Abstract
In this work, we report digital loop-mediated isothermal amplification (LAMP) or reverse-transcription LAMP (RT-LAMP) on a commercial membrane, without the need for complex chip fabrication or use of specialized equipment. Due to the pore size distribution, the theoretical error for digital LAMP on these membranes was analyzed, using a combination of Random Distribution Model and Multivolume Theory. A facile peel-off process was developed for effective droplet formation on the commercial track-etched polycarbonate (PCTE) membrane. Each pore functions as an individual nanoreactor for single DNA amplification. Absolute quantification of bacteria genomic DNA was realized with a dynamic range from 11 to 1.1 × 105 copies/μL. One-step digital RT-LAMP was also successfully performed on the membrane for the quantification of MS2 virus in wastewater. With the introduction of new probes, the positive pores can be easily distinguished from negative ones with 100 times difference in fluorescence intensities. Finally, the cost of a disposable membrane is less than $0.10/piece, which, to the best of our knowledge, is the most inexpensive way to perform digital LAMP. The membrane system offers opportunities for point-of-care users or common laboratories to perform digital quantification, single cell analysis, or other bioassays in an inexpensive, flexible, and simplified way.
Keywords: digital LAMP, membrane, microfluidic, droplets, nucleic acid, paper-based analytical device, PCR
Digital PCR (dPCR) has become a promising technologies for absolute quantification of nucleic acid without need of calibration curves.1 Conventional real-time PCR (qPCR) based on quantification cycles (Cq) is a relative quantification method, and the absolute concentration of target templates remains unknown until calibrated with standard samples. In contrast to qPCR, samples for dPCR quantification are first partitioned into numerous and separated droplets.2 Each droplet may contain one or no target molecule. These droplets function as isolated nanoreactors for template amplification, generating bright fluorescence for single-molecule counting. This “digital format” eliminates the kinetic variations of molecular amplification rates, therefore enabling precise, ultrasensitive, and rapid counting of target molecules.3 Meanwhile, dPCR also reduces device complexity, since only end-point readout is required (e.g., using a smartphone for imaging).4 Following dPCR, various digital isothermal amplification methods were also developed which only require isothermal incubation. Among them, loop-mediated isothermal amplification (LAMP) became the most popular one, as it is more rapid, sensitive, specific and robust than others.5
Micro/nanofluidics has emerged as a highly suitable platform for performing digital nucleic acid analysis, due to its ability of individual molecule manipulation and nanoscale fluidic control. Recently, many microfluidic droplet systems have been reported. For example, water in oil droplets generated by T-junction,6 flow focusing,7−9 centrifugation,10,11 SAMFS12,13 and XiE14,15 have been applied to dPCR or digital LAMP (dLAMP) analysis. In addition, dPCR can be achieved using microfluidic chips with a high density of polydimethylsiloxane (PDMS) or glass chambers. Sample partition is realized using valves,16 hydrophilic patterns,17,18 vacuum,19−21 pressure,2,22 SlipChip,23 gel,4,24,25 or self-digitization.26 Although many improvements have been made, these systems typically require elaborate chip fabrication, modification and complex fluidic control (e.g., pump, vacuum, and valve). More importantly, to avoid tedious washing/refabrication processes and potential contamination, a simple, low-cost, and disposable device is required that can be thrown away after single use.
Recently, we have fabricated an asymmetric membrane with micropores on one side and vertically aligned nanochannels on the other side for single-cell filtration and analysis.27 In this work, we performed a comprehensive study for digital LAMP on a piece of commercial membrane. Since the presence of pore size distribution, the theoretical error generated when using this commercial membrane for digital analysis was first simulated and investigated in detail, using a combination of the Random Distribution Model and Multivolume Theory. To completely remove the residual solution on the membrane surface while keeping sample partitioned inside each pore, a peel-off process was also developed based on asymmetric capillary force mechanism. Each pore functions as an individual nanoreactor for single DNA amplification. Absolute quantification of genomic DNA was realized from 11 to 1.1 × 105 copies/μL with a good correlation to the expected results. Next, one-step digital reverse-transcription LAMP (RT-LAMP) was also successfully performed on the membrane to quantify MS2 virus directly in wastewater. Since all the reported digital LAMP (including digital RT-LAMP) results have low fluorescence ratio between positive and negative droplets (usually 3–6), a new primer-dye-primer-quencher duplex fluorescent probe was adopted here, which could generate 100 times greater differences between positive and negative pores.
Experimental Section
Chemicals and Materials
All LAMP reagents were purchased from New England Biolabs (Ipswich, MA), and the primers were from Integrated DNA Technologies (Coralville, IA) unless otherwise mentioned. Calcein and MnCl2 were purchased from Sigma-Aldrich (St. Louis, MO). Culture media were obtained from ThermoFisher Scientific (San Jose, CA). Track-etched polycarbonate membranes were purchased from Sterlitech Corporation (Kent, WA) and GVS Filter Technology (USA). Sylgard 184 silicon elastomer kit consisting of a prepolymer base and a curing agent was obtained from Dow Corning (Midland, MI). The PDMS films were prepared by mixing their precursor and curing agent at a ratio of 10:1 and cured at 75 °C for 1.5 h.28
Bacteria Genomic DNA Extraction
Bacteria were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Escherichia coli (ATCC 10798) and Enterococcus faecalis (ATCC 19433) were cultivated in Luria–Bertani broth in the shaking incubator for ∼14 h at 37 °C. Salmonella Typhi (CVD 909) was cultivated in tryptic soy broth with 1 mg/L 2,3-dihydroxybenzoate in the incubator for ∼14 h at 35 °C. Genomic DNA extraction was performed using a commercial beads-beating tube (GeneRite, NJ), followed by heating at 95 °C to denature proteins. The precise concentration of genomic DNA was measured by QX200 droplet digital PCR system (Bio-Rad, Hercules, CA).
