Abstract
Previous studies have revealed that elevated expression of high mobility group A2 (HMGA2) is closely associated with the occurrence of pituitary adenomas (PAs). The expression of microRNA (miR)-16 is deregulated in PA tissues. Bioinformatics analysis has demonstrated that there is a complementary region between seed region of miR-16 and 3′-untranslated region (3′-UTR) of HMGA2 gene. In the present study, it was investigated whether miR-16 may regulate the expression of HMGA2 and whether it is involved in the pathogenesis of PAs. A total of 52 patients with PAs were recruited. Normal brain tissues obtained from 12 patients with traumatic brain injury were used as controls. The association between miR-16 and HMGA2 was validated using dual-luciferase reporter gene assay. HP75 cells were cultured in vitro and divided into the following groups: miR control, miR-16 mimic, small interfering RNA (si)-negative control, si-HMGA2 and miR-16 mimic+si-HMGA2 groups. The expression of miR-16 and HMGA2 in HP75 cells was determined using qRT-PCR and western blot. Cell proliferation was detected using the Cell Counting Kit-8 assay and apoptosis was detected using the TdT-UTP nick end labeling assay. Compared with normal pituitary tissues, the expression of miR-16 in PA tissues was significantly decreased, while the mRNA and protein levels of HMGA2 were significantly increased. miR-16 targeted the 3′-UTR of HMGA2 gene and regulated the expression of HMGA2. Transfection with siRNAs targeting HMGA2 and/or miR-16 mimics inhibited the expression of HMGA2 and the proliferative ability of HP75 cells, whereas it increased apoptosis of HP75 cells. The downregulation of miR-16 and upregulation of HMGA2 were involved in the pathogenesis of PAs. Thus, it is hypothesized that miR-16 inhibited the proliferation and promoted apoptosis of HP75 cells by inhibiting HMGA2 expression.
Keywords: microRNA-16, high mobility group A2, pituitary adenoma, proliferation, apoptosis
Introduction
Pituitary adenomas (PAs) are monoclonal tumors that originate from the anterior and posterior lobes of the pituitary gland and residual cells of the cranial pharyngeal epithelium (1). PA is a common intracranial tumor and accounts for ~15–20% of intracranial tumors, and its incidence ranks third among intracranial tumors and second among gliomas and meningiomas (2). High mobility group A2 (HMGA2) is a member of the HMGA family and a non-histone protein. HMGA2 is named after the high migration ability of HMGA2 on polyacrylamide gel electrophoresis (3). HMGA2 is a chromatin remodeling factor that contains AT-hook basic domains, which assign them the ability to bind the DNA minor groove of AT-rich DNA sequences (4). By changing the chromatin architecture and assembling transcriptional complexes on chromatin, HMGA2 may activate or impair the activity of transcriptional enhancers in various genes indirectly (5). In addition to serving as a transcriptional co-regulator, HMGA2 may also directly regulate the transcription activation and expression of various genes (6). It has been demonstrated that HMGA2 regulates various biological processes, including proliferation and cell cycle. Additionally, increased expression of HMGA2 is associated with the occurrence of various tumors, including prostate (7), colorectal (3), lung (4) and breast cancer (8), and serves an important function in the occurrence, progression and metastasis. Previous studies have revealed that increased expression of HMGA2 is associated with the occurrence of PA and prognosis, and the survival time of patients with PA (9–11). microRNA (miRNA) is an endogenous non-coding single-stranded RNA of 21–25 nucleotides in length, which may regulate the expression of target genes by inhibiting the translation or degradation of mRNAs (12) Previous studies have demonstrated that the expression of miR-16 is deregulated in PAs, and is associated with the occurrence of cancer and poor prognosis (13–15). Bioinformatics analysis indicated there is a complementary region between seed region of miR-16 and 3′-UTR of HMGA2 gene. In the present study, it was investigated whether miR-16 may regulate the expression of HMGA2 and whether it is involved in the pathogenesis of PAs.
