Abstract
mRNA therapeutics hold great promise for the treatment of human diseases. While incorporating naturally occurring modified nucleotides during synthesis has greatly increased their potency and safety, challenges in selective expression have hindered clinical applications. MicroRNA (miRNA)-regulated in vitro-transcribed mRNAs, called miRNA switches, have been used to control the expression of exogenous mRNA in a cell-selective manner. However, the effect of nucleotide modifications on miRNA-dependent silencing has not been examined. Here we show that the incorporation of pseudouridine, N1-methylpseudourdine, or pseudouridine and 5-methylcytidine, which increases translation, tends to decrease the regulation of miRNA switches. Moreover, pseudouridine and 5-methylcytidine modification enables one miRNA target site at the 3′ UTR to be as effective as four target sites. We also demonstrate that the effects of pseudouridine, pseudouridine and 5-methylcytidine, and N1-methylpseudourdine modification are miRNA switch specific and do not correlate with the proportion of modified nucleotides in the miRNA target site. Furthermore, modified miRNA switches containing seed-complementary target sites are poorly regulated by miRNA. We also show that placing the miRNA target site in the 5′ UTR of the miRNA switch abolishes the effect of nucleotide modification on miRNA-dependent silencing. This work provides insights into the influence of nucleotide modifications on miRNA-dependent silencing and informs the design of optimal miRNA switches.
Keywords: post-transcriptional regulation, synthetic biology, mRNA therapy, non-viral gene delivery
Introduction
mRNA therapeutics are becoming valuable tools for the treatment of a broad range of human diseases.1 Many preclinical and clinical trials are underway using in vitro-transcribed (IVT) mRNAs as cancer treatments,2 vaccines,3 and protein replacement therapies.4, 5 mRNA-based therapies represent a remarkable alternative to DNA-based therapies, with a wide range of advantages, including (1) increased safety: their non-replicative nature carries no risk for integration into the host genome, thus eliminating the chances of genomic alteration; (2) broader applications: exogenous mRNAs preclude the need for nuclear localization and allow rapid protein expression in any cell type, including non-dividing cells; and (3) streamlined production: cell-free systems enable reproducible, rapid, and cost-effective synthesis with stringent quality control. The inclusion of structural elements such as a 5′ cap, 5′ and 3′ UTRs, and a poly(A) tail has been shown to significantly improve the stability and translational efficiency of IVT mRNA.1 Additionally, codon engineering and the incorporation of chemically modified nucleotides that mimic the naturally occurring intracellular modifications have been shown to reduce immunogenicity and increase translation of IVT mRNA.6, 7
To date, 171 types of modified bases have been described, although most types are thought to be rare.8 Post-transcriptional modifications of RNA are frequent, conserved across species, and observed in different RNA classes. These modifications distinguish endogenous RNA molecules from invading viral or microbial RNA molecules. Pseudouridine (Ψ) is one of the most common nucleotide modifications in cellular RNA, and it is produced by the irreversible isomerization of uridine to Ψ.9, 10, 11 Hundreds of pseudouridylation sites have been identified in eukaryotic mRNA, and they have been shown to be dynamically modulated in response to environmental changes and stresses.10, 11, 12, 13 Ψ affects the stability of endogenous mRNA, indicating potential regulatory roles in mRNA metabolism.10, 11 Incorporating Ψ into IVT mRNA has been shown to increase translation and decrease immunogenicity.7, 14, 15, 16, 17 The benefits of incorporating Ψ in IVT mRNA are enhanced when Ψ is combined with other naturally modified nucleotides, such as 5-methylcytidine (m5C).18, 19 Other modifications, such as N1-methylpseudourdine (m1Ψ), have recently been shown to further improve translation and evasion of the innate immune system.20, 21 These insights have made IVT mRNA a viable therapeutic option. However, safe and effective mRNA therapeutics require cell-specific targeting to maximize the benefits and minimize off-target effects.
Cell-specific expression has been achieved using various strategies, including the incorporation of ligand-sensing aptamers22 or target sites for endogenous microRNA (miRNA).23, 24 The addition of artificial miRNA target sites into the 3′ UTR of the transgene expression vector allows for targeting by the RNA-Induced Silencing Complex (RISC) and leads to the silencing of the transgene in cells expressing the corresponding miRNA. We have previously employed this strategy using an adenoviral vector with target sites for miR-126-3p, an endothelial cell-specific miRNA, to selectively inhibit the proliferation of arterial vascular smooth muscle cells (VSMCs) and prevent restenosis while allowing re-endothelialization of the vessels after balloon injury in a rat carotid artery.25 Recently, IVT mRNAs that contain artificial miRNA target sites, called miRNA switches, were successfully used to achieve cell-specific expression of synthetic circuits, identify and eliminate undifferentiated induced stem cells, and achieve cell-selective genome editing using miRNA-responsive CRISPR-Cas9.26, 27, 28, 29
The intricate mechanisms of miRNA-mediated silencing have been the focus of extensive investigations. However, to date, the relative contributions of mRNA decay and translational repression to the overall silencing remain under debate. Moreover, the influence of chemically modified nucleotides on miRNA-dependent mRNA silencing has not been investigated. Since nucleotide modifications increase translation and stabilize the 2D and 3D RNA structures, thereby altering interactions within the RNA itself and with RNA-binding proteins,30 we sought to determine whether nucleotide modifications affect the regulation of miRNA switches. In this work we compared the effect of nucleotide modifications (Ψ, Ψ/m5C, and m1Ψ) on the performance of different miRNA switches in different cell types. We also tested the effect of the location and the complementarity of the miRNA target sites in the miRNA switches.
Results
Nucleotide Modifications Alter miRNA-Dependent Silencing of miRNA Switches
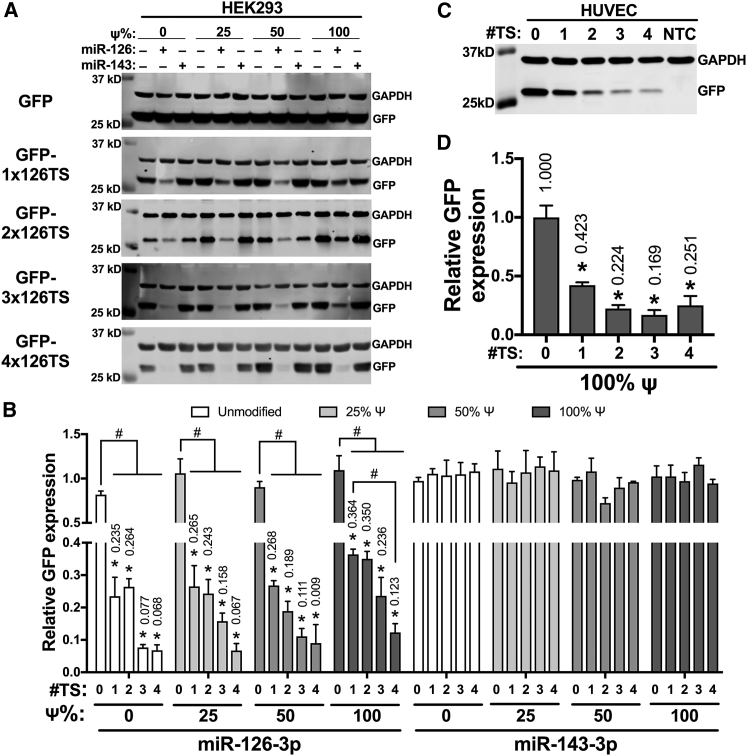
To create reporter miRNA switches, we introduced one to four copies of a 22-nt sequence fully complementary to the mature miR-126-3p at the 3′ UTR of IVT GFP mRNA (GFP-1x126TS, GFP-2x126TS, GFP-3x126TS, GFP-4x126TS). We used HEK293 cells, since they display minimal cytotoxicity following transfection with unmodified IVT mRNA due to the lack of expression of almost all Toll-like receptors (TLRs).16, 21, 31 Since HEK293 cells express very low levels of miR-126-3p compared to human umbilical vein endothelial cells (HUVECs) (Figure S1A), we used exogenous mimics to overexpress miR-126-3p in HEK293. To determine the optimal time needed for the loading of the miR-126-3p mimics into the RISC, HEK293 cells were transfected with miR-126-3p mimics simultaneously or 4, 8, or 24 h prior to transfection with unmodified GFP-4x126TS miRNA switches. miR-126-3p mimics reduced GFP expression to <37% (p < 0.05) at all time points compared to vehicle control, and the lowest GFP expression (16.3%) was seen in cells transfected with miR-126-3p mimics 24 h prior to the GFP-4x126TS miRNA switches (Figure S1B). Therefore, in all subsequent experiments involving HEK293 cells, we transfected miRNA mimics 24 h prior to the miRNA switch transfection.
