Abstract
Fat mass and obesity-associated (FTO) protein has been identified as a critical demethylase in regulating cellular mRNA stability by removing N6-methyladenosine (m6A) residues in mRNA. Even though the role of FTO in body energy metabolism has been well established, its role in cancer cell homeostasis remains unclear. In the present study, by using RNA interference, it was indicated that FTO is required for pancreatic cancer cell proliferation. Knockdown of FTO resulted in compromised proliferation of pancreatic cancer cells. Furthermore, DNA synthesis was compromised, followed by an increase in apoptosis in FTO small interfering RNA (siRNA)-treated cells. In terms of its underlying mechanism, FTO has been indicated to interact with MYC proto-oncogene, bHLH transcription factor and to enhance its stability by decreasing its m6A level. Therefore, the aforementioned observations indicate a novel mechanism for the regulation of pancreatic cancer cells by FTO, which may provide insight on pancreatic cancer treatment strategies.
Keywords: fat mass and obesity-associated protein, pancreatic cancer, proliferation
Introduction
Fat mass and obesity-associated (FTO) protein was first identified as a protein associated with obesity (1,2). Its overexpression is associated with obesity occurrence, whereas its underlying mechanisms remain unclear. As a member of AlkB-like DNA/RNA demethylase family, FTO contains a universal amino-terminal AlkB-like domain and a carboxy-terminal domain (3,4). In contrast to other family members, an extra loop in FTO, which competes with the unmethylated strand of the DNA duplex for binding to FTO, engages higher specificity to single-stranded RNA or DNA substrates (4). Importantly, a catalytic activity of FTO towards RNA has been previously identified (5). FTO contains a Fe(II) binding domain and substrate recognition domain, which catalyzes the removal of the methyl group on N6-adenosine in mRNA (5). As a prevalent internal modification of mRNA, N6-adenosine methylation (m6A) serves important roles in mRNA metabolism, including localization and stability. Oxidative demethylation by FTO represses the recognition of mRNA by YTH domain family protein for mRNA decay (6). The aforementioned observations indicate an important uncharacterized regulatory mechanism for gene expression.
By using a genetically engineered mouse model, the physiological functions of FTO have been extensively studied. Germline loss of FTO leads to perinatal lethality, growth retardation and reduced body weight, indicating that FTO is required for mammalian development (2,7). However, the underlying roles of FTO in other pathological processes, including the occurrence and development of cancer, remain unclear.
Treatment of pancreatic cancer remains inefficient, due to its high resistance to traditional therapeutic strategies, including chemotherapy, radiotherapy and surgical resection (8–10). Therefore, future studies to provide additional insight for the molecular and cellular mechanisms for the development of novel treatment strategies are required.
In the present study, the function of FTO in pancreatic cancer cell homeostasis was characterized to establish the association between them. Whether FTO participates in pancreatic cancer cell homeostasis through regulation of m6A modification was also studied. It was revealed that FTO was highly expressed in pancreatic cancer cells, and FTO knockdown resulted in compromised pancreatic cell proliferation and reduced DNA synthesis. Accordingly, mRNA methylation was accumulated. In conclusion, a previously unknown function of FTO in pancreatic cancer was uncovered, shedding light on a novel molecular target for pancreatic cancer treatment.
Materials and methods
Tissues sample collection
Fresh frozen tumor and normal tissue samples were obtained from surgical specimens of 2 patients at Jiangyin People's Hospital (Wuxi, China) between July 2014 and May 2015. One patient was a 52 year-old male and the other was a 43 year-old female. The present study had been reviewed and approved by the Institutional Review Board of Jiangyin People's Hospital, School of Medicine (Southeast Medical University College, Wuxi, China). Informed written consent was obtained from all patients, in accordance to the Declaration of Helsinki and its amendments.
Cell culture and transfection
Human pancreatic ductal cell (HPDE) and pancreatic cancer cell lines SW1990, PANC-1 and BXPC-3 were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences) at 37°C in a humidified atmosphere containing 5% CO2. For empty pcDNA 3.1 plasmid (cat. no. V79020; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or c-Myc-overexpressing pcDNA 3.1 plasmid (cat. no. V79020; Thermo Fisher Scientific, Inc.) transfection, 2×105 PANC-1 cells were seeded into 6-well plates overnight prior to transfection. A total of 2.5 µg expression plasmids or empty vectors were transfected into cells using Lipofectamine™ 2000 (cat. no. 11668027; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After 48 h, the transfected cells were harvested for further analysis.