Materials and methods
Reagents
The human pituitary adenoma cell line HP75 was provided by ScienCell Research Laboratories (USA). The HEK293 cell line was purchased from the American Type Culture Collection (Manassas, VA, USA). Dulbecco's modified Eagle's medium (DMEM) was purchased from Lonza Group, Ltd. (Basel, Switzerland). Streptomycin was purchased from Corning Incorporated (Corning, NY, USA). Horse serum, fetal bovine serum (FBS), An Opti-MEM, enhanced chemiluminesence (ECL) Western Blotting Substrate Kit, TRIzol and Lipofectamine® 2000 were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). The PrimeScript™ RT reagent kit and SYBR were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). miRNA nucleotide fragments and primers for polymerase chain reaction (PCR) were designed and synthesized by Guangzhou Ribobio (Guangzhou, China). Rabbit anti-HMGA2 antibody was purchased from GeneTex Inc. (cat no. GTX50799; Irvine, CA, USA). Mouse anti-histone H3.1 was purchased from Cell Signaling Technology, Inc. (cat no. 3638; Danvers, MA, USA). Horseradish peroxidase-conjugated secondary antibodies were purchased from Abcam (cat no. ab6721; Cambridge, UK). The TdT-UTP nick end labeling assay (TUNEL) Apoptosis Detection kit, radioimmunoprecipitation assay buffer and Antifade mounting medium were purchased from Jiangsu Beyotime Biotechnology Co., Ltd. (Jiangsu, China). Cell Counting Kit-8 (CCK-8) kit was purchased from Wuhan Boster Biological Technology Co., Ltd. (Wuhan, China). The Dual-Luciferase® Reporter Assay System and pGL3-promoter plasmid were purchased from Promega Corporation (Madison, WI, USA).
Patients
A total of 52 patients with PAs, including 27 males and 25 females, were recruited in Mudanjiang Forestry Hospital between March 2014 and May 2016. The average age of patients was 36.7 years (range, 31–56 years). Normal brain tissues from 16 patients with neurotrauma were collected during surgery and were used as controls. Among these, 9 patients were males and 7 patients were females. The average age of patients was 38.1 years (range, 33–57 years). All patients provided written informed consent and the study was approved by the Ethics Committee of Mudanjiang Forestry Hospital.
Cell culture
The human PA cell line HP75 was cultured in DMEM supplemented with 10% horse serum, 5% fetal bovine serum and 1% streptomycin, and cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. The culture medium was changed every 3 days. When cells reached 60–80% confluence, cells were used for subsequent experiments.
Dual-luciferase reporter assay
The HEK293 cell line genome was used as a template to amplify the full-length 3′-UTR fragment of the HMGA2 gene, cloned into the luciferase reporter vector pGL-3M and transformed into DH5α Escherichia coli competent cells. The positive clones were screened by colony PCR. Following sequencing, the positive clones were used for subsequent experiments. The HEK293 cell line was co-transfected with pGL3-HMGA2-3′-UTR (400 ng), miR-16 mimic/miR-control (25 nmol) and pRL-TK (25 ng) using Opti-MEM and Lipofectamine 2000. Luciferase assays were performed 48 h after transfection, The activity of firefly luciferase was measured and normalized to the corresponding Renilla luciferase activity.
Cell transfection
The sequences of siRNAs used in the present study were as follows: si-HMGA2, 5′-CAGCCUGAAUAACUUGAACTT-3′ (sense) and 5′-GUUCAAGUUAUUCAGGCUGTT-3 (anti-sense); si-negative control (NC), 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (anti-sense). HP75 cells were divided into the following groups: miR-control (scramble), miR-16 mimic, si-NC transfection group, si-HMGA2 group and miR-16 mimic+si-HMGA2 groups. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection, according the manufacturer's protocol. Briefly, HP75 cells were cultured in 100 mm dish at a density of 1.5–2×105/ml, Following cultivation for 24 h, the cells were transfected with 100 nmol/l miRNAs using Lipofectamine 2000. Cell were cultured for 72 h before subsequent experiments.