To establish the baseline for miRNA switch silencing, we first assessed GFP expression in HEK293 cells transfected with unmodified (0% Ψ) GFP mRNA containing zero to four copies of miR-126-3p fully complementary target sites at the 3′ UTR, in the presence of miR-126-3p mimics, miR-143-3p mimics, or vehicle control. In cells transfected with unmodified GFP-1x126TS mRNA, miR-126-3p mimics reduced GFP protein levels to 23.5% (p < 0.05) of vehicle control (Figures 1A and 1B). Increasing the number of miR-126-3p target sites from one to four decreased the expression of GFP protein levels from 23.5% to 6.8% in the presence of miR-126-3p mimics, whereas miR-143-3p mimics showed no difference in GFP expression (Figures 1A and 1B).
Figure 1.
Increasing the Number of miRNA Target Sites at the 3′ UTR of the miRNA Switches Counteracts the Impact of the Ψ Substitution
(A–D) Representative GFP and GAPDH immunoblots (A and C) and densitometric quantification (B and D) of HEK293 cells transfected with miR-126-3p or miR-134-3p mimic or vehicle, 24 h before transfection with the indicated GFP miRNA switch (A and B) or HUVECs transfected with the indicated 100% Ψ-modified GFP miRNA switches (C and D). GFP and GAPDH expressions were measured after 24 h. Data represent the mean ± SEM, normalized against GAPDH (B and D) and relative to vehicle controls (B) or untargeted GFP control (D). *p < 0.05 versus miR-143 control (B); versus GFP-treated cells (D). #p < 0.05 for the indicated comparisons. #TS, the number of miR-126-3p target sites in the 3′ UTR of the miRNA switch; NTC, non-transfected control. Numbers above bars indicate the mean values.
We then synthesized GFP mRNA with 25%, 50%, or 100% substitution of uridine for Ψ. Transfection of these Ψ-modified mRNAs into HEK293 cells showed >2-fold increase (p < 0.05) in GFP expression with 50% and 100% Ψ compared to unmodified (0% Ψ) GFP mRNA (Figure S2). We found that increasing Ψ substitutions, from 0% to 100%, reduced miR-126-3p-dependent silencing of GFP-1x126TS from 23.5% to 36.4% relative GFP expression (Figures 1A and 1B). Increasing the number of miR-126-3p target sites from one to four further decreased the expression of GFP protein levels to <13% at all percentages of Ψ substitution in the presence of miR-126-3p, but not miR-143-3p (Figures 1A and 1B). The increased silencing of GFP-4x126TS switches compared to GFP-1x126TS was seen at all levels of Ψ substitution, but it only reached statistical significance in the 100% Ψ-modified miRNA switches (p < 0.05).
We also assessed the ability of the endogenous miR-126-3p to silence unmodified (0% Ψ) or 100% Ψ-modified GFP miRNA switches containing zero to four copies of miR-126-3p fully complementary target sites at the 3′ UTR. HUVECs transfected with unmodified GFP mRNA displayed minimal GFP expression and a 3.4-fold (p < 0.05) increase in the number of dead cells compared to non-transfected controls (Figures S3A and S3B). However, HUVECs transfected with 100% Ψ-modified GFP mRNA displayed increased GFP expression and no difference in the number of dead cells compared to non-transfected controls (Figures S3A and S3B). Similar to HEK293 cells transfected with miR-126-3p mimics, HUVECs transfected with 100% Ψ-modified GFP-1x126TS showed 42.3% GFP expression compared to 100% Ψ-modified GFP mRNA (p < 0.05). Increasing the number of miR-126-3p target sites from one to four further decreased the expression of GFP to 25.1% but did not reach statistical significance (Figures 1C and 1D).
To determine whether the reduction in GFP expression is due to transcript cleavage or reduced translation, we analyzed GFP transcript levels of HUVECs 24 h after transfection with 100% Ψ-modified GFP-encoding miRNA switches, with zero, one, or four fully complementary target sites for miR-126-3p in the 3′ UTR. GFP-1x126TS mRNA levels were reduced to 59.8% of cells transfected with 100% Ψ-modified GFP mRNA. Increasing the number of miR-126-3p target sites to four did not further decrease GFP mRNA levels (Figure S4), whereas GFP protein levels continued to decrease with the additional target sites (Figures 1C and 1D). These results show that the additional fully complementary miRNA target sites do not increase the cleavage of miRNA switches but still contribute to the miRNA-mediated silencing by increasing translational repression.
Lastly, we assessed sponging of the endogenous miRNA by miRNA switches. HUVECs were transfected with 100% Ψ-modified GFP containing zero, one, or four miR-126-3p target sites at the 3′ UTR, and, after 24 h, the expressions of miR-126-3p and PIK3R2, a known miR-126-3p target, were determined by RT-PCR. We found that inclusion of one or four miR-126-3p target sites reduced endogenous miR-126-3p levels to ∼55% (p < 0.05) and increased the mRNA level of PIK3R2 ∼1.5-fold (p < 0.05). No difference was observed in the sponging of miR-126-3p or PIK3R2 expression between cells transfected with GFP-1x126TS and GFP-4x126TS (Figures S5A and S5B). These data indicate that miRNA switches that contain even one fully complementary miRNA target site reduce the availability of the endogenous cognate miRNA and impact the expression of its target genes.
