Abstract
BACKGROUND
Colorectal cancer (CRC) is the second most common cause of cancer death worldwide. It is broadly described that cyclooxygenase-2 (COX-2) is mainly overexpressed in CRC but less is known regarding post-translational modifications of this enzyme that may regulate its activity, intracellular localization and stability. Since metabolic and proteomic profile analysis is essential for cancer prognosis and diagnosis, our hypothesis is that the analysis of correlations between these specific parameters and COX-2 state in tumors of a high number of CRC patients could be useful for the understanding of the basis of this cancer in humans.
AIM
To analyze COX-2 regulation in colorectal cancer and to perform a detailed analysis of their metabolic and proteomic profile.
METHODS
Biopsies from both healthy and pathological colorectal tissues were taken under informed consent from patients during standard colonoscopy procedure in the University Hospital of Bellvitge (Barcelona, Spain) and Germans Trias i Pujol University Hospital (Campus Can Ruti) (Barcelona, Spain). Western blot analysis was used to determine COX-2 levels. Deglycosylation assays were performed in both cells and tumor samples incubating each sample with peptide N-glycosidase F (PNGase F). Prostaglandin E2 (PGE2) levels were determined using a specific ELISA. 1H high resolution magic angle spinning (HRMAS) analysis was performed using a Bruker AVIII 500 MHz spectrometer and proteomic analysis was performed in a nano-liquid chromatography-tandem mass spectrometer (nano LC-MS/MS) using a QExactive HF orbitrap MS.
RESULTS
Our data show that COX-2 has a differential expression profile in tumor tissue of CRC patients vs the adjacent non-tumor area, which correspond to a glycosylated and less active state of the protein. This fact was associated to a lesser PGE2 production in tumors. These results were corroborated in vitro performing deglycosylation assays in HT29 cell line where COX-2 protein profile was modified after PNGase F incubation, showing higher PGE2 levels. Moreover, HRMAS analysis indicated that tumor tissue has altered metabolic features vs non-tumor counterparts, presenting increased levels of certain metabolites such as taurine and phosphocholine and lower levels of lactate. In proteomic experiments, we detected an enlarged number of proteins in tumors that are mainly implicated in basic biological functions like mitochondrial activity, DNA/RNA processing, vesicular trafficking, metabolism, cytoskeleton and splicing.
CONCLUSION
In our colorectal cancer cohort, tumor tissue presents a differential COX-2 expression pattern with lower enzymatic activity that can be related to an altered metabolic and proteomic profile.
Keywords: Colon, Carcinoma, Cyclooxygenase, Prostaglandin, Proteomics, High resolution magic angle spinning
Core tip: Although increased levels of cyclooxygenase-2 (COX-2) have been broadly related to colorectal cancer (CRC), high variability exists among patients. In our cohort, the electrophoretic band profile of COX-2 was altered in tumor tissue due to COX-2 glycosylation. Furthermore, prostaglandin E2 levels in tumor tissue were significantly lower than in normal colonic mucosa. In order to further characterize these samples, we have also detected an altered metabolic and proteomic profile in the tumor samples compared to controls that can be associated with the presence of glycosylated COX-2. These findings contribute to delineate the post-translational complexity and heterogeneity of molecular signaling regulation in CRC progression.
INTRODUCTION
Colorectal cancer (CRC) is the second most common cause of cancer death worldwide. There were over 18.1 million new cases and 9.6 million deaths in 2018 according to the World Health Organization. Patient prognosis is based on several parameters like the clinical stage of the disease, histological type, tumor grade and the analysis of active oncogenes[1]. One of the targets for CRC therapy is cyclooxygenase-2 (COX-2), an inducible enzyme usually absent but transcriptionally upregulated upon neoplastic and pro-inflammatory insults[2,3]. In CRC, whereas cyclooxygenase 1 (COX-1) expression remains unaltered when comparing CRC cells and normal colonic mucosa, COX-2 is overexpressed in 40%-50% of benign polyps and in 80%-90% of adenocarcinomas[4,5]. Both enzymes catalyze a key step in the conversion of arachidonate (AA) to prostaglandin H2 (PGH2), the immediate substrate for a series of cell-specific prostaglandin (PG) and thromboxane synthases. In particular, PGE2 is partially pro-inflammatory (depending on the tissue and accumulation) and it has been related to CRC progression[6,7]. The potential tumor suppressor enzyme 15-prostaglandin dehydrogenase (15-PGDH), which catalyzes the degradation of PGE2, is usually down-regulated in colorectal adenoma and carcinoma cells[8,9]. In addition to this, experimental data have demonstrated that, not only the expression levels of COX-2 are important: post-translational modifications of this enzyme may also regulate its function and degradation[10]. COX-2 sequence contains five potential N-glycosylation sites, three of which are always glycosylated, one Asn580 in human and mouse that is glycosylated ≤ 50% of the times, and one that is never glycosylated[11]. The variability of glycosylation at Asn580 leads to the production of two distinct glycoforms of 72 and 74 kDa, respectively. In this context, it has been described that glycosylation of COX-2 can target the protein for proteolysis through the endoplasmic reticulum pathway and has been associated with alterations in the activity of the enzyme[12]. While the above-mentioned studies reported various COX-2 post-translational regulatory mechanisms, the biological and physiological significance of the post-translational regulation of COX-2 remains to be investigated.
For cancer prognosis and diagnosis, it also gains special relevance the analysis of alterations in both the metabolic and protein profile in tumor tissues of patients in contrast to the non-tumor adjacent regions. Nuclear magnetic resonance techniques, such as high resolution magic angle spinning (HRMAS) provide an in-depth analysis for the identification of distinctive metabolites in cancer and can be used to correlate changes in metabolic profiles with the severity of the disease and probable prognosis[13-15]. Although several recent studies have been carried out in CRC patients with nuclear magnetic resonance (NMR) techniques, they have been mainly focused on the improvement of screening (in serum, urinary tissue and fecal extracts) and early diagnosis of the disease[16-18]. Additionally, proteomic analyses have been effective in the identification of peptides present in different pathological contexts, including cancer[19-21]. Thus, the analysis of correlations between metabolic and molecular parameters in tumors of a high number of CRC patients could be useful for the understanding of the basis of this cancer in humans.
In this study, we have analyzed in samples from CRC patients the post-translational regulation of COX-2 together with a detailed analysis of both the metabolic and the proteomic profile of the tumor tissue compared to the non-tumor adjacent area to elucidate whether distinctive COX-2 post-translational modifications can be associated to a specific, altered metabolic profile in these CRC samples.
