Abstract
Extracellular vesicles (EVs) play key roles in cell biology and may provide new clinical diagnostics and therapies. However, it has proven difficult to develop protocols for their purification and characterization. One of the major barriers in the field has been a lack of convenient assays for their bioactivity. Developing assays has not been a trivial matter, because of the heterogeneity of EVs, the multiple activities they demonstrate, and the uncertainty about their modes of action. Therefore, it is likely that multiple assays for their activities are needed. One important assay will be for the anti-inflammatory activity observed in mice after administration of the small EVs commonly referred to as exosomes. We developed an assay for the anti-inflammatory activity of exosomes with a line of mouse macrophages. The assay makes it possible to rank different preparations of exosomes by their anti-inflammatory activity, and their ranking predicts their efficacy in suppressing LPS-stimulated inflammation in mice. The assay is convenient for comparing multiple samples and, therefore, should be useful in developing protocols for the purification and characterization of anti-inflammatory exosomes.
Keywords: in-process assay, bioactivity, extracellular vesicles
Graphical Abstract
Introduction
Extracellular vesicles (EVs) have attracted attention because of their roles in the biology of most organisms, ranging from some parasites and bacteria to essentially all the cells in multicellular organisms.1, 2, 3, 4, 5 Also, EVs have been pursued as plasma markers of disease progression and for their potential as therapeutic agents.3, 4 Special attention has been paid to the very small EVs (30 to 200 nm in diameter) that are referred to as exosomes and that appear to be derived from the multivesicular bodies found in the endomembrane system of most eukaryotic cells.2 As a consequence of their origin, exosomes are highly enriched in proteins involved in multivesicular endosome biogenesis (e.g., Alix and Tsg101) and tetraspanin membrane proteins (e.g., CD63 and CD81), which are widely used as exosome biomarkers.6 As reflected in a series of reviews,7, 8, 9, 10 research on EVs in general and on exosomes in particular has met a number of technological challenges. No consensus has emerged on the best protocol to isolate and purify exosomes in spite of numerous attempts with classical techniques such as centrifugation, precipitation, size exclusion chromatography, ion exchange chromatography, and immuno-purification. Similarly, no consensus on purification protocols has emerged from the application of advanced techniques developed for synthetic nanoparticles such as those based on hydrodynamic features combined with other properties.8, 10, 11 Also, there is no consensus on the compositions of the vesicles from the many reports and large databases that have been developed.12 Instead, the data reveal marked differences in the composition of similar EVs from different sources or different culture conditions. In addition, the reports and databases reveal thousands of proteins and other components that either co-purify or adhere to the surfaces of vesicles. Among the problems encountered is that there is no consensus as to how to count exosomes, because their small size is below the sensitivity of techniques for cell counting, and artifacts have been encountered in the use of techniques such as light scattering or nanoparticle tracking analysis.8, 9, 10
The technical challenges encountered with EVs, such as exosomes, would obviously benefit from convenient assays for their bioactivity. Unfortunately, few are available.13 A series of assays will probably be required, because exosomes from different sources have demonstrated to have multiple activities, including anti-inflammatory, anti-immune, anti-coagulant, pro-angiogenic, and pro-tumorigenic. Many of these activities were only demonstrated after injection of the exosomes into mice that were models for human diseases.14, 15, 16 Assays in mouse models, however, are time consuming and costly of mice and other materials, including exosomes. Also, if the exosomes are produced by human cells, the mice must be housed in facilities with special precautions to prevent the spread of human pathogens. Therefore, assays in mice are impractical for the multiple samples that must be assayed in a purification protocol.
One of the bioactivities observed with some exosome preparations obtained from mesenchymal stromal cells (MSCs) was to suppress inflammation in mouse models for traumatic brain injury17, 18, 19 and status epilepticus.20 Exosomes that are anti-inflammatory are of special interest as therapeutic agents, because inflammation is part of the pathology of a broad range of diseases, including obesity,21 diabetes,22 myocardial infarction,23 and Alzheimer’s disease.24 Anti-inflammatory exosomes may be particularly effective for treating CNS diseases, since they cross the blood barrier that is not breached effectively by any known drug.25, 26 Recently, we developed an assay with macrophages in vitro for the anti-inflammatory activity of exosomes. The assay made it possible to classify different preparations of exosomes produced with the same protocol by their anti-inflammatory activity. The classifications predicted the activity of the exosomes in suppressing lipopolysaccharide (LPS)-induced inflammation in mice.
