 Human milk, the best food for infants, is a dynamic and complex fluid that directly influences the immune system and microbiota establishment.
Human milk, the best food for infants, is a dynamic and complex fluid that directly influences the immune system and microbiota establishment.
Abstract
Human milk, the best food for infants, is a dynamic and complex fluid that directly influences the immune system and microbiota establishment. The protective role of human milk is well known although the mechanisms behind it still need to be uncovered. This study aimed to characterize the impact of human milk in the immature intestine of newborns by analyzing the global transcriptomic response of the FHs 74 int cell line (ATCC CCL-241). The expression of intestinal keratins and other genes with a well-annotated intestinal or epithelial function validated FHs 74 int derived from the fetal small intestine as a model of the intestinal epithelium of newborns. Cells exposed to skimmed human milk showed seventeen differentially expressed genes, most of them up-regulated, including four chemokine genes (CXCL1, CXCL2, CXCL3 and CXCL10) and other immune-related genes. qRT-PCR and ELISA analysis confirmed the microarray data and indicated a different pattern of expression upon milk exposure in FHs 74 int as compared to the adult tumorigenic Caco-2 cell line. The evaluation of the functional significance of these transcriptomic changes reveals that human milk exposure may contribute to the regulation of the inflammatory response in the intestine during the perinatal period, which is characterized by the immaturity of the immune system and a pro-inflammatory phenotype.
Introduction
Human milk (HM) has been recognized as the gold standard of infant nutrition. HM has also been recognized for its overall nutrient composition and also as a source of soluble factors that help protect the baby against infections, external insults and pathogenesis. Thus, breast-feeding is a key factor influencing infant health outcomes.1 In fact, a newborn's diet is not only important in preventing pathogenic conditions during the perinatal period but may also help prevent future health problems in early life, including allergies, atopy, chronic inflammation, obesity, diabetes, and autoimmune diseases.2
HM is unique in its ability to fulfill infant nutritional requirements, and its composition constantly adapts to neonatal requirements. HM contains bioactive components that directly influence the developing infant and shape the development of the intestinal microbiota, as well as immune system maturation. Beyond its nutritional composition, HM contains several nonspecific factors, such as lysozyme, lactoferrin, oligosaccharides and microbes, as well as several immune-related substances, mainly cytokines, chemokines and growth factors.3 This collection of molecules has mainly anti-inflammatory properties and is necessary for counteracting the pro-inflammatory phenotype of the newborn's immature immune system.4 The detailed roles of milk's specific bioactive molecules in modulating the newborn's immune system have been previously investigated in vitro and in vivo and are currently a subject of intense research for their translation to clinical applications. Transforming growth factor-β2 (TGF-β2) has been shown to modulate cytokine expression in intestinal epithelial cells (IECs), and the underlying molecular mechanisms have been elucidated.5 A TGF-β-supplemented formula has been successfully used for the treatment of pediatric Crohn's disease.6,7 Other milk bioactive compounds, such as HM oligosaccharides, peptides or HAMLET, have been used in clinical trials with different success rates.8 Only a few attempts have been made to describe these mechanisms from a more global and integrated point of view or employing diverse cellular models.9 Therefore, other models that are not based on animal cell lines or commonly used adult and carcinoma-derived cell lines and that closely resemble the characteristics of the newborn intestinal epithelium are required.
To characterize the functional role of HM as a whole complex food in the immature intestine of newborns, we have performed a global transcriptomic analysis using skimmed human milk (SHM) and the FHs 74 int fetal small intestine epithelial cell line. This cell line has previously been used to describe the pro-oxidant or proliferative effects of HM components on enterocytes and also as a model for Crytosporidium parvum infection.10–14 Despite these previous studies, the characteristics described for this cell line by the American Type Culture Collection (ATCC) include an absence of keratin expression, as determined by immunoperoxidase assay. Because keratins are important markers of epithelial origin in cells, our first goal was to validate the FHs 74 int cell line as a good model for the immature intestinal epithelium of newborns. Then, we investigated whether SHM has a functionally relevant transcriptomic effect on these cells.
Materials and methods
Human milk samples
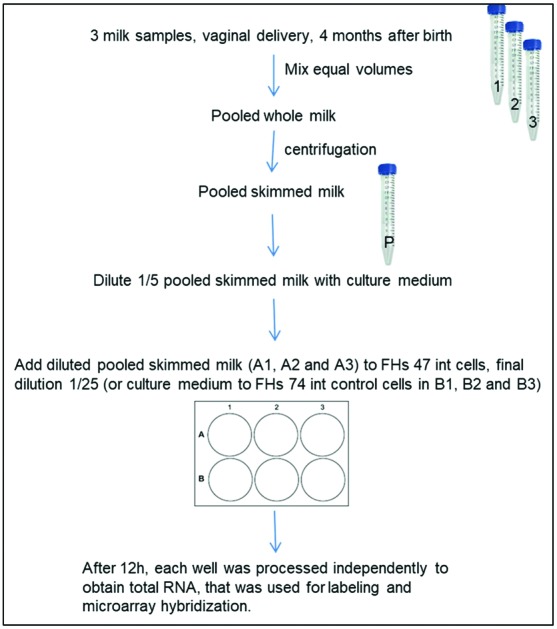
Human milk samples from healthy lactating mothers (n = 3) at 4 months after vaginal delivery were included in this study. A figure describing the overall study design is included as ESI S1.† All infants were in good health. Previous to sample collection, nipples and mammary areola were cleaned with soap and sterile water and soaked in chlorhexidine to reduce sampling of microorganisms residing on the skin. Milk samples were taken at the lactation moment with the infant, although the exact moment (beginning, during or the end) was not recorded. Although the composition may vary due to the moment of collection, the data on nutritional composition and bacterial load of the milk samples (ESI S2†) agree with previous data published by our group.15 The milk samples were collected in a sterile tube manually, discarding the first drops. All samples were frozen at –20 °C until further processing. Before sample collection, the mothers received oral and written information, and gave written informed consent to the protocol, which had been approved by the Ethics Committee of the Atencion Primaria Generalitat Valenciana (Spain), and the Bioethics Subcommittee of Consejo Superior de Investigaciones Científicas (CSIC). All experiments were carried out in accordance with relevant guidelines and regulations.
Cell culture
The FHs 74 int (ATCC CCL-241) and Caco-2 (ATCC HTB-37) cell lines were obtained from the ATCC. FHs 74 int was maintained in Hybri Care medium (ATCC 46X) reconstituted following ATCC instructions and supplemented with 10% (v/v) inactivated fetal bovine serum (Sigma Aldrich) and 45 ng mL–1 of epidermal growth factor (Corning). Cells from passage 3 were seeded onto 6-well plates (40 000 cells per well) and grown for six days until 90% confluence. The medium was changed every two days.
Caco-2 cells were cultured as previously described16 and seeded onto 6-well plates (700 000 cells per well). They were maintained in culture for eight days after seeding with frequent medium renewal.
