Abstract
Histone posttranslational modifications (PTMs) are key epigenetic marks involved in gene silencing or activation. Histone modifications impact chromatin organization and transcriptional processes through the changes in charge density between histones and DNA. They also serve as recognition and binding sites for specific binding proteins. His-tone tails and globular cores contain many basic amino acid residues, which are subject to various dynamic modifications, making the modification repertoire extremely diverse. Consequently, determination of histone PTM identity and quantity has been a challenging task. In recent years, mass spectrometry-based methods have proven useful in histone PTM characterization. This chapter provides a brief overview of these methods and describes the approach to analyze the PTMs of the histone variant CENP-A, essential for the cell cycle progression, when present in minute amounts from tumor and mammalian tissues. Because this method does not rely on antibody-based immunopurification, we anticipate that these tools could be readily adaptable to the investigation to other histone variants in a range of mammalian tissues and solid tumors.
1. INTRODUCTION
Histones are highly basic globular proteins that form nucleosomes, the basic fundamental repeating building blocks of DNA packaging into chromatin. DNA in chromatin is wrapped around histone octamers, consisting of histones H3, H4, H2A, and H2B, and the linker DNA between nucleosomes is bound to histone H1. During cell division, the metaphase chromosomes within the centromere region contain a specific histone H3 variant, CENP-A, which substitutes H3 within centromeric nucleosomes. CENP-A is present in less than 10% of total nucleosomes making its study in the native context challenging. Consequently, majority of the labs in the centromere field have resorted to the use of ectopically expressed tagged CENP-A, under the control of various promoters. However, there is no reliable data yet thoroughly investigating whether the presence of tags, commonly twice the size of a given histone and placed at the N- or C-terminus of the protein, interferes with nucleosome structure or dynamic processes such as assembly kinetics, strength of kinetochore formation, degradation, turnover, rate of transcription, or repair. Consequently, variability in nucleosomal structures containing CENP-A is still under intense investigation, with a range of groups using alternative tools and approaches arriving at disparate answers that still await reconciliation.
Extensive posttranslational modification (PTM) of histone tails has been long known to influence the chromatin state. Indeed, stably inherited covalent histone modifications are referred to as epigenetic marks (Holliday, 1990). More recently, the PTMs in the globular core of histone molecules and their diverse functions have been reported. PTMs such as acetylation, methylation, phosphorylation, and ubiquitination of many amino acids within histones have been known for decades (Allfrey, Faulkner, & Mirsky, 1964; Allfrey & Mirsky, 1964; Bonnet, Devys, & Tora, 2014; Jason, Moore, Lewis, Lindsey, & Ausio, 2002) to change the affinity of histones to chromatin and cause alterations in the complexes with histone-binding proteins, impacting transcription. The complexity of the combinations of histone modifications and their effects on the downstream events led to the proposal of the histone code hypothesis and methods to decipher the code (Dhall & Chatterjee, 2011; Jenuwein & Allis, 2001; Strahl & Allis, 2000; Turner, 2000). Multiple histone PTMs have been described (Zhao & Garcia, 2015). Changes in histone PTMs are absolutely necessary for many cellular processes, and aberrant modification of histones has been implicated as drivers of many diseases, including cancer, as recurrent mutations exist in many histone-modifying enzymes (for a comprehensive recent review, see Audia & Campbell, 2016). Specific to this review, changes in core domain CENP-A PTMs may also regulate centromere dynamics during cell cycle progression (Bui et al., 2012; Bui, Walkiewicz, Dimitriadis, & Dalal, 2013; Walkiewicz, Bui, Quenet, & Dalal, 2014).