Results
Standardization of the Assay in Murine RAW 264.7 Macrophages
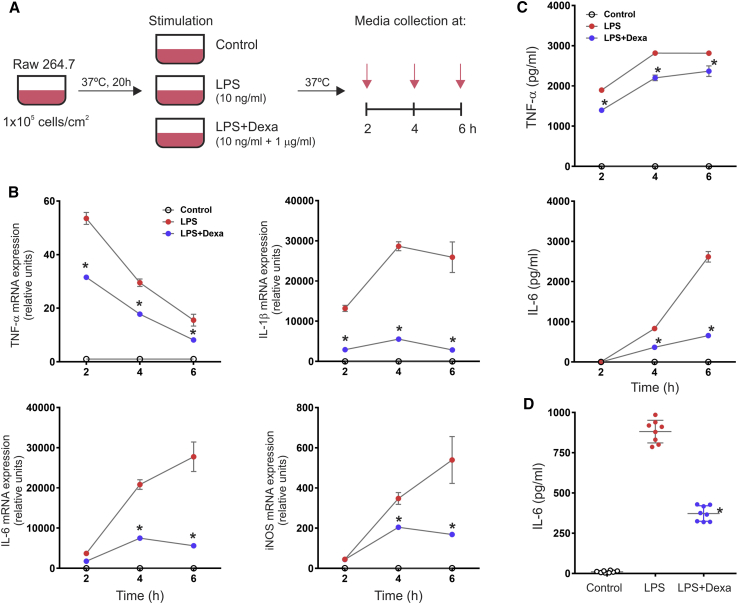
For the assay, we elected to use a cell line of mouse macrophages that have been commonly used in assays for anti-inflammatory activity and that can be used to generate a stable bank for repeated assays. It has been well established that LPS stimulation polarizes macrophages toward the M1 phenotype, which is characterized by the production of high levels of pro-inflammatory cytokines, such as tumor necrosis factor alpha (TNF-α) and interleukin (IL)-6.27 Thus, the assay is able to determine the anti-inflammatory activity of a molecule and/or compound based on its potency to inhibit the acquisition of the M1 phenotype in LPS-stimulated macrophages. For this purpose, we first standardized conditions for maximal recovery of viable macrophages from frozen vials to create a bank. We then found that the response to LPS varied with the plating density of the cells and that the exosomes we tested were ineffective if the cells were stimulated with doses higher than 10 ng/mL of LPS. Therefore, we standardized the assay for plating density of macrophages and the dose of purified LPS that stimulated the cells to a moderate level of cytokine expression, as judged by ready suppression by dexamethasone (Figures 1 and S1). We then examined the time course of the cytokine response to determine the optimal cytokine and the optimal time for the assay. Unstimulated cells showed almost undetectable levels of pro-inflammatory cytokines, indicating an uncommitted macrophage phenotype (M0) (Figure 1). As expected, macrophages responded to LPS by increased levels of mRNAs for TNF-α, IL-1β, IL-6, and iNOS that were effectively suppressed by dexamethasone (Figure 1B). The upregulation of these markers by LPS is in agreement with the acquisition of the M1-proinflammatory phenotype. ELISAs on medium confirmed increased secretion of TNF-α and IL-6 (Figure 1C). IL-1β was not detectable by ELISA, as LPS, per se, is not sufficient to trigger caspase-1-dependent IL-1β maturation and secretion in murine macrophages.28 For convenience and simplicity, we decided to use an ELISA for the readout of the in vitro macrophage assay. Results indicated that the assays for IL-6 at 4 h showed the optimal differences, considering that dexamethasone suppressed IL-6 production by about 60% at this time point (Figure 1C). In this experimental setting, containing only macrophages in culture, the secretion of IL-6 specifically represents the LPS-induced M1 polarization of macrophages. Repeated assays confirmed that the differences observed in the ELISAs for IL-6 at 4 h were reproducible (Figure 1D), and these conditions were selected for the assay. Therefore, we standardized the conditions for maximizing the recovery of viable macrophages from frozen stocks and for the expansion, seeding, and stimulation of the cells (Figure 2).
Figure 1.
Time Course Analysis of LPS-Stimulated Macrophages
(A) Diagram to depict the basic experimental design. Murine RAW 264.7 macrophage cells were seeded at a density of 1 × 105 cells per square centimeter and cultured for 20 h before stimulation with a dose of 10 ng/mL LPS with or without the addition of the anti-inflammatory corticosteroid dexamethasone (1 μg/mL). Cells and conditioned media were assayed at 2, 4, and 6 h after stimulation. (B) qRT-PCR assay for the expression of TNF-α, IL-1β, IL-6, and iNOS. Individual PCR reactions were normalized against the internal control GAPDH, and transcript levels in untreated macrophages were expressed as 1 U. Data represent mean ± SD of three independent experiments. *p < 0.0001 between the LPS and LPS+Dexa groups derived from one-way ANOVA after multiple comparisons, Tukey post hoc test. (C) TNF-α and IL-6 secretion in conditioned media assessed by ELISA. Data represent mean ± SD of an experiment run in quadruplicate. *p < 0.0001 between the LPS and LPS+Dexa groups derived from one-way ANOVA after multiple comparisons, Tukey post hoc test. (D) Reproducibility of the IL-6 ELISA in conditioned media at 4 h post-LPS stimulation. Data represent mean ± SD of eight independent experiments. *p < 0.0001 between the LPS and LPS+Dexa groups derived from one-way ANOVA after multiple comparisons, Tukey post hoc test.
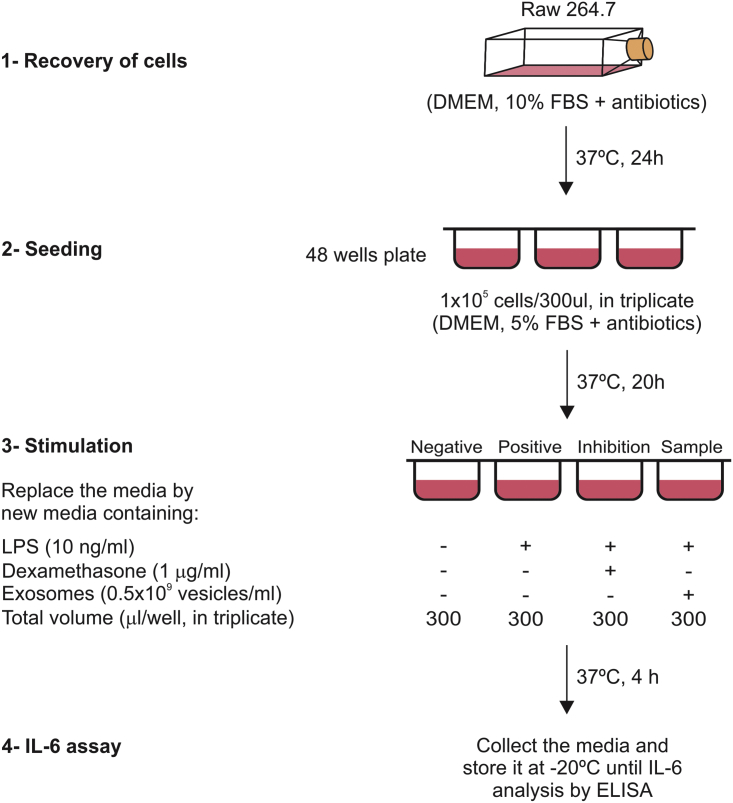
Figure 2.
In Vitro Macrophage Assay Design
Schema showing the different steps involved in the in vitro macrophage assay. First, macrophages (∼2 × 106 cells) were recovered from a frozen vial in 25 mL medium supplemented with 10% FBS and antibiotics. After 24 h of incubation at 37°C in 5% CO2, the cells were detached by gentle scraping with a cell scraper. Cells were counted and seeded in triplicate at a density of 1 × 105 cells per square centimeter in a volume of 300 μL DMEM containing 5% FBS plus antibiotics. The stimulation was performed by replacing the 300 μL culturing medium with a similar volume of medium alone (5% FBS and antibiotics) or containing LPS (10 ng/mL), LPS plus dexamethasone (1 μg/mL), or LPS plus exosomes (0.5 × 109 vesicles per milliliter). The supernatant was collected after 4 h of incubation at 37°C and kept frozen until interleukin determination by ELISA.