For the SHM exposition of cell cultures, equal volumes of the HM samples were equally pooled. This mixture was centrifuged at 1200 rpm at 4 °C for 20 minutes, and the upper fat layer was removed with a sterile Pasteur pipette under sterile conditions. This SHM pool was resuspended and diluted 1/5 with complete growth medium. Just before starting the incubation time, the medium was renewed in every well with 3 mL of fresh complete medium. 750 μL of complete medium was added to the control untreated wells, whereas 750 μL of the diluted SHM pool was added to the treated wells. Cells were maintained at 37 °C under a 5% CO2 atmosphere for 12 hours. Integrity of the cell culture was checked under an optical inverted microscope before and after milk treatment.
Tumor necrosis factor alpha (TNFα) at a final concentration of 10 ng mL–1 was added to the cell cultures alone for 6 hours or during the last 6 hours of SHM treatment, as described above.
RNA purification
Cell cultures were carefully washed 5 times with PBS and lysed in the well using the RNeasy Mini Kit (Qiagen) together with the DNase on column kit (Qiagen). Genomic DNA-free total RNA from the cells was quantified using a NanoDrop ND-1000 system (Thermo Fisher).
Microarray analysis
Agilent microarrays 4 × 44k (Agilent G4845A) including 27 958 biological features (60-mer oligonucleotides, AMADID 026652) were used. A two-color experiment was designed with four biological replicates of each set of conditions (control versus treated). In order to use the control samples for the evaluation of gene expression in the untreated FHs 74 int cell line, the dye-swap was avoided so the four control samples were labeled using the red dye Cy5 and the four treated samples were labeled in green (Cy3).
Total RNA integrity was checked before and after labeling using Bioanalyzer 2100 and the RNA 6000 Nano Kit (Agilent). 150 ng of total RNA samples were amplified and labeled with the Low Input Quick Amp Labeling Kit (Agilent). The Spike-In Two Color Kit (Agilent) was used for the evaluation of the efficiency of labeling and hybridization. Hybridization and washing of the slides were carried out with the Gene Expression Hybridization Kit (Agilent) and Gene Expression Wash Buffers (Agilent) following the Agilent protocol GeneExpression_two_Color_v6.9.1. After washing and drying, the slides were scanned in an Agilent G2565AA microarray scanner, at 5 μm resolution double pass scanning, as recommended. A dual scan extended dynamic range was used. Image files were extracted with Feature Extraction Software v9.5.1 (Agilent).
Differential expression and functional analysis
Raw data obtained from scanning of the arrays were analyzed using the Babelomics suite.17 The normalization of the raw data was carried out using the normexp background correction method.18,19 The differential expression of genes after SHM treatment was detected using the limma moderated t-test20 implemented in the Babelomics suite with a multiple test correction to avoid false positive discovery.21 We selected significant genes with an adjusted p-value smaller than 0.05, where a positive statistic value means higher expression in the treated group and a negative statistic value reflects lower gene expression in the treated group.
The Gene Ontology (GO) enrichment tool FatiGO22,23 was used to convert the up-regulated gene list into lists of functional terms using annotations in the GO database.24,25 A Fisher's exact test was applied to check for significant over-representation of functional terms in the list of up-regulated genes with respect to the rest of the genome. Multiple test correction to account for the multiple hypotheses tested (one for each functional term) was applied.21
Gene set enrichment analysis (GSEA) was carried out using the complete list of the analyzed genes (not only the up-regulated ones) ranked by the statistic value obtained in the class comparison using the limma moderated t-test. GSEA points out the cooperative behavior of genes using the complete list of ids without imposing any artificial threshold with an arbitrary p-value. Gene sets with common GO annotations that correlate with high or low values of the statistic in the ranked list of genes are displayed as the result. Statistical significance of GO term enrichment was quantified by the lor statistic (logarithm of odds ratio) and the corresponding adjusted p-value (selection p-value < 0.05). GO terms with a positive lor are enriched in gene sets included in the up-regulated part of the ranked list of genes; in contrast, GO terms with a negative lor are enriched in gene sets included in the down-regulated part of the ranked list of genes.
qRT-PCR and ELISA validation of gene expression
850 ng of total RNA from cell cultures were retro-transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) and used as a template for qRT-PCR. LightCycler 480 SYBR green I Master Mix (Roche) was used in a LightCycler 480 instrument using a 96-well plate format (Roche). The oligonucleotides used as primers for quantitative qRT-PCR are listed in Table 1. All the primers selected for this work were designed using the Primer-BLAST tool at the NCBI website26 and are separated by at least one intron on the corresponding genomic DNA. This option makes it easy to distinguish between amplification from mRNA and genomic DNA. We used ACTB and GAPDH genes as housekeeping reference genes. All the genes gave a single amplicon according to the melting curves of the amplified products carried out at the end of the amplification process. The Cp values were calculated for each amplification curve using the second derivative maximum method implemented in LightCycler Software release 1.5.1.62. Efficiencies of the PCR reactions were calculated for every amplification curve corresponding to a single well in the plate, using LinRegPCR Software version 2017.0.27 Mean efficiency for all the wells corresponding to the same gene in the same plate was used for further calculations.
Table 1. Oligonucleotide sequences used as primers for quantitative qRT-PCR.
| Gene | Sequence 5′ → 3′ | Ref. |
| ACTB-f | ATGCTATCACCTCCCCTGTGTG | 52 |
| ACTB-r | TTGTTACAGGAAGTCCCTTGCC | |
| GAPDH-f | GTCTCTCTCTTCCTCTTGTGCTCTTGC | 53 |
| GAPDH-r | TCCTCTGACTTCAACAGCGACACC | |
| FOS-f | CAGACTACGAGGCGTCATCC | This work |
| FOS-r | CGTGGGAATGAAGTTGGCAC | |
| ANGPTL4-f | GAAATCCAGCCTCAGGGGTC | This work |
| ANGPTL4-r | AAGTCCACTGAGCCATCGTG | |
| CXCL2-f | CTTGTCTCAACCCCGCATC | This work |
| CXCL2-r | CAGGAACAGCCACCAATAAGC | |
| CXCL3-f | AGCCACACTCAAGAATGGG | This work |
| CXCL3-r | GCAGGAAGTGTCAATGATACGC | |
| CXCL10-f | GTGGCATTCAAGGAGTACCTC | 54 |
| CXCL10-r | GCCTTCGATTCTGGATTCAGACA | |
| CDC69-f | CCTGGTCACCCATGGAAGTG | This work |
| CDC69-r | GCTGCTGACCTCTGTGTTTTTC | |
| PTX3-f | TCTCTGGTCTGCAGTGTTGG | 55 |
| PTX3-r | TGAAGAGCTTGTCCCATTCC | |
| IL6-f | GTGTGAAAGCAGCAAAGAGGC | 56 |
| IL6-r | TGCAGGAACTGGATCAGGACT | |
| IL8-f | ACTTCCAAGCTGGCCG | This work |
| IL8-r | CCTTGGGGTCCAGACAGAG |
Relative Expression Software Tool (REST 2009 v2.0.13) was used for analyzing gene expression using real-time amplification data. The software addresses issues surrounding the measurement of uncertainty in expression ratios by introducing randomization and bootstrapping techniques.28
The concentration of the protein products corresponding to differentially expressed genes was measured in the extracellular media of the cell cultures by ELISA (eBioscience) following the manufacturer's instructions.