MATERIALS AND METHODS
Human colon biopsies
Biopsies from both healthy and pathological colorectal tissues were taken under informed consent from patients during standard colonoscopy procedure in the University Hospital of Bellvitge (Barcelona, Spain) and Germans Trias i Pujol University Hospital (Campus Can Ruti) (Barcelona, Spain). The mean age of participants was 71 ± 15 years old. All procedures were approved by the Ethical Committee for Clinical Investigation of University Hospital of Bellvitge and University Hospital of Germans Trias i Pujol (Barcelona, Spain).
Cell culture
The colorectal adenocarcinoma cell line HT-29 was maintained in DMEM medium (Gibco) containing 100 U/mL penicillin, 100 μg/mL streptomycin and 10% heat-inactivated fetal bovine serum (FBS).
Deglycosylation assay in vivo for HT29 cells
Three hundred thousand cells were seeded per well on 6-well plates. When confluence was reached, cells were starved in serum-free media for 18 h. Then, deglycosylation was performed by treating for 24 h with 5 mmol/L glucosamine-hydrochloride (Sigma #G4875). To also evaluate PGE2 production, cells were then treated with 30 µmol/L arachidonic acid (Sigma #10931) for 30 min. Two hundred microliter of media was collected before and after treatment with arachidonic acid and plates were stored at -80 °C until processing.
Deglycosylation assay in vitro for tumor samples
Tumor samples were denaturalized by mixing 50 µg of total protein extract with 50 µL of 50 mmol/L NaH2PO4 buffer (pH 7.5) and 5 µL of “denaturing solution” containing 0.2% SDS and 1% 2-mercaptoethanol. After heating at 100 °C for 10 min and recovery of room temperature, deglycosylation was performed incubating each sample with 500 U/mL of PNGase F (Sigma #P7367) and 5 µL of 15% Triton X-100 for 18 h at 37 °C. Reaction was then stopped by boiling samples at 100 °C for 5 min.
Preparation of total protein cell extracts
Cells were homogenized in a medium containing 10 mmol/L Tris-HCl, pH 7.5; 1 mmol/L MgCl2, 1 mmol/L EGTA, 10% glycerol, 0.5% CHAPS and proteases and phosphatases inhibitors cocktails (Sigma). The extracts were vortexed for 30 min at 4 °C and after centrifuging for 15 min at 13000 rpm, the supernatants were stored at -20 °C. Protein levels were determined with Bradford reagent (Bio-Rad).
Preparation of total protein extracts from human tissue
Several milligrams of tissue were pulverized and then were homogenized in 10 mmol/L Tris-HCl, pH 7.5; 1 mmol/L MgCl2, 1 mmol/L EGTA, 10% glycerol, 0.5% CHAPS and proteases and phosphatases inhibitors cocktails (Sigma) at 4 °C using an Ultra-turrax dispersing instrument (Ika). The extracts were vortexed for 30 min at 4 °C and after centrifuging for 15 min at 13000 rpm, the supernatants were stored at -80 °C. Protein levels were determined with Bradford reagent (Bio-Rad).
Western blot analysis
Equal amounts of protein (20-40 μg) from each fraction obtained were loaded into a 10%-12% SDS-PAGE. Proteins were size fractionated, transferred to a PVDF membrane (Bio-Rad) and, after blocking with 5% non-fat dry milk, incubated with the corresponding antibody for COX-1 (sc-1752) or COX-2 (sc-1747) from Santa Cruz Biotechnology. Blots were normalized by the measurement of the amount for GAPDH (ThermoFisher Scientific #AM4300) and developed by ECL protocol and different exposition times were performed for each blot to ensure the linearity of the band intensities. Values of densitometry were determined using Image J software.
PGE2 measurement
Levels of PGE2 were determined in tumor and normal colonic mucosa samples and in the culture medium of HT29 cells using a specific Enzyme Immunoassay Kit (Arbor Assays, #K051-H1) following the manufacturer’s instructions.
1H HRMAS analysis
All 1H HRMAS experiments were carried out at 277.15 K on a Bruker AVANCE III 500 MHz spectrometer (Bruker Biospin, Germany) using a 1H probe with a sample spin rate of 4000 Hz. Each tissue sample was individually placed in D2O saline (99.2% 2H, Apollo Scientifc Limited, Stockport, United Kingdom) and inserted into a zirconium oxide rotor (50 µL) for all NMR acquisitions. In order to attenuate NMR signals of macromolecules, a Carr-Purcell-Meiboom-Gil (CPMG) spin-echo spectrum was collected for each sample with 2 pulses and TE = 36 ms and TR = 144 ms. An integration routine using Mnova software (Mestrelab Research, Santiago de Compostela, Spain) was used to determine the areas under the peaks (parts per million; ppm) after adjusting phase and baseline.
Proteomic analysis
Proteins from normal and tumor colon tissue were extracted using tissue homogenization with ceramic beads (MagNa Lyser Green Beads apparatus, Roche, Germany) in extraction buffer (50 mmol/L Tris-HCl pH 6.8; 4% SDS, 10 mmol/L DTT). Proteins were digested using the filter aided sample preparation (FASP) protocol as previously described[22] and the resulting peptides were analyzed by nano-liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) using a QExactive HF orbitrap mass spectrometer (Thermo Scientific). Peptides were injected onto a C18 reversed phase (RP) nano-column (75 µm I.D. and 50 cm, Acclaim PepMap100, Thermo Scientific) in buffer A [0.1% formic acid (v/v)] and eluted with a 240 min lineal gradient of buffer B [90% acetonitrile, 0.1% formic acid (v/v)]. MS runs consisted of enhanced FT-resolution spectra (140000 resolution) followed by data-dependent MS/MS spectra of the 15 most intense parent ions acquired along the chromatographic run. HCD fragmentation was performed at 27% of normalized collision energy. For peptide identification, the MS/MS spectra were searched with the SEQUEST HT algorithm implemented in Proteome Discoverer 2.1 (Thermo Scientific). The results were analyzed using the probability ratio method[23] and the false discovery rate (FDR) of peptide identification was calculated based on the search results against a decoy database using the refined method[24]. Peptide and scan counting were performed assuming as positive events those with a FDR equal or lower than 1%. Enrichment analyses were performed by using the DAVID functional annotation database (https://david.ncifcrf.gov/).
Statistical analysis
The values in graphs correspond to the means ± SD. Statistical analysis was performed by qualified personnel. Analysis was based on continuous variables, and statistical significance was assessed with the Student-t test for paired observations and the two-way ANOVA for multiple comparisons. Correlations between continuous variables were summarized with the Pearson coefficient (indicated in the corresponding figures).