RNA interference
FTO RNA interference (RNAi) (5′-GCACAAGCATGGCTGCTTA-3′), scramble control (5′-GCAACGACGGTCGTACTTA-3′) and pSilencer 3.1-H1 neo plasmid were purchased from Thermo Fisher Scientific, Inc. PANC1, SW1990 and BXPC-3 cells (~5×105) were seeded in 35-mm plates and cultured overnight at 37°C. Cells were subsequently transfected with the mixture of RNAi (50 nM) and Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Inc.), according to the manufacture's protocol.
MTT assay
Following FTO RNAi transfection for 12 h, PANC1, SW1990 and BXPC-3 cells were plated into 96-well plates for further culture at 37°C and samples were taken at day 2, 4 and 8. A total of 20 µl of MTT solution (5 mg/ml), dissolved in DMEM, was subsequently added into the cells and incubated for 2 h at 37°C. Precipitates were subsequently dissolved by dimethylsulfoxide for 20 min at room temperature. Absorbance was recorded at a wavelength of 540 nm using a microplate reader elx 800 (BioTek Instruments, Inc., Winooski, VT, USA).
Bromodeoxyuridine (BrdU) assay
For BrdU incorporation assay, PANC-1, SW1990 and BXPC-3 cells were plated in 8-well chamber slides at density of 3×104/ml. Following 48 h of transfection, cells were incubated with 10 µM BrdU for an additional 2 h. Cells were then fixed with 2% formaldehyde for 30 min at room temperature and permeabilized with 0.2% Triton X-100, followed by incubation with anti-BrdU primary antibody (1:1,000; cat. no. ab152095; Abcam, Cambridge, UK) overnight at 4°C. Cells were subsequently incubated with PE-Cy7-labeled mouse anti-rabbit secondary antibody (1:400; cat. no. sc-516721; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at room temperature. Images were acquired using a fluorescent microscope (magnification, ×400; Leica Microsystems, Inc., Buffalo Grove, IL, USA) and analyzed with ImageJ software (v1.8.0; National Institutes of Health, Bethesda, Maryland, USA).
TUNEL assay
For TUNEL assay, cells at a density of 3×104/ml were fixed with 2% formaldehyde for 30 min at room temperature and the TUNEL assay was performed according to the manufacturer's protocol (Click-iT® TUNEL Alexa Fluor® 488 Imaging assay; cat. no., C10245; Thermo Fisher Scientific, Inc.). Ten fields of view at ×400 magnification were observed using a fluorescent microscope (Leica Microsystems, Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RNA from PANC-1, SW1990 and BXPC-3 cells was isolated by TRIzol (Thermo Fisher Scientific, Inc.) and subsequently used for cDNA synthesis with High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. RT-qPCR was performed using StepOne/StepOnePlus™ Real-time PCR System with SYBR Green PCR Master mix (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The sequence of the FTO primers used in RT-qPCR were the following: Forward, 5′-ACTTGGCTCCCTTATCTGACC-3′ and reverse, 5′-TGTGCAGTGTGAGAAAGGCTT-3′. An optimal reaction was obtained with the following thermocycling conditions: Initial denaturation at 95°C for 10 min; 45 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for 1 min and elongation at 72°C for 1 min; and a final extension at 72°C for 10 min. β-actin mRNA was used as a control and the level of FTO mRNA expression was normalized to β-actin mRNA using the 2−ΔΔCq method (11). The sequence of the β-actin primers were as follows: Forward, 5′-CTCCATCATGAAGTGTGACGTT-3′ and reverse, 5′-ATCTCCTTCTGCATCCTGTCAG-3′. For amanitin treatment, a final concentration of 2.5 mg/l amanitin (cat. no., A2263; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to the culture medium and incubated at 37°C for 30, 60 and 120 min.