Data availability
The data sets generated during the current study are available in the ArrayExpress public repository at EMBL-EBI with accession number E-MTAB-6101 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6101).
Results
Transcriptomic characterization of the FHs 74int cell line
Agilent microarrays were used to design a two-color experiment in which the control samples were labeled with the red dye cyanine Cy5 as described in the Materials and methods section. The fluorescence signal that had been background corrected and obtained from the untreated control samples was used as an estimation of the relative expression levels of genes in FHs 74 int cells. This red processed signal (“rProcessedSignal” column in the data table obtained from the scanner; ArrayExpress number E-MTAB-6101) was used for further analysis.
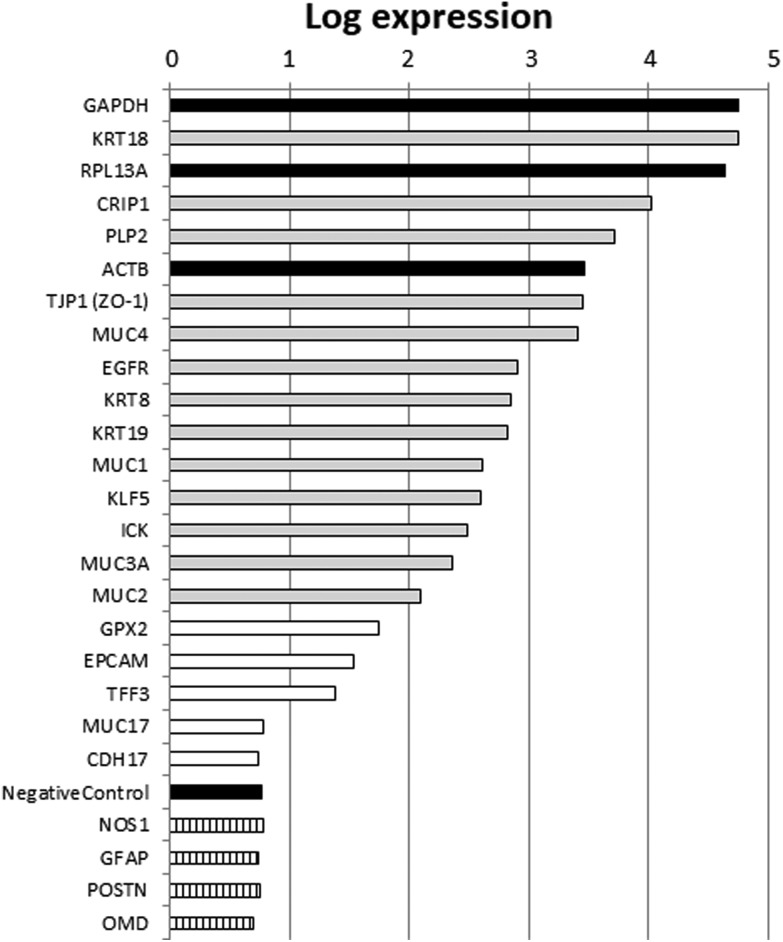
Based on the annotation of the probes represented in the array, we selected a list of genes with well-described intestinal or epithelial functions. We also represented in that list housekeeping genes with high expression levels (ACTB, GAPDH, RPL13A), non-intestinal expression controls and negative controls included in the array. The values of the red processed signal from that list of genes are presented in Fig. 1. Selected nervous or bone tissue marker genes, which were used as the non-intestinal expression controls, were expressed at levels similar to those of the negative controls included in the array. High expression levels comparable to those of the housekeeping genes were obtained for genes with well-annotated intestinal or epithelial functions, which corroborated the intestinal epithelial origin of FHs 74 int fetal cells. Therefore, these results suggest that the FHs 74 int cell line is a good model to use in determining HM's influence on transcriptomic changes in the intestinal epithelium of newborns.
Fig. 1. Relative expression for genes with well-annotated intestinal epithelium features in FHs 74 int untreated cells. Gene expression (average of four biological replicas) was estimated by red processed fluorescence signal from microarrays. Black bars indicate housekeeping genes or negative controls. Hatched bars indicate nervous (NOS1, GFAP) or bone (POSTN, OMD) tissue marker genes. Grey bars correspond to intestinal epithelium marker genes with high expression levels. White bars represent intestinal epithelium marker genes with lower gene expression.
Differential gene expression of fetal intestinal cells in response to skimmed human milk
Seventeen genes that were differentially expressed upon SHM treatment are listed in Table 2. Only one gene was down-regulated (POLR2L, RNA polymerase II polypeptide L), while the remaining sixteen were up-regulated. A functional enrichment analysis carried out using the up-regulated gene list revealed the significant over-representation of functional annotations, mainly those related to immune function (Table 3). Among these, Cell chemotaxis (GO:0060326) and Response to molecule of bacterial origin (GO:0002237) are the most significant.
Table 2. Genes differentially expressed (adjusted p-value < 0.05) upon skimmed human milk treatment in FHs 74 int fetal intestinal cells.
| Gene name | Description | Statistic | Adj. p-value | Regulation |
| ANGPTL4 | Angiopoietin-like 4, transcript variant 1, [NM_139314] | 28.6 | 0.004 | UP |
| CXCL3 | Chemokine (C–X–C motif) ligand 3, [NM_002090] | 18.1 | 0.012 | UP |
| FOS | FBJ murine osteosarcoma viral oncogene homolog, [NM_005252] | 17.9 | 0.012 | UP |
| CXCL10 | Chemokine (C–X–C motif) ligand 10, [NM_001565] | 17.5 | 0.012 | UP |
| ABCD3 | ATP-binding cassette, sub-family D (ALD), member 3, transcript variant 2, [NM_001122674] | 17.4 | 0.012 | UP |
| RGS18 | Regulator of G-protein signaling 18, [NM_130782] | 16.9 | 0.012 | UP |
| PTX3 | Pentraxin 3, long, [NM_002852] | 16.8 | 0.012 | UP |
| FBXL5 | F-box and leucine-rich repeat protein 5, transcript variant 1, [NM_012161] | 16.1 | 0.014 | UP |
| CD69 | CD69 molecule, [NM_001781] | 14.0 | 0.026 | UP |
| CXCL2 | Chemokine (C–X–C motif) ligand 2, [NM_002089] | 13.7 | 0.026 | UP |
| PDK4 | Pyruvate dehydrogenase kinase, isozyme 4, [NM_002612] | 13.7 | 0.026 | UP |
| C15orf48 | Chromosome 15 open reading frame 48, transcript variant 2, [NM_032413] | 13.4 | 0.026 | UP |
| PAQR5 | Progestin and adipoQ receptor family member V, transcript variant 1, [NM_001104554] | 12.1 | 0.043 | UP |
| CXCL1 | Chemokine (C–X–C motif) ligand 1 (melanoma growth stimulating activity, alpha), transcript variant 1, [NM_001511] | 11.8 | 0.043 | UP |
| IL6 | Interleukin 6, [NM_000600] | 11.7 | 0.043 | UP |
| POLR2L | Polymerase (RNA) II (DNA directed) polypeptide L, [NM_021128] | –11.9 | 0.043 | DOWN |
| HIST1H1A | Histone cluster 1, H1a, [NM_005325] | 11.6 | 0.044 | UP |
Table 3. Functional enrichment analysis carried out with the up-regulated gene list (adjusted p-value ≤ 0.05). Lor: logarithm of odds ratio.