RESULTS
Study of post-translational COX-2 modifications
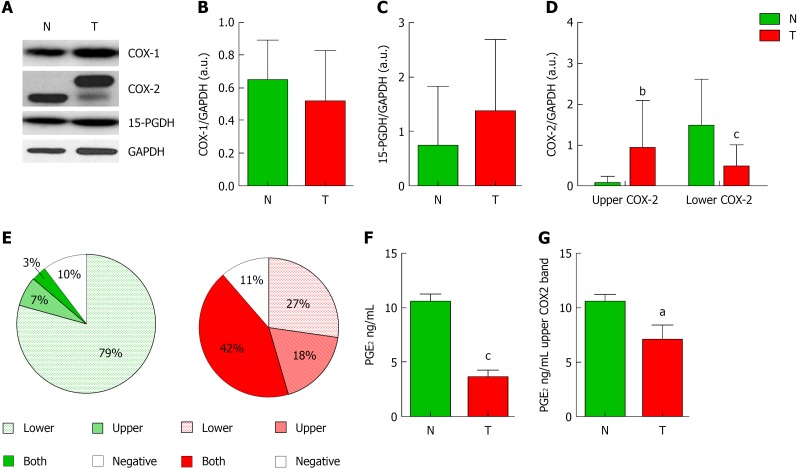
COX-2 seems to play an important role in multiple CRC cellular functions including apoptosis, cell invasiveness, and angiogenesis. We have analyzed both COX isoforms and 15-PGDH levels in 45 CRC patients whose data are detailed in Table 1. We first analyzed the levels of these proteins in tumors and the corresponding adjacent non-tumor tissues. In contrast to COX-1 or 15-PGDH, in which no significant differences were detected (Figure 1A-C), COX-2 exhibited a change in its protein expression profile in tumors presenting two bands instead of one, at approximately 66 and 72 kDa (Figure 1D). Regarding all non-tumor/tumor pairs included in this study, whereas non-tumor tissue predominantly exhibits the lower band (79% of cases) (Figure 1E), only a 27% of tumor samples had this band and 60% had the upper band or both. In fact, tumor samples have significantly more “upper COX-2” band and less “lower COX-2” band than their corresponding non-tumor tissue. To determine whether this modification can be related to the activity of the enzyme, we measured the content of PGE2 in both groups (Figure 1F). Our results show that tumor tissue had a significantly lower PGE2 content compared to their paired non-tumor regions. This situation also prevailed in tumors that only presented the upper band (Figure 1G).
Table 1.
Patient demographic and pathological data
| Age (median) | 75 (25-95) | |
| Sex | Male | 28 |
| Female | 17 | |
| Anatomical location | Distal | 15 |
| Proximal | 12 | |
| Unknown | 18 | |
| T stage | T1 | 1 |
| T2 | 19 | |
| T3 | 20 | |
| T4 | 5 | |
| N stage | N0 | 17 |
| N1 | 2 | |
| N2 | 7 | |
| Unknown | 19 |
Figure 1.
Analysis of cyclooxygenase-2 protein levels and prostaglandin E2 measurement. A: Representative image of western blot analysis of cyclooxygenase-1 (COX-1), cyclooxygenase-2 (COX-2) and 15-hydroxyprostaglandin dehydrogenase (15-PGDH) normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of tumor (T) or non-tumor (N) samples. B and C: Graphs represent normalized COX-1 (B) or 15-PGDH (C) bands in western blot in T vs N tissue. D: Graph represent normalized COX-2 protein levels corresponding to upper (Up) or lower (Down) band in western blot in T vs N tissue. E: Graphs represent the percentage of samples expressing lower, upper, both or none bands in western blot for COX-2. F: Levels of PGE2 in Tumor vs Non-tumor samples. G: Relative levels of prostaglandin E2 only in samples from colorectal cancer patients showing the upper band of COX-2 in western blot. n = 45. Graphs show mean ± SD. aP ≤ 0.05, bP ≤ 0.01 and cP ≤ 0.001 vs the non-tumor condition. COX-1: Cyclooxygenase-1; COX-2: Cyclooxygenase-2; 15-PGDH: 15-hydroxyprostaglandin dehydrogenase; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; T: Tumor; N: Non-tumor; PGE2: Prostaglandin E2.
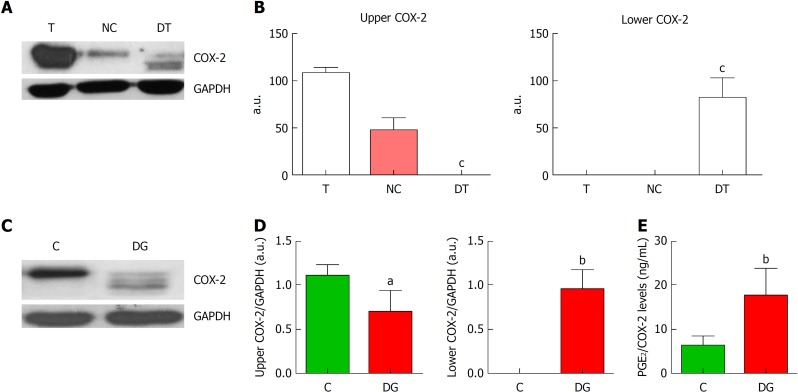
COX-2 glycosylation has been described as one of the main post-translational modifications that can affect the electrophoretic mobility of the protein. We have performed deglycosylation assays in several tumor samples and observed that in deglycosylated tumors (DT) COX-2 band appears at a lower molecular weight, while the upper initial band was missing (Figure 2A and B). These experiments were confirmed in vitro using the colon cancer cell line HT29 (Figure 2C). These cells presented only COX-2 upper band under basal conditions, but when treated for deglycosylation, several bands of lower molecular weight appeared along with the upper one that decreased (Figure 2D). These results indicate that COX-2 appears mainly in a glycosylated state in tumor cells and in tumor samples from CRC patients. Moreover, as it has been described that post-translational modifications can affect COX-2 activity, PGE2 was measured in cells before and after deglycosylation (Figure 2E). We obtained higher levels of PGE2 after deglycosylation indicating that the glycosylated-state of COX-2 appears to be less active than controls in both HT29 and in CRC samples.
Figure 2.