MS2 Culture
Coliphage MS2 virions (ATCC 15597-B1) were cultured with freshly growing E. coli-3000 (ATCC 15597) host suspensions in the Luria–Bertani media at 37 °C for 36 h. The propagated MS2 suspension was then centrifuged at 3000g for 4 min to remove the bacterial cells and debris. The supernatant, containing the MS2 virions, was further purified using a 0.2 μm syringe filter (GE Whatman, Pittsburgh, PA). MS2 concentration was quantified by double-agar-layer plaque assays to determine the titer of virus particles in plaque forming units per microliter (PFU/μL).29
dLAMP Assay
Here, 25 μL of dLAMP mix contained 1× isothermal buffer, 6 mM total MgSO4, 1.4 mM dNTP, 640 U/mL Bst 2.0 WarmStart polymerase, primer mix (1.6 μM FIB and BIP, 0.2 μM F3 and B3, 0.8 μM LF and LB), 1 mg/mL BSA, 50 μM calcein, 1 mM MnCl2, and 2.5 μL of template. The primers sequences are shown in Table S1. The selectivity of these certain primers toward different DNA samples has already been tested and published in previous works.30−32
dLAMP on Membrane
The PCTE membranes were used as received. For some commercial PCTE membranes with polyvinylpyrrolidone (PVP) coating, this hydrophilic coating needs to be removed, since it affects the LAMP process. PVP removal was accomplished by dipping the membranes in 10% acetic acid for 30 min, followed by heating to 140 °C for 60 min.33
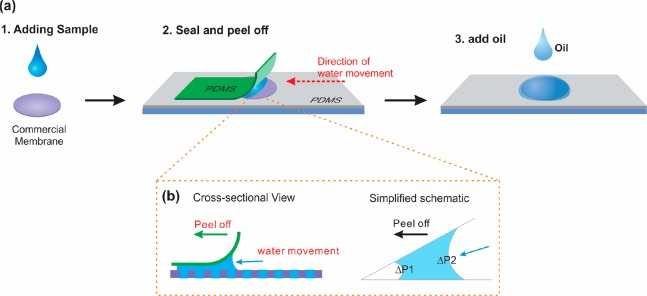
Digital LAMP on membrane is illustrated in Figure 1a. Here, 25 μL of LAMP mix was added on top of the membrane and then sealed between two pieces of PDMS film. Subsequently, the top PDMS film was peeled off, followed by adding mineral oil and a frame-seal (Bio-Rad, Hercules, CA) to cover the whole membrane. The membrane was incubated at 65 °C on a hot plate (MJ Research PTC-100, Watertown, MA) for 40 min. After amplification, images of the membranes were taken by a fluorescence microscope (Leica DMi8, Germany) using the 4× objective. Positive pores were counted using ImageJ (NIH, MD) software and calibrated by Poisson distribution. The total number of pores can be counted using ImageJ, since the negative pores also show a weak fluorescence, or estimated based on the membrane porosity (1 × 104 pores/cm2). Each sample was tested for a minimum of three times.
Figure 1.
(a) Schematic illustration of sample partition on the membrane. (b) Mechanism for excess sample removal.
qPCR and qLAMP
The 25 μL qPCR mix contained 1× PerfeCTa qPCR ToughMix (Quanta BioSciences Inc.), 0.25 μM forward primer, 0.25 μM reverse primer, 0.25 μM TaqMan probe, and 2.5 μL of DNA template. The primer was targeting the universal bacterial 16s rRNA gene and their sequences are listed in Table S2. Thermal cycling was performed with Eppendorf RealPlex2 (Hamburg, Germany). The initialization was 3 min at 95 °C, followed by 40 cycles of 15 s at 95 °C for denaturation and 30 s at 55 °C for annealing/extension.
25 μL qLAMP assay contained 1× WarmStart LAMP Mastermix, primer mix (1.6 μM FIB and BIP, 0.2 μM F3 and B3, 0.8 μM LF and LB), 1× self-contained dye, and 2.5 μL of template. The reaction was incubated at 65 °C using Eppendorf RealPlex2. Fluorescence intensity of the reaction was monitored every minute for 60 min.
MS2 Virus Quantification Using Probes
The assay for MS2 quantification also includes the RTx reverse transcriptase for one-step digital RT-LAMP. A primer-dye-primer-quencher duplex was used instead of calcein-Mn2+ indicators.4 The regular FIP primer was substituted with a fluorophore-labeled primer (5′FAM-FIP) and a complementary quencher primer (qFIP-3′IBFQ). The final 25 μL reaction for digital RT-LAMP contained 1× isothermal buffer, 6 mM total MgSO4, 1.4 mM dNTP, 640 U/mL Bst 2.0 WarmStart polymerase, 300 U/mL WarmStart RTx reverse transcriptase, primer mix (1.6 μM 5′FAM-FIB and BIP, 0.2 μM F3 and B3, 0.8 μM LF and LB), 3.2 μM quencher primer qFIP-3′IBFQ, 1 mg/mL BSA, and 2.5 μL of template. The primer sequences for MS2 RT-LAMP are also shown in Table S1.
Wastewater Samples
The wastewater was collected from the sedimentation and storage tank of a pilot scale solar-powered mobile toilet system located on the California Institute of Technology (Caltech) campus. Wastewater is composed of urine, feces, and hand-washing and toilet-flushing water. Cultured MS2 was spiked in with a final concentration of 100–1 × 105 PFU/μL. To eliminate the effect of large pollutants in the sample on the RT-LAMP process, a double-membrane system was used. A sacrificial commercial PCTE membrane with 1 μm pore size was placed on top of the 25 μm PCTE membrane for sample pretreatment. Sample was added to the composite membranes. After completely wetting, the sacrificial membrane was thrown away and the bottom membrane (25 μm pore size) was sealed and incubated at 65 °C, as described above, for digital analysis.
Characterization
Water static contact angle was measured using a contact angle goniometer equipped with an AmScope Microscope Camera model MU300. A drop of LAMP mix was placed on the surface of the membranes. The image was captured 10 s after the drop was placed, and then analyzed using ImageJ. Top-view and cross-sectional view SEM images were obtained with a ZEISS 1550VP field emission scanning electron microscope (FESEM). Before analysis, samples were sputtered with 10 nm Pd.