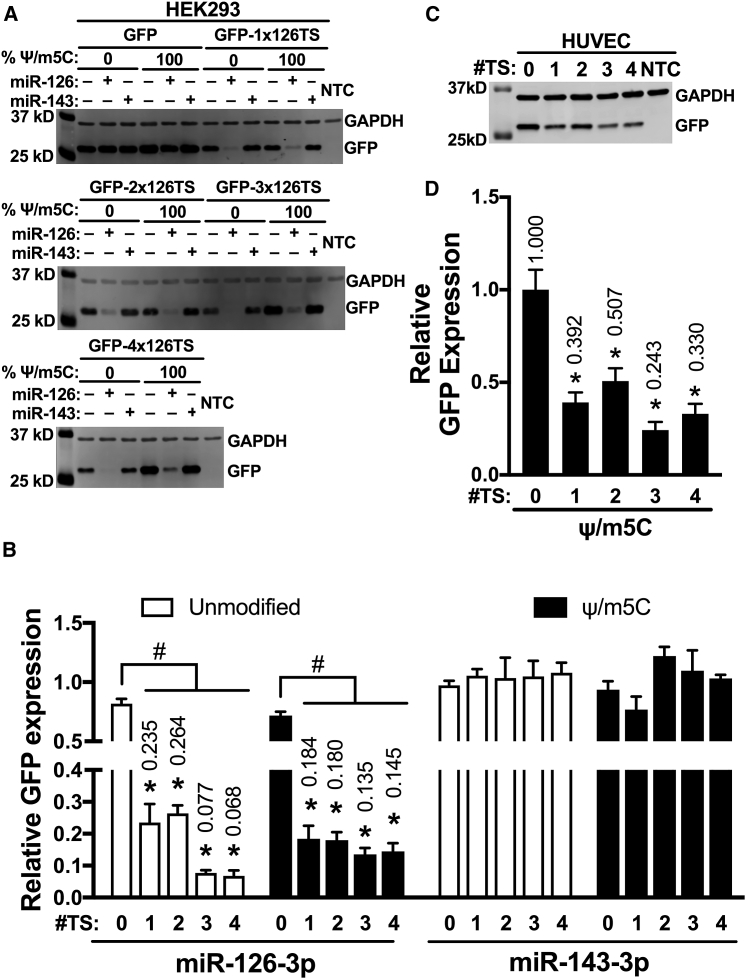
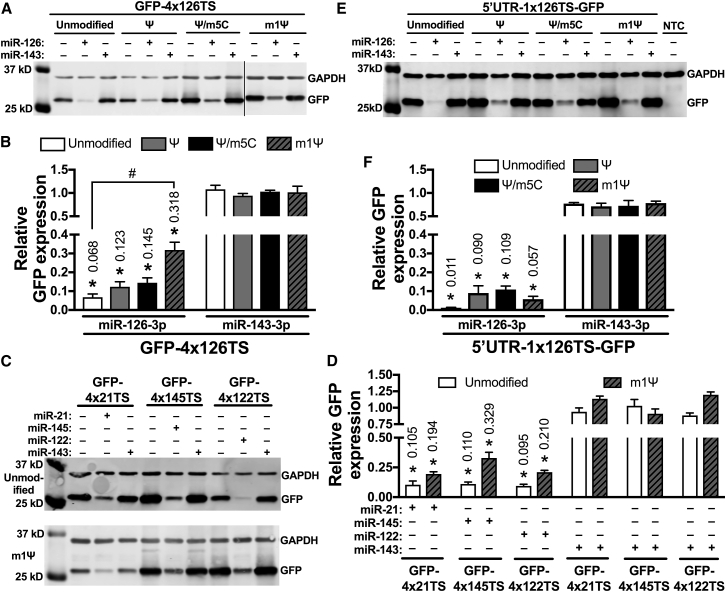
Recent studies have used combinations of modified nucleotides to further decrease the innate immune response to exogenous mRNA.18, 19 We therefore tested whether 100% substitution with both Ψ and m5C alters miRNA-dependent silencing of GFP miRNA switches that contained zero to four miR-126-3p fully complementary target sites at the 3′ UTR. HEK293 cells transfected with 100% Ψ/m5C GFP-1x126TS showed 18.4% GFP expression (p < 0.05), equivalent to unmodified GFP-1x126TS miRNA switches (23.5%). Similar to unmodified and Ψ-modified miR-126-3p switches, 100% Ψ/m5C-modified GFP-4x126TS produced 14.5% relative GFP expression (p < 0.05) (Figures 2A and 2B). However, unlike Ψ-modified miRNA switches, the silencing provided by one miR-126-3p target site in the Ψ/m5C-modified miRNA switches was equal to the silencing of miRNA switches with four target sites.
Figure 2.
Ψ/m5C Substitution Prevents Increased Silencing by Additional miR-126-3p Target Sites
(A–D) Representative GFP and GAPDH immunoblots (A and C) and densitometric quantification (B and D) of HEK293 cells transfected with miR-126-3p or miR-143-3p mimic or vehicle, 24 h before transfection with the indicated GFP miRNA switch (A and B) or HUVECs transfected with the indicated 100% Ψ/m5C-modified GFP miRNA switches (C and D). GFP and GAPDH expressions were measured after 24 h. Data represent the mean ± SEM, normalized against GAPDH (B and D) and relative to vehicle control (B) or untargeted GFP control (D). *p < 0.05 versus miR-143 control (B); versus GFP-treated cells (D). #p < 0.05 for the indicated comparisons. #TS, the number of miR-126-3p target sites in the 3′ UTR of the miRNA switch; NTC, non-transfected control. Numbers above bars indicate the mean values.
When the 100% Ψ/m5C-modified GFP mRNA was tested in HUVECs, we observed GFP expression and no difference in the number of dead cells compared to non-transfected controls (Figures S3A and S3B). HUVECs transfected with 100% Ψ/m5C GFP-1x126TS showed 39.2% GFP expression compared to GFP mRNA controls. As was observed in HEK293 cells, increasing the number of miR-126-3p target sites at the 3′ UTR produced no increase in the silencing (Figures 2C and 2D). These data demonstrate that, in Ψ/m5C-modified miR-126-3p switches, unlike unmodified or Ψ-modified miR-126-3p switches, inclusion of just one miR-126-3p target site at the 3′ UTR achieved the same miRNA-dependent silencing as four target sites.
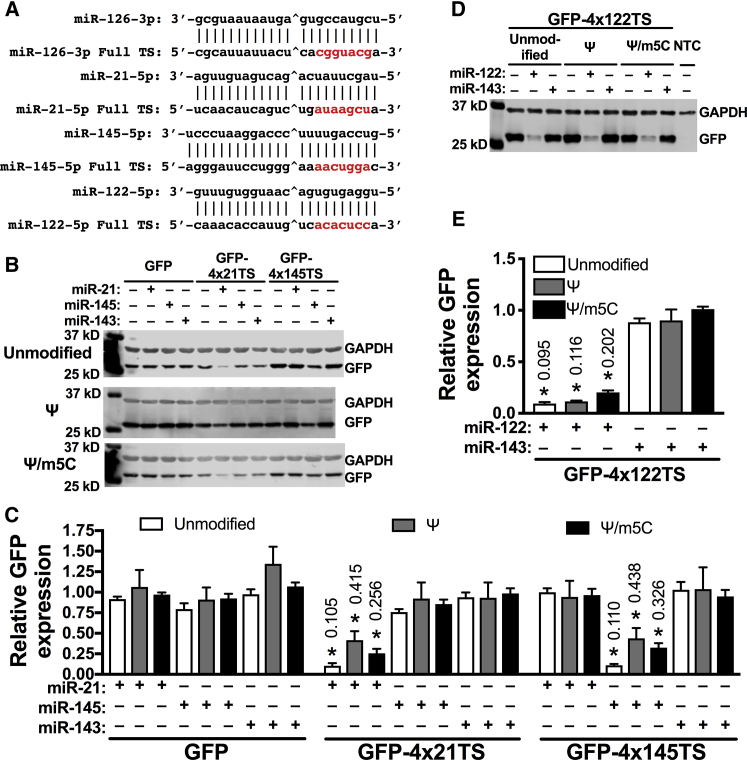
To further explore the influence of the modified nucleotides on the silencing of miRNA switches, we designed GFP mRNA containing four fully complementary target sites at the 3′ UTR for miR-21-5p, which contains six uridines and five cytosines (GFP-4x21TS), or miR-145-5p (GFP-4x145TS), which contains four uridines and four cytosines. In contrast, miR-126-3p contains six uridines and six cytosines (Figure 3A). HEK293 cells were transfected with unmodified, 100% Ψ-modified, or 100% Ψ/m5C-modified GFP, GFP-4x21TS, or GFP-4x145TS miRNA switches 24 h after transfection with miR-21-5p, miR-145-5p, or miR-143-3p mimic or vehicle control. In cells transfected with unmodified, Ψ-modified, or Ψ/m5C-modified GFP-4x21TS miRNA switches, only miR-21-5p mimic reduced GFP protein levels to 10.5%, 41.5%, and 25.6%, respectively, compared to vehicle control (p < 0.05), whereas miR-145-5p and miR-143-3p showed no inhibitory effect (Figures 3B and 3C).