Preparation of Exosomes for Assay
To test the assay, we used seven independent samples of exosomes prepared under the same protocol. In brief, we used bone marrow-derived MSCs from a single donor, and the same preparation at a low density of passage 4 or 5.29 The MSCs were expanded to about 70% confluency in medium containing fetal bovine serum (FBS) and then conditioned in a chemically defined, protein-free medium.18 The exosomes secreted into the conditioned medium were concentrated and purified with an anion exchange column.
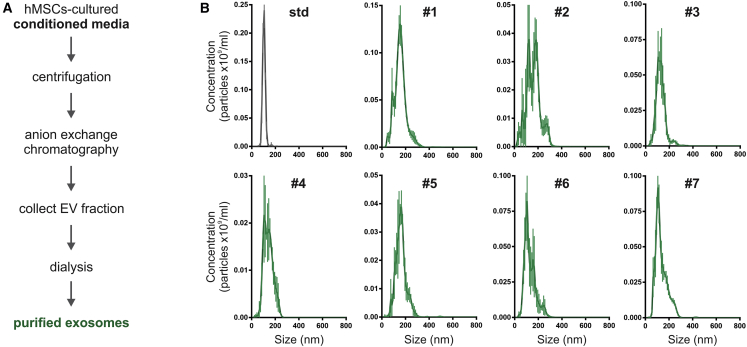
The size and number of the exosomes in the seven separate preparations were assessed by nanoparticle tracking analysis (NTA). To ensure reproducibility of the NTA data, the settings on the instrument were maintained as described in Materials and Methods. In addition, the instrument was calibrated before each assay with a stock solution of silica microspheres that were about the same size (100-nm diameter) and about the same refractive index as the exosomes.30 The mean and mode diameters of the seven preparations ranged from 99 to 162 nm (Figure 3; Table 1). The same assays suggested that the concentrations of exosomes varied from 13.1 to 51.2 × 109/mL. However, as is generally recognized,8, 10 there are limitations to data obtained on exosomes by NTA and all other available procedures. The NTA profile of sizes (Figure 3) showed that the nanoparticles in the samples were not homogeneous, as indicated by the comparison to the profile of the microspheres. The data for concentrations probably included nanoparticles of cellular origin that were not exosomes, exosomes of varying sizes, disrupted exosomes, or fused exosomes.31 Nevertheless, the diameter of the nanoparticles included in the 90th percentile (D90), when ranked from the smallest to largest, were below 200 nm for most of the isolated samples (Table 1), implying that they contained mainly exosomes. Furthermore, exosomes have a defined protein signature according to recent proteomic data collected in the ExoCarta database.32 In this context, the isolated exosomes were positive for human-specific CD63 and CD81, negative for 11 other epitopes found on human MSCs,18 and positive for Tsg101 and Alix (D.-k.K. and D.J.P., unpublished data). The identification of this selective subset of proteins further supports the hypothesis that exosome samples were highly purified.
Figure 3.
Isolation and Characterization of Different Preparations of Exosomes
(A) Schema of the procedure to isolate exosomes from MSC-conditioned media. Exosomes were collected in chemically defined protein-free media during 48 h and then were concentrated and purified by using anion exchange chromatography. The elution fractions containing exosomes were pooled and dialyzed against PBS. (B) Characterization of exosomes by nanoparticle tracking analysis (NTA). Representative size distribution profiles of the different exosome preparations used in this study. See Table 1 for concentration (particles per milliliter) and size (in nanometers) of exosome samples according to NTA measures.
Table 1.
Characteristics of the Different Exosome Preparations Used in This Study
| Exosome Preparation | NTA Analysis |
Protein (μg/mL) | CD63 (OD U/mL)a | |||
|---|---|---|---|---|---|---|
| Concentration (Particles × 109/mL) | Size (in Nanometers) |
|||||
| Mean | Mode | D90 | ||||
| #1 | 30.3 | 155.1 | 151.8 | 206.4 | 105 | 3.21 |
| #2 | 25.0 | 160.9 | 137.0 | 230.5 | 62 | 1.95 |
| #3 | 19.5 | 134.6 | 123.1 | 183.0 | 86 | 1.80 |
| #4 | 13.1 | 141.4 | 131.0 | 186.2 | 34 | 1.00 |
| #5 | 14.1 | 161.9 | 155.7 | 212.3 | 44 | 1.50 |
| #6 | 40.8 | 130.3 | 99.2 | 183.2 | 110 | 3.15 |
| #7 | 51.2 | 134.3 | 106.4 | 198.2 | 111 | 4.26 |
Values relative to level for sample #4. OD, optical density.
Efficacy of the Assay of the Anti-inflammatory Activity in Culture
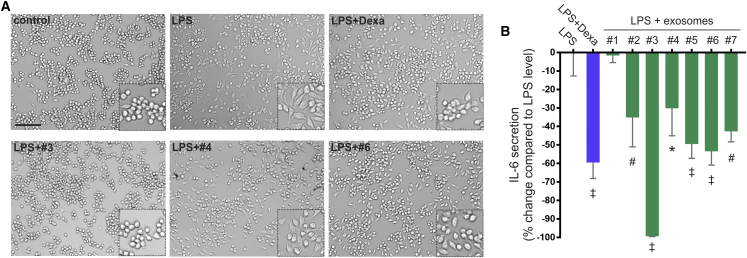
Preliminary dose-response tests with five of the preparations indicated that maximal effects in the assay were obtained with an exosome dose of approximately 0.5 × 109 vesicles per milliliter (Figure S2). To simplify the assay, this dose was used in further comparisons. The effects of the LPS in converting unstimulated M0 cells to an M1-like phenotype were apparent from visual inspection of the cultures, in which the cells with rounded morphology of M0 macrophages flattened into a pancake-like morphology of M1 macrophages (Figure 4A). Also, the effects of dexamethasone and the exosomes on inhibiting the acquisition of the M1-like morphology were apparent. For convenience, we expressed the results in terms of how much the exosomes reduced the LPS-stimulated levels of IL-6 (Figure 4B). The results indicated that five of the seven preparations had about the same anti-inflammatory activity. One (#1) was inactive. One (#3) had more activity than any of the others and completely inhibited the LPS-stimulated expression of IL-6; i.e., it was 100% effective on the scale used in Figure 4B. These differences in the anti-inflammatory activity of the preparations can be attributed to several factors, but the stability of exosomes through the purification process and storage conditions may be a critical aspect. A recent review article summarized current strategies for EV preservation, which are primarily based on data obtained by measuring particle numbers, size, and aggregation.33 Unfortunately, little is known about the best conditions to preserve the biological activity of exosomes, mainly because biological tests are lacking.