RNA immunoprecipitation
A total of two 10-cm plates of PANC-1 cells (selected as representative cells) were crosslinked by 1% paraformaldehyde for 10 min at room temperature. Cells at a density of ~2×107/ml were subsequently neutralized with 2 mg/ml glycine and lyzed in cell lysis buffer (150 mM KCl, 10 mM HEPES pH 7.6, 2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, RNase inhibitor) on ice for 30 min. Cell lysate were subsequently sonicated (20 KHz; 5s sonication, 5s pause, 12 cycles) at 4°C and precleaned by Dynabeads® Protein G (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. FTO antibody (1:50; cat. no. ab92821; Abcam) and control IgG (1:50; catalog no. ab188776; Abcam) were incubated with the sonicated lysates overnight at 4°C. Dynabeads® Protein G was subsequently incubated with the lysate for an additional 2 h at 4°C. Immunoprecipitated complex were subsequently washed with standard Chromatin immunoprecipitation (cat. no., 17-295, Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocols. TRIzol was subsequently added into the supernatant to extract immunoprecipitated RNA.
Western blot analysis
Cells were harvested in lysis buffer (100 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40 and protein inhibitor cocktail) and centrifuged at 1,000 × g for 3 min at 4°C to collect the supernatants. Samples were separated with 12% Mini-PROTEAN® TGX™ Precast Gels (cat. no., 4561093; Bio-Rad Laboratories, Inc., Hercules, CA, USA) and 50 µg total protein determined with BCA assay (cat. no. 23235; Thermo Fisher Scientific, Inc.) was loaded into each lane. The gel was subsequently transferred to Immobilon®-P polyvinylidene difluoride Membrane (cat. no., IPVH00010; EMD Millipore, Billerica, MA, USA). The membrane was subsequently blocked with 5% Blotting-Grade Blocker (cat. no., 1706404; Bio-Rad Laboratories, Inc.) dissolved in Tris-buffered saline containing Tween-20 (PBST) at room temperature for 2 h. The blots were incubated with antibodies against FTO (cat. no., sc-271713; Santa Cruz Biotechnology, Inc.), GAPDH (cat. no., sc-47724; Santa Cruz Biotechnology, Inc.) or anti-FLAG-M2 (cat. no., F1804; Sigma-Aldrich; Merck KGaA) diluted at 1:1,000 with blocking buffer overnight at 4°C, followed by washing with PBST and horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (cat. no., sc-2005; Santa Cruz Biotechnology, Inc.) diluted at 1:5,000 with blocking buffer for 30 min at room temperature. Following washing with PBST, protein bands were visualized by enhanced chemiluminescence (cat. no., 7003; Cell Signaling Technology, Inc., Danvers, MA, USA) and analyzed with ImageJ software (v1.8.0; National Institutes of Health).
Dot blot
mRNA was isolated with Magnetic mRNA Isolation kit (New England BioLabs, Inc., Ipswich, MA, USA), according to the manufacturer's protocols. 200 ng total mRNA was spotted onto a nitrocellulose membrane (Thermo Fisher Scientific, Inc.), which was subsequently crosslinked by ultraviolet exposure. The membrane was subsequently incubated with 5% non-fat milk and blocked for 30 min at room temperature. Anti-m6A antibody (cat. no. 202 111; 1:1,000; Synaptic Systems, Goettingen, Germany) was incubated with the membrane overnight at 4°C. HRP-conjugated rabbit secondary antibody (cat. no. sc-2357; 1:10,000; Santa Cruz Biotechnology) was incubated with the membrane at room temperature for 2 h. Subsequently, the membrane was used for further chemiluminescence analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Comparisons between 2 groups were analyzed using an unpaired Student's t-test. Data are presented as the mean ± standard error of mean. Comparisons between 3 or more were analyzed by one-way analysis of variance followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference
Results
Overexpression of FTO in pancreatic cancer cells
To determine the expression profile of FTO in pancreatic cancer cells, the expression levels in normal pancreatic epithelial cells HPDE and pancreatic cancer cell lines SW1990, PANC-1 and BXPC-3 were examined. Higher mRNA expression levels of FTO were observed in SW1990, PANC-1 and BXPC-3 cells compared with expression level of HPDE (Fig. 1A). Accordingly, the protein expression level of FTO was upregulated in SW1990, PANC-1 and BXPC-3 compared with that in HPDE cells (Fig. 1B). Increased FTO expression was also observed in pancreatic tumors (Fig. 1C).