Figure 3.
Silencing of Ψ- and Ψ/m5C-Modified miRNA Switches Is Independent of the U or C Content
(A) Sequences of miR-126-3p, miR-21-5p, miR-145-5p, and miR-122-5p and their complementary target sites. ˆThe site of Argonaute cleavage. Red bases are complementary to the seed sequence. (B–E) Representative immunoblots (B and D) and densitometric quantification (C and E) of HEK293 cells transfected with the indicated miRNA mimics or vehicle control 24 h before transfection with the indicated miRNA switches. GFP and GAPDH expressions were measured after 24 h. Data represent the mean ± SEM, normalized against GAPDH and relative to vehicle control. *p < 0.05 versus miR-143 control. NTC, non-transfected control. Numbers above bars indicate the mean values.
Similarly, in cells transfected with unmodified, Ψ-modified, or Ψ/m5C-modified GFP-4x145TS mRNA, only miR-145-5p mimic reduced GFP protein levels to 11.0%, 43.8%, and 32.6%, respectively, compared to vehicle control (p < 0.05), whereas miR-21-5p and miR-143-3p showed no inhibitory effect (Figures 3B and 3C). Although the silencing of Ψ- or Ψ/m5C-modified GFP-4x21TS and GFP-4x145TS by miR-21-5p and miR-145-5p mimics, respectively, was less effective compared to unmodified miRNA switches, it did not reach statistical significance (Figures 3B and 3C).
We also designed GFP mRNA containing four fully complementary target sites at the 3′ UTR for miR-122-5p, which contains 4 uridines and 9 cytosines (Figure 3A), and we found no difference in GFP silencing of unmodified (9.5%), 100% Ψ-modified (11.6%), or 100% Ψ/m5C-modified (20.2%) GFP-4x122TS miRNA switch by miR-122-5p mimic (Figures 3D and 3E). Taken together, these data indicate that Ψ and Ψ/m5C modifications tend to decrease the silencing of the miRNA switches in comparison to the unmodified miRNA switches.
Regulation of Ψ- and Ψ/m5C-Modified miRNA Switches Is Cell Type Dependent
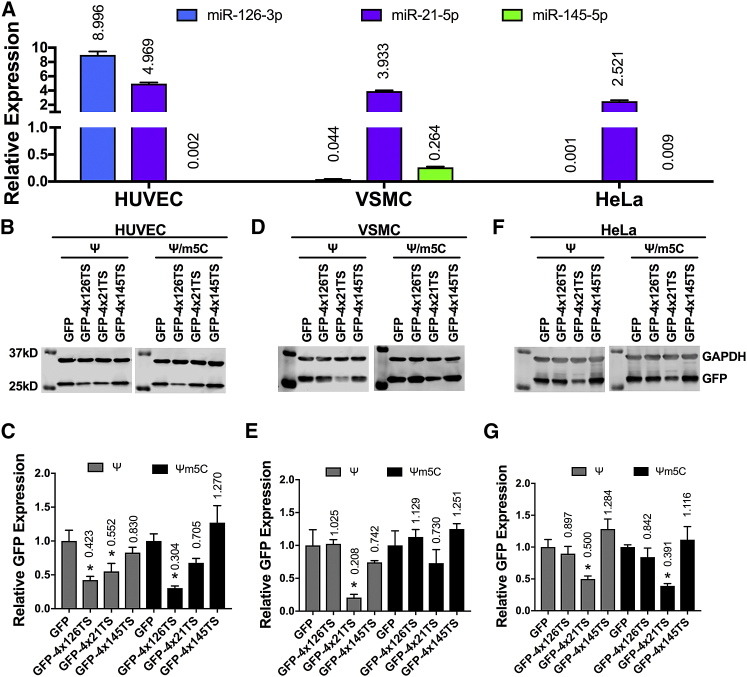
We also tested these miRNA switches in cell types that endogenously express the cognate miRNAs to confirm the effects we observed using miRNA mimics in HEK293 cells. We used HUVECs and VSMCs, which express high levels of miR-126-3p and miR-145-5p, respectively, in addition to miR-21-5p. We also used HeLa cells, which express high levels of miR-21-5p, but neither miR-126-3p nor miR-145-5p (Figure 4A). HUVECs transfected with Ψ-modified and Ψ/m5C-modified GFP-4x126TS switches showed a similar reduction in GFP expression, to 42.3% and 30.4%, respectively, compared to their respective GFP mRNA-transfected controls (p < 0.05). However, Ψ-modified GFP-4x21TS miRNA switch reduced GFP expression to 55.2% of Ψ-modified GFP-transfected cells (p < 0.05), while Ψ/m5C-modified GFP-4x21TS expression was reduced to only 70.5% of Ψ/m5C-modified GFP-transfected controls, which did not reach statistical significance. Lastly, HUVECs transfected with Ψ- or Ψ/m5C-modified GFP-4x145TS showed no decrease in GFP expression compared to GFP controls (Figures 4B and 4C).
Figure 4.
Silencing of Ψ- and Ψ/m5C-Modified miRNA Switches by Endogenous miRNA
(A) Real-time PCR analysis of the indicated miRNA expression in HUVECs, VSMCs, and HeLa cells normalized to U18. (B–G) Representative immunoblots of GFP and GAPDH expressions (B, D, and F) and densitometric quantification (C, E, and G) of HUVECs (B and C), VSMCs (D and E), and HeLa (F and G) cells transfected with the indicated miRNA switch. GFP and GAPDH expressions were measured after 24 h. Data represent the mean ± SEM, normalized against GAPDH and relative to GFP-transfected control. *p < 0.05 versus GFP-transfected cells. Numbers above bars indicate the mean values.
In VSMCs, transfection of either Ψ- or Ψ/m5C-modified GFP-4x126TS switch did not decrease GFP expression compared to Ψ- or Ψ/m5C-modified GFP mRNA control. Ψ-modified GFP-4x21TS switch produced 20.8% GFP expression relative to Ψ-modified GFP mRNA control (p < 0.05), while Ψ/m5C modification exhibited 73.0% GFP expression relative to the Ψ/m5C-modified GFP mRNA control. Interestingly, Ψ-modified GFP-4x145TS switch exhibited very little silencing (74.2% relative GFP expression, p > 0.05) compared to the Ψ-modified GFP mRNA-transfected control, and Ψ/5mC-modified GFP-4x145TS switch showed no silencing at all (Figures 4D and 4E). No statistically significant differences in GFP expression were observed in the Ψ/5mC-modifed GFP-4x21TS and GFP-4x145TS compared to the Ψ-modified switches.
In HeLa cells, Ψ-modified GFP-4x21TS switch produced 50.0% relative GFP expression, while Ψ/5mC-modified GFP-4x21TS switch was further reduced to 39.1% relative GFP expression. No silencing was seen with Ψ- and Ψ/m5C-modified GFP-4x126TS and GFP-4x145TS switches (Figures 4F and 4G). These data indicate that the silencing of Ψ- and Ψ/m5C-modified miRNA switches may be influenced by the abundance of the cognate miRNA and/or cell type.
