|---|---|---|---|---|---|
| RUNX1 | T cell acute lymphoblastic leukemia versus normal | 2.554 | 4.42E−31 | 15.793 | Haferlach Leukemia 221 |
| pro-B acute lymphoblastic leukemia versus normal | 2.559 | 8.65E−10 | 9.076 | Haferlach Leukemia 221 | |
| acute myeloid leukemia versus normal | 4.907 | 9.93E−5 | 5.119 | Stegmaier Leukemia22 | |
| T cell acute lymphoblastic leukemia versus normal | 2.464 | 3.65E−33 | 14.045 | Haferlach Leukemia21 | |
| acute myeloid leukemia versus normal | 2.105 | 3.64E−24 | 12.67 | Haferlach Leukemia21 | |
| B cell acute lymphoblastic leukemia versus normal | 2.342 | 1.47E−26 | 12.136 | Haferlach Leukemia21 | |
| pro-B acute lymphoblastic leukemia versus normal | 2.161 | 4.98E−14 | 8.586 | Haferlach Leukemia21 | |
| chronic lymphocytic leukemia versus normal | 3.066 | 1.21E−4 | 5.253 | Haslinger Leukemia23 | |
| acute myeloid leukemia versus normal | 2.128 | 1.00E−3 | 4.402 | Valk Leukemia24 | |
| T cell acute lymphoblastic leukemia versus normal | 2.735 | 6.54E−6 | 7.092 | Andersson Leukemia25 | |
| B cell childhood acute lymphoblastic leukemia versus normal | 3.126 | 0.01 | 4.377 | Coustan-Smith Leukemia26 | |
| T cell childhood acute lymphoblastic leukemia versus normal | 5.366 | 2.00E−3 | 6.246 | Coustan-Smith Leukemia26 | |
| RUNX2 | acute myeloid leukemia versus normal | 2.89 | 6.00E−3 | 3.178 | Stegmaier Leukemia22 |
| T cell prolymphocytic leukemia versus normal | 2.371 | 2.10E−2 | 2.298 | Durig Leukemia27 | |