Bioinformatics analysis
The potential direct common target genes of miR-16 were predicted using miRNA databases, including TargetScan Human 7.2 (16), miRanda (17), and miRDB (18). Intersecting the results of these three databases demonstrated that the HMGA2 gene was a prediction target gene.
Reverse transcription quantitative (RT-q)PCR
Total RNA was isolated from cells using TRIzol reagent. RNA was reverse-transcribed into cDNA using PrimeScript™ RT Reagent kit, according to the manufacturer's protocol. qPCR was performed using Bio-Rad CFX96 quantitative PCR system and SYBR, according to the manufacturer's protocol. The following primers were used: miR-16, 5′-AACCCGUAGAUCUUGGAUCCUG-3′ (forward) and 5′-CAAGAUCAUCUACGGUUUGGGU-3′ (reverse); U6, 5′-ATTGGAACGATACAGAGAAGATT-3′ (forward) and 5′-GGAACGCTTCACGAATTTG-3′ (reverse); HMGA2, 5′-ACCCAGGGGAAGACCCAAA-3′ (forward) and 5′-CCTCTTGGCCGTTTTTCTCCA-3′ (reverse); β-actin, 5′-GAACCCTAAGGCCAAC-3′ (forward) and 5′-TGTCACGCACGATTTCC-3′ (reverse). PCR was conducted as follows: Initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 sec and annealing at 60°C for 30 sec. All the samples were assessed by relative quantification (2−ΔΔCq method) (19).
Western blot analysis
Cells were lysed using radioimmunoprecipitation assay buffer and centrifuged at 10,000 × g for 10 min. Proteins (40 µg) were separated by SDS-PAGE (10% gels) and then transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% non-fat milk for 60 min, followed by incubation with primary antibodies against HMGA2 (dilution, 1:500) and histone H3.1 (dilution, 1:1,000, serving as loading control) overnight at 4°C. Membranes were then washed with 0.1% Tween-20 in PBS (PBST) three times. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000) for 60 min at room temperature and then washed with PBST three times. Immunoreactive bands were visualized by an ECL Western Blotting Substrate kit. X-ray film was used to analyze the image and intensity of bands via ImageJ software (version 1.51b; National Institutes of Health, Bethesda, MA, USA).
CCK-8 assay
Cells were seeded in 96-well cell plates at a density of 1×104 cells/well for 24 h. Cells were cultured for additional 24, 48, 72 and 96 h at 37°C in a humidified atmosphere containing 5% CO2. Then, CCK-8 reagent (10 µl) was added to each well and cells were cultured for 4 h at 37°C in a humidified atmosphere containing 5% CO2. The absorbance values were read at 450 nm wavelength using a microplate reader.
TUNEL assay
Cells were washed with PBS and fixed with 4% paraformaldehyde for 60 min. PBS containing 0.1% Triton X-100 was added and cells were incubated on ice for 2 min and washed twice with PBS. Then, 50 µl of TUNEL solution was added and cells incubated for 60 min at 37°C in dark, washed twice with PBS. Antifade mounting medium were added onto the cells and the fluorescein isothiocyanate-labeled TUNEL-positive cells were imaged under a fluorescent microscope. The cells with green fluorescence were defined as apoptotic cells. A total of 5 fields of each sample were observed.