| GO term | Gene names | Lor | Adj. p-value |
| Response to cold (GO:0009409) | FOS, CXCL10 | 6.55 | 0.0076 |
| Positive regulation of leukocyte chemotaxis (GO:0002690) | IL6, CXCL10 | 5.48 | 0.0465 |
| Cell chemotaxis (GO:0060326) | IL6, CXCL10, CXCL2, CXCL3, CXCL1 | 5.25 | 0.0000 |
| Leukocyte chemotaxis (GO:0030595) | IL6, CXCL10, CXCL3 | 5.03 | 0.0054 |
| Response to molecule of bacterial origin (GO:0002237) | IL6, FOS, CXCL10, CXCL2 | 4.99 | 0.0002 |
| Response to lipopolysaccharide (GO:0032496) | IL6, FOS, CXCL10 | 4.70 | 0.0076 |
| Regulation of angiogenesis (GO:0045765) | IL6, CXCL10, ANGPTL4 | 4.69 | 0.0076 |
| Regulation of vasculature development (GO:1901342) | IL6, CXCL10, ANGPTL4 | 4.63 | 0.0076 |
Gene set enrichment analysis (GSEA) was carried out on the complete gene list to determine whether immune-related functional annotation enrichment could be confirmed by a complementary analysis. The results from GSEA extended the diversity of immune-related functional terms that were significantly enriched. Twenty-six terms corresponding to the molecular function ontology and six terms corresponding to the cellular component ontology were significantly enriched, while the terms corresponding to the biological process ontology included 1528 enriched annotations. The complete list of enriched Gene Ontology (GO) terms and the corresponding gene sets are listed in ESI S3.† Among all the enriched GO terms resulting from GSEA, selections were made based on the significance and relationship to immune function, among other criteria. The selected items (Table 4) confirmed that SHM addition to FHs 74 int cells in culture produced changes in gene expression that could regulate immune system function.
Table 4. Selected GO terms significantly enriched in GSEA. Statistical significance of enrichment is quantified by the lor statistic (logarithm of odds ratio) and the corresponding adjusted p-value (selection p-value < 0.05).
| GO term | Gene set/terms in the genome | Lor | Adj. p-value |
| CXCR3 chemokine receptor binding (GO:0048248) | 5/6 | 0.32 | 0.000001 |
| Negative regulation of tumor necrosis factor-mediated signaling pathway (GO:0010804) | 10/24 | 0.24 | 0.000246 |
| Positive regulation of interleukin-6 secretion (GO:2000778) | 6/11 | 0.23 | 0.007230 |
| Interleukin-6 production (GO:0032635) | 78/161 | 0.11 | 0.022169 |
| Negative regulation of SMAD protein complex assembly (GO:0010991) | 5/17 | –0.61 | 0.000009 |
| Negative regulation of pathway-restricted SMAD protein phosphorylation (GO:0060394) | 10/30 | –0.73 | 0.000000 |
Effect of TNFα stimulation after skimmed human milk exposure in FHs 74 int cells
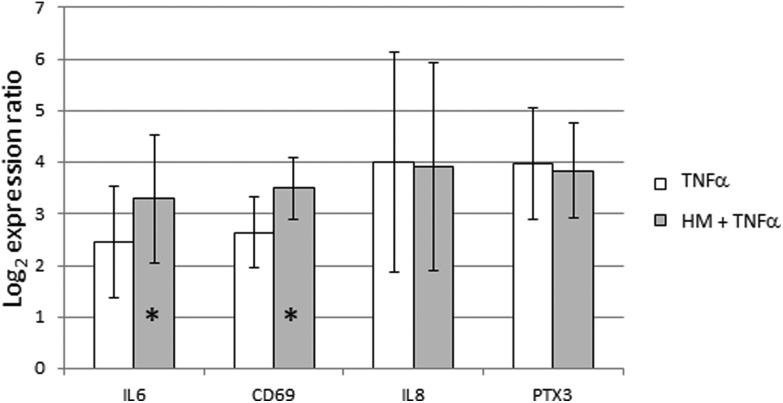
The GO term Negative regulation of tumor necrosis factor-mediated signaling pathway (GO:0010804) was significantly enriched in GSEA with a lor value of 0.24. This result prompted us to characterize the likely protective effect of SHM on the intestinal epithelium. TNFα stimulation was performed alone or during the last 6 hours of SHM addition to FHs 74 int cells. All the genes that had been previously shown to be induced by SHM addition were also induced by TNFα alone. The pre-treatment with SHM did not reduce the level of gene expression induced by TNFα alone. Indeed, IL6 and CD69 induction by TNFα was more intense after the SHM pre-treatment. However, this intensifying effect on gene expression was not observed for IL8 or PTX3 genes (Fig. 2).
Fig. 2. Effect of TNFα stimulation of gene expression after SHM exposure in FHs 74 int cells. Expression ratios of IL6, CD69, IL8 and PTX3 immune-related genes are represented. The addition of TNFα to a final concentration of 10 ng mL–1 was carried out for 6 hours, alone or during the last 6 hours of SHM treatment (SHM pre-treatment). Bars represent mean of the log2 expression ratio, and lines represent standard deviation. * p-Value < 0.05.