| RUNX3 | chronic adult T cell leukemia and lymphoma versus normal | 2.783 | 3.90E−5 | 4.949 | Choi Leukemia28 |
In Stegmaier Leukemia’s dataset,22 RUNX2 is overexpressed in AML with a fold change of 2.89, and RUNX2 is overexpressed in T cell childhood acute lymphoblastic leukemia with a fold change of 2.371 of Durig Leukemia’s dataset27 (Table 1).
In Choi Leukemia’s dataset,28 RUNX3 is overexpressed in chronic adult T cell leukemia and lymphoma with a fold change of 2.783 (Table 1).
Relationship between the mRNA Levels of RUNXs and the Clinicopathological Parameters of Patients with Leukemia
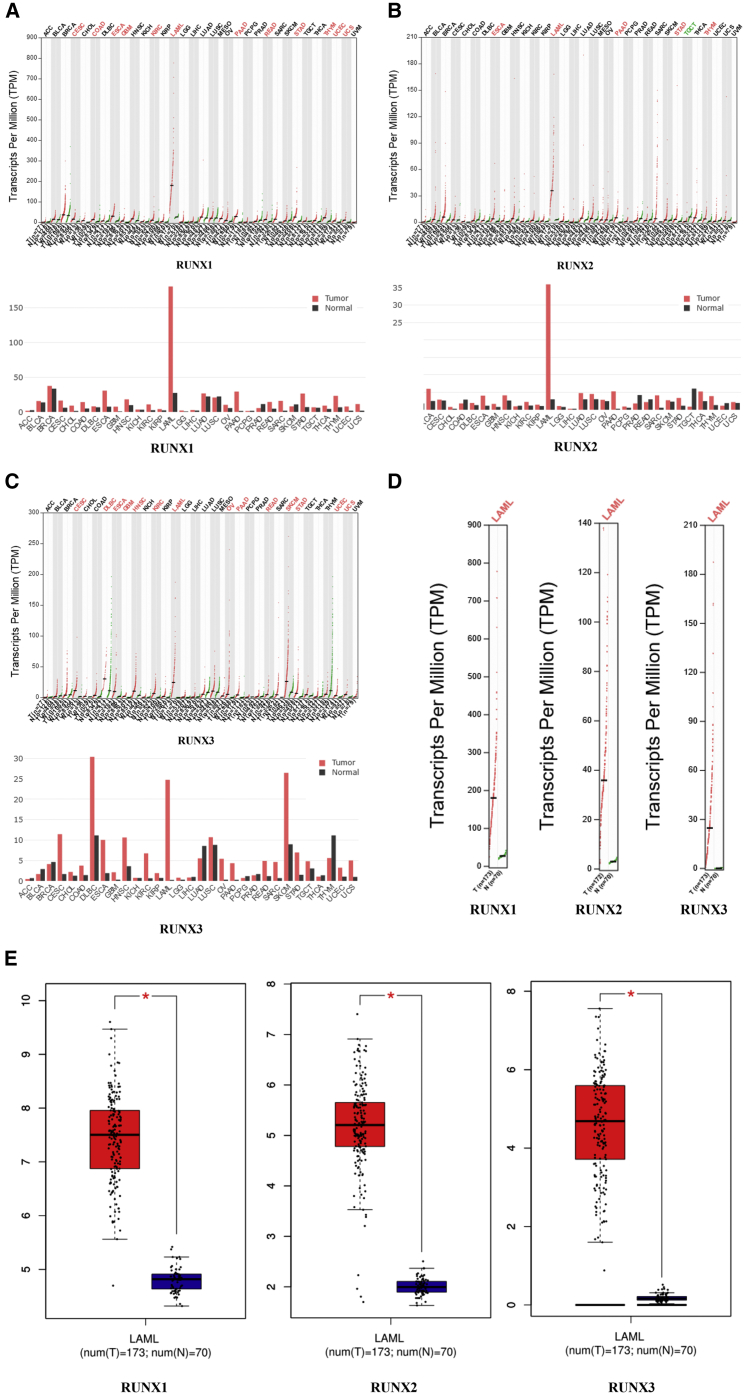
Using the GEPIA (Gene Expression Profiling Interactive Analysis) dataset (http://gepia.cancer-pku.cn/), we compared the mRNA expression of RUNX factors between leukemia and normal blood samples. The results indicated that the expression levels of RUNX1, RUNX2, and RUNX3 were all higher in leukemia than in normal blood samples (Figures 2A–2E).
Figure 2.
The Expression of RUNXs in Leukemia (GEPIA)
(A) The expression of RUNX1 in pan-cancer. (B) The expression of RUNX2 in pan-cancer. (C) The expression of RUNX3 in pan-cancer. (D–E) The expression of RUNXs in LAML.
RUNX Translational Factors Expression in Cell Lines of Leukemia
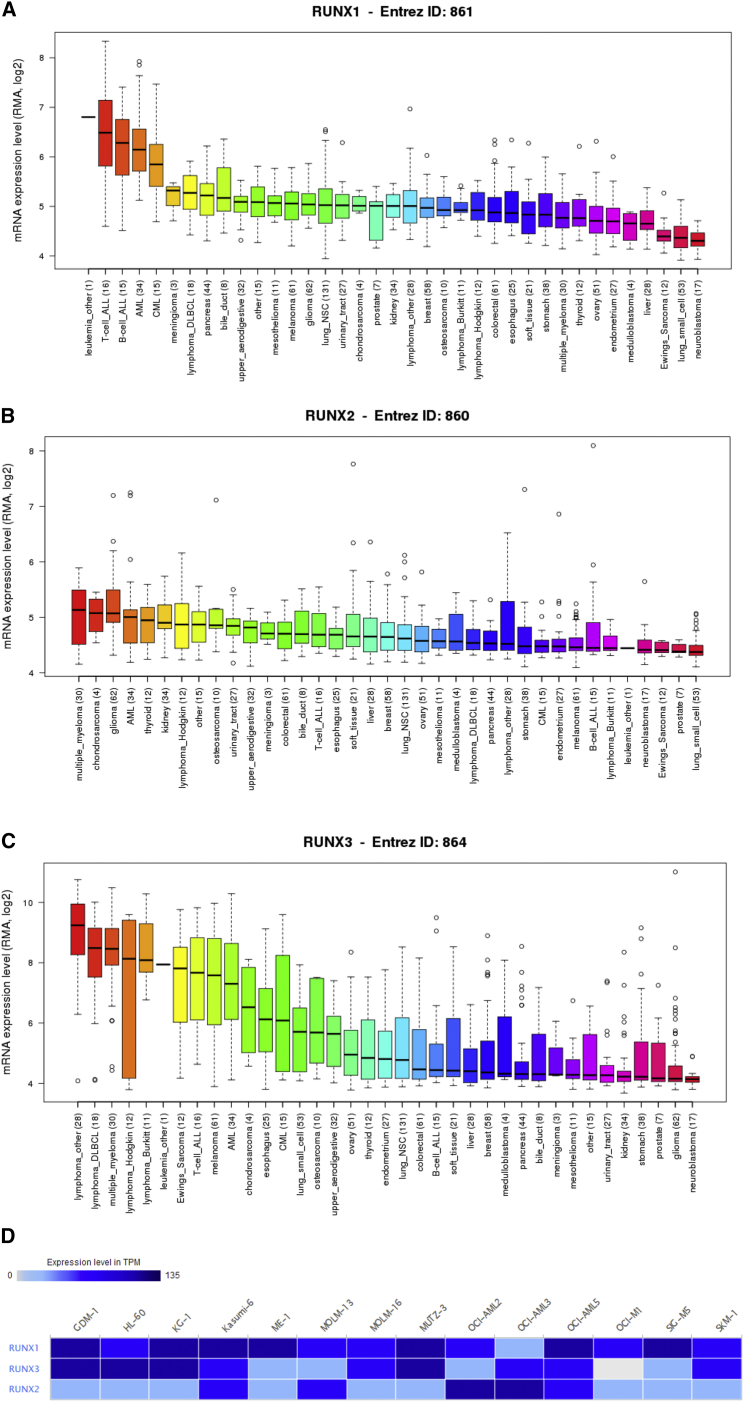