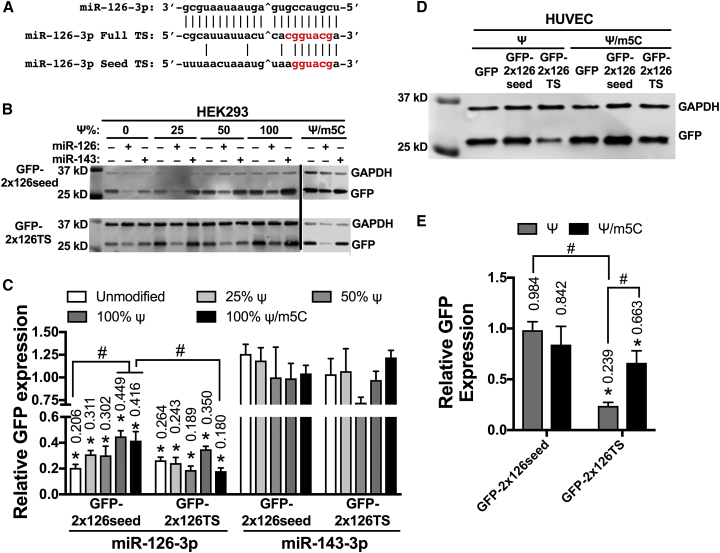
Silencing of Modified miRNA Switches Is Affected by the Degree of Complementarity of the miRNA Target Sites
To determine whether the miRNA-dependent silencing of modified mRNA is affected by the degree of complementarity of target sites, we designed a GFP-encoding mRNA that included two 7-mer-A1 target sites complementary to the seed region of miR-126-3p in the 3′ UTR (GFP-2x126seed), and we compared it to GFP-2x126TS (Figure 5A). Two target sites were purposely selected to ensure that we could detect any increase or decrease in the ability of miRNA to silence the miRNA switch. In HEK293 cells transfected with unmodified GFP-2x126seed or GFP-2x126TS, miR-126-3p mimics decreased GFP protein levels to 20.6% or 26.4%, respectively, compared to vehicle controls (Figures 5B and 5C). Increasing the percentage of Ψ from 0% to 100% reduced the silencing of GFP-2x126seed by miR-126-5p mimics from 20.6% to 44.9% (p < 0.05), in contrast to GFP-2x126TS, which exhibited relatively equal GFP expression at all percentages of Ψ substitution (Figures 5B and 5C). Cells transfected with Ψ/m5C-modified GFP-2x126seed also exhibited impaired miRNA-dependent silencing compared to unmodified GFP-2x126seed miRNA switches (41.6% versus 20.6%, p < 0.05) and to Ψ/m5C-modifed GFP-2x126TS (41.6% versus 18.0%, p < 0.05) (Figures 5B and 5C).
Figure 5.
miRNA Target Site Complementarity Affects the Silencing of Ψ- and Ψ/m5C-Modified miRNA Switches
(A) Sequences of mature miR-126-3p and fully complementary and 7-mer-A1 seed target sites. ˆThe site of Argonaute cleavage. Red bases are complementary to the seed sequence. (B–E) Representative immunoblots (B and D) and densitometric quantification (C and E) of HEK293 cells transfected with miR-126-3p or miR-143-3p mimic or vehicle control 24 h before transfection with the indicated miRNA switch (B and C) or HUVECs transfected with the indicated miRNA switches (D and E). GFP and GAPDH expressions were measured after 24 h. Data represent the mean ± SEM, normalized against GAPDH and relative to vehicle control (C) or GFP-transfected cells (E). *p < 0.05 versus miR-143 control (C); versus GFP-treated cells (E). #p < 0.05 for the indicated comparisons. Numbers above bars indicate the mean values.
Strikingly, when 100% Ψ- or 100% Ψ/m5C-modified GFP-2x126seed miRNA switches were transfected into HUVECs, we observed no decrease in GFP expression compared to 100% Ψ- or 100% Ψ/m5C-modified GFP mRNA; whereas 100% Ψ- and 100% Ψ/m5C-modified GFP-2x126TS reduced GFP expression to 23.9% and 66.3% (p < 0.05), respectively (Figures 5D and 5E). The silencing of Ψ-modified GFP-2x126TS was significantly stronger than the silencing of Ψ-modified GFP-2x126seed (p < 0.05), but no difference was observed between the Ψ/m5C-modified miRNA switches. Interestingly, 100% Ψ/m5C-modified GFP-2x126TS was significantly less silenced than 100% Ψ-modified GFP-2x126TS (Figures 5D and 5E), an effect that was not seen when using GFP-4x126TS miRNA switches in the same cells (Figures 4B and 4C). These data indicate that nucleotide modifications largely inactivate miRNA switches containing partial complementarity to the mature miRNA sequence.
m1Ψ Modification Affects the Silencing of miRNA Switches
Recently, it has been reported that mRNAs containing m1Ψ modification outperformed the Ψ- and Ψ/m5C-modified mRNA platforms.20, 21 Therefore, we compared the silencing of GFP-4x126TS containing 100% m1Ψ substitutions to unmodified, 100% Ψ-modified, and 100% Ψ/m5C-modified miRNA switches in HEK293 cells. In the presence of miR-126-3p mimics, m1Ψ-modified GFP-4x126TS exhibited 31.8% relative GFP expression (p < 0.05) and was significantly different than unmodified GFP-4x126TS that showed only 6.8% GFP expression relative to vehicle controls (Figures 6A and 6B).
Figure 6.
Silencing of Modified miRNA Switches Is Affected by the Location of the miRNA Target Sites
(A–F) Representative immunoblots (A, C, and E) and densitometric quantification (B, D, and F) of HEK293 cells transfected with the indicated miRNA mimics or vehicle control 24 h before transfection with the indicated miRNA switches. GFP and GAPDH expressions were measured after 24 h. Data represent the mean ± SEM, normalized against GAPDH and relative to vehicle control. *p < 0.05 versus miR-143-treated control. #p < 0.05 for the indicated comparisons. NTC, non-transfected control. Numbers above bars indicate the mean values.
To test whether modified nucleotides alter the silencing of miRNA switches beyond the 24-h time point, HEK293 cells were transfected with vehicle or miR-126-3p or miR-143 mimic 24 h before transfection with unmodified, 100% Ψ-modified, Ψ/m5C-modified, or m1Ψ-modified GFP-4x126TS together with 100% Ψ/m5C-modifed RFP mRNA for transfection control. The percentages of GFP- and RFP-positive cells were determined every 24 h for 5 days by flow cytometry. At day 1, cells transfected with unmodified GFP-4x126TS together with miR-143 mimic or vehicle control were ∼30% GFP and RFP positive. After 5 days, the percentages of GFP- and RFP-positive cells decreased to 4% and 15%, respectively (Figure S6A). In contrast, cells transfected with Ψ-, Ψ/m5C-, or m1Ψ-modified GFP-4x126TS together with miR-143 mimic or vehicle control were >70% GFP positive at day 1, which decreased to 24%, 27.4%, and 54%, respectively, at day 5, whereas the percentage of RFP-positive cells was ∼60% at day 1 and decreased to 6% by day 5 with all modifications (Figures S6B–S6D). Cells transfected with 100% Ψ/m5C RFP mRNA alone exhibited a similar pattern of expression with ∼65% RFP-postive cells at day 1 that decreased to 6% by day 5 (Figure S6E).