Statistical analysis
Data were analyzed using SPSS software (version 18.0; SPSS, Inc., Chicago, IL, USA). Data are expressed as the mean ± standard deviation. Results were analyzed using Student's t-test when only 2 groups were compared. One-way analysis of variance followed by Bonferroni post hoc test was used to examine differences between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Decreased expression of miR-16 decreased and increased expression of HMGA2 in PA tissues
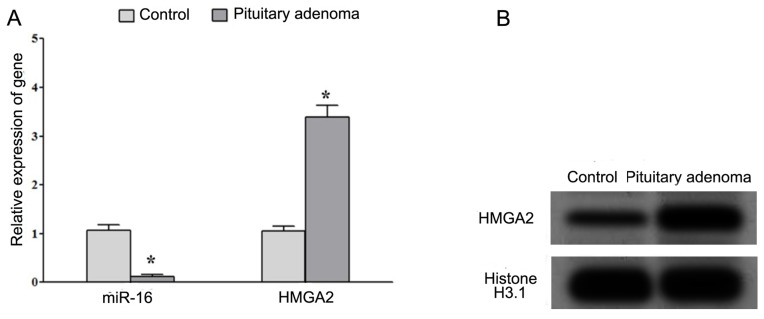
RT-qPCR analysis demonstrated that the expression of miR-16 was significantly decreased whereas the expression of HMGA2 mRNA was significantly increased in patients with PAs compared with that in normal pituitary tissues (Fig. 1A). Western blot analysis indicated that the protein expression of HMGA2 was increased in patients with PAs compared with that in normal pituitary tissues (Fig. 1B). These results suggested that the decreased expression of miR-16 and increased expression of HMGA2 may be associated with the development of PA.
Figure 1.
Decreased expression of miR-16 and elevated expression of HMGA2 in PA tissues. (A) The expression levels of miR-16 and HMGA2 in pituitary adenoma tissues and normal pituitary tissues were assessed using reverse transcription-quantitative polymerase chain reaction. (B) The expression of HMGA2 protein in pituitary adenoma tissues and normal pituitary tissues was assessed using western blot analysis. *P<0.05. miR, microRNA; HMGA2, high mobility group A2; PA, pituitary adenoma.
Regulation of HMGA2 by miR-16
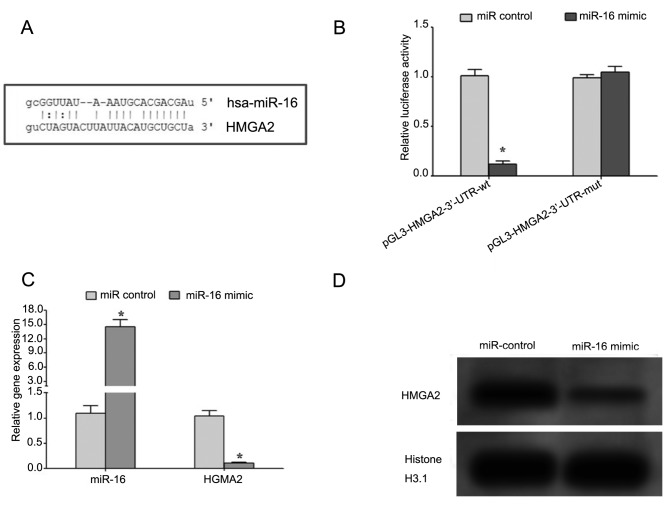
Bioinformatics analysis revealed that there is a complementary region between seed region of miR-16 and 3′-UTR of HMGA2 gene (Fig. 2A). Dual-luciferase reporter assay demonstrated that miR-16 mimic significantly decreased the relative luciferase activity in the wild-type but not mutant constructs (Fig. 2B), suggesting that miR-16 may target the 3′-UTR region of HMGA2 mRNA and regulate the expression of HMGA2. Additionally, the introduction of miR-16 mimic in HP75 cells led to decrease in gene and protein expression levels of HMGA2, thus confirming that miR-16 may regulate the expression of HMGA2 (Fig. 2C and D).
Figure 2.
miR-16 regulates the expression of HMGA2. (A) Bioinformatic analysis revealed that there is a complementary region between seed region of miR-16 and 3′-UTR of HMGA2 gene. (B) Dual luciferase reporter assay was performed to detect the interaction between miR-16 and HMGA2. (C) The expression of miR-16 and HMGA2 was assessed using reverse transcription-quantitative polymerase chain reaction. (D) The expression of HMGA2 was assessed using western blot analysis. *P<0.05. miR, microRNA; HMGA2, high mobility group A2; hsa, homo sapiens; wt, wild-type; mut, mutant.