By assembling the Cancer Cell Line Encyclopedia (CCLE), we have expanded the process of detailed annotation of preclinical human cancer models (https://www.broadinstitute.org/ccle). We found that RUNX1, RUNX2, and RUNX3 were all highly expressed in cell lines of leukemia (Figures 3A–3C). Moreover, the European Bioinformatics Institute (EMBL-EBI) bioinformatics website (https://www.ebi.ac.uk/gxa/home) was also used to test the expression of RUNX translational factors in leukemia cell lines, and results indicated that RUNX1, RUNX2, and RUNX3 were increased in most cell lines of leukemia (Figure 3D).
Figure 3.
The Expression of RUNXs in Leukemia Cell Lines (CCLE and EMBL-EBI)
(A) The expression of RUNX1 in leukemia cell lines, analyzing by CCLE. (B) The expression of RUNX2 in leukemia cell lines, analyzed by CCLE. (C) The expression of RUNX3 in leukemia cell lines, analyzed by CCLE. (D) The expression of RUNXs in leukemia cell lines, analyzed by EMBL-EBI.
The Prognostic Values of RUNXs in Leukemia
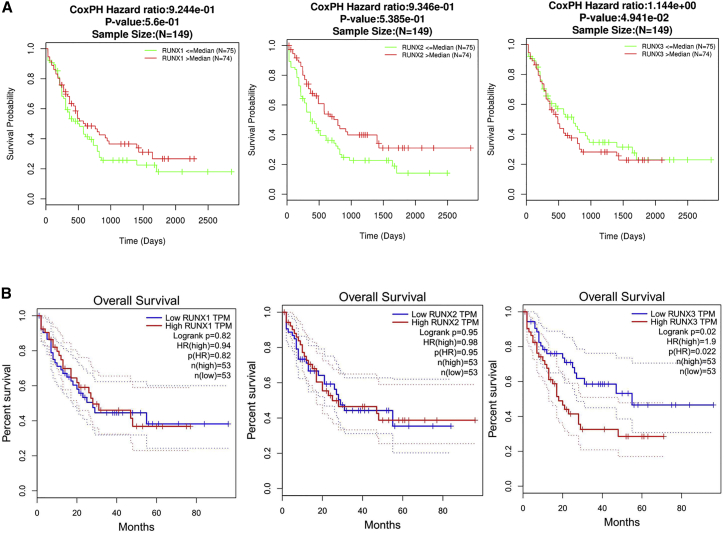
Nest, we investigated prognosis analysis for RUNX1, RUNX2, and RUNX3 using LinkedOmics and GEPIA databases in leukemia. In particular, decreased RUNX1 and RUNX2 were associated with poor overall survival (OS) in leukemia, but with no significance, in the LinkedOmics dataset (Figure 4A). Interestingly, there were no differences for RUNX1 and RUNX2 in prognostic analysis by GEPIA databases (Figure 4B). Nevertheless, increased RUNX3 was associated with poor OS in leukemia (Figures 4A and 4B).