Fetal and adult intestinal epithelial cells display different gene expression responses to skimmed human milk
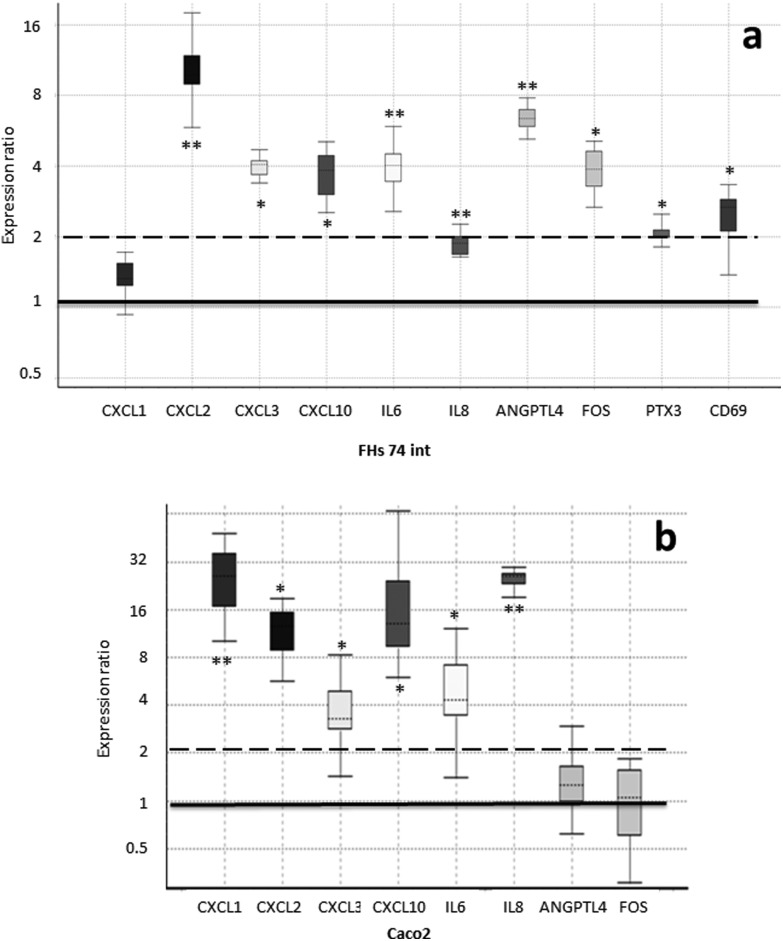
In order to confirm the array expression data and to compare the expression response to SHM on the parts of fetal and adult IECs, we analyzed the expression levels of genes in the FHs 74 int and Caco-2 cell lines upon SHM treatment via quantitative real-time PCR (qRT-PCR). For this purpose, we employed those genes that were responsible for a single enrichment of functional terms (Table 3). This list was further extended to include IL8 (interleukin-8), CD69 transmembrane receptor and PTX3 (pentraxin 3) due to their important roles in immune response. The expression fold change as compared to the untreated controls is represented in Fig. 3.
Fig. 3. Expression of genes which were responsible for the enrichment of functional terms was analyzed by qRT-PCR. The fold change expression of the SHM-treated cells compared to the untreated controls is represented in a box–whisker plot. The box represents the interquartile range (middle 50% of the observations), the dotted line represents the sample median and the whiskers represent the outer 50% of the observations. The black bar is set at an expression ratio of 1 (no change) and the dashed bar represents the threshold for fold change greater than 2. a. Fetal IECs from the FHs 74 int cell line. b. Adult IECs from the Caco-2 cell line. * p-Value < 0.05, ** p-value < 0.01.
The upregulation of all these genes, except for CXCL1 (C–X–C motif chemokine ligand 1), was confirmed via RT-qPCR in FHs 74 int cells upon SHM treatment, with a fold change greater than 2, except for IL8, which was still significantly up-regulated with a fold change of 1.9. Significantly higher concentrations of IP-10 (23 vs. 9 pg mL–1, p value < 0.0001), the protein product of the CXCL10 (C–X–C motif chemokine ligand 10) gene, and secreted IL-6 (interleukin-6) (333 vs. 242 pg mL–1, p value < 0.05) were measured by ELISA in extracellular media from cultures of SHM-treated versus non-treated FHs 74 int cells. However, adult IECs from the Caco-2 cell line displayed a different pattern of gene expression upon SHM treatment: CD69 and PTX3 expression could not be detected, while FOS1 (FBJ murine osteosarcoma viral oncogene homolog) and ANGPTL4 (angiopoietin-like 4) expression did not change significantly. Remarkably, in contrast to FHs 74 int cells, IL8 and CXCL1 (C–X–C motif chemokine ligand 1) are significantly upregulated with a fold change of 25, while IL6, CXCL2 (C–X–C motif chemokine ligand 2), CXCL3 (C–X–C motif chemokine ligand 3) and CXCL10 were up-regulated after SHM exposure in a similar way in both cell lines (Fig. 3).
Discussion
The FHs 74 int cell line has previously been used as a model for normal non-transformed IECs.10–12,29,30 The description of this cell line in the ATCC indicates that keratins could not be detected in these cells via immunoperoxidase assay and that they have an epithelial-like morphology (characteristics available from ; https://www.lgcstandards-atcc.org/products/all/CCL-241.aspx#characteristics). In addition to this, FHs 74 int cells do not form tight monolayers, as we could appreciate based on our experiments. Other human fetal non-transformed small intestine primary epithelial cells that have previously been characterized do not form tight junctions or polarize.31 For all these reasons, we wanted to validate the FHs 74 int cell line as an adequate model for the intestinal epithelium of newborns, investigating its general expression profile. To our knowledge, global transcriptome analysis is not publicly available for this cell line, so our work represents the first FHs 74 int global transcriptome derived using microarray data.
We took advantage of the two-color Agilent technology and used the red processed signal corresponding to the untreated controls as an estimate of the expression values of the genome. Selected nervous or bone tissue marker genes, namely NOS1 (nitric oxide synthase 1), GFAP (glial fibrillary acidic protein), POST (periostin) and OMD (osteomodulin), which were used as non-intestinal expression controls, were expressed at as low a level as the negative controls included in the array. Among the selected genes with a well-annotated intestinal or epithelial function, KRT18, KRT8 and KRT19, which code for the principally expressed keratins in the intestinal epithelium,32 were highly expressed, which would stand in contrast to the previous data of the ATCC regarding the detection of keratins in these cells. In this respect, we have measured mRNA levels, but we have not measured protein levels. This may explain the differences observed in the expression data of keratins and should be taken into account for further analysis. The expression of TJP-1 (tight junction protein 1), which codes for ZO-1, a protein involved in cytoplasmic plaque assembly in the apical junction complex,33 is well detected in the intestinal epithelium, and it was also identified in FHs 74 int cells. Defects in the forming of a tight monolayer in FHs 74 int cells may be due to the low expression levels of other integral components of tight junctions (e.g., claudin expression levels were low) or lower expression levels of other intercellular adhesion proteins, such as EPCAM (epithelial cell adhesion molecule) or CDH17 (cadherin 17). Decreased epithelial barrier function in preterm infants and newborns up to 6 months of age has been previously described,34,35 which would invalidate the FHs 74 int cell line's use in nutrient or drug transport studies. The expression of the genes coding for mucins (MUC1, MUC2, MUC3A, and MUC4) was also detected to various extents. However, the expression of MUC17, a mucin highly present in adult small intestine tissue,36 is not detected in FHs 74 int fetal cells. High expression levels were also detected for EGFR (epidermal growth factor receptor) and KLF5 (Kruppel-like factor 5). This transcription factor has been ascribed to functional annotation GO:0030033, which is related to the assembly of microvilli, a characteristic cellular structure of enterocytes.
Other genes that were highly expressed in FHs 74 int cells were PLP2, CRIP1 and ICK. The expression pattern detected was in agreement with the small intestine expression pattern detected in publicly available RNA-seq experiments.36 Taking into account the expression patterns of all the intestinal epithelium marker genes analyzed, we conclude that although FHs 74 int cells should not be employed in transport studies due to their inability to form tight monolayers, the expression data support the use of FHs 74 int cells as a model to investigate HM's influence on the transcriptome in the intestinal epithelium of newborns.