Results and Discussion
Sample Partition on Membrane
The track-etched membrane is a type of commercial membranes which contains a high density of micro/nanopores with uniform pore sizes, ranging from 10 nm to 30 μm.34 In this work, we chose membranes with a nominal pore size of 25 μm for experiments. Figure 2a shows a photograph of the commercial PCTE membrane (1.3 cm diameter), which is transparent and flexible. The membrane has a smooth surface, and contains a high density of cylindrical pores with an average pore size of 25 ± 1 μm and a thickness of 27 ± 1 μm, as confirmed by top-view and cross-sectional view scanning electron microscopy (SEM) images (Figure 2b,c). Samples can be partitioned on the membrane, as illustrated in Figure 1a (see Experimental Section for details). The original PCTE membrane without PVP coating displays a contact angle of 45 ± 3° for LAMP solutions (Figure 2d). After adding 25 μL of LAMP mix onto a membrane, the pores become easily wetted due to the capillary forces, without the need of vacuum. The wetted PCTE membrane was then sealed between two PDMS films to remove the residue solution on the membrane surface. However, for solutions with low surface tension (e.g., containing enzymes, proteins), there was always a thin liquid film present between the top PDMS and membrane upper surface (see Movie S1). We found this residual liquid could be removed completely by peeling off the top PDMS film (see Movie S1). The removal mechanism is attributed to the asymmetric capillary force formed when peeling off PDMS, as illustrated in Figure 1b.35 Since the pressure difference at the air–water interface on one side (ΔP1) was always larger than that on the other side (ΔP2), water tends to be taken away by the PDMS films, even though the liquid showed a very low contact angle. After PDMS was peeled off, a drop of mineral oil was added to prevent evaporation. As shown in the fluorescence image (Figure 2e), the sample was indeed partitioned into each pore successfully. All the pores (100%) were filled up with the solutions. About 1.7 × 104 droplets of ∼13 pL in volume were formed on a commercial 1.3 cm diameter membrane within 1 min. The droplets formed in each pore were uniform, as confirmed by the size and fluorescence distribution results (Figure 2f,g). However, since the pores on the membranes were produced by random heavy ion irradiation and subsequent track etching, there is a probability that two or more pores will overlap, if they are close to each other. The overlapped pores have almost double volume (see Figure S1), resulting in an error for the digital nucleic acid quantification. These overlapped pores could be excluded when calculating the results, since single pores can be easily distinguished from the overlapped pores, using common software (e.g., ImageJ). However, we prefer to include all the results for simple calculation, and the resulting error for DNA quantification will be discussed in the following. The droplet size, could be easily changed by using membranes with different pore sizes or thickness. As shown in Figure S2, a high density of droplets of 900 fL and 25 pL in volume were also successfully formed on commercial membranes with nominal 14 and 30 μm pore size, respectively. We did not test membranes with smaller pore sizes, due to the resolution limitation of the fluorescence microscope.
Figure 2.
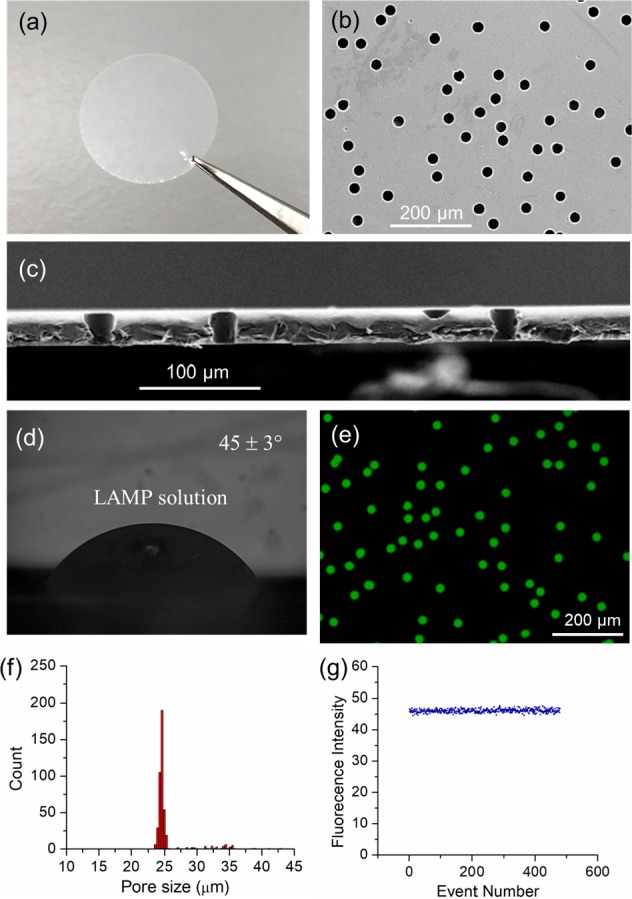
Images of the commercial PCTE membranes. (a) Photograph. (b) Top-view SEM image. (c) Cross-sectional view SEM image. (d) Static contact angle of LAMP solution on the membrane. (e) Fluorescence images of membrane when filled with fluorescent solution. (f) Pore size distribution. (g) Fluorescence intensity distribution.
Error Analysis
As mentioned above, the overlapped pores would cause an error in the nucleic acid quantification. Herein, we analyze this error, using a combination of the Random Distribution Model and Multivolume Theory. In single-volume systems, where all droplets have identical volumes, the number of DNA molecules in a given experiment can be precisely calculated by the Poisson distribution,36
| 1 |
where c is the DNA concentration, b is the number of positive droplets, n is the total number of droplets, and v is the droplet volume.
The track-etched membranes are produced by heavy ion irradiation and subsequent track etching. Since the irradiation process is random, there is a probability that two or more pores will overlap, if they are close to each other. This probability increases as the porosity increases. The possible number of different kinds of pores on the membrane, ni (i = 1 denotes single pores, 2 for two overlapped pores, etc.), can be estimated, by the following formula based on the total number of pores (ntotal) and porosity (f),37
| 2 |
The overlapped pores have a maximum probability being tangent at the pore edges. Thus, for a simplified calculation, we assume that the overlapped pores have same volumes (vi), and are equal to the sum of each pore (iv1). Therefore, the pores with different volumes can be analyzed separately.