In the presence of miR-126-3p mimic, cells transfected with unmodified GFP-4x126TS were only 4% GFP positive at day 1, whereas Ψ-, Ψ/m5C-, or m1Ψ-modified GFP-4x126TS transfected cells were 22%, 18.7%, and 47% GFP positive, respectively. The percentage of GPF-positive cells decreased to <2% at day 5 in all miR-126-3p mimic-treated groups (Figures S6A–S6D). These data show that the nucleotide modifications, particularly m1Ψ, increase the translation and extend the duration of protein expression, thereby reducing the silencing efficiency of the miRNA switches.
We also tested whether 100% m1Ψ substitution affects silencing of the other miRNA switches in HEK293 cells, and we found that m1Ψ-modified GFP-4x21TS, GFP-4x145TS, and GFP-4x122TS exhibited 19.4%, 32.9%, and 21.0% GFP expression, respectively, in the presence of their cognate miRNA mimic (p < 0.05). Unmodified GFP-4x21TS, GFP-4x145TS, and GFP-4x122TS showed 10.5%, 11.0%, and 9.5% relative GFP expression, respectively. Although the m1Ψ-modified miRNA switches tended to be less effectively silenced by the cognate miRNA than the unmodified miRNA switches, these differences did not reach statistical significance (Figures 6C and 6D).
Addition of miRNA Target Sites at the 5′ UTR of the miRNA Switches Counteracts the Impact of Nucleotide Modification
A recent reports has shown that placing miRNA target sites in the 5′ UTR of the miRNA switches produced stronger miRNA-dependent silencing.28 To test whether the addition of miRNA target sites at the 5′ UTR could counteract the impact of nucleotide modifications on the silencing of miRNA switches, we designed GFP-encoding miRNA switches with one fully complementary target site for miR-126-3p in the 5′ UTR of human β-globin (5′ UTR-1x126TS-GFP). In HEK293 cells transfected with unmodified 5′ UTR-1x126TS-GFP, miR-126-3p reduced GFP expression to 1.1% in the presence of miR-126-3p mimics (Figures 6E and 6F). The suppression of GFP was equivalent to that of the four miR-126-3p target sites in the 3′ UTR (Figures 6A and 6B). Moreover, cells transfected with 100% Ψ-, 100% Ψ/m5C-, and 100% m1Ψ-modifed 5′ UTR-1x126TS-GFP switches also showed significant silencing by miR-126-3p compared to miR-143-3p controls (9.0%, 10.9%, and 5.7%, respectively; p < 0.05). These data indicate that placing one fully complementary miRNA target site at the 5′ UTR of miRNA switches eliminates the impact of m1Ψ modification seen when using four target sites at the 3′ UTR.
Discussion
miRNAs have increasingly been used to build regulatory circuits in synthetic biology. miRNA switches have been recently used to enable cell-specific expression of IVT mRNA. Nucleotide modifications are commonly incorporated in these IVT mRNA to decrease immunogenicity and increase translation. In this study, we sought to investigate the influence of modified nucleotides on the performance of miRNA switches. We found that the incorporations of Ψ and m1Ψ, which increase translation, tend to decrease the miRNA-dependent regulation of miRNA switches, while Ψ/m5C modification enables one miRNA target site at the 3′ UTR to regulate the miRNA switch as effectively as four target sites. We also demonstrated that the effects of Ψ, Ψ/m5C, and m1Ψ modifications are sequence dependent and are not correlated with the proportion of modified nucleotides in the miRNA target site. Furthermore, modified miRNA switches containing seed-complementary target sites are poorly regulated by miRNA, while placing the miRNA target site in the 5′ UTR makes the miRNA-dependent silencing largely insensitive to nucleotide modification (Table S2).
The relative contribution of the endonuclease cleavage and/or suppression of translation in the dynamics of miRNA-mediated mRNA silencing has not been fully elucidated, but, ultimately, both pathways contribute to the decrease in protein expression. To explore the differences in miRNA-dependent silencing between unmodified and modified miRNA switches, we measured changes at the protein level, which also accounts for the increase in mRNA translation seen with the modifications. We explored different methods for measuring protein expression (flow cytometry and immunoblotting), and we carefully quantified and pooled the data when possible. Although immunoblotting is not the most sensitive method of measuring protein expression, it was sufficient for our initial assessments. Further studies that use more sensitive assays are necessary to pinpoint the exact mechanisms by which nucleotide modification alters miRNA-dependent silencing.
We also took advantage of the fact that HEK293 cells lack expression of most TLRs16, 21, 31 and tolerate transfection with unmodified IVT mRNA. Although HEK293 cells displayed minimal toxicity when transfected with unmodified IVT mRNA, we noticed lower expression of Ψ/m5C-modifed RFP mRNA when co-transfected with unmodified GFP-4x126TS (Figure S6A). This trans-repression is likely due to the presence of other pattern recognition receptors such as protein kinase R (PKR), which phosphorylates the eukaryotic translation initiation factor eIF2a and leads to translation inhibition.32 Previous studies have performed similar experiments in HEK293 cells to show that the incorporation of modified nucleotides in IVT mRNA reduces PKR activation and 2′-5′-oligoadenylate synthetase and RNase L activities.14, 15, 16
Here we found that 100% Ψ substitution doubled the expression of IVT mRNA compared to unmodified mRNA and slightly reduced miRNA-mediated silencing of all the tested miRNA switches but did not reach statistical significance. Similar results were also observed with m1Ψ modification, which is known to produce even more protein expression than Ψ modification, and it significantly reduced the miRNA-dependent silencing of miR-126-3p switches. These data are in line with previous reports showing that incorporation of Ψ or m1Ψ in IVT mRNA enhanced translational efficiency.20, 21, 33
Moreover, alterations in mRNA translation initiation, for example, tethering of the translation factors eIF-4E or eiF-4G to an mRNA, have been shown to confer resistance to miRNA-induced repression.34 We also found that the increased silencing of Ψ-modified miRNA switches with multiple fully complementary target sites at the 3′ UTR was due to an increase in translational repression rather than transcript degradation. Moreover, miRNA switches drastically reduced the availability of the endogenous miRNA. The miRNA sponging was similar when using miRNA switches with one or four fully complementary target sites. Previous studies using plasmids or viral vectors have reported no sponging activity even when using four fully complementary target sites.24 The differences in miRNA sponging might be due to the temporal expression between the two delivery systems. Plasmids and viral vectors that are gradually transcribed and transported to the cytoplasm are less likely to sponge the miRNA, whereas transfection of miRNA switches can immediately bind and potentially saturate the available miRNA due to rapid delivery of a high number of transcripts into the cytoplasm. However, because miRNA switches are transient, the sponging caused by the miRNA switches is temporary.