Figure 4.
The Prognostic Value of mRNA Level of RUNX Factors in Leukemia Patients (LinkedOmics and GEPIA)
(A) The prognostic value of mRNA level of RUNX factors in leukemia patients, analyzed by LinkedOmics. (B) The prognostic value of mRNA level of RUNX factors in leukemia patients, analyzed by GEPIA.
The Prognostic Values of RUNXs in Leukemia
Nest, we investigated prognosis analysis for RUNX1, RUNX2, and RUNX3 using LinkedOmics and GEPIA databases in leukemia. In particular, decreased RUNX1 and RUNX2 were associated with poor OS in leukemia, but with no significance, in the LinkedOmics dataset (Figure 4A). Interestingly, there were no differences for RUNX1 and RUNX2 in prognostic analysis by GEPIA databases (Figure 4B). Nevertheless, increased RUNX3 was associated with poor OS in leukemia (Figures 4A and 4B). Hence highly expressed RUNX1 is a prognostic factor for leukemia.
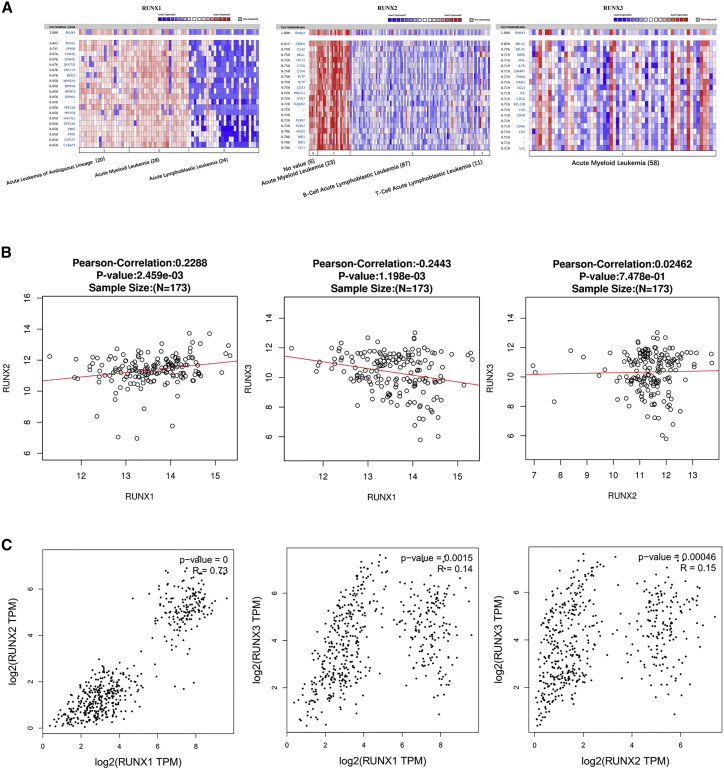
Co-expressed Genes of RUNXs, and the Correction between RUNXs in Leukemia
Genes co-expressed with RUNX1 were analyzed in Armstrong Leukemia,29 and we found RUNX1 was positively corrected with EP400, FAM3C, KDM4C, ZNF75D, ZNF177, BICD1, MAP2K4, ZBTB48, HDAC9, SEPHS1, and VPS13D. Genes co-expressed with RUNX2 were analyzed in Andersson Leukemia,25 and we found RUNX2 was positively corrected with CEBPA, GGA2, MGLL, TAF15, CTSA, PLTP, CD33, PMS2L4, ATG7, TGOLN2, and P2RY2. Genes co-expressed with RUNX3 were analyzed in Raponi Leukemia 2,30 and we found RUNX3 was positively corrected with ARL4C, SIRPG, MAL, IL7R, GIMAP5, PVRIG, TIAM1, CCL4, ITK, CD3G, and BCL11B (Figure 5A). We also analyzed the association among RUNX1, RUNX2, and RUNX3 by the LinkedOmics database and found that RUNX1 was positively corrected with RUNX2 (R = 0.2288, p < 0.05), RUNX1 was negatively corrected with RUNX3 (R = −0.2443, p < 0.05), and there was no significant correction between RUNX2 and RUNX3 (Figure 5B). Furthermore, we verified it using the GEPIA dataset; surprisingly, RUNX1 was both positively corrected with RUNX2 (R = 0.73, p < 0.05) and RUNX3 (R = 0.14, p < 0.05), and RUNX2 was positively corrected with RUNX3 (R = 0.15, p < 0.05) (Figure 5C). Hence RUNX1 has a positive correlation with RUNX2 in leukemia.