Overexpression of miR-16 promotes apoptosis and inhibits proliferation of HP75 cells
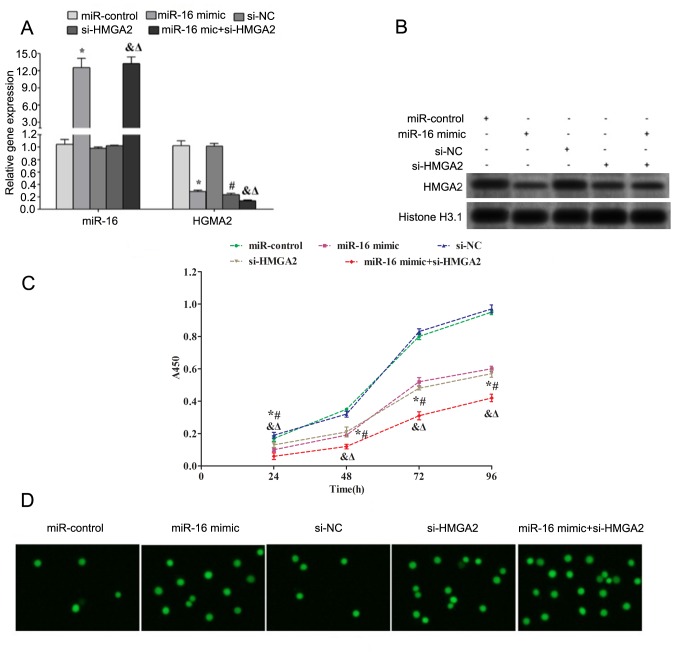
Transfection with miR-16 mimic and/or si-HMGA2 downregulated the gene and protein expression levels of HMGA2 in HP75 cells (Fig. 3A and B). Cell proliferation was assessed using a CCK-8 assay. The results demonstrated that transfection with miR-16 and/or si-HMGA2 significantly decreased the proliferation of HP75 cells after 24, 48, 72 and 96 h (Fig. 3C). Additionally, apoptosis of HP75 cells was assessed using TUNEL assay. The results demonstrated that transfection with miR-16 and/or si-HMGA2 increased the number of apoptotic cells (Fig. 3D).
Figure 3.
Overexpression of miR-16 promotes apoptosis but inhibits proliferation of HP75 cells. (A) The expression levels of miR-16 and HMGA2 in HP75 cells following different transfections [miR-control (scramble), miR-16 mimic, si-NC, si-HMGA2 and miR-16 mimic+si-HMGA2 transfection groups] were assessed using reverse transcription-quantitative polymerase chain reaction. (B) The expression of HMGA2 was assessed using western blot analysis. Histone H3 was used as a nuclear loading control. (C) Cell proliferation was assessed using Cell Counting Kit-8. (D) Apoptosis was assessed using TdT UTP nick end labeling assay. The cells with green fluorescence were defined as apoptotic cells. *P<0.05 vs. miR-control; #P<0.05 vs. si-NC; &P<0.05 vs. miR-16 mimic+ si-HMGA2 vs. miR-control; ΔP<0.05 vs. si-NC. miR, microRNA; HMGA2, high mobility group A2; si, small interfering RNA; NC, negative control.
Discussion
PA may lead to a series of endocrine symptoms, including acromegaly, decreased sexual function, headache, vision loss, visual field defect, hypothalamic syndrome, cerebrospinal fluid rhinorrhea and other symptoms of adjacent local tissue compression. Invasive PAs (IPAs) account for ~30% of PAs and are large tumors with infiltration of adjacent tissue. Patients with IPAs exhibit an increased postoperative recurrence rate of 50% and poor prognosis (20). Therefore, future studies investigating the underlying molecular mechanisms are required for the diagnosis and treatment of PA.