Figure 4.
Efficacy of the In Vitro Macrophage Assay to Test the Anti-inflammatory Activity of Exosomes
(A) Representative phase-contrast images showing changes in the morphology of RAW264.7 macrophages after stimulation with LPS with or without the concomitant addition of exosomes (scale bar represents 100 μm). Inserts indicate a higher magnification image. (B) ELISA for IL-6 in conditioned media of LPS-stimulated macrophages. Seven exosome preparations were tested for their anti-inflammatory activity using a dose of 0.5 × 109 vesicles per milliliter. Dexamethasone and active exosome preparations significantly reduced IL-6 secretion. Results are expressed as percentage of change of IL-6 secretion in conditioned media relative to LPS-stimulated levels. Data represent mean ± SD of three independent experiments. *p < 0.05; #p < 0.01; ‡p < 0.001, against the LPS group derived from one-way ANOVA after multiple comparisons, Tukey post hoc test.
To better characterize the effect of exosomes in LPS-stimulated macrophages, we also determined the mRNA expression of iNOS and arginase-1 (Arg1) at 4 h. We found that exosomes significantly decreased the upregulation of iNOS by 45% in LPS-stimulated cells (1.00 ± 0.26 versus 0.55 ± 0.16 relative units; LPS versus LPS + exosomes, respectively, p < 0.009). The specific M2-macrophage marker Arg1 was undetectable in our assay conditions. These results infer that exosomes act by suppressing the polarization of M0 macrophages toward the M1 phenotype by LPS stimulation.
The Assay as a Predictor of Anti-inflammatory Activity In Vivo
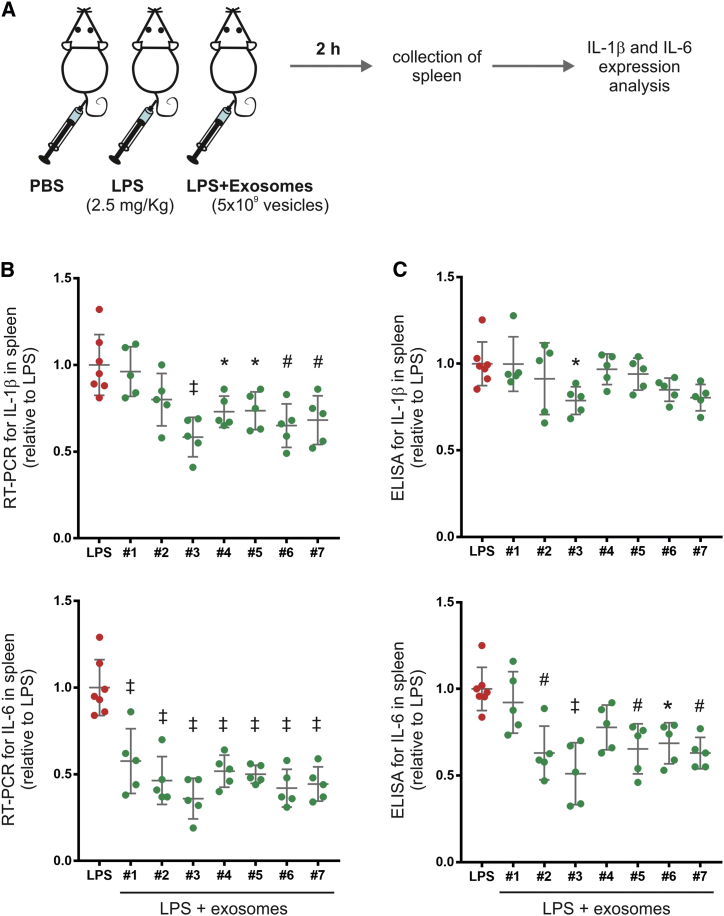
To assess the predictive efficacy of the assay, the same seven preparations were tested in mice in which a moderate degree of inflammation was induced with LPS and the inflammatory response followed by expression of IL-1β and IL-6 in spleen (Figure 5). These pro-inflammatory cytokines peaked at 2 h after LPS administration (Figure S3); thus, this time point was used to test the anti-inflammatory potential of the different exosome preparations. The intravenous (i.v.) infusion of exosomes (about 5 × 109 vesicles per mouse) was more effective in suppressing the upregulation of IL-6 than IL-1β in spleen (Figure 5), possibly because LPS-stimulated levels were lower (Figure S3).
Figure 5.
Anti-inflammatory Effects of Exosomes in a Mouse Model of LPS-Induced Acute Systemic Inflammation
(A) Schematic diagram of the experimental study design. Mice were separated into different groups receiving PBS, LPS (2.5 mg/kg), or LPS and exosomes (5 × 109 vesicles per mouse). The same seven independent exosome preparations tested in the macrophage assay were evaluated in vivo. The anti-inflammatory effect of the exosomes was determined by assessing their potential to inhibit the upregulation of IL-1β and IL-6 in spleen samples collected after 2 h of LPS administration. (B) qRT-PCR assay for the expression of IL-1β and IL-6 in spleen samples. PCR reactions were normalized using GAPDH, and transcript levels in LPS-treated mice were expressed as 1 U. Dots represent individual mice, and the lines represent the mean and SD. *p < 0.05; #p < 0.01; ‡p < 0.001, between LPS and LPS + exosome groups derived from one-way ANOVA after multiple comparisons, Tukey post hoc test. (C) Quantitation of the pro-inflammatory cytokines IL-1β and IL-6 in spleen samples by ELISA. Results are expressed as relative units taking cytokine levels in LPS-treated mice as 1 U. Dots represent individual mice, and the lines represent the mean and SD. *p < 0.05; #p < 0.01; ‡p < 0.001, between LPS and LPS + exosome groups derived from one-way ANOVA after multiple comparisons, Tukey post hoc test.