Figure 5.
Co-expressed Genes of RUNXs, and the Correction between RUNXs in Leukemia (ONCOMINE, LinkedOmics, and GEPIA)
(A) Co-expressed genes of RUNXs in leukemia, analyzed by ONCOMINE. (B) The correction between RUNXs in leukemia, analyzed by LinkedOmics and GEPIA.
Discussion
RUNX factors dysregulation has been reported in many cancers.1, 8, 12, 13, 15, 17, 18, 31, 32 The present study is the first to explore the mRNA expression and prognostic (OS) values of different RUNX factors in leukemia. We hope that our findings will contribute to available knowledge, improve treatment designs, and enhance the accuracy of prognosis for patients with leukemia.
The three RUNX family members are lineage-specific master regulators, which also have important, context-dependent roles in carcinogenesis as either tumor suppressors or oncogenes.33 RUNX1 plays an essential role in the development of normal hematopoiesis34 and is generally considered a tumor suppressor in myeloid neoplasms.35 In malignant lymphoma with leukemia (MLL) fusion leukemia, RUNX1 expression is downregulated through degradation by MLL fusion proteins.10 RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis.36 Moreover, Goyama et al.8 reported that RUNX1 promotes survival of AML cells. Thus, RUNX1 has been treated as a beneficial tumor suppressor for myeloid leukemogenesis. In our study, ONCOMINE datasets and GEPIA datasets revealed that the expression of RUNX1 was higher in human leukemia than in normal tissues. CCLE and EMBL-EBI databases also showed that RUNX1 was highly expressed in human leukemia cell lines. Using GEPIA and LinkedOmics datasets, we determined the prognostic value of RUNX1 in patients with leukemia. A low RUNX1 expression was associated with poor OS in leukemia, but with no significance.
Some evidence also points to a role for RUNX2 specifically in bone metastasis in advanced breast and prostate cancer.37 RUNX2 regulates the expression of genes intimately associated with tumor progression, invasion, and metastasis. Li et al.12 reported that RUNX2 promoted breast cancer bone metastasis by increasing integrin alpha5-mediated colonization. Schnerch and colleagues38 showed that RUNX2 was upregulated in AML in a patient with an inherent RUNX2 haploinsufficiency and CCD. Runx2 was also reported to induce AML in cooperation with Cbfbeta-SMMHC in mice.13 Hence RUNX2 might act as an oncogene in leukemia. In our report, the expression of RUNX2 in leukemia tissues was higher than that in normal tissues. RUNX2 was also highly expressed in human leukemia cell lines, as implicated by CCLE and EMBL-EBI databases. But to our surprise, decreased RUNX1 expression was shown to associate with poor OS in leukemia, with no significance as well.