Figure 1.
Expression of FTO in pancreatic cancer cells. (A) Relative expression level of FTO in pancreatic cancer cells. (B) Protein levels of FTO in pancreatic cancer cells. (C) Immunostaining of FTO in pancreatic tumor tissue and peri-cancer tissue (magnification, ×400). *P<0.05 and **P<0.01 vs. HPDE. FTO, fat mass and obesity-associated protein; HPDE human pancreatic ductal cell; Ctrl, control.
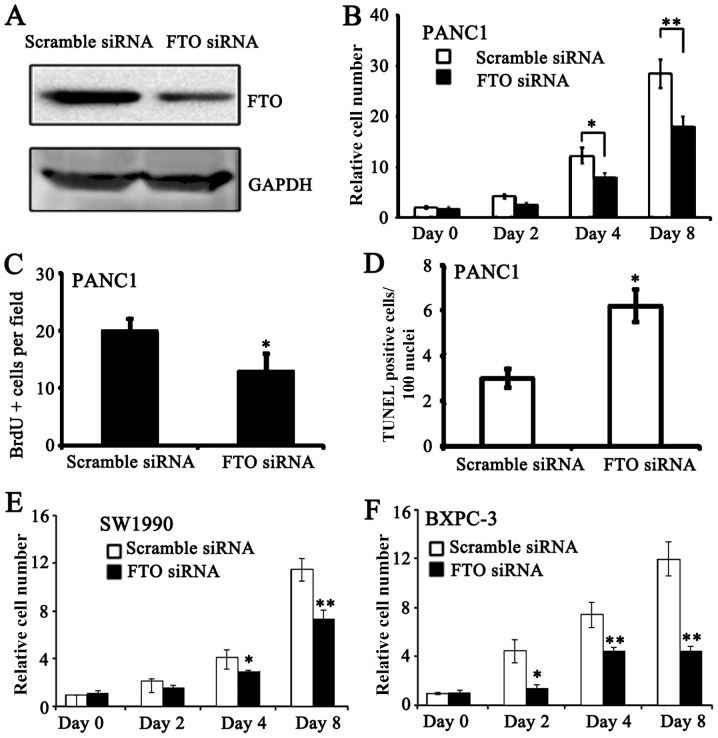
FTO knockdown compromises pancreatic cancer cell proliferation
To examine the roles of FTO in pancreatic cancer cells, RNAi was used to knockdown the expression levels of FTO in PANC-1 cells. FTO protein expression was successfully knockdown in PANC-1 cell lines (Fig. 2A). FTO siRNA-treated PANC-1 cells exhibited reduced proliferation compared with scramble siRNA-treated PANC-1 cells at days 4 and 8 (Fig. 2B). Accordingly, DNA synthesis was reduced in FTO siRNA-treated PANC-1 cells (Fig. 2C). Furthermore, apoptosis was increased in FTO knockdown PANC-1 cells (Fig. 2D). To verify the results from the PANC-1 cells, proliferation in SW1990 and BXPC-3 in response to treatment using FTO siRNA was examined. FTO knockdown lead to similar cytotoxic effects on the proliferation of SW1990 and BXPC-3 cells for 8 days (Fig. 2E and F). These data indicate that FTO may regulate proliferation in pancreatic cancer cells by modulating cell cycle progression.
Figure 2.
FTO regulates pancreatic cancer cell proliferation. (A) Knockdown efficiency of FTO siRNA in PANC-1 cells. (B) Proliferation rate of PANC-1 cells treated with FTO or control siRNA. (C) Quantitative BrdU incorporation analysis of FTO siRNA-treated PANC-1 cells. (D) Quantitative TUNEL analysis of FTO siRNA-treated PANC-1 cells. Proliferation rate of (E) SW1990 and (F) BXPC-3 cells. *P<0.05 and **P<0.01 vs. scramble siRNA. FTO, fat mass and obesity-associated protein; HPDE human pancreatic ductal cell; siRNA, small interfering RNA.