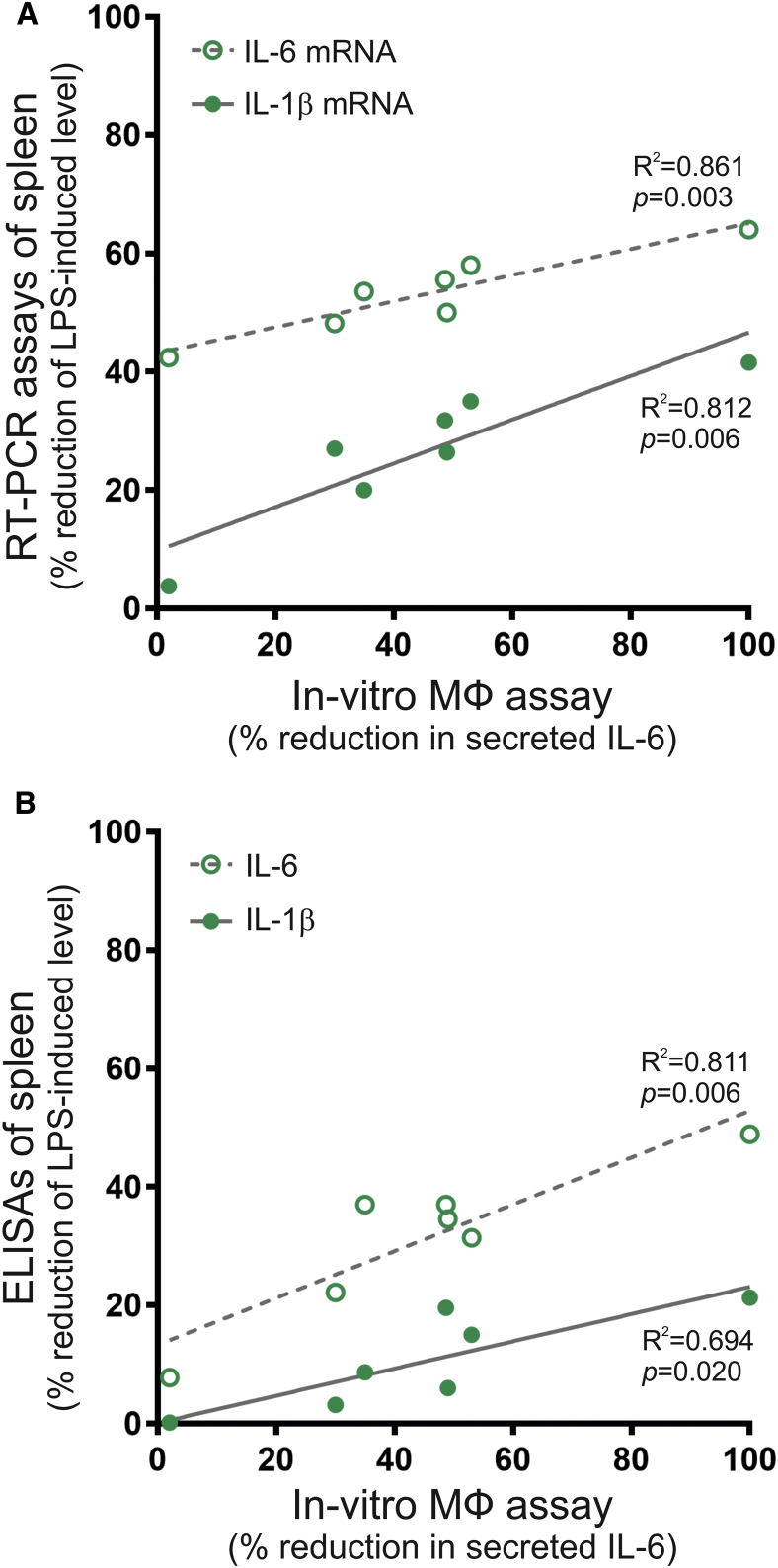
To determine the predictive potential of the in vitro macrophage assay, we plotted the suppression of IL-6 secretion in the macrophage assay against the reduction of IL-1β and IL-6 in the spleen of LPS-treated mice using data from Figures 4B and 5B. The values for anti-inflammatory activity (percentage of reduction in secreted IL-6) from the in vitro assay correlated with the suppression of mRNA levels in spleen for IL-6 (r2 = 0.861, p = 0.003) and for IL-1β (r2 = 0.812, p = 0.006) (Figure 6A). Furthermore, the assay also showed a positive correlation with the reduction of IL-1β (r2 = 0.694, p = 0.020) and IL-6 (r2 = 0.811, p = 0.006) contents in the spleen of mice receiving exosomes after LPS administration (Figure 6B). These data imply that IL-6 determination in the macrophage assay may be useful for estimating the anti-inflammatory potential of exosome preparations in vivo.
Figure 6.
Correlation between the Anti-inflammatory Activity of Exosomes in the In Vitro Macrophage Assay and in the Mouse Model of LPS-Induced Inflammation
(A) Correlation of the reduction in secreted IL-6 in the macrophage assay with the reduction of IL-1β or IL-6 mRNA expression in the spleen of LPS-treated mice. (B) Correlation of the reduction in secreted IL-6 in the macrophage assay with the reduction of IL-1β or IL-6 in the spleen of LPS-treated mice. Pearson coefficient of determination (r2) and the value of probability, p, are shown for each analysis.
Discussion
The in vitro assay described here can assess multiple samples of exosomes for one of their important activities in vivo, the ability to suppress inflammation both systemically and in the brain.18, 19, 20 Therefore, the assay has many obvious potential applications. One is to develop more rigorous protocols for the isolation and purification of anti-inflammatory exosomes. Another is to identify the best conditions for storage or transport without loss of activity. Still another is the identification of active components in the exosomes by detailed comparisons of those that have anti-inflammatory activity with those that do not.