For a certain input DNA template concentration c, the total number of positive pores can be obtained by calculating the number of positive pores separately for each kind of pore, using the Poisson distribution,
| 3 |
The measured concentration of template, cx, can be calculated based on the total number of positive pores (btotal) and the total number of pores (ntotal),
| 4 |
where vx is the pore volume we treated. The relative error between measured concentration (cx) and input concentration (c) is defined as
| 5 |
Hence, by introducing eqs 2–4 into eq 5, the relative error can be expressed as
| 6 |
Using a Taylor series, eq 6 can be simplified as
| 7 |
Equation 7 gives the relationship between relative error and input DNA concentration (c).
In the first case, we assume that the membrane is a single-volume system, and all the pores have the same pore volume of vx = v1. In other words, we assume all pores are single pores without overlap. According to membrane images, porosity (f) was measured to be 0.04, and v1 was measured to be 12.2 pL, respectively. The relationship between the relative error and the input DNA concentration, c, is shown in Figure 3 (blue line). About 8% error were observed when the input DNA concentration was below 1 × 104 copies/μL, while the error drops to 0% at higher DNA concentrations. Indeed, the large error generated at low DNA concentrations is mainly attributed to the volume error, since we assume all the overlapped pores to be the single pores.
Figure 3.
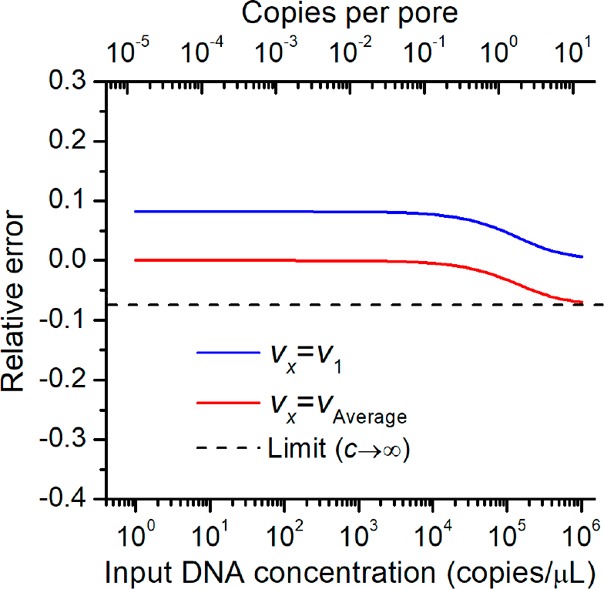
Relationship between the relative error and the input DNA concentration.
In the second case, we assume the membrane is a single-volume system, and all the pores have an average pore volume.
| 8 |
Using the average volume for DNA quantification, the relationship between the relative error and the input DNA concentrations is plotted in Figure 3 (red line). Nearly 0% error will occur at target DNA concentrations below 1 × 104 copies/μL, while approaching −3% at 1 × 105 copies/μL. Considering the dynamic range (10–105 copy/μL) of our membrane system, using an average pore volume for the calculation of DNA concentrations is more reliable. As all pores have the same depths (membrane thickness), the average pore volume can be calculated using the average pore size.
In this study, we used the nominal pore size for calculation of pore volume. The nominal pore size (25 μm) as provided by the manufacturer was measured by air flow or average bubble point techniques, which was indeed the average pore size (25 μm). Therefore, the resulting digital nucleic acid detection on these commercial membranes would has a relative error less than 3%, which is acceptable for nucleic acid quantification. If needed, this small error can be further eliminated by calibrating the results with red line in Figure 3 automatically.
Performance of dLAMP on Membrane (mdLAMP)
LAMP is an isothermal amplification method with fast amplification rates and excellent specificity. Therefore, it is more suitable for nucleic acid detection.5 We first perform real-time LAMP (qLAMP) in tube for DNA quantification, based on time threshold (Tt) values. The Tt is defined as the time required for the fluorescence of the LAMP solution to exceed a given value, similar to Cq in PCR. A smaller Tt value means higher concentration of target templates in the solution. As shown in Figure S3, even samples with same DNA concentration show quite different Tt values each time when qLAMP were performed. The quantification results can differ by 2 orders of magnitude. This poor quantification performance may be attributed to the different amplification rates of the template molecules in different environments, resulting in a variable Tt value each time.38 This issue can be fully addressed by mdLAMP, since only end-point counting is required. Digital LAMP on the membrane was validated using extracted E. coli genomic DNA. The DNA sample was first mixed with LAMP reagents, and partitioned on the membrane. During isothermal incubation at 65 °C, each pore functioned as an isolated nanoreactor for single DNA amplification. The use of mineral oil and frame-seal prevented the evaporation of droplets inside pores. To achieve a rapid and robust amplification on the membrane, the concentration of each component in the LAMP mix, such as Bst, Bovine serum albumin (BSA), Mg2+, and betaine, was optimized, as illustrated in Figure S4. Figure 4a shows a typical fluorescence image of a membrane after mdLAMP. The pores containing target DNA exhibited bright fluorescence, while those without target DNA showed a weak background signal. This result demonstrates that the commercial PCTE membrane is an excellent biocompatible material that allows effective amplification of single nucleic acids in small isolated pores, without need for additional surface modifications. Even when two pores were very close to each other (Figure 4b,c), cross-contamination was not observed, conforming perfect pore isolation in our system.
Figure 4.

Fluorescence images of the membrane after mdLAMP. The red circles denote two adjacent pores with perfect isolation.