HMGA2 is a non-histone chromatin protein that is involved in the regulation of chromatin. HMGA2 is involved in biological processes, including cell proliferation, differentiation, cell cycle and migration. Under physiologic conditions, HMGA2 is not expressed or minimally expressed in normal tissues. Abnormal expression of HMGA2 is detected in various types of cancer, including prostate (7), colorectal (3), lung (4) and breast cancer (8). Increased expression of HMGA2 is associated with the occurrence, progression and poor prognosis of cancer. Shi et al (7) revealed that HMGA2 may be involved in the occurrence of prostate cancer by regulating cell proliferation, apoptosis, epithelial mesenchymal transition and invasion. Wu et al (21) demonstrated that upregulation of HMGA2 may be used as a biomarker of breast cancer progression and poor prognosis. Borrmann et al (22) reported that HMGA2 negatively regulated the activity of the Nucleotide excision repair cross complementing gene 1 (ERCC1) gene promoter. Additionally, HMGA2 inhibited the nucleotide excision repair pathway. Thus, HMGA2 may be involved in the occurrence of tumor by regulating DNA damage and repair. Previous studies (9–11) have demonstrated that elevated expression of HMGA2 may be associated with the occurrence of PA, the survival and prognosis of patients with PA. Previous studies (13–15) revealed that the expression of miR-16 was deregulated in PA tissues. Bioinformatics analysis indicated there is a complementary region between seed region of miR-16 and 3′-UTR of HMGA2 gene. In the present study, it was investigated whether miR-16 may regulate the expression of HMGA2 and whether it is involved in the pathogenesis of PAs.
The results of present study demonstrated that the expression level of miR-16 was decreased, whereas the expression level of HMGA2 was increased in PA tissues compared with normal pituitary tissues. These results suggest that the decreased expression of miR-16 may serve a function in upregulating the expression of HMGA2 and promoting the occurrence of PA. D'Angelo et al (10) demonstrated that the expression level of HMGA2 in tissues from patients with growth hormone-secreting PAs was significantly increased compared with that in normal pituitary glands. Oruckaptan et al (20) revealed that HMGA2-transgenic mice have an increased incidence of PA compared with normal wild-type mice. Fedele et al (11) demonstrated that overexpression of HMGA2 may lead to the development of prolactin or growth hormone adenomas in 85% of transgenic mice within 6 months. In the present study, it was demonstrated that the expression level of HMGA2 in tumor tissues from patients with PA was significantly increased compared with that in normal pituitary tissues, thus confirming the results from a previous study (10). Amaral et al (13) demonstrated the expression level of miR-16 in tumor tissues from patients with PA was significantly decreased compared with normal tissues. Bottoni et al (14) revealed that the expression level of miR-16 was significantly decreased in PA tissues compared with normal pituitary tissues. Renjie and Haiqian (15) demonstrated that the expression level of miR-16 was significantly decreased in PA cell lines compared with that in control. Additionally, the expression level of miR-16 was significantly increased in patients with IPA compared with that in patients with non-invasive PAs (15), suggesting that decreased expression of miR-16 may be involved in the pathogenesis of PA. The present study demonstrated that the expression level of miR-16 in PA tissues was significantly decreased compared with that in normal pituitary tissues, thus confirming the results from previous studies (13,14). The dual-luciferase reporter assay demonstrated that transfection of miR-16 mimic decreased the relative luciferase activity, and the protein and gene expression level of HMGA2 in HP75 cells, suggesting that miR-16 may target HMGA2. Additionally, upregulation of miR-16 or transfection of si-HMGA2 significantly decreased the expression level of HMGA2 in HP75 cells. Additionally, transfection with miR-16 and/or si-HMGA2 significantly decreased the proliferation but