The purification and characterization of most biomaterials have required convenient assays for either their structure or their function. However, the stringency of the assays and criteria for purification have, of necessity, decreased with the complexity of the biomaterial, ranging from small molecules to proteins, and to cells. The purity of most biologically important small molecules can readily be defined by their atomic compositions and formulas for chemical functions. The purity of proteins cannot be defined by the same criteria. Instead, the goal is “homogeneity,” defined as recovery as a single component through multiple fractionation procedures. Cells present a higher order of complexity, and there is, as yet, no way to define any of them in terms of their thousands of components that can change in response to environmental cues. Exosomes are simpler than cells, but serious challenges have been encountered in efforts to define them in terms of structure and the few convenient assays available for their function.
In the past years, the potential application of exosomes as biomarkers of disease and therapeutic agents has attracted a lot of research interest.34 Recently, a bioassay to measure the plasmin generation capacity of leukocyte-derived extracellular vesicles isolated from human plasma demonstrated prognostic value in the survival of septic shock patients.13 Here, we developed an in vitro assay that measures the anti-inflammatory potential of MSC-derived exosomes based on their ability to suppress the acquisition of the M1 phenotype in LPS-stimulated macrophages. The mechanisms mediating the anti-inflammatory effect of exosomes are still unknown, but they are more likely related to a complex combination of factors than to the activity of a specific molecule or enzyme. Interestingly, we found that the anti-inflammatory activity of exosomes in the in vitro assay positively correlates with their activity in vivo. Hence, the most effective preparations in vitro showed the highest suppression of IL-1β and IL-6 in the spleen of mice subjected to LPS-induced systemic inflammation. The fact that macrophages are critical elements in the defense against bacterial infections and the fact that both IL-1β and IL-6 secretion in spleen are mainly mediated by Toll-like receptor 4 (TLR4)-expressing monocytes and macrophages may explain the positive correlation observed with the in vitro macrophage assay. Whether the classification of different exosome preparations in terms of their anti-inflammatory potential in vitro is also associated with the therapeutic effect in other inflammatory diseases needs to be further studied. In this context, the in vitro assay developed here is similar to assays used extensively in the past to screen for drugs that suppress inflammation.35 It shares with them many of the same limitations, such as the inability to simulate fully the complex inflammatory responses seen in vivo. The assay also has several specific limitations. One is the difficulty of ensuring that the same numbers of exosomes are assayed in comparing different samples, given the current limitations of assaying exosomes.8, 10 Another limitation is that it was difficult in the assay to establish the usual pharmacological criterion of an effective dose for a 50% response. Still another limitation is the need to standardize a bank of macrophages, their response to LPS, and then their response to exosomes. The initial efforts are by trial and error, in part because the mode of action of exosomes on macrophages is still unknown. Once standardized, however, the assay is reproducible and, as illustrated here, predicts some of the effects of anti-inflammatory exosomes in vivo.
Materials and Methods
Reagents and Cell Culture
High-glucose DMEM with GlutaMAX Supplement and sodium pyruvate, alpha-minimum essential medium (MEM) eagle medium, L-glutamine, and the antibiotics (penicillin-streptomycin) were obtained from GIBCO (Life Technologies, Rockville, MD, USA). FBS and LPS from Escherichia coli serotype 0111:B4 (Sigma, L4391) were purchased from Atlanta Biologicals (Lawrenceville, GA, USA) and Sigma (St. Louis, MO, USA), respectively. Dexamethasone was from Spectrum Chemicals (Gardena, CA, USA). Chemically defined Chinese hamster ovary (CHO) medium (CD-CHO Medium; catalog #10743-002), HT Supplement, MEM Vitamin Solution, MEM Non-Essential Amino Acids solution, PBS (pH 7.4), Trypan Blue (0.4%), and Trypsin-EDTA (0.25%/1 mM) were obtained from Invitrogen (Carlsbad, CA, USA). D-(+)-glucose was from Sigma (St. Louis, MO, USA). Tissue culture flasks (75 cm2), 5-STACK multi-layer chambers (CellSTACK, 5 layers, 3,180-cm2 cell growth area), and 15-cm and 48-well tissue culture-treated plates were from Corning (Corning, NY, USA). Mouse TNF-α, IL-1β, and IL-6 DuoSet ELISA kits were purchased from R&D Systems (Minneapolis, MN, USA). Bradford reagent was purchased from Bio-Rad (Hercules, CA, USA).
Cells
RAW 264.7 cells, a murine macrophage-like cell line, were acquired from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were cultured in DMEM supplemented with 5% (v/v) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2. Human MSCs were obtained from normal, healthy donors with informed consent under procedures approved by the Scott & White and Texas A&M institutional review boards (Center for Distribution; https://medicine.tamhsc.edu/centers/irm/index.html). MSCs were cultured as reported previously.18
Extracellular Vesicle Production and Exosome Purification
Human MSC-derived exosomes were produced and purified as described previously.18 Briefly, MSCs (500 cells per square centimeter) were plated in 15-cm-diameter tissue culture plates or 5-STACK multi-layer chambers in complete culture medium (CCM). After 4–6 days (70% confluence cells), the medium was removed, the cell layer was washed, and the cells were cultured in a chemically defined protein-free medium that consisted of CD-CHO Medium supplemented with L-glutamine, HT Supplement, D-[+]-glucose, non-essential amino acids, and vitamins.18 After 6 h, the medium was changed, the conditioned medium recovered from 6 to 48 h was centrifuged to remove cellular debris at 17,000 × g for 10 min at room temperature, and the supernatant was stored at −80°C or used directly to isolate the exosomes. For the isolation of exosomes, the conditioned medium was applied directly at room temperature to a column containing the anion exchange resin (Q Sepharose Fast Flow, GE Healthcare, Chicago, IL, USA) that had been equilibrated with 50 mM NaCl in 50 mM phosphate buffer (pH 7.5). The column resin was washed with 100 mM NaCl in 50 mM phosphate buffer (pH 7.5) and then eluted with 500 mM NaCl in 50 mM phosphate buffer (pH 7.5). Exosome fractions (20–30 mL) were pooled, dialyzed against PBS (molecular weight cut-off [MWCO] of 300 kDa), and stored at −20°C.