Deglycosylation assays of cyclooxygenase-2. A: Representative images of western blot of cyclooxygenase-2 (COX-2) protein levels corresponding to tumor tissue (T), negative control (NC) and deglycosylated tumor tissue (DT). B: Graphs correspond to upper or lower COX-2 bands quantifications. n = 4. C: Representative images of western blot of COX-2 protein levels in HT29 cells under basal conditions (C) or deglycosylated (DG). D: Graphs correspond to upper or lower COX-2 bands quantifications. n = 8. E. Relative levels of prostaglandin E2 (PGE2) normalized by total COX-2 band obtained in the corresponding western blot analysis. n = 11. Graphs show mean ± SD. aP ≤ 0.05, bP ≤ 0.01 and cP ≤ 0.001 vs the glycosylated condition. COX-2: Cyclooxygenase-2; T: Tumor; NC: Negative control; DT: Deglycosylated tumor; C: Cells under basal conditions; DG: Deglycosylated; PGE2: Prostaglandin E2.
Analysis of the metabolic profile of CRC samples
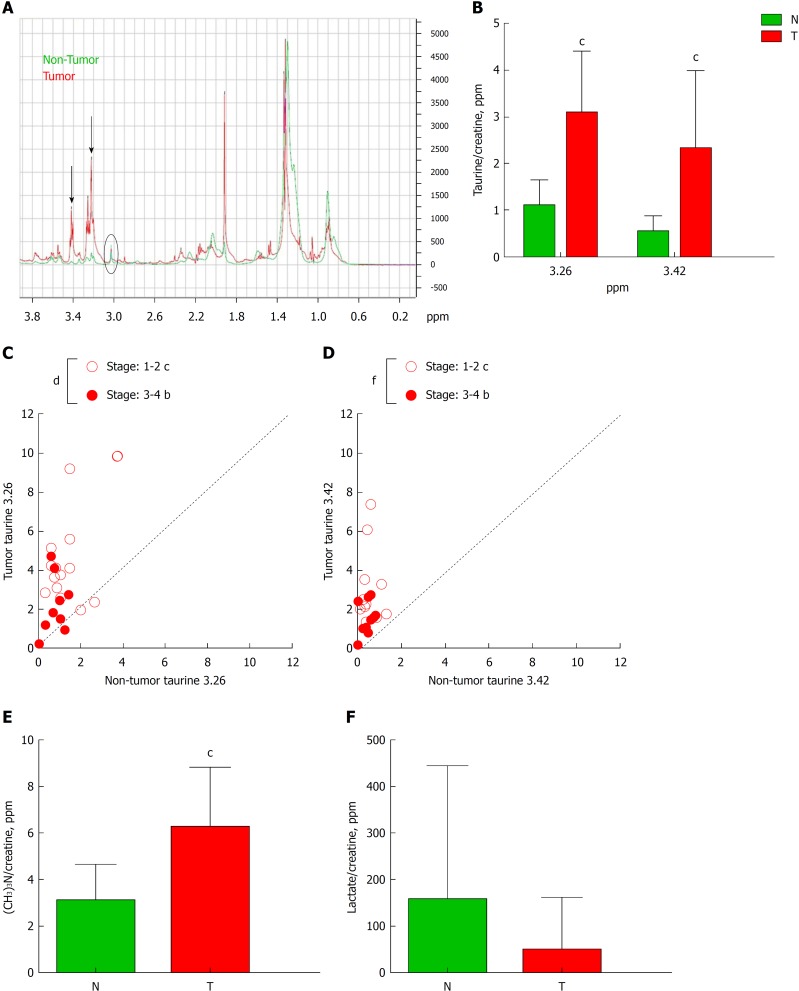
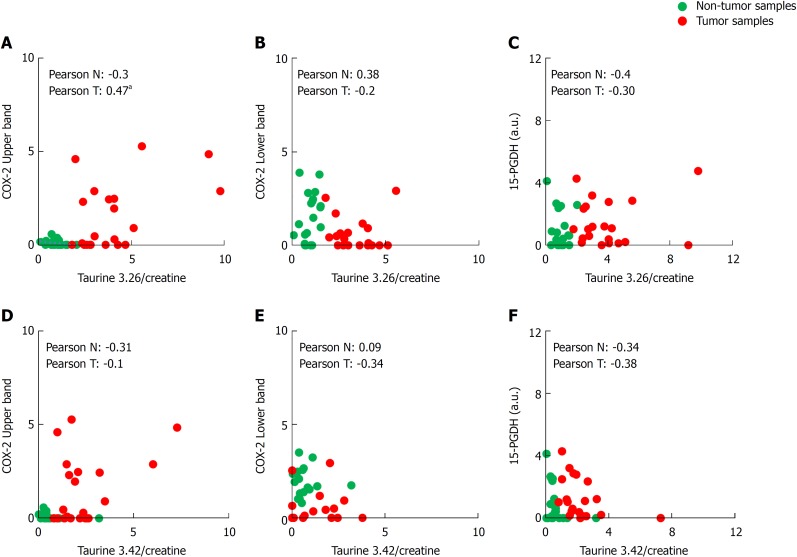
Another important aspect to consider in CRC is the metabolic profile of the patient, which may reflect the general state of the organism. We have done a comparative study in non-tumor vs. tumor colorectal tissues from the same patient by HRMAS (representative obtained profile shown in Figure 3A), demonstrating that, in tumors, there is an altered metabolic profile with significantly higher presence of taurine (both peaks at δ ppm 3.26 and 3.42) (Figure 3B). We then analyzed whether these differences in taurine for both peaks could be sex- or tumor stage-related. We found a significant correlation between the levels of both peaks of taurine 3.26 and 3.42 and the tumor stage, being higher in stage 1-2 when compared to stage 3-4 (Figure 3C and D). However, we did not find any significant difference between sexes (Supplementary Figure 1). Moreover, in HRMAS analysis we also detected significantly higher levels of phosphocholine [(CH3)3N] in tumors (peak at δ ppm 3.22; Figure 3E). Other metabolites, such as lactate (Figure 3F), acetate and glycine did not show significant difference (Supplementary Figure 2A-B). To further investigate if there is a correlation between taurine levels and the presence of COX-2 (both bands) and 15-PGDH, we performed a broad statistical analysis calculating Pearson’s correlation coefficient between these variables (Figure 4A-F). Interestingly, only the levels of taurine at 3.26 peak significantly correlates with the levels of the upper COX-2 band (Figure 4A).
Figure 3.
High resolution magic angle spinning analysis of tumor tissue. A: Schematic diagram of high resolution magic angle spinning spectra of tumor tissue (red line) and non-tumor tissue (blue line). Black arrows indicate two peaks corresponding to taurine δ ppm 3.26 and 3.42. B: Graph represents the quantification of both taurines normalized by creatine of tumor (T) and non-tumor (N) tissue. C and D: Correlation graphs of taurine 3.26 (C) and taurine 3.42 (D) in T vs N subdivided depending on the stage of the Tumor: 1-2 vs 3-4. E and F: Graphs represent the quantification of phosphocholine (E) or lactate (F) in T vs N. n = 29. Graphs show mean ± SD. bP ≤ 0.01, cP ≤ 0.001 of T vs N and dP ≤ 0.05, fP ≤ 0.001 between stages in tumors. T: Tumor; N: Non-tumor.