2. MASS SPECTROMETRY-BASED METHODS OVERVIEW
Histone modifications have been extensively studied in many biological systems with use of biochemical methods, mainly based on modification-specific antibodies. Chromatin immunoprecipitation (ChIP) approaches routinely exploit the available antibody repertoire. Combining the ChIP with next-generation sequencing enabled genome-wide mapping of histone modification status (Barski et al., 2007; Johnson, Mortazavi, Myers, & Wold, 2007; Mikkelsen et al., 2007). However, these methods require the prior knowledge of the potential modification and suffer from critical issues shared by all antibody-based techniques, such as antibody availability, batch-to-batch variability, cross-reactivity, dependence on the surrounding amino acid sequence, and/or protein structure. Thus, antibody-based immunopurifications do not allow for an unbiased interrogation of multiple histone PTMs at once, a solution for which is provided by mass spectrometry-based methods for comprehensive analysis of histone PTMs, which have been extensively described previously (Huang, Lin, Garcia, & Zhao, 2015; Zheng, Huang, & Kelleher, 2016). Briefly, for high-throughput, multihistone analyses, three analytical techniques, called Bottom Up, Middle Down, and Top Down, are established. Bottom Up and Middle Down analyses require digestion of histones by proteolytic enzymes and subsequent analysis of the peptides by tandem mass spectrometry (MS/ MS). Top Down analysis uses no proteases and whole histone molecules with modification are detected by MS/MS (for an excellent recent review, see Zheng et al., 2016).
The analysis of peptides in Bottom Up mode, when the histones are digested with trypsin, for complex histone mixtures requires derivatization by propionylation of free amines to prevent the appearance of very short tryptic peptides due to high occurrence of lysine residues within histones (Garcia et al., 2007). Here, we present an alternative combinatorial method we have used successfully for the qualitative studies of PTMs of a single his-tone variant, CENP-A. This method first separates all histones and their modified version, at very high resolution, on Triton acid urea (TAU) gels, bands of which can then be excised directly and analyzed using Bottom Up approach without derivatization, using trypsin and/or chymotrypsin and subsequent nano-LC–MS/MS.
3. PROTOCOL
3.1. Preparation of Chromatin-Bound Histones and Separation of Histones on TAU Gels
Unlike traditional SDS-PAGE gels, TAU gels separate not only by size but also by charge and hydrophobicity (Shechter, Dormann, Allis, & Hake, 2007), making TAU gels the ideal method of separation of histones for downstream mass spectrometric analysis. Indeed, histone variants and his-tone modifications have been studied extensively for almost 30 years, on TAU gels by workers such as Waterborg, Annunziato, and Hake (Ryan & Annunziato, 2001; Shechter et al., 2007; Waterborg & Matthews, 1984). Here, we provide a detailed protocol in which we adapted chromatin-bound histone purification on TAU gels for subsequent use with mass spectrometry. The first nine steps of the protocol are therefore useful for examining only chromatin-bound histone variants and modifications.
Prior to separating the histones on the TAU, five T175 flask of human cells (HeLa, SW480, etc.) are grown to about 80–90% confluency. Before starting, cool down centrifuge to 4°C.
3.1.1. Prepping Cells and Extracting Histones With High Salt
Trypsinize and collect cells into a 50-mL tube with DMEM containing FBS to neutralize the trypsin.
Centrifuge cells at 230 rcf for 5 min at 4°C (from this point on, “centrifuge” will refer to centrifuge at 230 rcf for 5 min at 4°C, unless otherwise noted).
Wash with PBS and centrifuge.
Wash with cold PBS+0.1% Tween (PBS-T) and centrifuge. During spin, prepare 10 mL of cold TM2 solution (20 mM Tris, pH 8.0; 2 mM MgCl2) supplemented with 0.5% NP40 or NP40 substitute, per sample prep. Vortex for several minutes to ensure the NP40 detergent has completely dissolved.
Remove the PBS-T supernatant, flick tube to break up cell pellet, and add the TM2+NP40 mix. Gently rotate a few times to break up pellet and leave on ice for 2 min. The cells are now lysed and nuclei are left behind.
Centrifuge nuclei.
Perform a wash with cold TM2 and centrifuge.
Remove TM2 supernatant and to the 50-mL tube, add 1 g (10%) hydroxylapatite, 10 mL of 350 mM NaCl PBS (PBS already has ~150 mM NaCl, so add an additional 200 mM NaCl), 2 mM EDTA, Roche cOmplete protease inhibitor cocktail (Cat #050556489001), and a small stir bar.