Nanoparticle Tracking Analysis
The concentration and size distribution of isolated exosomes were measured with a NanoSight LM14 instrument (Malvern Instruments, Malvern, UK). Data were analyzed with Nanoparticle Tracking Analysis (NTA) software (v.3.2). The analysis settings were optimized and kept constant between samples. Readings were taken in triplicate by capturing video during 45 s at 30 frames per second (fps), at a camera level set to 13. Silica microspheres were used as a size standard (100 nm, Polysciences). Purified exosome samples were diluted 25–50 times with sterile and filtered PBS yielding particle concentrations in the region of 0.5–1.5 × 109 particles per milliliter in accordance with the manufacturer’s recommendations.
In Vitro Macrophage Assay
Frozen cells (∼2 × 106 cells per vial) were washed in 10 mL previously warm (37°C) DMEM supplemented with FBS (10%) and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin). Cells were centrifuged at 450 × g during 10 min, resuspended in 1 mL medium, and cultured in a T75 flask containing 25 mL of the same medium for 24 h at 37°C in 5% CO2. For the macrophage assay, cells were detached by gentle scraping with a cell scraper, and they were collected, centrifuged (10 min at 450 × g), and resuspended in DMEM containing 5% of FBS and antibiotics. After that, cells were counted and seeded at a density of 1 × 105 cells per square centimeter using 300 μL medium (5% of FBS plus antibiotics) per well in a 48-well plate. After 20 h of incubation, the macrophages were stimulated by replacing the 300 μL culturing medium with a similar volume of medium alone (5% FBS and antibiotics) or containing LPS (10 ng/mL), LPS plus dexamethasone (1 μg/mL), or LPS in combination with exosomes (0.5 × 109 vesicles per milliliter). All the different conditions were prepared in a total volume of 1 mL and tested in triplicates. In all cases, the dose of exosomes was loaded using a volume that was not higher than 10% of the total volume. The conditioned medium was collected after 4 h of incubation at 37°C and kept frozen until assayed for cytokines by ELISA.
LPS Mouse Model
All animal protocols were approved by the Texas A&M Animal Care and Use Committee. Male BALB/c mice (10–12 weeks old) were used for the experiments. We used a mouse model of endotoxemia that consists of an i.v. injection of LPS. Vasodilation was achieved by immersing the tail into warm water for 1 min. For i.v. administration, LPS (2.5 mg/kg) was injected via tail vein using a 28G needle in a total volume of 250 μL, either alone or in combination with exosomes (approximately 5 × 109 vesicles). Control mice were injected with saline. Mice were euthanized at 2 h after injection, and the spleens were aseptically removed, snap frozen, and stored at −80°C until use. A preliminary experiment using 12 mice was carried out to determine the best time point (0, 2, or 6 h) for the detection of IL-1β and IL-6 in spleen samples (Figure S3).
qRT-PCR Analysis
Total RNA was extracted from cells or tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as described by the manufacturer. One microgram of RNA was reverse-transcribed into cDNA using random primers and MultiScribe reverse transcriptase (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems, Foster City, CA, USA). qRT-PCR was performed by using TaqMan Gene Expression Assays for genes of interest (Thermo Fisher Scientific) (Table S1). Samples were assayed in triplicate using the TaqMan Fast Advanced Master Mix on an Applied Biosystems Sequence Detection System (SDS7900HT) with the following conditions: 1 cycle for 2 min at 95°C and 40 cycles at 95°C for 1 s and 60°C for 20 s. Results were analyzed using the relative quantification (2−ΔΔCt) method by normalizing the threshold cycle (Ct) values against the reference gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH).36
Quantitation of TNF-α, IL-1β, and IL-6 by ELISA
Conditioned media from the macrophage assays were analyzed for TNF-α, IL-1β, and IL-6 by ELISA following the manufacturer’s recommendations (R&D Systems). Spleen samples were homogenized in ice-cold 50 mM potassium phosphate buffer (pH 7.4) containing 0.1% Triton X-100 and protease inhibitors (cOmplete Protease Inhibitor Cocktail, Roche). Homogenates were clarified by centrifugation (15 min at 10,000 × g) and stored at −80°C until use. Samples were diluted in PBS (1 mg protein per milliliter) and assayed for IL-1β and IL-6 by ELISA following the manufacturer’s recommendations (R&D Systems). Protein concentrations were determined using the Bradford micromethod assay (Bio-Rad).
Statistical Analysis
Continuous variables were expressed as mean ± SD. One-way ANOVA with post hoc Tukey’s test was used for intergroup comparisons. The correlation between the anti-inflammatory activity observed in the in vitro macrophage assay and in the LPS mouse model was calculated by the Pearson coefficient of determination (r2) and the p value. A value of p < 0.05 was considered statistically significant.
Author Contributions
N.P. designed the study; performed experiments; collected, analyzed, and interpreted data; and prepared the figures. R.H.L., E.-H.B., D.-k.K., and Q.L. performed experiments and collected, analyzed, and interpreted data. D.J.P. and G.Y. designed the study, analyzed and interpreted data, coordinated and supervised the project, and wrote the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported in part by NIH grant P40OD011050 held by D.J.P. This work was also supported by a PIP-2015-2017 grant (11220150100188CO) from CONICET and by a Bilateral Cooperation Program (PCB-I, 2250160100022CO) NIH-CONICET held by G.Y. N.P. and G.Y. are staff researchers of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina.
Footnotes
Supplemental Information includes three figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.12.003.
Contributor Information
Darwin J. Prockop, Email: prockop@medicine.tamhsc.edu.
Gustavo Yannarelli, Email: gyannarelli@favaloro.edu.ar.