increased apoptosis of HP75 cells. Renjie and Haiqian (15) reported that miR-16 may regulate the proliferation, migration and invasion of pituitary tumor cells by inhibiting the expression of sex determining region Y-box 5, and overexpression of miR-16 inhibited the proliferation of PA cell lines, GH3 and MMQ and attenuated their invasive ability. In the present study, overexpression of miR-16 significantly attenuated the malignant biological behavior of HP75 cells, thus confirming the findings from Renjie and Haiqian (15). Fedele et al (23) demonstrated that histone deacetylase (HDAC) 2 interacted with retinoblastoma protein (pRB) and substituted HDAC1 in the phosphorylated pRB/E2F transcription factor 1 complex to enhance E2F1 acetylation and transcription activity, and promote cells to enter S phase. Thus, HMGA2 may be involved in the occurrence of PA. De Martino et al (24) confirmed that HMGA2 binds to the promoter region of the G2/mitotic-specific cyclin-B2 (CCNB2) gene to upregulate the expression of CCNB2 and promote cell mitosis and proliferation, thus serving a function in the occurrence of PAs. Additionally, the association between HMGA2 and apoptosis has been demonstrated by previous studies (25,26). In the present study, si-HMGA2 interference and miR-16 overexpression inhibited cell proliferation and promoted apoptosis by inhibiting the expression of HMGA2, which in turn may regulate cell cycle, mitosis proliferation and apoptosis. The molecular mechanism underlying the function of HMGA2 in PAs needs to be further identified.
In summary, the results of the present study demonstrated that the expression of miR-16 was decreased and the expression of HMGA2 was increased in PA tissues. miR-16 may inhibit the proliferation of HP75 cells and promote apoptosis by inhibiting the expression of HMGA2.
Acknowledgements
The authors would like to thank Mudanjiang Medical University (Mudanjiang, China) for providing assistance in the development of related experiments, Dean Li Xiaoxia and Dean Liu Fenghai (School of Public Health, Mudanjiang Medical University, Mudanjiang, China) for giving guidance on the design of relevant experiments.
Funding
The present study was supported by the Scientific Research Project of Heilongjiang Provincial Health Department (grant no. 2012-290), the Science and Technology Research Project of Mudanjiang Medical University (grant nos. ZS201523 and ZS201526) and the Key Laboratory of Mudanjiang Center for Disease Control and Prevention.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
NA, YN and YW made substantial contributions to conception and design. YL and HZ were responsible for the analysis and interpretation of data. CF and JX made substantial contributions to acquisition and analysis of data. NA, CF and JX were involved in drafting the manuscript and revising it critically for important intellectual content. NA agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients provided written informed consent and the study was approved by the Ethics Committee of Mudanjiang Forestry Hospital.
Patient consent for publication
All patients provided consent for the publication of this manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Leone V, Langella C, D'Angelo D, Mussnich P, Wierinckx A, Terracciano L, Raverot G, Lachuer J, Rotondi S, Jaffrain-Rea ML, et al. Mir-23b and miR-130b expression is downregulated in pituitary adenomas. Mol Cell Endocrinol. 2014;390:1–7. doi: 10.1016/j.mce.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Esposito F, De Martino M, D'Angelo D, Mussnich P, Raverot G, Jaffrain-Rea ML, Fraggetta F, Trouillas J, Fusco A. HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle. 2015;14:1471–1475. doi: 10.1080/15384101.2015.1021520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siahmansouri H, Somi MH, Babaloo Z, Baradaran B, Jadidi-Niaragh F, Atyabi F, Mohammadi H, Ahmadi M, Yousefi M. Effects of HMGA2 siRNA and doxorubicin dual delivery by chitosan nanoparticles on cytotoxicity and gene expression of HT-29 colorectal cancer cell line. J Pharm Pharmacol. 2016;68:1119–1130. doi: 10.1111/jphp.12593. [DOI] [PubMed] [Google Scholar]