RUNX3 is a master regulator of gene expression in major developmental pathways, and has been implicated in a multitude of cancers where it can function as a tumor suppressor or oncogene.16, 17 It is shown to be a tumor suppressor in a variety of cancers including gastric, colon, and breast cancers.16, 18 However, RUNX3 overexpression is frequently observed and is well correlated with malignant behaviors in head and neck cancer.17 In addition, RUNX3 is frequently highly expressed in the nuclei of ovarian cancer cell lines and plays an oncogenic role in ovarian cancer.16 Further, Selvarajan et al.19 reported overexpressed RUNX3 in NKTL had functional oncogenic properties. Therefore, it is inconclusive on the definite function of RUNX3 in diverse cancers. In our report, we demonstrated that the expression of RUNX3 in leukemia was higher than that in normal tissues. But to our surprise, a higher RUNX3 expression was significantly correlated with poor OS in patients with leukemia, which seemed consistent with the role of RUNX3 as an oncogene.
In this study, we systemically analyzed the expression and prognostic value of RUNXs in leukemia and provided a thorough understanding of the heterogeneity and complexity of the molecular biological properties of leukemia. Our results indicated that the increased expression of RUNX1, RUNX2, and RUNX3 in leukemia might play an important role in leukemia tumorigenesis. High RUNX3 expression could also serve as a molecular marker to identify high-risk subgroups of patients with leukemia. Our findings suggested that RUNX3 was a potential therapeutic target for leukemia, and transcriptional RUNX1 and RUNX2 were potential prognostic markers for the improvement of leukemia survival and prognostic accuracy.
Materials and Methods
Ethics Statement
This study was approved by the Academic Committee of Wuhan University and conducted according to the principles expressed in the Declaration of Helsinki. All of the datasets were retrieved from the published literature, so it was confirmed that all written informed consent was obtained.
ONCOMINE Analysis
ONCOMINE gene expression array datasets (https://www.oncomine.org/), an online cancer microarray database, was used to analyze the transcription levels of the RUNX family in different cancers. The mRNA expressions of the RUNX family in clinical cancer specimens were compared with that in normal controls, using a Student’s t test to generate a p value. The cutoff of p value and fold change were defined as 0.01 and 2, respectively.
GEPIA Dataset
GEPIA (Gene Expression Profiling Interactive Analysis) is a newly developed interactive web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects, using a standard processing pipeline (http://gepia.cancer-pku.cn/). GEPIA provides customizable functions such as tumor or normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis, and dimensionality reduction analysis.39
LinkedOmics Dataset
LinkedOmics (http://www.linkedomics.orglogin.php) is a new and unique tool in the software ecosystem for disseminating data from large-scale cancer omics projects. It uses preprocessed and normalized data from the Broad TCGA Firehose and Clinical Proteomic Tumor Analysis (CPTAC) data portal to reduce redundant efforts, and focused on the discovery and interpretation of attribute associations, and thus complements existing cancer data portals.40
CCLE Dataset
The CCLE ( https://www.broadinstitute.org/ccle) project is a collaboration between the Broad Institute and the Novartis Institutes for Biomedical Research and its Genomics Institute of the Novartis Research Foundation to conduct a detailed genetic and pharmacologic characterization of a large panel of human cancer models, to develop integrated computational analyses that link distinct pharmacologic vulnerabilities to genomic patterns and to translate cell line integrative genomics into cancer patient stratification.41 The CCLE provides public access to genomic data, analysis, and visualization for about 1,000 cell lines. The RUNX family’s expression in cancer cell lines is verified by the CCLE dataset.
EMBL-EBI Dataset
EMBL-EBI (https://www.ebi.ac.uk) has provided free and open access to a range of bioinformatics applications for sequence analysis since 1998.42 The RUNX family’s expression in AML cell lines is verified by the EMBL-EBI dataset.
Author Contributions
Conceptualization, C.-C.S.; Investigation, C.-C.S., S.-J.L., and D.-J.L.; Writing – Original Draft, C.-C.S., S.-J.L., and D.-J.L.; Writing – Review & Editing, C.-C.S., S.-J.L., G.L., Z.-L.C., Q.Z., and D.-J.L.; Visualization, C.-C.S., S.-J.L., G.L., and D.-J.L.; Supervision, C.-C.S.; Funding Acquisition, C.-C.S.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study is funded by the National Natural Science Foundation of China (grant 81802285 to C.-C.S.), Fundamental Research Funds for the Central Universities (grants 2015305020202 and 2042018kf0025 to C.-C.S.), China Postdoctoral Science Foundation (grant 2017M620340 to C.-C.S.), National Postdoctoral Program for Innovative Talents (grant BX201700178 to C.-C.S.), and Health Commission of Hubei Province scientific research project (grant WJ2019Q039 to C.-C.S.). It was also supported by Hubei Province Key Laboratory of Occupational Hazard Identification and Control, Wuhan University of Science and Technology (grant OHIC2017Y02 to C.-C.S.).