Figure 4.
Biostatistical analysis of the correlation between the presence of cyclooxygenase-2 bands in western blot and both taurine peaks. A-F: Statistical analysis was performed searching the correlation coefficient between the presence of upper or lower cyclooxygenase-2 band or 15-hydroxyprostaglandin dehydrogenase in western blot and the taurine peaks (3.26 and 3.42 δ ppm) levels of 29 Tumor/Non-tumor pairs of samples. The Pearson’s correlation coefficient is shown in each figure for non-tumor or tumor sample. aP ≤ 0.05. COX-2: Cyclooxygenase-2; 15-PGDH: 15-hydroxyprostaglandin dehydrogenase.
Proteomic analysis of CRC samples
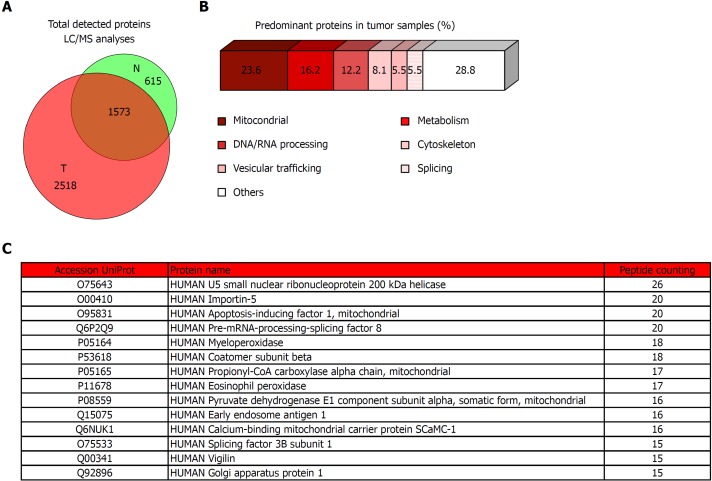
To complete the molecular analysis of our CRC cohort, we performed a proteomic shotgun analysis (Figure 5). Our results shown that tumor tissues have a higher number of identified proteins, detecting more than 4000 proteins in tumors of which 1573 are shared with non-tumor adjacent tissue but 2518 are exclusive (Figure 5A). To evaluate the implications of these proteins, we then selected only the exclusive proteins from tumors and established a threshold for the score of “peptide counting” above 5, resulting in a total of 271 proteins that we have classified according to their biological functions (Figure 5B). We have detected that most of tumor exclusive proteins have mitochondrial activity (23.6%) and the rest were mainly implicated in metabolism (16.2%), DNA/RNA processing (12.2%), and cytoskeleton (8.1%), vesicular trafficking (5.5%) and splicing (5.5%) among others pathways. A simplified list of the main proteins detected in tumor vs. normal mucosa, containing those with a score of “peptide counting” over 15, is shown in Figure 5C. All of them are included in the biological functions previously cited.
Figure 5.
Proteomic analysis of tumor tissue of colorectal cancer patients and the corresponding non-tumor adjacent tissue. A: Venn diagram with specific and overlapped proteins. 2188 and 4091 proteins were identified (FDR < 1%) in non-tumor and tumor tissue, respectively. Among them, 1573 were identified in both tissues, and 615 and 2518 were unique in non-tumor and tumor tissue, respectively. B: Enrichment analysis of specific proteins identified in tumor tissue by mass spectrometry performed with proteins that were identified with 5 or more peptides (271 most abundant proteins). Most of these proteins had mitochondrial and metabolic function, or were involved in processes related to nucleic acids processing, cytoskeleton, vesicular trafficking, or splicing. C: Specific tumor tissue proteins identified with 15 or more peptides in liquid chromatography-mass spectrometry analyses. T: Tumor; N: Non-tumor; LC/MS: Liquid chromatography-mass spectrometry.
DISCUSSION
Cancer includes several complex diseases that can affect different organs in the body. The recent report about global burden of cancer “GLOBOCAN 2018” estimates that it causes more than 9 millions of deaths per year worldwide[25]. Many medical and research efforts have been made trying to improve the treatments against this complex disease. Nevertheless, it is still necessary to study in depth the molecular, metabolic and proteomic features of cancer tissue to find the remaining answer of cancer problematic.
At the molecular level, numerous studies have related increased levels of COX-2 and derived prostanoids (mainly PGE2) to cancer development[26]. Thus, elevated COX-2 expression has been associated to larger cancer size, greater depth of cancer invasion and reduced patient survival[27,28]. Due to these findings, COX-2 was proposed as a potential molecular predictor for CRC progression. However, its predictive relevance is controversial because other studies have been unable to identify a significant correlation between CRC patient outcome and COX-2 expression mainly due to the high variability in COX-2 levels among CRC patients[29]. In our study, when we analyzed by Western blot the COX-2 levels in tumor tissues from CRC patients, we observed significantly higher levels of total protein and also a change in the expression profile compared to non-tumor colonic mucosa, comprising several bands. Although most reports in the literature refer to only one homogenous COX-2 protein band in animal tissue, some groups have reported the presence of several bands corresponding to COX-2 in tumors[30,31]. They proposed that this multiplicity of bands can be associated to post-translational modifications of the enzyme, mainly due to COX-2 glycosylation which can be related to several pathologies such as lung adenocarcinoma[32]. Interestingly, we detected non-glycosylated COX-2 protein in the majority of non-tumor mucosa samples, while glycosylated COX-2 protein was mainly detected in tumor tissue from CRC patients. These findings were confirmed by deglycosylation assays performed both in tumor tissue from CRC patients and in HT29 cells. Moreover, it is known that PGE2 is the most abundant prostaglandin in human colon and the levels of PGE2 are usually increased in colorectal neoplasia compared to normal tissue, which demonstrates its tumor expansion and metastatic functions. Interestingly, in our tumor samples we detected less PGE2 compared to normal colonic mucosa. These results may be due to the presence of glycosylated COX-2, as previous work pointed out that post-translational modifications such as glycosylation of COX-2 can lead to a decrease in both protein levels and activity. In fact, the levels of PGE2 were significantly lower in tumors that only present the upper COX-2 band, corresponding to those with a glycosylated state of the protein. Moreover, in spite of the high variability of the samples, we have also detected a tendency towards an increase in 15-PGDH content in tumors, while levels of EP4 receptors remained unaltered (data not shown). Thus, low levels of PGE2 detected in tumor samples of our CRC cohort can be associated not only to COX-2 protein alterations, but also to an inhibitory scenario of the downstream signaling pathways.