Similar to Ψ-modified mRNA, Ψ/m5C-modified mRNAs are also known to enhance protein expression and reduce immunogenicity.21 However, in contrast to Ψ-modified miRNA switches, the addition of one miRNA target site to the 3′ UTR produced miRNA-dependent silencing of Ψ/m5C-modified miR-126-3p switches as effectively as when four target sites were used. Moreover, this effect was detected not only in HEK293 cells but also occurred in HUVECs. The proportion of m5C in the target sites of the tested miRNA switches did not correlate with the miRNA-dependent silencing efficiency, and the silencing efficiency also varied in different cell types used in this work. Nucleotide modification alters the secondary or tertiary structure of the mRNA,35 which may alter accessibility of the miRNA target sites in a switch-specific manner. In fact, Ψ modification has been reported to reduce protein binding to consensus sequences in mRNA.36 The nucleotide modifications may also affect the kinetics of miRNA switch shuttling in or out of p-bodies.37, 38 Further studies are necessary to explore the interaction of miRNA target site sequence and the changes in miRNA-dependent silencing caused by nucleotide modifications, especially because previous reports have shown enrichment of m5C at the site of Argonaute binding39 and differences in protein expression from modified mRNA in different cell types.18
The design of miRNA target sites and its location in a miRNA switch can have a large impact on the capacity for miRNA-mediated silencing. Our results showed that nucleotide modifications have a larger effect on the silencing of miRNA switches that utilize target sites complementary to the seed sequence of the mature miRNA. Because the majority of miRNA target sites in animals utilize seed-complementary target sites,40, 41 post-transcriptional modification of mRNAs may provide an additional level of control over gene expression by tuning miRNA activity. An important feature of the miRNA:mRNA interaction is the thermally stable base pairing between the miRNA 5′ end (residues 2–7) and the mRNA target. In addition, efficient endonuclease cleavage by Argonaute 2 requires base pairing at the site of cleavage, between bases 10 and 11.42, 43, 44 Previous reports have shown that miRNA-mediated silencing primarily occurs though direct cleavage of the targeted mRNA by Argonaute 2 when bound to a fully complementary target site.34, 45, 46 However, the silencing of partial complementary target genes occurs by translational repression and/or mRNA decay in a manner independent of endonucleolytic cleavage. Binding of the RISC has been show to recruit several proteins, including GW182, which can mediate poly(A)-binding protein displacement, recruit translational repressors, and/or dissociate eIF4A from the cap-binding complex eIF4F.34, 47
Additionally, we show that placing one fully complementary miRNA target site at the 5′ UTR eliminates the influence of the nucleotide modifications seen with the four target sites at the 3′ UTR of the miRNA switches. The increase in silencing efficiency when using miRNA target sites at the 5′ UTR was most pronounced in the m1Ψ-modified miR-126-3p switches. These data are in agreement with previous work that showed that modified miRNA switches with four miRNA target sites at the 3′ UTR are less effective than one target site at the 5′ UTR.28 Silencing of miRNA switches by miRNA target sites in the 3′ UTR and the 5′ UTR has been shown to involve deadenylation and cap-dependent translation inhibition.48 However, the enhanced silencing of the modified miRNA switches with one fully complementary miRNA target site at the 5′ UTR indicates that there may be another mechanism triggered by miRNA target sites in the 5′ UTR. One potential explanation is that direct Argonaute 2-mediated cleavage of the targeted miRNA switch upstream of the start codon may be more effective in silencing due to the removal of initiation factors that are bound to the 5′ cap, whereas cleavage at the 3′ UTR promotes transcript degradation by removing the stabilizing poly(A) tail. It is also possible that RISC binding at the 5′ UTR interferes with translation machinery through steric hindrance of ribosome assembly.49 Furthermore, a miRNA target site near the start codon may be more accessible to the RISC, as it has less secondary structure than other regions of mRNA.50 Further studies are needed to fully explain the enhanced activity of miRNA target sites at the 5′ UTR.
Based on the data presented here, several important considerations must be taken into account when designing effective modified miRNA switches. First, it is crucial to ensure that the selected miRNA is expressed in a cell-specific manner. Second, the endogenous expression levels of the selected miRNA must be abundant. Third, use a fully complementary miRNA target site and not miRNA seed sequences. Fourth, including multiple miRNA target sites increases the miRNA-mediated suppression of the miRNA switch. Fifth, placing the miRNA target sites at the 5′ UTR will result in effective miRNA-dependent silencing and eliminate the reduction in silencing caused by nucleotide modification; however, switches with miRNA target sites at the 5′ UTR can increase translation in some instances,51 and, therefore, they should be tested to ensure switch activity. Lastly, the dose of miRNA switches should be fine-tuned to prevent saturation of endogenous miRNA.
The present study is of broad significance given the dynamic changes in the naturally occurring modified nucleotides in all cellular mammalian RNA. The impact of these modifications on miRNA-mediated silencing provides a new mechanism of gene regulation at the epitranscriptome level, which has not been explored. This work provides insights into the influence of the naturally occurring nucleotide modifications on miRNA-dependent silencing, and it informs the design of optimal miRNA switches. Furthermore, given that modified nucleotides increase translation, they can serve as a novel tool to elucidate the relative contribution of the endonuclease cleavage and/or suppression of translation in the dynamics of miRNA-mediated mRNA silencing.
Materials and Methods
Constructs and In Vitro Transcription
GFP constructs for the generation of in vitro transcription templates were designed using the pLL3.7 plasmid, a gift from Luk Parijs (Addgene plasmid 11795). The 3′ UTR of human β-globin (132 bp) was amplified using HeLa genomic DNA as a template, with forward and reverse primer oligonucleotides 1 and 2 (Table S1), and EcoRI digested and cloned into the pLL3.7 plasmid. BamHI, PacI, and SphI restriction sites were included upstream of the β-globin 3′ UTR in oligonucleotide 1 to facilitate the insertion of miRNA target sites at the 3′ UTR.
To construct GFP miRNA switches with one or two copies of the fully complementary target sites for miR-126-3p (GFP-1x126TS or GFP-2x-126TS), we annealed oligonucleotides 3 and 4 or oligonucleotides 5 and 6, respectively, and we ligated them into the plasmid using the BamHI restriction site added to the GFP 3′ UTR. To construct GFP with three or four copies of the fully complementary target sites for miR-126-3p (GFP-3x126TS or GFP-4x126TS) at the 3′ UTR, we annealed oligonucleotides 7 and 8, containing two copies of miR-126-3p fully complementary target sites, and we ligated them into the GFP-1x126TS or GFP-2x126TS plasmids using the PacI restriction site. Likewise, to construct GFP with two miR-126-3p seed target sites (GFP-2x126seed) at the 3′ UTR, we annealed oligonucleotides 9 and 10 that contained two copies of regions complementary to the seed sequence of miR-126-3p, and we introduced them into the GFP plasmid using the BamHI restriction site. The same strategy was used to construct GFP-4x21TS, GFP-4x145TS, and GFP-4x122TS plasmids by inserting four target sites for miR-21-5p (oligonucleotides 11–14), miR-145-5p (oligonucleotides 15–18), or miR-122-5p (oligonucleotides 19–22), respectively, using the BamHI and PacI sites (Table S1).
For in vitro transcription, templates were generated by PCR using forward primer 23 (containing a T7 promoter, 5′ UTR of human β-globin and the first 26 bases of GFP) and reverse primer 24 (starting at the end of the human β-globin 3′ UTR). To generate a template that contained one copy of the fully complementary target sites for miR-126-3p at the 5′ UTR of GFP (5′ UTR-1x126TS-GFP), PCR was performed using forward primer 25 (containing a T7 promoter, 5′ UTR of human β-globin, one miR-126-3p fully complementary target sequence, and the first 26 bases of GFP) and reverse primer 24. In vitro transcription was performed, per the manufacturer’s protocol, using the HiScribe T7 High Yield RNA synthesis kit (New England Biolabs) followed by ammonium acetate precipitation, as previously described.52 Pseudouridine-5′-Triphosphate (TriLink Biotechnologies, N-1019), 5-Methylcytidine-5′-Triphosphate (TriLink Biotechnologies, N-1014), N1-methylpseudouridine-5′-Triphosphate (TriLink Biotechnologies, N-1081), and 2-Thiouridine-5′-Triphosphate (TriLink Biotechnologies, N-1032) were substituted during IVT to generate modified mRNA. The RNA was then capped using the ScriptCap m7G Capping System (CellScript) and tailed using the A-Plus Poly(A) Polymerase Tailing Kit (CellScript). Following poly(A) tailing, the mRNA was precipitated using 1× vol of 5 M ammonium acetate and re-suspended in dH2O.