Contributor Information
Cheng-Cao Sun, Email: chengcaosun@whu.edu.cn.
De-Jia Li, Email: lodjlwhu@sina.com.
References
- 1.Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 2.Adya N., Castilla L.H., Liu P.P. Function of CBFbeta/Bro proteins. Semin. Cell Dev. Biol. 2000;11:361–368. doi: 10.1006/scdb.2000.0189. [DOI] [PubMed] [Google Scholar]
- 3.Huang G., Shigesada K., Ito K., Wee H.J., Yokomizo T., Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 6.Osato M., Ito Y. Increased dosage of the RUNX1/AML1 gene: a third mode of RUNX leukemia? Crit. Rev. Eukaryot. Gene Expr. 2005;15:217–228. doi: 10.1615/critreveukargeneexpr.v15.i3.40. [DOI] [PubMed] [Google Scholar]
- 7.Speck N.A., Gilliland D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 8.Goyama S., Schibler J., Cunningham L., Zhang Y., Rao Y., Nishimoto N., Nakagawa M., Olsson A., Wunderlich M., Link K.A. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Invest. 2013;123:3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob B., Osato M., Yamashita N., Wang C.Q., Taniuchi I., Littman D.R., Asou N., Ito Y. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood. 2010;115:1610–1620. doi: 10.1182/blood-2009-07-232249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang G., Zhao X., Wang L., Elf S., Xu H., Zhao X., Sashida G., Zhang Y., Liu Y., Lee J. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood. 2011;118:6544–6552. doi: 10.1182/blood-2010-11-317909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto F., Kanegane H., Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum. Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- 12.Li X.Q., Lu J.T., Tan C.C., Wang Q.S., Feng Y.M. RUNX2 promotes breast cancer bone metastasis by increasing integrin α5-mediated colonization. Cancer Lett. 2016;380:78–86. doi: 10.1016/j.canlet.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Kuo Y.H., Zaidi S.K., Gornostaeva S., Komori T., Stein G.S., Castilla L.H. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q.L., Ito K., Sakakura C., Fukamachi H., Inoue Ki., Chi X.Z., Lee K.Y., Nomura S., Lee C.W., Han S.B. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 15.Brenner O., Levanon D., Negreanu V., Golubkov O., Fainaru O., Woolf E., Groner Y. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc. Natl. Acad. Sci. USA. 2004;101:16016–16021. doi: 10.1073/pnas.0407180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C.W., Chuang L.S., Kimura S., Lai S.K., Ong C.W., Yan B., Salto-Tellez M., Choolani M., Ito Y. RUNX3 functions as an oncogene in ovarian cancer. Gynecol. Oncol. 2011;122:410–417. doi: 10.1016/j.ygyno.2011.04.044. [DOI] [PubMed] [Google Scholar]
- 17.Kudo Y., Tsunematsu T., Takata T. Oncogenic role of RUNX3 in head and neck cancer. J. Cell. Biochem. 2011;112:387–393. doi: 10.1002/jcb.22967. [DOI] [PubMed] [Google Scholar]
- 18.Huang B., Qu Z., Ong C.W., Tsang Y.H., Xiao G., Shapiro D., Salto-Tellez M., Ito K., Ito Y., Chen L.F. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor α. Oncogene. 2012;31:527–534. doi: 10.1038/onc.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selvarajan V., Osato M., Nah G.S.S., Yan J., Chung T.H., Voon D.C., Ito Y., Ham M.F., Salto-Tellez M., Shimizu N. RUNX3 is oncogenic in natural killer/T-cell lymphoma and is transcriptionally regulated by MYC. Leukemia. 2017;31:2219–2227. doi: 10.1038/leu.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sealfon S.C., Chu T.T. RNA and DNA microarrays. Methods Mol. Biol. 2011;671:3–34. doi: 10.1007/978-1-59745-551-0_1. [DOI] [PubMed] [Google Scholar]