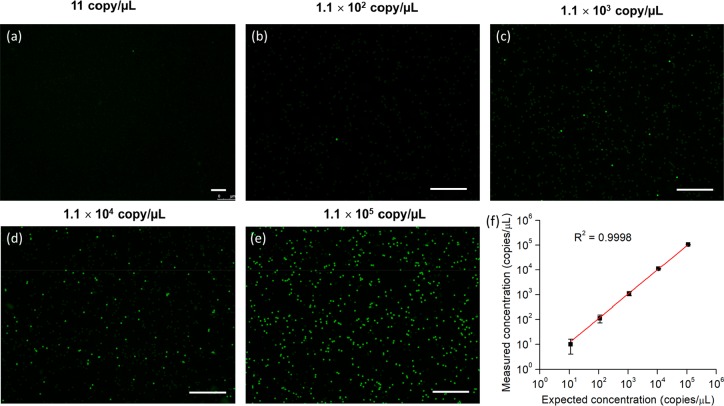
To test the quantitative performance of the mdLAMP, a series of genomic DNA solutions with final concentrations ranging from 11 to 1.1 × 105 copies/μL was used. Figure 5a–e shows the end-point fluorescence images of the mdLAMP results. With increasing concentration of target DNA in the samples, more positive pores were observed on the membranes. The template concentration can be calculated by direct counting of the positive pores and apply the Poisson distribution. The relationship between measured concentration and expected concentration is shown in Figure 5f. The actual concentration was analyzed and verified using the Bio-Rad QX200 droplet digital PCR (ddPCR) system. The measured concentrations correspond very well to the expected concentrations (R2 = 0.9998), demonstrating the excellent reliability of this method for the absolute quantification of nucleic acids. Besides, mdLAMP was also successfully performed on the membrane with a smaller pore size (14 μm), as shown in Figure S5. The increased pore density (5 × 104>) and reduced pore volume (only 900 fL) could help to improve single-molecule detection efficiency, reduce contamination, increase dynamic range, and enhance precision.2 While assays with E. coli served as a model, we also applied the membrane for the successful detection of specific DNA from Enterococcus faecalis and Salmonella Typhi, as shown in Figure S6. It should be noted that the disposable membrane could be thrown away after single use, avoiding cross-contamination and tedious washing/refabrication processes. All results presented above illustrate that mdLAMP offers a simple, low-cost, and precise method for the absolute quantification of nucleic acids.
Figure 5.
(a–e) End-point fluorescence images of the membrane after mdLAMP with a series concentration of genomic DNA. All the scale bars are 0.5 mm. (f) Comparison of measured DNA concentrations to the expected concentrations.
MS2 Virus Quantification via One-Step Digital RT-LAMP with New Probes
One-step digital RT-LAMP for virus/RNA detection is still challenging and seldom reported.23 Here, the membrane system was further applied to the quantification of MS2 virus via digital RT-LAMP (mdRT-LAMP). MS2 virus is a icosahedral, positive-sense single-stranded RNA that infects the bacterium E. coli and other members of the Enterobacteriaceae.39 The conventional plaque assay quantification method requires complicated double-agar layers, and long incubation times (>36 h) for the growth of host bacteria and target virus. To detect MS2 virus by membrane system, 8 U/mL Warmstart RTx reverse transcriptase was included in the sample solution. During 65 °C incubation, the reverse transcription and LAMP amplification was proceeding simultaneously within each pore. One disadvantage of digital LAMP or RT-LAMP is the low fluorescence ratio (usually 3–6) between positive and negative droplets, making it difficult to distinguish them.7,14,23,26,40 To address this issue, a primer-dye-primer-quencher duplex, was adopted here instead of conventional calcein-Mn2+ indicators.4 In this case, the forward internal primer (FIP) is labeled with a fluorophore at the 5′ prime end (5′ FAM-FIP), and its complementary primer is labeled with a quencher (Iowa Black FQ) at the 3′ end (qFIP-3′IBFQ). Before LAMP amplification, the probe fluorescence is suppressed by the complementary primer with a quencher. During the reaction, 5′ FAM-FIPs were released and incorporated into the LAMP amplicons, generating bright fluorescence. In contrast, excess unincorporated 5′ FAM-FIPs are quenched again by the complementary quencher primer qFIP-3′ IBFQs. The resulting fluorescence image for mdRT-LAMP is shown in Figure 6a, exhibiting at least 100 times higher intensity in positive pores in comparison to negative ones. In fact, the negative pores had nearly no fluorescence, and can hardly be distinguished from the background. Poisson distribution analysis can still be performed, as the total number of pores here can be calculated based on the membrane porosity. It should be noted that, although no fluorescence was observed for negative ones, all these pores were still filled up with samples as checked by bright-field microscope.
Figure 6.
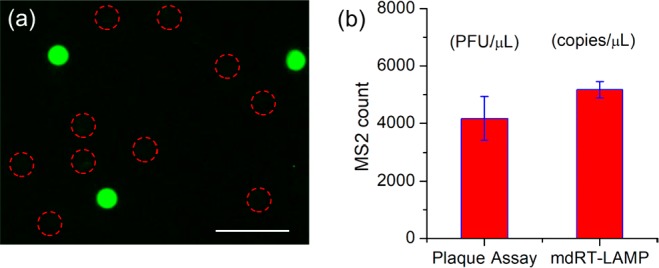
(a) Fluorescence image of membrane after mdRT-LAMP analysis of MS2. The red circles indicate the position of negative pores. The scale bar is 100 μm. (b) Concentration of MS2 detected in wastewater using plaque assay and mdRT-LAMP.
Measurement of MS2 Virus in Wastewater
To test the performance on complex water samples, the membrane was applied for MS2 detection directly in wastewater. The wastewater sample was collected from the storage tank of a pilot-scale solar-powered mobile toilet system at the California Institute of Technology (Caltech). More details about the conditions of the toilet system were reported in previous studies.41,42 Since the wastewater consists of various urine, feces, particles, bacteria, and hand-washing/toilet-flushing water, the real-time RT-LAMP was strongly inhibited, as shown in Figure S7. However, the concentration of MS2 spiked inside the wastewater sample could be precisely quantified using the mdRT-LAMP system. One advantage of membrane system is the simple capacity of sample pretreatment by combining different membranes. In order to eliminate the impact of large pollutants in the wastewater on the RT-LAMP process, a sacrificial membrane was introduced. The sacrificial PCTE membrane with 1 μm pore size was placed on top of the original 25 μm membrane. After adding the human wastewater sample (premixed with RT-LAMP reagents), both membranes became wetted due to the capillary force. However, the large particles, feces, and bacteria were retained by the top sacrificial membrane with 1 μm pore, while the smaller virus particles and LAMP reagents can pass through and be partitioned into the underlying membrane with 25 μm pore size. The bottom 25 μm pore membrane was then sealed and incubated at 65 °C, as described above, for digital analysis. As shown in Figure 6b, 5100 ± 300 copies/μL of MS2 was detected in the wastewater sample, a slightly larger concentration than that determined by a traditional plaque assay. This discrepancy is most likely due to the fact that the plaque assay is a functional measurement rather than a measurement of the absolute quantity of viral particles. It has been reported that more than one viral particle may infect a single host bacterium, and produce only one plaque-forming unit.43 In contrast, the mdRT-LAMP is an absolute quantification method that detect target virus at single-molecule level and resolution.