Previous studies comparing the metabolic content of healthy and cancerous colon biopsies by HRMAS obtained from the same patient have demonstrated that adenocarcinomas are characterized by higher levels of taurine, glutamate, aspartate and lactate among others[18,33]. Accordingly, we have detected higher levels of taurine (both peaks) and phosphocholine in tumor samples whose peaks were missing in normal colonic mucosa. In contrast to the broadly described increase in the literature[34,35], lactate levels in our cohort showed a decreasing trend in tumors that did not reach statistical significance due to the high variability between patients. Although some authors have proposed lactate as a non-invasive biomarker to determine malignancy for some kind of tumors[36], further analyses in larger populations will be needed to establish the value of serum lactate to determine the response to therapy or early recurrence in CRC.
Furthermore, in the last years several proteomic studies of tissue samples obtained from CRC patients has allowed to compare the protein profile between tumor and the adjacent healthy mucosa[19,37]. It has been particularly effective in the discovery of prognostic tumor markers. However, these proteomic studies in tumors have also some disadvantages, mainly due to the limited availability of sample and the cellular heterogeneity of tumors. Our proteomic study revealed that tumor tissue has significantly more protein load and higher heterogeneity than its corresponding non-tumor pair. This fact has been recently described in several cancer cell lines, where it has been related to genomic aberrations on protein networks underlying physiological cellular activities[38]. Among these proteins, our study highlights some implicated in basic biological functions such as mitochondrial activity, processing of DNA and RNA, vesicular trafficking, cytoskeleton, metabolism and splicing. These findings are in agreement with previously published data in CRC and in other tumors[19,39,40]. Particularly, in our cohort of CRC patients, four proteins are missing in healthy mucosa but are highly expressed in tumor with a peptide counting more than 20. Interestingly, two of them are implicated in splicing, another specific hit is importin-5, which is a nuclear transport protein and the other is the apoptotic factor AIF-1. Recent studies have revealed multiple ways by which splicing is pathologically altered to promote the initiation and/or maintenance of cancer[41,42]. Furthermore, the importins may also play an important role in cancer by transporting key mediators of oncogenesis across the nuclear membrane in cancer cells. In fact, these proteins are usually overexpressed in multiple tumors including melanoma, pancreatic, breast, colon, gastric, prostate, esophageal, lung cancer and lymphomas[43]. Apoptotic pathways dysregulation has a direct relationship with cancer development[44]. In particular, it has been described that the pro-apoptotic factor AIF-1 shows higher expression in colorectal tumors[45]. In addition to this, it should be noted that among the more significantly upregulated proteins we found neutrophil-associated proteins such as myeloperoxidase and eosinophil peroxidase, which are involved in inflammation. This result is consistent with what we expected since cancer has a strong inflammatory component, which is known to play a significant role in cancer initiation, promotion and progression.
Our results relate by the first time post-translational COX-2 modifications with a metabolic and proteomic profile and can be useful in building new multivariate classifiers looking for a robust cross-talk among molecular, metabolic and proteomics data to improve the knowledge on CRC, thus contributing to establish new protocols for the diagnosis, prognosis and therapeutics for this cancer.
ARTICLE HIGHLIGHTS
Research background
Colorectal cancer (CRC) is the second most common cause of cancer death worldwide. It is broadly described that cyclooxygenase-2 (COX-2) is mainly overexpressed in CRC but very low is known about the nature and effects of the post-translational modifications of this enzyme within the tumor environment. Using metabolic and proteomic profile analyses which are essential for cancer prognosis and diagnosis, our hypothesis is that integrating these parameters with the study of COX-2 post-translational modifications in a high number of CRC patients may provide new insights on the implications of post-translational modifications for CRC prognosis and therapeutics.
Research motivation
Many medical and research efforts have been made trying to better evaluate CRC progression. Nevertheless, it is still necessary to study in depth the molecular, metabolic and proteomic features to unravel the mechanisms leading to CRC progression and to provide a rationale for a personalized therapeutic approach.
Research objectives
The aim of this study was to analyze the regulation of COX-2 in samples from patients with colorectal cancer and to perform a detailed analysis of both the metabolic profile and the proteomic content of the tumor tissue compared to the non-tumor adjacent area.
Research methods
Both normal and tumor tissue obtained from CRC patients were processed and the protein levels of COX-2 were determined. Deglycosylation assays were performed in both cells and tumor samples before measuring prostaglandin E2 (PGE2) levels. Moreover, metabolic and proteomic profile in both samples types were carried out to complete the study.
Research results
Tumor tissue of colorectal cancer patients of our cohort presents an altered COX-2 protein expression profile, which correspond to a glycosylated state of the protein. This was associated to a lesser PGE2 production in tumors. Moreover, high resolution magic angle spinning (HRMAS) analysis indicated that tumor tissue exhibits increased levels of certain metabolites as taurine and phosphocholine and lower levels of lactate. Moreover, we detected an enlarged number of proteins in tumors that are mainly implicated in basic biological functions. Due to the high variability between patients, it will be necessary to analyze a large number of samples in order to achieve sufficient statistical power to find a biomarker that will be applicable in the future as a tool to improve both the diagnosis and prognosis of CRC patients.
Research conclusions
In our colorectal cancer cohort, tumor tissue presents a differential COX-2 expression pattern compared to non-tumor and a lower activity of this enzyme. Moreover, this tissue showed an altered metabolic and proteomics profile that can be correlated to post-translational COX-2 modifications. The analysis of correlations between metabolic and molecular parameters in tumors of a high number of CRC patients could be useful for the understanding of the basis of this cancer in humans.
Research perspectives
Our results relate by the first time post-translational COX-2 modifications with a metabolic and proteomic profile and can be useful in building new multivariate classifiers looking for a robust cross-talk among molecular, metabolic and proteomics data to improve the knowledge on CRC contributing to establish new protocols for the diagnosis, prognosis and therapeutics for this cancer. However, more samples should be analyzed in the future in order to discriminate a robust biomarker that can be useful in CRC diagnosis and prognosis.
ACKNOWLEDGEMENTS
Authors would like to thank Verónica Terrón Arcos and Natalia Canales Bueno for their excellent technical assistance.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
Institutional review board statement: Colorectal cancer biopsies were obtained from 45 patients under informed consent, and were used in accordance with the procedures approved by Clinical Investigation ethics committees of the Germans Trias i Pujol Hospital (Badalona, Spain) and Bellvitge Hospital (Barcelona, Spain).