Tightly cap the 50-mL tube, flip it upside down, place into a small container, and let it stir overnight on a magnetic stirrer at 4°C.
Next day, centrifuge the slurry at 500 rcf, remove supernatant, and perform two washes with 350 mM NaCl PBS. At this stage, chromatin-bound nucleosomal histones are still bound to DNA, whereas most weakly associated/nonhistone proteins will be removed. For preassembly histones inside the nucleus, we recommend following steps 1–10, saving this soluble “0.35 M” fraction at step 10, and reentering the protocol by concentrating the soluble fraction down using spin columns at step 17. Alternatively, using chromatin-free nuclear extracts might be preferred, but these will be enriched in several hundred other nuclear factors.
Add 6 mL of 2 M NaCl PBS (PBS already has ~150 mM NaCl, so add an additional 1.85 M NaCl) and place on magnetic stirrer at 4°C for 2–4 h. A fraction of histones will be released into the supernatant.
Centrifuge the slurry at 500 rcf and save the 2 M NaCl PBS supernatant, which contains the histones. Store at 4°C.
To the hydroxylapatite slurry, add 6 mL of 2.8 M NaCl PBS (PBS already has ~150 mM NaCl, so add an additional 2.65 M NaCl) and place on magnetic stirrer overnight at 4°C. As the majority of histones will be released from the DNA during this incubation, this step is critical and should not be shortchanged.
On the next day, centrifuge the 2.8 M NaCl PBS slurry at 500 rcf.
Pool the 2 M and 2.8 M supernatants into a 15-mL tube and centrifuge at 900 rcf to sediment residual hydroxylapatite material.
Transfer the supernatant (leaving the sedimented hydroxylapatite behind) to a clean 15-mL tube.
Concentrate samples by centrifuging in Amicon Ultra Centrifugal Filter units (3K MW cutoff, Millipore Cat #UFC800396) at 2000 rcf at 4°C. Continue spinning and adding additional sample containing histones until samples have concentrated to ~200 μL.
Dialyze sample overnight by using Slide-A-Lyzer Dialysis Cassettes (ThermoScientific, Waltham, MA, Cat #66373) in low-salt buffer(0.5 × PBS) at 4°C in a beaker with a stir bar. Make sure dialysis is done on a magnetic stirrer.
Histone samples are now ready to be loaded and ran on a TAU gel.
3.1.2. Prepping TAU Gels and Running Samples
To a 250-mL vacuum filter flask, add 12 g urea, 5.6 mL 40% acrylamide, 1.25 mL glacial acetic acid, 125 μL TEMED, 0.5 mL 0.3 M Triton X, 7.6 mL dH2O (to 25 mL).
Gently stir without introducing bubbles in a warm water bath (~37°C) to aid dissolution of the urea.
Degas for 10 min with a vacuum and hose attached to the flask. In the meantime, assemble the BioRad PROTEAN II xi cell according to manufacturer’s recommendations (alternatively, to your own manufacturer’s recommendations).
Add 300 μL 10% APS to the flask containing the contents in step 1, gently swirl and pipette once up and down with a 10-mL pipette tip, and transfer in between the assembled glass plates, leaving ~3 cm from the top to allow room for the stacking gel and comb.
Cover the top of the resolving gel with a layer of butanol by pipetting from one side and letting it level out. This prevents drying of the gel, which normally takes ~30 min.
Prepare stacking gel by adding the following components to a 250-mL vacuum filter flask: 4.8 g urea, 1.2 mL 40% acrylamide, 500 μL glacial acetic acid, 100 μL TEMED, 200 μL 0.3 M Triton X-100, 4.5 mL dH2O.
Gently stir flask in a water bath to dissolve urea crystals and degas for 10 min.
Take the assembled glass plates with the solidified TAU resolving gel, and wash off the butanol and unpolymerized acrylamide. Vacuum any residual water in between the glass plates.
Reassemble the gel on the stand, add 250 μL 10% APS to the stacking gel mix in step 6, swirl the flask to mix, and add in between the glass plates. Fill to the rim and gently assemble the comb. Allow to polymerize for ~30 min.
Gently remove the comb and tape or any sealant used at the base of glass plates, and assemble the unit into the running buffer container.