Comparison with Other Digital Systems
Compared with commercial Bio-Rad digital PCR systems, which are bulky and expensive (∼$80,000), the mdLAMP system simplified the whole machine into a small commercial membrane (∼$0.10), showing potential application in point-of-care detection. Compared with recently developed digital system using droplets or microchips, the membrane system is more simple and facile. First, lab users do not need to enter the cleanroom for complex chip fabrication. Second, the price for a single membrane is less than $0.10, which is much cheaper than most microfluidic chips (∼$10). Third, there is no requirement for syringe pump, centrifuge, vacuum, or other specialized equipment, while other digital systems usually included these. The disadvantage of the current membrane system is the high detection limit for nucleic acids (10 copies/μL) due to the small droplet size formed in the membrane. However, this disadvantage can be easily addressed by combining sample filtration when we modify DNA-binding sites in the pore of membrane, like chitosan and aptamer.44
Conclusion
Herein, we report digital LAMP directly on a commercial membrane without using any specialized equipment or complex chip fabrication procedures. Approximately 104 uniform picoliter droplets could be generated on a single membrane within 1 min. Theoretical calculations using a Random Distribution Model and Multivolume Theory revealed that a maximum relative error of 3% would be generated when using these membranes for digital analysis. Absolute quantification of E. coli, E. faecalis, and Salmonella Typhi DNA was successfully achieved in a dynamic range from 11 to 1.1 × 105 copies/μL. In addition, a double-membrane system was applied for the detection of MS2 virus particles in wastewater, with final quantification results slightly higher than those obtained by a traditional plaque assay. By using a new fluorescence probe, the positive pores can be easily distinguished from negative pores with at least 100 times difference in fluorescence intensities.
In the future, membranes could be directly sealed by an adhesive tape to increase the system simplicity. Since many membranes have been developed for DNA extraction, purification, or concentration (i.e., Qiagen DNA extraction kits),44 it would be powerful to combine all these features and digital detection into one piece of membrane. Besides, the mdLAMP can also be integrated with paper-based analytical devices (μPAD) for complex sample manipulation and subsequent detection.45−47 The herein presented lab-on-membrane (LOM) system could be further applied to pathogen quantification, gene sequencing, immunoassays, and single cell analysis. We believe this simple, novel, low-cost, and disposable lab-on-membrane system will provide a great platform for users without expertise in microfluidics.
Acknowledgments
The authors acknowledge the financial support provided by the Bill and Melinda Gates Foundation (Grant No. OPP1111252).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications Web site. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.8b01419.
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Hindson C. M.; Chevillet J. R.; Briggs H. A.; Gallichotte E. N.; Ruf I. K.; Hindson B. J.; Vessella R. L.; Tewari M. Absolute Quantification by Droplet Digital PCR Versus Analog Real-Time PCR. Nat. Methods 2013, 10, 1003–1005. 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyries K. A.; Tropini C.; Vaninsberghe M.; Doolin C.; Petriv O. I.; Singhal A.; Leung K.; Hughesman C. B.; Hansen C. L. Megapixel Digital PCR. Nat. Methods 2011, 8, 649–651. 10.1038/nmeth.1640. [DOI] [PubMed] [Google Scholar]
- Baker M. Digital PCR Hits Its Stride. Nat. Methods 2012, 9, 541–544. 10.1038/nmeth.2027. [DOI] [Google Scholar]
- Huang X.; Lin X.; Urmann K.; Li L.; Xie X.; Jiang S.; Hoffmann M. R. A Smartphone Based in-Gel Loop Mediated Isothermal Amplification (gLAMP) System Enables Rapid Coliphage Ms2 Quantification in Environmental Waters. Environ. Sci. Technol. 2018, 52, 6399–6407. 10.1021/acs.est.8b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N.; Mori Y.; Kanda H.; Notomi T. Loop-Mediated Isothermal Amplification (LAMP) of Gene Sequences and Simple Visual Detection of Products. Nat. Protoc. 2008, 3, 877–882. 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Lin X.; Hu X.; Bai Z.; He Q.; Chen H.; Yan Y.; Ding Z. A Microfluidic Chip Capable of Switching W/O Droplets to Vertical Laminar Flow for Electrochemical Detection of Droplet Contents. Anal. Chim. Acta 2014, 828, 70–79. 10.1016/j.aca.2014.04.023. [DOI] [PubMed] [Google Scholar]
- Rane T. D.; Chen L.; Zec H. C.; Wang T. H. Microfluidic Continuous Flow Digital Loop-Mediated Isothermal Amplification (LAMP). Lab Chip 2015, 15, 776–782. 10.1039/C4LC01158A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak R.; Zeng Y.; Shuga J.; Venugopalan G.; Fletcher D. A.; Smith M. T.; Mathies R. A. Single-Cell Multiplex Gene Detection and Sequencing with Microfluidically Generated Agarose Emulsions. Angew. Chem., Int. Ed. 2011, 50, 390–395. 10.1002/anie.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. D.; Luo K.; Chang W. H.; Lee G. B. A Microfluidic Chip Capable of Generating and Trapping Emulsion Droplets for Digital Loop-Mediated Isothermal Amplification Analysis. Lab Chip 2018, 18, 296–303. 10.1039/C7LC01004D. [DOI] [PubMed] [Google Scholar]