Conflict-of-interest statement: MAP is cofounder and equity holder of Aniling, a biotech company with no interests in this work. MAP lab has received research funding from Celgene. The rest of the authors declare no conflict of interest.
Data sharing statement: No additional data is available.
Manuscript source: Invited manuscript
Peer-review started: October 17, 2018
First decision: December 5, 2018
Article in press: January 10, 2019
P- Reviewer: Leon J, Liagre B, Temraz S S- Editor: Ma RY L- Editor: A E- Editor: Yin SY
Contributor Information
Patricia Prieto, Department of Metabolism and Physiopathology of Inflammatory Diseases, Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
Rafael I Jaén, Department of Metabolism and Physiopathology of Inflammatory Diseases, Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
Daniel Calle, Laboratorio de Imagen Médica, Hospital Universitario Gregorio Marañón, Madrid 28007, Spain.
María Gómez-Serrano, Laboratorio de Proteómica Cardiovascular, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
Estefanía Núñez, Laboratorio de Proteómica Cardiovascular, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
María Fernández-Velasco, Instituto de Investigación Sanitaria del Hospital Universitario la Paz (IdiPaz), Madrid 28046, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
Paloma Martín-Sanz, Department of Metabolism and Physiopathology of Inflammatory Diseases, Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
Sergio Alonso, Programa de Medicina Predictiva y Personalizada del Cáncer (PMPPC), Fundación Instituto de investigación en ciencias de la salud Germans Trias i Pujol, Ctra Can Ruti, Badalona 08916, Spain.
Jesús Vázquez, Laboratorio de Proteómica Cardiovascular, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain.
Sebastián Cerdán, Department of Metabolism and Physiopathology of Inflammatory Diseases, Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Madrid 28029, Spain.
Miguel Ángel Peinado, Programa de Medicina Predictiva y Personalizada del Cáncer (PMPPC), Fundación Instituto de investigación en ciencias de la salud Germans Trias i Pujol, Ctra Can Ruti, Badalona 08916, Spain.
Lisardo Boscá, Department of Metabolism and Physiopathology of Inflammatory Diseases, Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Enfermedades Cardiovasculares (Ciber-CV), Instituto de Salud Carlos III (ISCIII), Madrid 28029, Spain. lbosca@iib.uam.es.
References
- 1.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu B, Qu L, Yan S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. 2015;15:106. doi: 10.1186/s12935-015-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pang LY, Hurst EA, Argyle DJ. Cyclooxygenase-2: A Role in Cancer Stem Cell Survival and Repopulation of Cancer Cells during Therapy. Stem Cells Int. 2016;2016:2048731. doi: 10.1155/2016/2048731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrández A, Prescott S, Burt RW. COX-2 and colorectal cancer. Curr Pharm Des. 2003;9:2229–2251. doi: 10.2174/1381612033454036. [DOI] [PubMed] [Google Scholar]
- 6.Baker N, O’Meara SJ, Scannell M, Maderna P, Godson C. Lipoxin A4: anti-inflammatory and anti-angiogenic impact on endothelial cells. J Immunol. 2009;182:3819–3826. doi: 10.4049/jimmunol.0803175. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology. 2015;149:1884–1895.e4. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backlund MG, Mann JR, Holla VR, Buchanan FG, Tai HH, Musiek ES, Milne GL, Katkuri S, DuBois RN. 15-Hydroxyprostaglandin dehydrogenase is down-regulated in colorectal cancer. J Biol Chem. 2005;280:3217–3223. doi: 10.1074/jbc.M411221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myung SJ, Rerko RM, Yan M, Platzer P, Guda K, Dotson A, Lawrence E, Dannenberg AJ, Lovgren AK, Luo G, Pretlow TP, Newman RA, Willis J, Dawson D, Markowitz SD. 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc Natl Acad Sci USA. 2006;103:12098–12102. doi: 10.1073/pnas.0603235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexanian A, Sorokin A. Cyclooxygenase 2: protein-protein interactions and posttranslational modifications. Physiol Genomics. 2017;49:667–681. doi: 10.1152/physiolgenomics.00086.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sevigny MB, Graham K, Ponce E, Louie MC, Mitchell K. Glycosylation of human cyclooxygenase-2 (COX-2) decreases the efficacy of certain COX-2 inhibitors. Pharmacol Res. 2012;65:445–450. doi: 10.1016/j.phrs.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Sevigny MB, Li CF, Alas M, Hughes-Fulford M. Glycosylation regulates turnover of cyclooxygenase-2. FEBS Lett. 2006;580:6533–6536. doi: 10.1016/j.febslet.2006.10.073. [DOI] [PubMed] [Google Scholar]
- 13.Begley JK, Redpath TW, Bolan PJ, Gilbert FJ. In vivo proton magnetic resonance spectroscopy of breast cancer: a review of the literature. Breast Cancer Res. 2012;14:207. doi: 10.1186/bcr3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beloueche-Babari M, Chung YL, Al-Saffar NM, Falck-Miniotis M, Leach MO. Metabolic assessment of the action of targeted cancer therapeutics using magnetic resonance spectroscopy. Br J Cancer. 2010;102:1–7. doi: 10.1038/sj.bjc.6605457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haris M, Yadav SK, Rizwan A, Singh A, Wang E, Hariharan H, Reddy R, Marincola FM. Molecular magnetic resonance imaging in cancer. J Transl Med. 2015;13:313. doi: 10.1186/s12967-015-0659-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan KW, Nordenstam J, Lauwers GY, Rothenberger DA, Alavi K, Garwood M, Cheng LL. Metabolomic characterization of human rectal adenocarcinoma with intact tissue magnetic resonance spectroscopy. Dis Colon Rectum. 2009;52:520–525. doi: 10.1007/DCR.0b013e31819c9a2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIntyre DJ, Howe FA, Ladroue C, Lofts F, Stubbs M, Griffiths JR. Can localised (19)F magnetic resonance spectroscopy pharmacokinetics of 5FU in colorectal metastases predict clinical response? Cancer Chemother Pharmacol. 2011;68:29–36. doi: 10.1007/s00280-010-1438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y, Xu T, Huang J, Zhang L, Xu S, Xiong B, Wang Y, Tang H. Tissue Metabonomic Phenotyping for Diagnosis and Prognosis of Human Colorectal Cancer. Sci Rep. 2016;6:20790. doi: 10.1038/srep20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Chaver P, Otero-Estévez O, Páez de la Cadena M, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Proteomics for discovery of candidate colorectal cancer biomarkers. World J Gastroenterol. 2014;20:3804–3824. doi: 10.3748/wjg.v20.i14.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzozie A, Nanni P, Staiano T, Grossmann J, Barkow-Oesterreicher S, Shay JW, Tiwari A, Buffoli F, Laczko E, Marra G. Sorbitol dehydrogenase overexpression and other aspects of dysregulated protein expression in human precancerous colorectal neoplasms: a quantitative proteomics study. Mol Cell Proteomics. 2014;13:1198–1218. [Google Scholar]