We treated the cells that had been stored frozen from the collection time until the day of the experiment with SHM. Although the concentration of bioactive proteins in SHM may not vary during the prolonged freezing period,37,38 the well-known denaturing effect of freeze-thawing could influence the bioactivity of the SHM samples used in our study. In addition, fat-soluble/associated molecules with potential bioactivity (milk fat globule membrane proteins, glycosphingolipids, phospholipids) may have been reduced through the skimming process. Taking into account the storage and processing conditions of the HM samples, the addition of SHM to FHs 74 int cells in culture resulted in significant changes in the expression levels of 17 genes, mostly those related to immune function (Table 2). The up-regulation of genes responsible for a single enrichment of functional terms (Table 3) was confirmed via qRT-PCR in FHs 74 int cells upon SHM treatment (Fig. 3). We also included IL8, CD69 and PTX3 because they have important roles in immune response. In the case of IL6, which was up-regulated with a fold change of 4 in FHs 74 int cells, this expression pattern agrees with the GSEA functional enrichment of the GO terms Positive regulation of interleukin-6 production (GO:0032755) and Positive regulation of interleukin-6 secretion (GO:2000778), with lor values of 0.16 and 0.23, respectively. This result links the up-regulation of IL6 gene expression with the increased expression of genes related to IL6 production and secretion in FHs 74 int cells after SHM addition. The secreted IL-6 measured in the extracellular culture media indicated that protein levels were also higher after SHM exposure, which also validates the results of GO functional terms enriched in GSEA.
Genes that are up-regulated upon SHM treatment, such as PTX3 and CD69, are involved in immune-related functions. PTX3 acts as a fluid phase pattern recognition molecule, participating in the opsonization of pathogens during innate immune response39 and also as a regulator of inflammation by diminishing leukocyte recruitment at sites of inflammation under pathological conditions.40 The up-regulation of the PTX3 expression after SHM treatment may contribute to the attenuation of the pro-inflammatory phenotype of immature IECs in newborns. The promoters of human and murine PTX3 genes have potential binding sites for many transcription factors, including AP-1 (activator protein 1). Remarkably, the gene coding for the FOS protein, which participates in AP-1 formation, is also up-regulated upon milk treatment, along with its potential target, PTX3, in FHs 74 int cells in culture (Table 2).
The CD69 gene also contains elements that are responsive to transcription factor AP-1, which is formed by the FOS and JUN proteins.41 The up-regulation of FOS1 gene expression after milk treatment may contribute to the up-regulation of CD69 gene expression in IECs. The CD69 gene codes for a membrane receptor that is expressed in T-cells. CD69 plays a complex role in immunity depending on the context. In addition to its role in tissue damage processes under various inflammatory conditions, it has been proposed to function as an activation factor of various leukocyte subsets. On the other hand, in humans, CD69 negatively regulates immune reactivity, and its deficiency results in an enhanced risk of autoimmune and chronic inflammatory diseases.41 The up-regulated expression of CD69 in FHs 74 int cells after SHM treatment may reflect an anti-inflammatory effect and points to immature human IECs, in addition to T-cells, but not adult enterocytes like Caco-2, as producers of CD69. There are cases of proteins usually associated with immune cells that have also a role in other cell types. As an example, TLRs are membrane receptors involved both in the development and differentiation of T-cell subsets42 and also in the recognition of molecules of pathogenic origin in the membrane of IECs.43 In other cases, CD26/DPP4 (dipeptidyl peptidase 4), which is a molecule involved in the activation of T-cells,44 has also been detected in exosomes originating from IECs.45 The role of CD69 expression in immature human enterocytes requires further investigation.
We wanted to determine whether gene expression changes were reflecting any significant enrichment of specific functions. For this purpose, we used two different types of analysis. We carried out the enrichment of GO terms within the significantly up-regulated genes, and we also investigated which functions were enriched by using gene set enrichment analysis (GSEA) to reveal gene sets that although they did not significantly change their expression after SHM addition, did display cooperative behavior and would be responsible for enrichment in common functional terms. Both kinds of analysis provided complementary information about targeted functions upon SHM exposure, many of them related to immunity.
Despite the fact that only sixteen genes were significantly up-regulated, the functional enrichment analysis carried out within this up-regulated gene list pointed to their close functional relationships. It showed the significant enrichment of GO terms related to immune function, such as Response to molecule of bacterial origin (GO:0002237), which is a direct parent of Response to lipopolysaccharide (GO:0032496). IL6, FOS, CXCL2 and CXCL10 are the up-regulated genes responsible for this functional enrichment. In addition to IL6, CXCL2 and CXCL10, CXCL1 and CXCL3 are also the up-regulated genes responsible for the significant functional enrichment of the term Cell chemotaxis (GO:0060326), which is a direct parent of Leukocyte chemotaxis (GO:0030595) and Positive regulation of leukocyte chemotaxis (GO:0002690). Therefore, the SHM treatment of human fetal IECs results in the up-regulated expression of genes functionally related to Response to molecule of bacterial origin and also to Positive regulation of leukocyte chemotaxis. As mentioned above, PTX3 can act locally to dampen excessive leukocyte recruitment, and the gene expression-dependent PTX3 production has also been proposed as a negative feedback loop acting systemically.40 One explanation for this is that HM would allow the regulation of the pro-inflammatory phenotype in the intestine of newborns5 and, secondarily, may contribute to immunotolerance towards the milk microbiota.
Although not related to the immune system, other significantly up-regulated genes have functions that are relevant to intestinal physiology or nutritional sensing. The most significant difference in gene expression corresponded to ANGPTL4 (6-fold up-regulation, Fig. 3). When we carried out a functional enrichment analysis with the up-regulated gene list, ANGPTL4 was responsible for the significant enrichment of the GO terms Regulation of angiogenesis (GO:0045765) and Regulation of vasculature development (GO:1901342), together with IL6 and CXCL10 (Table 3). ANGPTL4 is also involved in nutrient sensing when it is produced as a peptide hormone in the intestinal tract. This factor has been shown to be involved in the regulation of lipid homeostasis and has been proposed as a circulating mediator between the gut microbiota and fat storage in adipose tissue. It has been shown that the expression of ANGPTL4 in several human colonic cell lines can be stimulated by specific probiotic strains of bacteria.46,47 These results are in agreement with the fact that in our experimental setup, the HM samples were simply skimmed. Therefore, the microbial component of HM48 was present during incubation with FHs 74 int cells.