- Schuler F.; Trotter M.; Geltman M.; Schwemmer F.; Wadle S.; Dominguez-Garrido E.; Lopez M.; Cervera-Acedo C.; Santibanez P.; von Stetten F.; et al. Digital Droplet PCR on Disk. Lab Chip 2016, 16, 208–16. 10.1039/C5LC01068C. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Liao P.; Zhang F.; Jiang M.; Zhu Y.; Huang Y. Centrifugal Micro-Channel Array Droplet Generation for Highly Parallel Digital PCR. Lab Chip 2017, 17, 235–240. 10.1039/C6LC01305H. [DOI] [PubMed] [Google Scholar]
- Liu W. W.; Zhu Y.; Feng Y. M.; Fang J.; Fang Q. Droplet-Based Multivolume Digital Polymerase Chain Reaction by a Surface-Assisted Multifactor Fluid Segmentation Approach. Anal. Chem. 2017, 89, 822–829. 10.1021/acs.analchem.6b03687. [DOI] [PubMed] [Google Scholar]
- Liu W. W.; Zhu Y.; Fang Q. Femtomole-Scale High-Throughput Screening of Protein Ligands with Droplet-Based Thermal Shift Assay. Anal. Chem. 2017, 89, 6678–6685. 10.1021/acs.analchem.7b00899. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Xu P.; Luo J.; He H.; Du W. Absolute Quantification of H5-Subtype Avian Influenza Viruses Using Droplet Digital Loop-Mediated Isothermal Amplification. Anal. Chem. 2017, 89, 745–750. 10.1021/acs.analchem.6b03328. [DOI] [PubMed] [Google Scholar]
- Xu P.; Zheng X.; Tao Y.; Du W. Cross-Interface Emulsification for Generating Size-Tunable Droplets. Anal. Chem. 2016, 88, 3171–3177. 10.1021/acs.analchem.5b04510. [DOI] [PubMed] [Google Scholar]
- Ottesen E. A.; Hong J. W.; Quake S. R.; Leadbetter J. R. Microfluidic Digital PCR Enables Multigene Analysis of Individual Environmental Bacteria. Science 2006, 314, 1464–1467. 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- Bao L.; Rezk A. R.; Yeo L. Y.; Zhang X. Highly Ordered Arrays of Femtoliter Surface Droplets. Small 2015, 11, 4850–5. 10.1002/smll.201501105. [DOI] [PubMed] [Google Scholar]
- Yen T. M.; Zhang T.; Chen P.-W.; Ku T.-H.; Chiu Y.-J.; Lian I.; Lo Y.-H. Self-Assembled Pico-Liter Droplet Microarray for Ultrasensitive Nucleic Acid Quantification. ACS Nano 2015, 9, 10655–10663. 10.1021/acsnano.5b03848. [DOI] [PubMed] [Google Scholar]
- Yeh E. C.; Fu C. C.; Hu L.; Thakur R.; Feng J.; Lee L. P. Self-Powered Integrated Microfluidic Point-of-Care Low-Cost Enabling (Simple) Chip. Sci. Adv. 2017, 3, e1501645 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.; Xu Y.; Qiu L.; Ma C.; Yu B.; Song Q.; Jin W.; Jin Q.; Liu J.; Mu Y. A Scalable Self-Priming Fractal Branching Microchannel Net Chip for Digital PCR. Lab Chip 2017, 17, 1655–1665. 10.1039/C7LC00267J. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Yan S.; Zhang X.; Ma P.; Du W.; Feng X.; Liu B. F. Monte Carlo Modeling-Based Digital Loop-Mediated Isothermal Amplification on a Spiral Chip for Absolute Quantification of Nucleic Acids. Anal. Chem. 2017, 89, 3716–3723. 10.1021/acs.analchem.7b00031. [DOI] [PubMed] [Google Scholar]
- Men Y.; Fu Y.; Chen Z.; Sims P. A.; Greenleaf W. J.; Huang Y. Digital Polymerase Chain Reaction in an Array of Femtoliter Polydimethylsiloxane Microreactors. Anal. Chem. 2012, 84, 4262–4266. 10.1021/ac300761n. [DOI] [PubMed] [Google Scholar]
- Sun B.; Shen F.; McCalla S. E.; Kreutz J. E.; Karymov M. A.; Ismagilov R. F. Mechanistic Evaluation of the Pros and Cons of Digital Rt-LAMP for Hiv-1 Viral Load Quantification on a Microfluidic Device and Improved Efficiency Via a Two-Step Digital Protocol. Anal. Chem. 2013, 85, 1540–1546. 10.1021/ac3037206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Zhang D.; Zhang H.; Guan Z.; Song Y.; Liu R.; Zhu Z.; Yang C. Microwell Array Method for Rapid Generation of Uniform Agarose Droplets and Beads for Single Molecule Analysis. Anal. Chem. 2018, 90, 2570–2577. 10.1021/acs.analchem.7b04040. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Southard K. M.; Zeng Y. Digital PCR Using Micropatterned Superporous Absorbent Array Chips. Analyst 2016, 141, 3821–3831. 10.1039/C6AN00164E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansen A.; Herrick A. M.; Dimov I. K.; Lee L. P.; Chiu D. T. Digital LAMP in a Sample Self-Digitization (Sd) Chip. Lab Chip 2012, 12, 2247–2254. 10.1039/c2lc21247a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.; Huang X.; Zhu Y.; Urmann K.; Xie X.; Hoffmann M. R. Asymmetric Membrane for Digital Detection of Single Bacteria in Milliliters of Complex Water Samples. ACS Nano 2018, 12, 10281–10290. 10.1021/acsnano.8b05384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.; Zhang B.; Yang Q.; Yan F.; Hua X.; Su B. Polydimethysiloxane Modified Silica Nanochannel Membrane for Hydrophobicity-Based Molecular Filtration and Detection. Anal. Chem. 2016, 88, 7821–7827. 10.1021/acs.analchem.6b01866. [DOI] [PubMed] [Google Scholar]