Fill the chamber and let the rest flow into the buffer container with TAU running buffer (65 mL glacial acetic acid, 1.3 mL concentrated Triton X-100, and dH2O up to 1300 mL, made in a 2-L graduated cylinder with a stir bar on a magnetic stirrer for an hour).
Using a needle and syringe, pipette up and down several times in each well to clear it of any unpolymerized acrylamide debris.
Pipette 30 μL of the Cysteamine Pre-run solution (3.84 g urea, 0.57 g cysteamine or β-mercaptoethanol, 430 μL glacial acetic acid, 160 μL0.3 M Triton X-100, a pinch of pyronin Y (for tracking)), add water to a total of 8 mL into each lane, and electrophorese at a constant 15 mA for 16–22 h. Be sure to reverse the polarity as the acetic acid running conditions will reverse the positive and negative poles.
Make a fresh batch of TAU running buffer as in step 11.
Next day, replace the TAU running buffer from the upper chamber and replace with the new buffer from step 14 and let that flow into the running buffer container. Getting rid of the previous buffer from the running container is not necessary.
Use a needle and syringe to clean out the wells as in step 12.
Dissolve sample(s) in 2× Sample Running Dye (9.6 g urea, 750 μL β-mercaptoethanol, 750 μL glacial acetic acid, a pinch of pyronin Y (for tracking), add water to a total of 15 mL), and load samples into the wells
Run gel at a constant 15 mA for 4–5 h (traditional 20-cm long L-TAU) or 9 h (double long TAU, dLτ). Be sure to reverse the polarity.
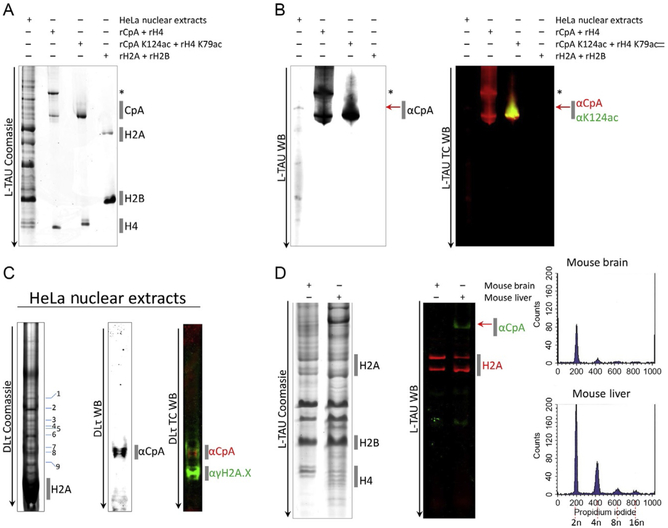
Once the run is complete, the gel can be stained with Coomassie Brilliant Blue after fixing for 15 min with 50% methanol, 5% acetic acid, and 45% dH2O. If recombinant proteins are available, run them alongside the histones extracted from the nuclei as a reference (see Fig. 1). Modifications such as acetylations and phosphorylations shift bands higher. If a Western is desired, please proceed to Section 3.1.3.
Fig. 1.
Separating histones from cells and tissues on traditional long Triton acid urea (L-TAU) and Double Long TAU gels (dLτ). (A) Nuclear extracts from HeLa cells were extracted and ran on an L-TAU alongside recombinant (r) histones expressed in bacteria, and the gel was stained with Coomassie Brilliant Blue to reveal CENP-A and other canonical histone bands. (B) Replicate gel was transferred to nitrocellulose for Western detection using antibodies against CENP-A or CENP-A K124ac. (C) Resolving the gel further by running on a double long TAU (dLτ). The majority of the histones ran off the gel, resulting in further separation of the CENP-A band into modified species running above histone H2A and γH2A.X histone bands. (D) Histones from mouse brain and liver were extracted (Dalal, 2003; Stein, Dalal, & Fleury, 2002) and ran on an L-TAU gel and Western probing against histone H2A and mouse CENP-A. Fluorescence-activated cell sorting (FACS) analysis confirmed the difference in ploidy among the two tissues (2n, 4n, 8n, or 16n). “*” Denotes CENP-A with a 6His tag and red arrow denotes native/endogenous CENP-A. TC, two-color; WB, Western blot.