- 21.Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G.T., Béné M.C., De Vos J., Hernández J.M., Hofmann W.K., Mills K.I. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stegmaier K., Ross K.N., Colavito S.A., O’Malley S., Stockwell B.R., Golub T.R. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat. Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 23.Haslinger C., Schweifer N., Stilgenbauer S., Döhner H., Lichter P., Kraut N., Stratowa C., Abseher R. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J. Clin. Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 24.Valk P.J., Verhaak R.G., Beijen M.A., Erpelinck C.A., Barjesteh van Waalwijk van Doorn-Khosrovani S., Boer J.M., Beverloo H.B., Moorhouse M.J., van der Spek P.J., Löwenberg B., Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 25.Andersson A., Ritz C., Lindgren D., Edén P., Lassen C., Heldrup J., Olofsson T., Råde J., Fontes M., Porwit-Macdonald A. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 26.Coustan-Smith E., Song G., Clark C., Key L., Liu P., Mehrpooya M., Stow P., Su X., Shurtleff S., Pui C.H. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2011;117:6267–6276. doi: 10.1182/blood-2010-12-324004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dürig J., Bug S., Klein-Hitpass L., Boes T., Jöns T., Martin-Subero J.I., Harder L., Baudis M., Dührsen U., Siebert R. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32) Leukemia. 2007;21:2153–2163. doi: 10.1038/sj.leu.2404877. [DOI] [PubMed] [Google Scholar]
- 28.Choi Y.L., Tsukasaki K., O’Neill M.C., Yamada Y., Onimaru Y., Matsumoto K., Ohashi J., Yamashita Y., Tsutsumi S., Kaneda R. A genomic analysis of adult T-cell leukemia. Oncogene. 2007;26:1245–1255. doi: 10.1038/sj.onc.1209898. [DOI] [PubMed] [Google Scholar]
- 29.Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., Korsmeyer S.J. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 30.Raponi M., Harousseau J.L., Lancet J.E., Löwenberg B., Stone R., Zhang Y., Rackoff W., Wang Y., Atkins D. Identification of molecular predictors of response in a study of tipifarnib treatment in relapsed and refractory acute myelogenous leukemia. Clin. Cancer Res. 2007;13:2254–2260. doi: 10.1158/1078-0432.CCR-06-2609. [DOI] [PubMed] [Google Scholar]
- 31.Fukamachi H., Ito K. Growth regulation of gastric epithelial cells by Runx3. Oncogene. 2004;23:4330–4335. doi: 10.1038/sj.onc.1207121. [DOI] [PubMed] [Google Scholar]
- 32.Levanon D., Bettoun D., Harris-Cerruti C., Woolf E., Negreanu V., Eilam R., Bernstein Y., Goldenberg D., Xiao C., Fliegauf M. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chimge N.O., Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32:2121–2130. doi: 10.1038/onc.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurokawa M. AML1/Runx1 as a versatile regulator of hematopoiesis: regulation of its function and a role in adult hematopoiesis. Int. J. Hematol. 2006;84:136–142. doi: 10.1532/IJH97.06070. [DOI] [PubMed] [Google Scholar]
- 35.Krauth M.T., Eder C., Alpermann T., Bacher U., Nadarajah N., Kern W., Haferlach C., Haferlach T., Schnittger S. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome. Leukemia. 2014;28:1449–1458. doi: 10.1038/leu.2014.4. [DOI] [PubMed] [Google Scholar]
- 36.Schnittger S., Dicker F., Kern W., Wendland N., Sundermann J., Alpermann T., Haferlach C., Haferlach T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 37.Pratap J., Lian J.B., Stein G.S. Metastatic bone disease: role of transcription factors and future targets. Bone. 2011;48:30–36. doi: 10.1016/j.bone.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnerch D., Lausch E., Becker H., Felthaus J., Pfeifer D., Mundlos S., Engelhardt M., Schwabe M., Wäsch R. Up-regulation of RUNX2 in acute myeloid leukemia in a patient with an inherent RUNX2 haploinsufficiency and cleidocranial dysplasia. Leuk. Lymphoma. 2014;55:1930–1932. doi: 10.3109/10428194.2013.855310. [DOI] [PubMed] [Google Scholar]
- 39.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43(W1):W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]