- Kropinski A. M.; Mazzocco A.; Waddell T. E.; Lingohr E.; Johnson R. P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Methods Mol. Biol. 2009, 501, 69–76. 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- Hill J.; Beriwal S.; Chandra I.; Paul V. K.; Kapil A.; Singh T.; Wadowsky R. M.; Singh V.; Goyal A.; Jahnukainen T.; et al. Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Common Strains of Escherichia Coli. J. Clin. Microbiol. 2008, 46, 2800–2804. 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F.; Yan M.; Du P.; Chen C.; Kan B. Rapid and Sensitive Salmonella Typhi Detection in Blood and Fecal Samples Using Reverse Transcription Loop-Mediated Isothermal Amplification. Foodborne Pathog. Dis. 2015, 12, 778–786. 10.1089/fpd.2015.1950. [DOI] [PubMed] [Google Scholar]
- Kato H.; Yoshida A.; Ansai T.; Watari H.; Notomi T.; Takehara T. Loop-Mediated Isothermal Amplification Method for the Rapid Detection of Enterococcus Faecalis in Infected Root Canals. Oral Microbiol. Immunol. 2007, 22, 131–135. 10.1111/j.1399-302X.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- Cheng I. F.; Martin C. R. Ultramicrodisk Electrode Ensembles Prepared by Incorporating Carbon Paste into a Microporous Host Membrane. Anal. Chem. 1988, 60, 2163–2165. 10.1021/ac00170a038. [DOI] [Google Scholar]
- Lin X.; Yang Q.; Ding L.; Su B. Ultrathin Silica Membranes with Highly Ordered and Perpendicular Nanochannels for Precise and Fast Molecular Separation. ACS Nano 2015, 9, 11266–11277. 10.1021/acsnano.5b04887. [DOI] [PubMed] [Google Scholar]
- Ma X.; Liu Q.; Xu D.; Zhu Y.; Kim S.; Cui Y.; Zhong L.; Liu M. Capillary-Force-Assisted Clean-Stamp Transfer of Two-Dimensional Materials. Nano Lett. 2017, 17, 6961–6967. 10.1021/acs.nanolett.7b03449. [DOI] [PubMed] [Google Scholar]
- Kreutz J. E.; Munson T.; Huynh T.; Shen F.; Du W.; Ismagilov R. F. Theoretical Design and Analysis of Multivolume Digital Assays with Wide Dynamic Range Validated Experimentally with Microfluidic Digital PCR. Anal. Chem. 2011, 83, 8158–8168. 10.1021/ac201658s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.; Anderson J.; Ho W.; Petzny W. Model Pores of Molecular Dimension: The Preparation and Characterization of Track-Etched Membranes. Biophys. J. 1972, 12, 990–1007. 10.1016/S0006-3495(72)86139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P.; Tangomo M.; Hibbs J.; Bonetti E. J.; Boehme C. C.; Notomi T.; Perkins M. D.; Schrenzel J. Robustness of a Loop-Mediated Isothermal Amplification Reaction for Diagnostic Applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- van Duin J.Single-Stranded RNA Bacteriophages. In The Bacteriophages; Springer: 1988; pp 117–167. [Google Scholar]
- Zhu Q.; Gao Y.; Yu B.; Ren H.; Qiu L.; Han S.; Jin W.; Jin Q.; Mu Y. Self-Priming Compartmentalization Digital LAMP for Point-of-Care. Lab Chip 2012, 12, 4755–63. 10.1039/c2lc40774d. [DOI] [PubMed] [Google Scholar]
- Cho K.; Qu Y.; Kwon D.; Zhang H.; Cid C. m. A.; Aryanfar A.; Hoffmann M. R. Effects of Anodic Potential and Chloride Ion on Overall Reactivity in Electrochemical Reactors Designed for Solar-Powered Wastewater Treatment. Environ. Sci. Technol. 2014, 48, 2377–2384. 10.1021/es404137u. [DOI] [PubMed] [Google Scholar]
- Huang X.; Qu Y.; Cid C. A.; Finke C.; Hoffmann M. R.; Lim K.; Jiang S. C. Electrochemical Disinfection of Toilet Wastewater Using Wastewater Electrolysis Cell. Water Res. 2016, 92, 164–172. 10.1016/j.watres.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfson K. J.; Avena L. E.; Beadles M. W.; Staples H.; Nunneley J. W.; Ticer A.; Dick E. J. Jr.; Owston M. A.; Reed C.; Patterson J. L.; et al. Particle-to-Pfu Ratio of Ebola Virus Influences Disease Course and Survival in Cynomolgus Macaques. J. Virol. 2015, 89, 6773–81. 10.1128/JVI.00649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlappi T. S.; McCalla S. E.; Schoepp N. G.; Ismagilov R. F. Flow-through Capture and in Situ Amplification Can Enable Rapid Detection of a Few Single Molecules of Nucleic Acids from Several Milliliters of Solution. Anal. Chem. 2016, 88, 7647–7653. 10.1021/acs.analchem.6b01485. [DOI] [PubMed] [Google Scholar]
- Liu M.; Hui C. Y.; Zhang Q.; Gu J.; Kannan B.; Jahanshahi-Anbuhi S.; Filipe C. D.; Brennan J. D.; Li Y. Target-Induced and Equipment-Free DNA Amplification with a Simple Paper Device. Angew. Chem., Int. Ed. 2016, 55, 2709–2713. 10.1002/anie.201509389. [DOI] [PubMed] [Google Scholar]
- Truong A. S.; Lochbaum C. A.; Boyce M. W.; Lockett M. R. Tracking the Invasion of Small Numbers of Cells in Paper-Based Assays with Quantitative PCR. Anal. Chem. 2015, 87, 11263–11270. 10.1021/acs.analchem.5b02362. [DOI] [PubMed] [Google Scholar]
- Boehle K. E.; Gilliand J.; Wheeldon C. R.; Holder A.; Adkins J. A.; Geiss B. J.; Ryan E. P.; Henry C. S. Utilizing Paper-Based Devices for Antimicrobial-Resistant Bacteria Detection. Angew. Chem., Int. Ed. 2017, 56, 6886–6890. 10.1002/anie.201702776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.