3.1.3. TAU Westerns
Prior to transferring the gel, it needs to be equilibrated to remove excess Triton X. Equilibrate the gel twice for 20 min each with TAU equilibrating solution (50 mM glacial acetic acid; 0.5% SDS), under gentle shaking conditions.
Next, equilibrate the gel in transfer buffer (1 × Tris–glycine transfer buffer containing 25 mM Tris–HCl, pH 8.0; 192mM glycine; 20% ethanol) with gentle shaking, twice at 20 min each.
Assemble the filter paper, membrane and gel sandwich with the midi-format Transfer-Blot Turbo Transfer pack, and transfer with the preset BioRad’s high-molecular-weight (MW) setting (2.5 A, 25 V) for a total of at least 15 but no more than 20 min using the Transfer-Blot Turbo Transfer system.
Disassemble the sandwich and wash the membrane with 1 × PBS with gentle shaking to remove excess ethanol for 2 min.
Block the membrane with membrane blocking buffer (5% nonfat dry milk solubilized in 1× PBS) at room temperature for 30 min.
Pour off the membrane blocking buffer and perform a quick rinse with PBS-T.
Assemble into a hybridization bag and add the hybridization buffer (3% BSA solubilized in PBS-T) containing the desired amount of antibody against the protein of interest.
Wash the membrane twice with PBS-T and proceed with secondary antibody incubation using hybridization buffer as well. Follow manufacturer’s conditions for detection of the secondary antibody. For the detection of human CENP-A, a 1:2000 dilution with the CENP-A antibody (AbCam, Cambridge, MA, Cat #ab13939) is recommended with overnight incubation at 4°C (see Fig. 1).
3.2. In-Gel Digest of TAU Gel Bands
Transfer the gel to an unused Petri dish or onto the clean Saran wrap surface, preferably in the sterile environment (e.g., a biosafety cabinet). The gel handling and all the subsequent steps should be performed with gloved hands to reduce contamination with keratins or dust (see Note 1). Cut out gel bands of interest with a fresh scalpel, taking as little excess polyacrylamide as possible, and place them inside separate, labeled 1.5-mL microcentrifuge tubes.
Cut the gel bands into approximately 1-mm cubes using a scalpel or micropoint tweezers.
Depending on the amount of protein loaded, the gel band can be kept whole for the proteolytic digest or split in half for a digest with an additional protease.
Destain the gel bands with approximately 100 μL of 1:1 100 mM ammonium bicarbonate (NH4HCO3)/acetonitrile for 30 min. Vortex occasionally.
Add 500 μL of acetonitrile and vortex occasionally for 10 min. Gel pieces will be more opaque and should be significantly destained. Complete destaining is not necessary.
Use a centrifuge to cluster gel cubes together at the bottom of the tube and then remove the supernatant with a pipette.
Add 50 μL of dithiothreitol (DTT) (Sigma-Aldrich, St. Louis, MO, Cat #D5545–5G) (DTT is dissolved in 100 mM NH4HCO3) to a final concentration of 10 mM. The gel pieces should be completely covered with DTT solution (see Note 2).
Incubate at 56°C for 30 min.
Chill the tubes to room temperature and add 500 μL of acetonitrile to shrink the gel pieces. After 10 min, the gel pieces should be more opaque. Remove the supernatant.
Add 50 μL of freshly made iodoacetamide (IDA; Sigma-Aldrich, St. Louis, MO, I1149–5G) in 100 mM NH4HCO3 to a final concentration of 55 mM (see Note 2). The gel pieces should be covered with the IDA solution.
Incubate at room temperature for 20 min in the dark.
Add 500 μL of acetonitrile and vortex occasionally for 10 min to shrink the gel pieces. Remove the supernatant.