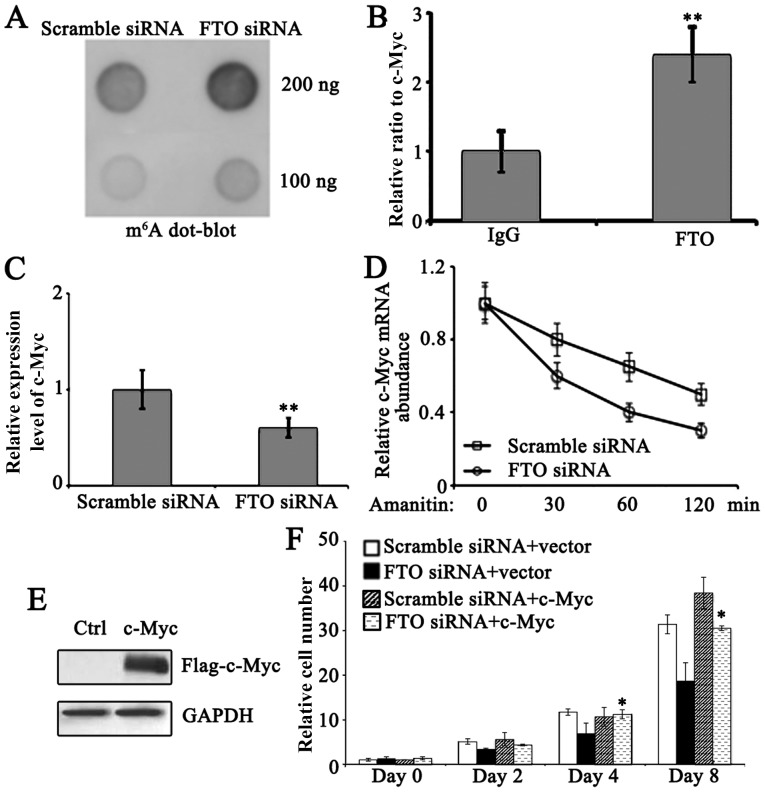
FTO regulates mRNA m6A levels
To examine the underlying mechanisms of FTO in the proliferation and cell cycle of pancreatic cancer cells, the m6A levels in PANC-1 cells were determined. FTO knockdown resulted in increased m6A expression levels compared with scramble siRNA-treated PANC-1 cells (Fig. 3A), indicating FTO serves important roles in the regulation of m6A expression levels in PANC-1 cells. Since c-Myc is a critical mediator in regulating cell entry into S phase of cell cycle (12), further investigation on whether FTO may interact with c-Myc expression and regulate its metabolism in PANC-1 cells was carried out. By using RNA immunoprecipitation, which enriches FTO-interacting RNA, with FTO antibody in PANC-1 cells, it was observed that FTO interacts with c-Myc expression (Fig. 3B). c-Myc expression was reduced following FTO knockdown (Fig. 3C). To test the stability of c-Myc expression, amanitin, a specific inhibitor of RNA polymerase II, was used to inhibit mRNA de novo synthesis in PANC-1 cells. The degradation of existent c-Myc expression was subsequently monitored. As expected, enhanced loss of c-Myc expression was detected in FTO knockdown PANC-1 cells, indicating its stability was compromised in FTO deficient PANC-1 cells (Fig. 3D). These data strongly suggest that c-Myc is a primary target for FTO-mediated cell cycle regulation in pancreatic cancer cells. To verify this, c-Myc was overexpressed in FTO knockdown PANC-1 cells (Fig. 3E). To verify its significance in FTO-regulated proliferation, c-Myc was overexpressed in FTO knockdown cells and it was indicated that c-Myc significantly restored proliferation (Fig. 3F). Therefore, c-Myc is suggested to serve a major role in FTO-mediated proliferation.
Figure 3.