- 21.O’Dwyer D, Ralton LD, O’Shea A, Murray GI. The proteomics of colorectal cancer: identification of a protein signature associated with prognosis. PLoS One. 2011;6:e27718. doi: 10.1371/journal.pone.0027718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiśniewski JR, Ostasiewicz P, Mann M. High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J Proteome Res. 2011;10:3040–3049. doi: 10.1021/pr200019m. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Bartolomé S, Navarro P, Martín-Maroto F, López-Ferrer D, Ramos-Fernández A, Villar M, García-Ruiz JP, Vázquez J. Properties of average score distributions of SEQUEST: the probability ratio method. Mol Cell Proteomics. 2008;7:1135–1145. doi: 10.1074/mcp.M700239-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Navarro P, Vázquez J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J Proteome Res. 2009;8:1792–1796. doi: 10.1021/pr800362h. [DOI] [PubMed] [Google Scholar]
- 25.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 26.Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 27.Sinicrope FA, Gill S. Role of cyclooxygenase-2 in colorectal cancer. Cancer Metastasis Rev. 2004;23:63–75. doi: 10.1023/a:1025863029529. [DOI] [PubMed] [Google Scholar]
- 28.Elder DJ, Baker JA, Banu NA, Moorghen M, Paraskeva C. Human colorectal adenomas demonstrate a size-dependent increase in epithelial cyclooxygenase-2 expression. J Pathol. 2002;198:428–434. doi: 10.1002/path.1232. [DOI] [PubMed] [Google Scholar]
- 29.Peng L, Zhou Y, Wang Y, Mou H, Zhao Q. Prognostic significance of COX-2 immunohistochemical expression in colorectal cancer: a meta-analysis of the literature. PLoS One. 2013;8:e58891. doi: 10.1371/journal.pone.0058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asting AG, Farivar A, Iresjö BM, Svensson H, Gustavsson B, Lundholm K. EGF receptor and COX-1/COX-2 enzyme proteins as related to corresponding mRNAs in human per-operative biopsies of colorectal cancer. BMC Cancer. 2013;13:511. doi: 10.1186/1471-2407-13-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crew TE, Elder DJ, Paraskeva C. A cyclooxygenase-2 (COX-2) selective non-steroidal anti-inflammatory drug enhances the growth inhibitory effect of butyrate in colorectal carcinoma cells expressing COX-2 protein: regulation of COX-2 by butyrate. Carcinogenesis. 2000;21:69–77. doi: 10.1093/carcin/21.1.69. [DOI] [PubMed] [Google Scholar]
- 32.Cao C, Gao R, Zhang M, Amelio AL, Fallahi M, Chen Z, Gu Y, Hu C, Welsh EA, Engel BE, Haura EB, Cress WD, Wu L, Zajac-Kaye M, Kaye FJ. Role of LKB1-CRTC1 on glycosylated COX-2 and response to COX-2 inhibition in lung cancer. J Natl Cancer Inst. 2014;107:358. doi: 10.1093/jnci/dju358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirnezami R, Jiménez B, Li JV, Kinross JM, Veselkov K, Goldin RD, Holmes E, Nicholson JK, Darzi A. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann Surg. 2014;259:1138–1149. doi: 10.1097/SLA.0b013e31829d5c45. [DOI] [PubMed] [Google Scholar]
- 34.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71:6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 35.San-Millán I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2017;38:119–133. doi: 10.1093/carcin/bgw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih CC, Lee TS, Tsuang FY, Lin PL, Cheng YJ, Cheng HL, Wu CY. Pretreatment serum lactate level as a prognostic biomarker in patients undergoing supratentorial primary brain tumor resection. Oncotarget. 2017;8:63715–63723. doi: 10.18632/oncotarget.18891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang JJ, Liu Y, Zheng Y, Lin F, Cai GF, Yao XQ. Comparative proteomics analysis of colorectal cancer. Asian Pac J Cancer Prev. 2012;13:1663–1666. doi: 10.7314/apjcp.2012.13.4.1663. [DOI] [PubMed] [Google Scholar]
- 38.Roumeliotis TI, Williams SP, Gonçalves E, Alsinet C, Del Castillo Velasco-Herrera M, Aben N, Ghavidel FZ, Michaut M, Schubert M, Price S, Wright JC, Yu L, Yang M, Dienstmann R, Guinney J, Beltrao P, Brazma A, Pardo M, Stegle O, Adams DJ, Wessels L, Saez-Rodriguez J, McDermott U, Choudhary JS. Genomic Determinants of Protein Abundance Variation in Colorectal Cancer Cells. Cell Rep. 2017;20:2201–2214. doi: 10.1016/j.celrep.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao JJ, Zhi X, Wang Y, Zhang Z, Hao Z, Ye R, Tang Z, Qian F, Wang Q, Zhu J. Comprehensive Proteomic Characterization of the Human Colorectal Carcinoma Reveals Signature Proteins and Perturbed Pathways. Sci Rep. 2017;7:42436. doi: 10.1038/srep42436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethi MK, Thaysen-Andersen M, Kim H, Park CK, Baker MS, Packer NH, Paik YK, Hancock WS, Fanayan S. Quantitative proteomic analysis of paired colorectal cancer and non-tumorigenic tissues reveals signature proteins and perturbed pathways involved in CRC progression and metastasis. J Proteomics. 2015;126:54–67. doi: 10.1016/j.jprot.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 41.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahipal A, Malafa M. Importins and exportins as therapeutic targets in cancer. Pharmacol Ther. 2016;164:135–143. doi: 10.1016/j.pharmthera.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 44.Plati J, Bucur O, Khosravi-Far R. Apoptotic cell signaling in cancer progression and therapy. Integr Biol (Camb) 2011;3:279–296. doi: 10.1039/c0ib00144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobanoglu B, Ceyran AB, Simsek M, Şenol S. Immunohistochemical analysis of Bax and AIF in colorectal tumors. Int J Clin Exp Med. 2015;8:16071–16076. [PMC free article] [PubMed] [Google Scholar]