Add enough protease to cover gel pieces. The trypsin solution is 13 ng/μL Sequencing Grade Trypsin (Promega, Madison, WI, Cat #V5111) in 10 mM NH4HCO3 with 10% (v/v) acetonitrile. It can take up to 30 min for the gel pieces to absorb trypsin due to its slow diffusion speed relative to the rest of the aqueous solution. The chymotrypsin solution (Promega, Madison, WI, Cat #V1061) is 13 ng/μL in 50 mM NH4HCO3.
The gel pieces should be fully saturated with the protease solution. Add more protease solution if needed. After 45–60 min, the gel pieces should be checked to make sure they are still completely covered with liquid. If not, add enough NH4HCO3 (10 mM) solution (50 mM NH4HCO3 if chymotrypsin is used) to cover the gel pieces.
Incubate gel pieces overnight: for trypsin, at 37°C; for chymotrypsin, at 25°C.
Add 100 μL of 1:2 (v/v) 5% formic acid/acetonitrile to extract peptides from gel pieces. Extract supernatant using gel-loading pipette tips to avoid pipetting gel debris, which could cause a physical blockage in the LC–MS/MS system.
Vacuum centrifuge the peptide extraction to dryness if the samples are to be stored in a freezer. If the samples will be promptly analyzed via mass spectrometry, then vacuum centrifuge long enough to remove acetonitrile from the extracted peptide solution. Evaporating the peptide extraction long enough to reduce the volume from 100 μL to approximately 10 μL (to a sample injection volume) should be sufficient because acetonitrile will evaporate faster than water.
3.3. Liquid Chromatography Mass Spectrometry
Prepare 1 L of 0.1% (v/v) formic acid (Buffer A) and 1 L of acetonitrile with 0.1% (v/v) formic acid (Buffer B). Use only LC–MS grade solvents.
Analyze samples using a 60 min liquid chromatography gradient of 3–35% Buffer B (acetonitrile containing 0.1% formic acid). Wash with 5-column volumes of 80% Buffer B and then reequilibrate with 10-column volumes of 97% Buffer A.
Trypsin-digested samples should be analyzed with a discovery method that uses data-dependent analysis (DDA) and collision-induced dissociation (CID). Chymotrypsin-digested samples should utilize a DDA method with an electron transfer dissociation (ETD) data-dependent decision tree.
3.4. Data Analysis
Use a database search program to analyze the data files. The searches should be against a CENP-A only database so that many dynamic modifications can be searched within a minimal search space. Chemical modifications added during sample preparation should be added as fixed modifications. There are several dynamic modifications of interest for CENP-A: phosphorylation (STY), acetylation (K), methylation (KR), dimethylation (KR), trimethylation (K), ubiquitination (K). If IDA was used for protein alkylation, then ubiquitin matches could be chemically rather than biologically induced (Nielsen et al., 2008). Alternative reduction and alkylation methods are available as mentioned in Note 2 (Nielsen et al., 2008).
Remove false discovery rate filters when looking at search results. Single gel band runs may not contain sufficient number of spectra for statistical models to be effective at removing random matches.
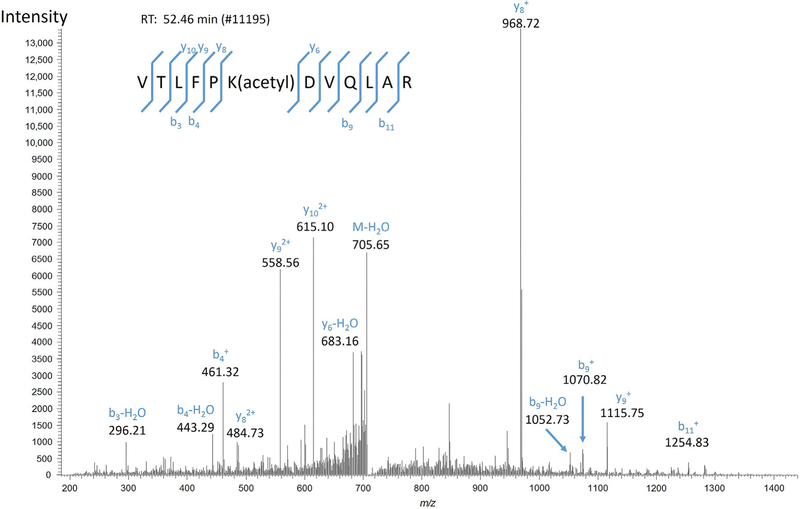
Manually validate CENP-A matches using the criteria given by Tabb, Friedman, and Ham (2006). High-confidence matches will have high-intensity fragment peaks accounted for as members of b and y or c and z ion fragment series for CID and ETD spectra, respectively. The presence of several unmatched intense peaks lowers the confidence of a match, and it is especially important that the base peak is matched (Fig. 2).