Since immunity seemed to play a central role in the functional response of the intestinal epithelium to SHM, we therefore used GSEA to shed light on the hypothesis that SHM attenuates the pro-inflammatory phenotype of the FHs 74 int cell line. Out of all the enriched GO terms resulting from GSEA, we selected the terms in view of their significance and relation to immune function, among other criteria. Remarkably, CXCR3 chemokine receptor binding (GO:0048248) is the most significant annotation of the molecular function ontology, with a statistical value of 0.32. The gene set responsible for this functional enrichment (CXCL9, CXCL10, CXCL11, CXCL13 and PF4, Table 4) accounts for five of the six genes annotated for this function in the human genome. The up-regulation of CXCL10 was confirmed by qRT-PCR and ELISA. CXCR3 engagement by CXCL10 produces a Th1-type inflammation loop that amplifies effector responses, but it has also been proposed to recruit T regulatory cells into peripheral tissues generating suppressive T cell responses. One potential explanation for this result is that SHM enhances the expression of chemokines in FHs 74 int cells, with these chemokines being ligands of CXCR3 in T-cells, thus leading to the regulation of T-cell function and trafficking.49 Since the GO term Negative regulation of tumor necrosis factor-mediated signaling pathway (GO:0010804) was significantly enriched in GSEA we wanted to characterize the likely protective effect of SHM on the intestinal epithelium. However, when we pre-treated FHs 74 int cells with SHM and then stimulated them with TNFα, we could not detect a reduction in gene expression. While an intensifying effect was detected on IL6 and CD69 expression, this additive effect was not observed in the case of IL8 or PTX3 expression, which could be interpreted as a certain containment effect of SHM on the expression of these genes.
Remarkably, the expression patterns of the selected functionally relevant genes were found to be different when we compared human fetal non-transformed normal small intestine epithelial cells (FHs 74 int cell line) with human adult colonic adenocarcinoma epithelial cells (Caco-2 cell line). In the Caco-2 cells, CD69 and PTX3 expression could not be detected, while FOS1 and ANGPTL4 expression did not change significantly. In contrast to FHs 74 int cells, IL8 and CXCL1 (C–X–C motif chemokine ligand 1) are significantly upregulated with a fold change of 25 in Caco-2 (Fig. 3). Differences between the FHs 74 int and colon cancer cell lines have previously been reported.50,51 In this case, the authors observed that the anti-proliferative effects of thymoquinone on the Caco-2 or HT-29 cell lines were not observed in the normal intestine FHs 74 int cell line. In relation to differences in the gene expression profile between normal and tumorigenic intestinal cell lines, in our study, fetal FHs 74 int cells and adult Caco-2 cells displayed different transcriptional profiles of immune-related genes after incubation with SHM. Other authors have reported that in primary fetal human IECs and also in the fetal H4 cell line, IL-1β (interleukin-1β) induced a significant increase in IL-8 secretion, which could be attenuated by the addition of TGF-β2. The adult untransformed IEC line NCM460 also displayed an IL-1β-induced increase in IL-8 secretion, but unlike immature fetal IECs, the mature enterocytes were unaffected by TGF-β2.5 Our results reinforce the need for suitable cellular models in order to obtain biologically relevant data regarding the role of HM in the physiology of the newborn intestinal epithelium. In addition to the tumorigenic versus normal origin and maturity (fetal versus adult), other factors such as the developmental stage may also be relevant to the detected gene expression response. Caco-2 cell cultures form a differentiated epithelium after achieving confluency, while the slow-growing FHs 74 int cells had reached 90% confluency at the initiation of our experiment.
In conclusion, we have demonstrated that FHs 74 int cells can be an appropriate model of the immature intestine. In contrast to the response displayed in adult tumorigenic cell lines, the incubation of fetal intestinal cells with SHM results in the increased expression of immune-related genes that may contribute to the regulation of the inflammatory response in the intestine during the perinatal period, which is characterized by the immaturity of the immune system and a pro-inflammatory phenotype. Further research is needed to dissect the contributions of the many active components of HM (e.g., cytokines, hormones, oligosaccharides and bacteria) to the observed effects. This will help in constructing a clearer picture of the impact of breast-feeding on the functionality and development of the newborn intestine.
Author contributions
J. V. G.-A. and M. C. C. both planned the experiments and wrote the manuscript, and both accepted the final version of the manuscript. J. V. G.-A. performed the experiments and the data analyses.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Material
Acknowledgments
This research has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (ERC starting grant, n° 639226). J. V. Gimeno-Alcañiz and M. C. Collado would also like to acknowledge the AGL2015-707487-P grant from the Spanish Government's Ministry of Economy and Competitiveness (MINECO). The authors also acknowledge Dr Lorena Latorre from the genomics facility of the Institute for Plant Molecular and Cell Biology (IBMCP-CSIC) for the training in the analysis of microarray results and for revising the Materials and methods section of the manuscript.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8fo01107a
References
- Gomez-Gallego C., Garcia-Mantrana I., Salminen S., Collado M. C. Semin. Fetal Neonatal Med. 2016;21:400–405. doi: 10.1016/j.siny.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Guaraldi F., Salvatori G. Front. Cell. Infect. Microbiol. 2012;2:94. doi: 10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Santaella M., Mira-Pascual L., Martinez-Arias E., Khodayar-Pardo P., Ros G., Martinez-Costa C. Nutrients. 2015;7:8577–8591. doi: 10.3390/nu7105415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg D. S., Walker W. A. Pediatr. Res. 2007;61:2–8. doi: 10.1203/01.pdr.0000250274.68571.18. [DOI] [PubMed] [Google Scholar]
- Rautava S., Nanthakumar N. N., Dubert-Ferrandon A., Lu L., Rautava J., Walker W. A. Neonatology. 2011;99:192–201. doi: 10.1159/000314109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A. S., Whitten K. E., Lemberg D. A., Clarkson C., Vitug-Sales M., Jackson R., Bohane T. D. J. Gastroenterol. Hepatol. 2006;21:1609–1614. doi: 10.1111/j.1440-1746.2006.04294.x. [DOI] [PubMed] [Google Scholar]
- Fell J. M. E., Paintin M., Arnaud-Battandier F., Beattie R. M., Hollis A., Kitching P., Donnet-Hughes A., Macdonald T. T., Walker-Smith J. A. Aliment. Pharmacol. Ther. 2001;14:281–289. doi: 10.1046/j.1365-2036.2000.00707.x. [DOI] [PubMed] [Google Scholar]