Cell Culture and Transfection
HEK293 (Stratagene) and HeLa (ATCC) cells were cultured in DMEM with 10% fetal bovine serum (FBS). HUVECs and VSMCs (Lonza) were cultured in EGM-2 Endothelial Cell Growth Media or SMGM-2 Smooth Muscle Cell Growth Media (Lonza), respectively. All cells were maintained in sub-confluent densities to allow cell division throughout the course of the experiments. Experiments using HUVECs and VSMCs were carried out in cells between 3 and 7 passages. Cells were plated in 24-well plates (HEK293, HeLa, and VSMC, 5 × 104/well; HUVEC, 1 × 105/well) on the day prior to the first transfection. miRIDIAN miRNA mimics and hairpin inhibitors (Dharmacon) were transfected at a final concentration of 200 nM using DharmaFECT 4 Transfection Reagent (Dharmacon). Lipofectamine 2000 (Thermo Fisher Scientific) was used for mRNA transfections. Media were changed after 4 h to remove transfection reagent from HUVECs, VSMCs, and HeLa cells. Cells were collected 24 h following mRNA transfection.
Cytotoxicity
Cytotoxicity was analyzed 24 h after transfection of miRNA switches by adding propidium iodide (PI) directly to wells without removing the culture media. PI was added to the transfected wells to a final concentration of 4 μM. Fluorescence microscopy images were captured after a 30-min incubation. The number of dead cells was calculated from 4 fields/well using ImageJ.
Immunoblotting
Immunoblot analyses were performed as previously described.53, 54 Protein lysates were fractionated based on size by SDS-PAGE, and they were transferred to nitrocellulose membrane before immunoprobing using rabbit antibodies against GAPDH (1:1,000, Cell Signaling Technology) or GFP (1:1,000, Life Technologies), followed by IRDye 680 donkey anti-rabbit immunoglobulin G (IgG) secondary antibodies (1:5,000, LI-COR). Blots were imaged using the Odyssey Infrared Imaging System (LI-COR) and quantified using Image Studio software (LI-COR). GFP expression was normalized to GAPDH, which was used as a loading control.
miRNA Isolation and qRT-PCR
Total cellular RNA was isolated from cultured cells using miRNeasy mini kit (QIAGEN), according to the manufacturer’s protocol, and stored at −80°C in RNase-free water. Reverse transcription was performed on 50 ng total RNA using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) and olidgo-d(T) or miRNA-specific RT primers (Thermo Fisher Scientific). qPCR was performed using TaqMan MicroRNA Assays (Thermo Fisher Scientific) for 40 cycles on a QuantStudio 3 (Applied Biosystems). Relative expression of miRNAs and mRNA was calculated in comparison to the small nucleolar RNA (snoRNA) U18 or endogenous GAPDH, respectively, using the 2−ΔCt method.
Statistical Analysis
Data were analyzed using two-tailed Student’s t test with Holm-Sidak correction, one-way ANOVA with Tukey’s post hoc test, or two-way ANOVA with Tukey’s post hoc test using GraphPad Prism 7.0 software. p < 0.05 was considered statistically significant. All data are reported as mean ± SEM of at least 3 independent experiments unless stated otherwise.
Author Contributions
J.L., J.V., and H.T.-J. designed the research, performed the experiments, and analyzed the data. J.C. and E.F.M. performed experiments and edited the paper. J.L. and H.T.-J. prepared figures and wrote the paper.
Conflicts of Interest
The authors have no potential conflicts of interest.
Acknowledgments
This work was supported by the NIH to H.T.J. (grants R00HL109133 and R01HL128411), the American Heart Association to J.C. (grant 15PRE25850019), and the TriLink Biotechnologies Research Award.
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.12.007.
Supplemental Information
References
- 1.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 2.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 3.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 6.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamma T., Ferré-D’Amaré A.R. Pseudouridine synthases. Chem. Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovejoy A.F., Riordan D.P., Brown P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 14.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchida S., Kataoka K., Itaka K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics. 2015;7:137–151. doi: 10.3390/pharmaceutics7030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki S., Fujita Y., Nagaike T., Tomita K., Saito H. Synthetic mRNA devices that detect endogenous proteins and distinguish mammalian cells. Nucleic Acids Res. 2017;45:e117. doi: 10.1093/nar/gkx298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown B.D., Venneri M.A., Zingale A., Sergi Sergi L., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 24.Brown B.D., Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 25.Santulli G., Wronska A., Uryu K., Diacovo T.G., Gao M., Marx S.O., Kitajewski J., Chilton J.M., Akat K.M., Tuschl T. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J. Clin. Invest. 2014;124:4102–4114. doi: 10.1172/JCI76069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirosawa M., Fujita Y., Parr C.J.C., Hayashi K., Kashida S., Hotta A., Woltjen K., Saito H. Cell-type-specific genome editing with a microRNA-responsive CRISPR-Cas9 switch. Nucleic Acids Res. 2017;45:e118. doi: 10.1093/nar/gkx309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015;33:839–841. doi: 10.1038/nbt.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miki K., Endo K., Takahashi S., Funakoshi S., Takei I., Katayama S., Toyoda T., Kotaka M., Takaki T., Umeda M. Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell. 2015;16:699–711. doi: 10.1016/j.stem.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Parr C.J., Katayama S., Miki K., Kuang Y., Yoshida Y., Morizane A., Takahashi J., Yamanaka S., Saito H. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci. Rep. 2016;6:32532. doi: 10.1038/srep32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdörfer B., Giese T., Endres S., Hartmann G. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 32.Balachandran S., Roberts P.C., Brown L.E., Truong H., Pattnaik A.K., Archer D.R., Barber G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 33.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 35.Davis D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaidyanathan P.P., AlSadhan I., Merriman D.K., Al-Hashimi H.M., Herschlag D. Pseudouridine and N6-methyladenosine modifications weaken PUF protein/RNA interactions. RNA. 2017;23:611–618. doi: 10.1261/rna.060053.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., 3rd, Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez J., Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haley B., Zamore P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 44.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 46.Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 47.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moretti F., Thermann R., Hentze M.W. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang B., Yanez A., Novina C.D. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl. Acad. Sci. USA. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu W., Xu Y., Xie X., Wang T., Ko J.H., Zhou T. The role of RNA structure at 5′ untranslated region in microRNA-mediated gene regulation. RNA. 2014;20:1369–1375. doi: 10.1261/rna.044792.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ørom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Totary-Jain H., Naveh-Many T., Riahi Y., Kaiser N., Eckel J., Sasson S. Calreticulin destabilizes glucose transporter-1 mRNA in vascular endothelial and smooth muscle cells under high-glucose conditions. Circ. Res. 2005;97:1001–1008. doi: 10.1161/01.RES.0000189260.46084.e5. [DOI] [PubMed] [Google Scholar]
- 53.Totary-Jain H., Sanoudou D., Ben-Dov I.Z., Dautriche C.N., Guarnieri P., Marx S.O., Tuschl T., Marks A.R. Reprogramming of the microRNA transcriptome mediates resistance to rapamycin. J. Biol. Chem. 2013;288:6034–6044. doi: 10.1074/jbc.M112.416446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Totary-Jain H., Sanoudou D., Dautriche C.N., Schneller H., Zambrana L., Marks A.R. Rapamycin resistance is linked to defective regulation of Skp2. Cancer Res. 2012;72:1836–1843. doi: 10.1158/0008-5472.CAN-11-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.