Peptide matches that have high numbers of dynamic modifications must be heavily scrutinized. Each additional dynamic modification included in a search increases the chance for random peptide matches. Search algorithms provide a score reflecting the overall quality of a match. Matches with Mascot ion scores below 30, and SEQUEST XCorrs below 2.5 should be manually validated. The SEQUEST XCorr is not normalized by peptide length so long peptides need to have higher XCorrs to be considered confident matches. Long protein cleavage products can result from missed cleavages or if a particular protein has large regions without a cleavage site.
Preliminary CENP-A matches should then be searched against a database of common lab contaminants and histones, including CENP-A. Individual spectra can be submitted to reduce search time. Spectra that still match to CENP-A in this validation search represent confident matches.
Fig. 2.
High-confidence spectral match. This confident spectrum match has its high-intensity peaks, including the base peak m/z 986.72, matched to members of the b and y ions series for the identified CENP-A peptide VTLFPK(acetyl)DVQLAR.
5. CONCLUDING REMARKS
Analysis of histone modifications presents unique challenges, and because of the high biological significance of histone mark patterns, it is a rapidly evolving field, offering new advances especially for the analysis of the complex mixtures of histones. This protocol summarizes the method that can be used for the detailed analysis of the modifications on single species of chromatin-bound histones with a combination of the TAU gel separation and mass spectrometry.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research program of NIAID, NIH, and of the NCI/CCR/NIH.
4. NOTES
Note 1: Exclusion list for common laboratory contaminants
Proteomic experiments frequently sustain some sort of contamination from ubiquitous laboratory contaminants. Any undesired proteins present in a sample will be digested into peptides, and those peptides will be subsequently selected and fragmented by the mass spectrometer. Contaminants that originate from sample handling, like keratins, can be very highly abundant and mask proteins of interest. It is possible to avoid wasting scan cycles on contaminants by using a mass-exclusion list.
A contaminants database, such as the common Repository of Adventitious Proteins (cRAP), is available from the website of The Global Proteome Machine (http://www.thegpm.org/crap). Each entry of the cRAP database has a Uniprot Accession ID number that can be entered into an in silico digest tool such as MS-Digest (http://prospector.ucsf.edu/prospector/html/instruct/digestman.htm) or PeptideMass (http://web.expasy.org/peptide_mass/). The output from these tools is a mass list of tryptic peptides, which can then be used to build an exclude list. When the mass spectrometer detects masses from the exclude list, they will not be selected for fragmentation.
Note 2: Alkylation method for ubiquitin analyses
The ubiquitination of proteins is a widely studied signaling modification. Mass spectrometry is frequently used to identify and map this modification. Duringatrypsindigestofaubiquitinatedprotein,theubiquitinmodification is cleaved, and a diglycine (MW 114.042927 Da) tag remains on any lysines that were the site of ubiquitination (Denis, Vasilescu, Lambert, Smith, & Figeys,2007).Unfortunately, protein cysteine alkylation using IDA can lead to the addition of a 2-acetamidoacetamide modification that has the same atomic composition as the diglycine that is characteristic of ubiquitination (Nielsen et al., 2008). This makes the biological modification and the chemical artifact nearly indistinguishable by mass spectrometry. A replacement alkylating agent, 2-chloroacetamide (Sigma-Aldrich, St. Louis, MO, Cat #C0267), can be used instead of IDA to avoid the diglycine artifact. Chloroacetamide can be used at concentrations of 10–55 mM, and the cysteine alkylation of samples using chloroacetamide should be performed at room temperature (21°C) in the dark.