Supplemental Information
References
- 1.Quesenberry P.J., Aliotta J., Deregibus M.C., Camussi G. Role of extracellular RNA-carrying vesicles in cell differentiation and reprogramming. Stem Cell Res. Ther. 2015;6:153. doi: 10.1186/s13287-015-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yáñez-Mó M., Siljander P.R., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.György B., Hung M.E., Breakefield X.O., Leonard J.N. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang B., Yeo R.W., Tan K.H., Lim S.K. Focus on extracellular vesicles: therapeutic potential of stem cell-derived extracellular vesicles. Int. J. Mol. Sci. 2016;17:174. doi: 10.3390/ijms17020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosa-Fernandes L., Rocha V.B., Carregari V.C., Urbani A., Palmisano G. A perspective on extracellular vesicles proteomics. Front Chem. 2017;5:102. doi: 10.3389/fchem.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coumans F.A.W., Brisson A.R., Buzas E.I., Dignat-George F., Drees E.E.E., El-Andaloussi S., Emanueli C., Gasecka A., Hendrix A., Hill A.F. Methodological guidelines to study extracellular vesicles. Circ. Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 8.Li P., Kaslan M., Lee S.H., Yao J., Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mateescu B., Kowal E.J., van Balkom B.W., Bartel S., Bhattacharyya S.N., Buzás E.I., Buck A.H., de Candia P., Chow F.W., Das S. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tkach M., Kowal J., Théry C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20160479. doi: 10.1098/rstb.2016.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Freitas D., Kim H.S., Fabijanic K., Li Z., Chen H., Mark M.T., Molina H., Martin A.B., Bojmar L. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018;20:332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Deun J., Mestdagh P., Agostinis P., Akay Ö., Anand S., Anckaert J., Martinez Z.A., Baetens T., Beghein E., Bertier L., EV-TRACK Consortium EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017;14:228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 13.Cointe S., Harti Souab K., Bouriche T., Vallier L., Bonifay A., Judicone C., Robert S., Armand R., Poncelet P., Albanese J. A new assay to evaluate microvesicle plasmin generation capacity: validation in disease with fibrinolysis imbalance. J. Extracell. Vesicles. 2018;7:1494482. doi: 10.1080/20013078.2018.1494482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai R.C., Arslan F., Lee M.M., Sze N.S., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. (Amst.) 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Zhu W., Huang L., Li Y., Zhang X., Gu J., Yan Y., Xu X., Wang M., Qian H., Xu W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012;315:28–37. doi: 10.1016/j.canlet.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Tan C.Y., Lai R.C., Wong W., Dan Y.Y., Lim S.K., Ho H.K. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res. Ther. 2014;5:76. doi: 10.1186/scrt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Chopp M., Meng Y., Katakowski M., Xin H., Mahmood A., Xiong Y. Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J. Neurosurg. 2015;122:856–867. doi: 10.3171/2014.11.JNS14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D.K., Nishida H., An S.Y., Shetty A.K., Bartosh T.J., Prockop D.J. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc. Natl. Acad. Sci. USA. 2016;113:170–175. doi: 10.1073/pnas.1522297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y., Ye Y., Su X., He J., Bai W., He X. MSCs-derived exosomes and neuroinflammation, neurogenesis and therapy of traumatic brain injury. Front. Cell. Neurosci. 2017;11:55. doi: 10.3389/fncel.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long Q., Upadhya D., Hattiangady B., Kim D.K., An S.Y., Shuai B., Prockop D.J., Shetty A.K. Intranasal MSC-derived A1-exosomes ease inflammation, and prevent abnormal neurogenesis and memory dysfunction after status epilepticus. Proc. Natl. Acad. Sci. USA. 2017;114:E3536–E3545. doi: 10.1073/pnas.1703920114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox A.J., West N.P., Cripps A.W. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015;3:207–215. doi: 10.1016/S2213-8587(14)70134-2. [DOI] [PubMed] [Google Scholar]
- 22.Lontchi-Yimagou E., Sobngwi E., Matsha T.E., Kengne A.P. Diabetes mellitus and inflammation. Curr. Diab. Rep. 2013;13:435–444. doi: 10.1007/s11892-013-0375-y. [DOI] [PubMed] [Google Scholar]
- 23.van Zuylen V.L., den Haan M.C., Geutskens S.B., Roelofs H., Fibbe W.E., Schalij M.J., Atsma D.E. Post-myocardial infarct inflammation and the potential role of cell therapy. Cardiovasc. Drugs Ther. 2015;29:59–73. doi: 10.1007/s10557-014-6568-z. [DOI] [PubMed] [Google Scholar]
- 24.Serpente M., Bonsi R., Scarpini E., Galimberti D. Innate immune system and inflammation in Alzheimer’s disease: from pathogenesis to treatment. Neuroimmunomodulation. 2014;21:79–87. doi: 10.1159/000356529. [DOI] [PubMed] [Google Scholar]
- 25.Tajes M., Ramos-Fernández E., Weng-Jiang X., Bosch-Morató M., Guivernau B., Eraso-Pichot A., Salvador B., Fernàndez-Busquets X., Roquer J., Muñoz F.J. The blood-brain barrier: structure, function and therapeutic approaches to cross it. Mol. Membr. Biol. 2014;31:152–167. doi: 10.3109/09687688.2014.937468. [DOI] [PubMed] [Google Scholar]
- 26.Pardridge W.M. Delivery of biologics across the blood-brain barrier with molecular trojan horse technology. BioDrugs. 2017;31:503–519. doi: 10.1007/s40259-017-0248-z. [DOI] [PubMed] [Google Scholar]
- 27.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaidt M.M., Ebert T.S., Chauhan D., Schmidt T., Schmid-Burgk J.L., Rapino F., Robertson A.A., Cooper M.A., Graf T., Hornung V. Human monocytese an alternative inflammasome pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Sekiya I., Larson B.L., Smith J.R., Pochampally R., Cui J.G., Prockop D.J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner C., Ferreira Y.J., Dragovic R.A., Redman C.W., Sargent I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles. 2013;2:19671. doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordin J.Z., Lee Y., Vader P., Mäger I., Johansson H.J., Heusermann W., Wiklander O.P., Hällbrink M., Seow Y., Bultema J.J. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine (Lond.) 2015;11:879–883. doi: 10.1016/j.nano.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Simpson R.J., Kalra H., Mathivanan S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles. 2012;1:18374. doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusuma G.D., Barabadi M., Tan J.L., Morton D.A.V., Frith J.E., Lim R. To protect and to preserve: novel preservation strategies for extracellular vesicles. Front. Pharmacol. 2018;9:1199. doi: 10.3389/fphar.2018.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 35.Schultze J.L., Schmieder A., Goerdt S. Macrophage activation in human diseases. Semin. Immunol. 2015;27:249–256. doi: 10.1016/j.smim.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.