- 4.Zhuo HC, Song YF, Ye J, Lai GX, Liu DL. MicroRNA-154 functions as a tumor suppressor and directly targets HMGA2 in human non-small cell lung cancer. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15028173. doi: 10.4238/gmr.15028173. [DOI] [PubMed] [Google Scholar]
- 5.Zhao XP, Zhang H, Jiao JY, Tang DX, Wu YL, Pan CB. Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J Transl Med. 2016;14:26. doi: 10.1186/s12967-016-0777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X, Dai M, Li Q, Wang Z, Lu Y, Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer. 2017;8:501–510. doi: 10.1111/1759-7714.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Z, Wu D, Tang R, Li X, Chen R, Xue S, Zhang C, Sun X. Silencing of HMGA2 promotes apoptosis and inhibits migration and invasion of prostate cancer cells. J Biosci. 2016;41:229–236. doi: 10.1007/s12038-016-9603-3. [DOI] [PubMed] [Google Scholar]
- 8.Zou Q, Wu H, Fu F, Yi W, Pei L, Zhou M. RKIP suppresses the proliferation and metastasis of breast cancer cell lines through up-regulation of miR-185 targeting HMGA2. Arch Biochem Biophys. 2016;610:25–32. doi: 10.1016/j.abb.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 9.D'Angelo D, Esposito F, Fusco A. Epigenetic mechanisms leading to overexpression of HMGA proteins in human pituitary adenomas. Front Med (Lausanne) 2015;2:39. doi: 10.3389/fmed.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Angelo D, Palmieri D, Mussnich P, Roche M, Wierinckx A, Raverot G, Fedele M, Croce CM, Trouillas J, Fusco A. Altered microRNA expression profile in human pituitary GH adenomas: Down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab. 2012;97:E1128–E1138. doi: 10.1210/jc.2011-3482. [DOI] [PubMed] [Google Scholar]
- 11.Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, et al. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–3198. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- 12.Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma H, Yu D, Maitikabili A, Xiao H, Zhang C, et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget. 2016;7:21825–21839. doi: 10.18632/oncotarget.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amaral FC, Torres N, Saggioro F, Neder L, Machado HR, Silva WA, Jr, Moreira AC, Castro M. MicroRNAs differentially expressed in ACTH-secreting pituitary tumors. J Clin Endocrinol Metab. 2009;94:320–323. doi: 10.1210/jc.2008-1451. [DOI] [PubMed] [Google Scholar]
- 14.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, Degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 15.Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356:568–578. doi: 10.1016/j.canlet.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Oruckaptan HH, Senmevsim O, Ozcan OE, Ozgen T. Pituitary adenomas: Results of 684 surgically treated patients and review of the literature. Surg Neurol. 2000;53:211–219. doi: 10.1016/S0090-3019(00)00171-3. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Zhang S, Shan J, Hu Z, Liu X, Chen L, Ren X, Yao L, Sheng H, Li L, et al. Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett. 2016;376:284–292. doi: 10.1016/j.canlet.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Borrmann L, Schwanbeck R, Heyduk T, Seebeck B, Rogalla P, Bullerdiek J, Wisniewski JR. High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCC1 and modulate its activity. Nucleic Acids Res. 2003;31:6841–6851. doi: 10.1093/nar/gkg884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 24.De Martino I, Visone R, Wierinckx A, Palmieri D, Ferraro A, Cappabianca P, Chiappetta G, Forzati F, Lombardi G, Colao A, et al. HMGA proteins up-regulate CCNB2 gene in mouse and human pituitary adenomas. Cancer Res. 2009;69:1844–1850. doi: 10.1158/0008-5472.CAN-08-4133. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Xu G, Liu H, Li T. MicroRNA-490-3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589:3148–3153. doi: 10.1016/j.febslet.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Cai J, Shen G, Liu S, Meng Q. Downregulation of HMGA2 inhibits cellular proliferation and invasion, improves cellular apoptosis in prostate cancer. Tumour Biol. 2016;37:699–707. doi: 10.1007/s13277-015-3853-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.