REFERENCES
- Allfrey VG, Faulkner R, & Mirsky AE (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, & Mirsky AE (1964). Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science, 144(3618), 559 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- Audia JE, & Campbell RM (2016). Histone modifications and cancer. Cold Spring Harbor Perspectives in Biology, 8(4), a019521 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, … Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell, 129(4), 823–837. 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Devys D, & Tora L (2014). Histone H2B ubiquitination: Signaling not scrapping. Drug Discovery Today: Technologies, 12, e19–e27. 10.1016/j.ddtec.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, … Dalal Y (2012). Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell, 150(2), 317–326. 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Walkiewicz MP, Dimitriadis EK, & Dalal Y (2013). The CENP-A nucleosome: A battle between Dr. Jekyll and Mr. Hyde. Nucleus, 4(1), 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y (2003). Signals in DNA that influence chromatin structure in vitro and in vivo. PurdueUniversity Press. PhD dissertation. [Google Scholar]
- Denis NJ, Vasilescu J, Lambert JP, Smith JC, & Figeys D (2007). Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics, 7(6), 868–874. 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- Dhall A, & Chatterjee C (2011). Chemical approaches to understand the language of his-tone modifications. ACS Chemical Biology, 6(10), 987–999. 10.1021/cb200142c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, & Hunt DF (2007). Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nature Protocols, 2(4), 933–938. 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R (1990). DNA methylation and epigenetic inheritance. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 326(1235), 329–338. [DOI] [PubMed] [Google Scholar]
- Huang H, Lin S, Garcia BA, & Zhao Y (2015). Quantitative proteomic analysis of histone modifications. Chemical Reviews, 115(6), 2376–2418. 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason LJ, Moore SC, Lewis JD, Lindsey G, & Ausio J (2002). Histone ubiquitination: A tagging tail unfolds? Bioessays, 24(2), 166–174. 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, & Allis CD (2001). Translating the histone code. Science, 293(5532),1074–1080. 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, & Wold B (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science, 316(5830), 1497–1502. 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, … Bernstein BE (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature, 448(7153), 553–560. 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, & Mann M (2008). Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nature Methods, 5(6), 459–460. 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- Ryan CA, & Annunziato AT (2001). Separation of histone variants and post-translationally modified isoforms by triton/acetic acid/urea polyacrylamide gel electrophoresis. Current Protocols in Molecular Biology. 10.1002/0471142727.mb2102s45. chapter 21, Unit 21.2. [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, & Hake SB (2007). Extraction, purification and analysis of histones. Nature Protocols, 2(6), 1445–1457. 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Stein A, Dalal Y, & Fleury TJ (2002). Circle ligation of in vitro assembled chromatin indicates a highly flexible structure. Nucleic Acids Research, 30(23), 5103–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, & Allis CD (2000). The language of covalent histone modifications. Nature,403(6765), 41–45. 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tabb DL, Friedman DB, & Ham AJ (2006). Verification of automated peptide identifications from proteomic tandem mass spectra. Nature Protocols, 1(5), 2213–2222. 10.1038/nprot.2006.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (2000). Histone acetylation and an epigenetic code. Bioessays, 22(9), 836–845. . [DOI] [PubMed] [Google Scholar]
- Walkiewicz MP, Bui M, Quenet D, & Dalal Y (2014). Tracking histone variant nucleosomes across the human cell cycle using biophysical, biochemical, and cytological analyses. Methods in Molecular Biology, 1170, 589–615. 10.1007/978-1-4939-0888-2_34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg JH, & Matthews HR (1984). Fluorography of polyacrylamide gels containing tritium. Methods in Molecular Biology, 1, 147–152. 10.1385/0-89603-062-8:147. [DOI] [PubMed] [Google Scholar]
- Zhao Y, & Garcia BA (2015). Comprehensive catalog of currently documented histone modifications. Cold Spring Harbor Perspectives in Biology, 7(9), a025064 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Huang X, & Kelleher NL (2016). Epiproteomics: Quantitative analysis of histone marks and codes by mass spectrometry. Current Opinion in Chemical Biology, 33, 142–150. 10.1016/j.cbpa.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]