FTO regulates m6A levels in PANC-1 cells. (A) m6A expression levels of control and FTO siRNA-treated PANC-1 cells. (B) FTO interaction with c-Myc expression level. **P<0.01. vs. IgG. (C) c-Myc expression levels in control and FTO siRNA-treated PANC-1 cells. **P<0.01. vs. scramble siRNA. (D) c-Myc expression levels in control and FTO siRNA-treated PANC-1 cells in the presence and absence of amanitin. Cells were collected for analysis at time points indicated in the graph. (E) PANC-1 cells were treated with empty pcDNA 3.1 plasmid or c-Myc-pcDNA 3.1 plasmid and the expression of c-Myc protein was evaluated by western blotting. (F) Proliferation of FTO siRNA treatment with or without c-Myc overexpression in PANC-1 cells. *P<0.05 scramble siRNA + vector vs. scramble siRNA + c-Myc. c-Myc, MYC proto-oncogene, BHLH transcription factor; FTO, fat mass and obesity-associated protein; siRNA, small interfering RNA; Ctrl, control.
Discussion
mRNA carries genetic information between the DNA and the protein, and the underlying mechanisms for mRNA generation and decay have been extensively studied. Similar to DNA, mRNA can also be modified (13–15). One of its most documented internal modifications is in its m6A sites (16,17). Until recently, the functions of this modification remained unclear. m6A has been reported to decrease mRNA stability and lead to its degradation. Once m6A is demethylated, mRNA is stabilized and relocated into the cytosol for protein translation (6). These observations indicate a novel mechanism for regulation of gene expression. However, its physiological and pathological significance remain unclear. In the present study, the existence of the aforementioned mechanisms in pancreatic cancer cells was examined. It was indicated that FTO, a primary demethylase in vivo, was overexpressed in pancreatic cancer cells compared with normal pancreatic epithelial cells, implying that it is involved in pancreatic cancer development and progression. The knockdown of FTO resulted in a compromised proliferation in pancreatic cancer cells, as well as an increased in apoptosis. These observations demonstrated FTO is required for the proliferation of pancreatic cancer cells. In addition, regulating m6A modification in mRNA may affect pancreatic cancer homeostasis.
The regulation of mRNA stability is critical for gene expression in cancer cells (18). In the past decade, a central role of microRNA in mRNA decay has been established (19,20). In the present study, it was revealed that m6A demethylation is an important missing mechanism for the regulation of mRNA stability in pancreatic cancer cells. In addition, it was demonstrated that the stability of c-Myc mRNA was compromised upon FTO knockdown, which subsequently repressed DNA synthesis. These data indicated an endogenous target for FTO to regulate pancreatic cancer cell proliferation.
A number of limitations existed in the present study. Due to a lack of knowledge of the accurate m6A sites on c-Myc mRNA, an in vitro demethylation assay, combining FTO recombinant protein and methylated c-Myc, was not possible in the present study. Therefore, further investigation is required, as it would assist to identify the direct effects of FTO on c-Myc. In addition, c-Myc was only determined as a primary target of FTO, while other cell cycle-associated proteins have yet to be identified. Further studies will focus on other genes regulated by FTO in pancreatic cancer cells. Given the roles of FTO in energy metabolism, it is possible that metabolism-associated mRNAs were controlled by FTO for pancreatic cancer homeostasis. Therefore, further investigation is required.
In conclusion, it was indicated that mRNA stability regulated by FTO serves important roles in pancreatic cancer cells, suggesting an underlying mechanism for the development and progression of pancreatic cancer. The data of the present study suggest a novel possible therapeutic target for the treatment of pancreatic cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JZ and XT designed the experiment. XT and SL performed the experiments with cancer cell lines. DC and ZZ performed histological experiments with tissue samples. All authors contributed in the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study had been reviewed and approved by the Institutional Review Board of Jiangyin People's Hospital, School of Medicine, Southeast University (Wuxi, China). Informed written consent was obtained from all patients, in accordance to the Declaration of Helsinki and its amendments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, Rüther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 3.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, Cheng W, Wang J, Feng Y, Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 5.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayraktar S, Rocha-Lima CM. Advanced or metastatic pancreatic cancer: Molecular targeted therapies. Mt Sinai J Med. 2010;77:606–619. doi: 10.1002/msj.20217. [DOI] [PubMed] [Google Scholar]
- 9.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 10.Loos M, Kleeff J, Friess H, Buchler MW. Surgical treatment of pancreatic cancer. Ann NY Acad Sci. 2008;1138:169–180. doi: 10.1196/annals.1414.024. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 14.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 15.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 16.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 17.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 20.Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–140. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.