- Hill D. R., Newburg D. S. Nutr. Rev. 2015;73:463–476. doi: 10.1093/nutrit/nuv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais M., Fortier M., Pouliot Y., Gauthier S. F., Boutin Y., Asselin C., Lessard M. Br. J. Nutr. 2015;113:200–211. doi: 10.1017/S0007114514003481. [DOI] [PubMed] [Google Scholar]
- Elisia I., Tsopmo A., Friel J. K., Diehl-Jones W., Kitts D. D. J. Nutr. 2011;141:1417–1423. doi: 10.3945/jn.111.139451. [DOI] [PubMed] [Google Scholar]
- Takeda T., Sakata M., Minekawa R., Yamamoto T., Hayashi M., Tasaka K., Murata Y. J. Endocrinol. 2004;181:449–457. doi: 10.1677/joe.0.1810449. [DOI] [PubMed] [Google Scholar]
- Varughese E. A., Bennett-Stamper C. L., Wymer L. J., Yadav J. S. J. Microbiol. Methods. 2014;106:47–54. doi: 10.1016/j.mimet.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Varughese E. A., Kasper S., Anneken E. M., Yadav J. S. PLoS One. 2015;10:e0142219. doi: 10.1371/journal.pone.0142219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl-Jones W. L., Askin D. F., Friel J. K. Breastfeed. Med. 2007;2:209–218. doi: 10.1089/bfm.2007.0017. [DOI] [PubMed] [Google Scholar]
- Boix-Amorós A., Collado M. C., Mira A. Front. Microbiol. 2016;7:492. doi: 10.3389/fmicb.2016.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäuerl C., Pérez-Martínez G., Yan F., Polk D. B., Monedero V. J. Mol. Microbiol. Biotechnol. 2011;19:231–241. doi: 10.1159/000322233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R., Salavert F., Garcia-Garcia F., Carbonell-Caballero J., Bleda M., Garcia-Alonso L., Sanchis-Juan A., Perez-Gil D., Marin-Garcia P., Sanchez R., Cubuk C., Hidalgo M. R., Amadoz A., Hernansaiz-Ballesteros R. D., Aleman A., Tarraga J., Montaner D., Medina I., Dopazo J. Nucleic Acids Res. 2015;43:W117–W121. doi: 10.1093/nar/gkv384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., Smyth G. K. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Silver J. D., Ritchie M. E., Smyth G. K. Biostatistics. 2009;10:352–363. doi: 10.1093/biostatistics/kxn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. J. R. Stat. Soc. Series B, Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Al-Shahrour F., Arbiza L., Dopazo H., Huerta-Cepas J., Minguez P., Montaner D., Dopazo J. BMC Bioinf. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F., Diaz-Uriarte R., Dopazo J. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology C. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., van den Hoff M. J., Moorman A. F. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarwal A., Agarwal R., Singh R. P. Mol. Carcinog. 2017;56:499–514. doi: 10.1002/mc.22512. [DOI] [PubMed] [Google Scholar]
- Waseem T., Duxbury M., Ashley S. W., Robinson M. K. Peptides. 2014;52:113–121. doi: 10.1016/j.peptides.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Claud E. C., Savidge T., Walker W. A. Pediatr. Res. 2003;53:419–425. doi: 10.1203/01.PDR.0000050141.73528.AD. [DOI] [PubMed] [Google Scholar]
- Majumdar D., Tiernan J. P., Lobo A. J., Evans C. A., Corfe B. M. Int. J. Exp. Pathol. 2012;93:305–318. doi: 10.1111/j.1365-2613.2012.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N. N., Eddington N. D., Fasano A. Adv. Drug Delivery Rev. 2006;58:15–28. doi: 10.1016/j.addr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Dall'Asta C., Florio P., Lammardo A. M., Prandi B., Mazzeo T., Budelli A., Pellegrini N. Int. J. Food Sci. Nutr. 2015;66:409–415. doi: 10.3109/09637486.2015.1042846. [DOI] [PubMed] [Google Scholar]
- Poquet L., Wooster T. J. Mol. Nutr. Food Res. 2016;60:1876–1895. doi: 10.1002/mnfr.201500883. [DOI] [PubMed] [Google Scholar]
- Duff M. O., Olson S., Wei X., Garrett S. C., Osman A., Bolisetty M., Plocik A., Celniker S. E., Graveley B. R. Nature. 2015;521:376–379. doi: 10.1038/nature14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksu T., Atalay Y., Türkyılmaz C., Gülbahar Ö., Hirfanoğlu I. M., Demirel N., Önal E., Ergenekon E., Koç E. J. Matern.-Fetal Neonat. Med. 2015;28:1799–1802. doi: 10.3109/14767058.2014.968844. [DOI] [PubMed] [Google Scholar]
- Chang J.-C., Chen C.-H., Fang L.-J., Tsai C.-R., Chang Y.-C., Wang T.-M. Pediatr. Neonatol. 2013;54:360–366. doi: 10.1016/j.pedneo.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Mantovani A., Valentino S., Gentile S., Inforzato A., Bottazzi B., Garlanda C. Ann. N. Y. Acad. Sci. 2013;1285:1–14. doi: 10.1111/nyas.12043. [DOI] [PubMed] [Google Scholar]
- Deban L., Russo R. C., Sironi M., Moalli F., Scanziani M., Zambelli V., Cuccovillo I., Bastone A., Gobbi M., Valentino S., Doni A., Garlanda C., Danese S., Salvatori G., Sassano M., Evangelista V., Rossi B., Zenaro E., Constantin G., Laudanna C., Bottazzi B., Mantovani A. Nat. Immunol. 2010;11:328–334. doi: 10.1038/ni.1854. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Amaro R., Cortes J. R., Sanchez-Madrid F., Martin P. Trends Mol. Med. 2013;19:625–632. doi: 10.1016/j.molmed.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B., Sun T., Yu X.-H., Yang Y.-X., Yeo A. E. T. Clin. Dev. Immunol. 2012;2012:836485. doi: 10.1155/2012/836485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M., Rossi O., Meijerink M., van Baarlen P. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li Y., Gong Y., Yu N., Zhang Y., You R., Qu C., Lu G., Huang Y., Gao Y., Gao Y., Guo X. Int. Immunopharmacol. 2018;54:280–285. doi: 10.1016/j.intimp.2017.11.024. [DOI] [PubMed] [Google Scholar]
- Van Niel G., Raposo G., Candalh C., Boussac M., Hershberg R., Cerf–Bensussan N., Heyman M. Gastroenterology. 2001;121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- Alex S., Lichtenstein L., Dijk W., Mensink R. P., Tan N. S., Kersten S. Histochem. Cell Biol. 2014;141:383–391. doi: 10.1007/s00418-013-1157-y. [DOI] [PubMed] [Google Scholar]
- Aronsson L., Huang Y., Parini P., Korach-André M., Håkansson J., Gustafsson J.-Å., Pettersson S., Arulampalam V., Rafter J. PLoS One. 2010;5:e13087. doi: 10.1371/journal.pone.0013087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann H., Rodríguez J. M., Salminen S., Szajewska H. Br. J. Nutr. 2014;112:1119–1128. doi: 10.1017/S0007114514001949. [DOI] [PubMed] [Google Scholar]
- Groom J. R., Luster A. D. Exp. Cell Res. 2011;317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Najjar N., Chatila M., Moukadem H., Vuorela H., Ocker M., Gandesiri M., Schneider-Stock R., Gali-Muhtasib H. Apoptosis. 2010;15:183–195. doi: 10.1007/s10495-009-0421-z. [DOI] [PubMed] [Google Scholar]
- Itani W., Geara F., Haykal J., Haddadin M., Gali-Muhtasib H. Radiat. Oncol. 2007;2:1. doi: 10.1186/1748-717X-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn F., Robbrecht P., De Paepe A., Speleman F., Vandesompele J. Nucleic Acids Res. 2006;34:D684–D688. doi: 10.1093/nar/gkj155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monjazeb A. M., High K. P., Connoy A., Hart L. S., Koumenis C., Chilton F. H. Carcinogenesis. 2006;27:1950–1960. doi: 10.1093/carcin/bgl023. [DOI] [PubMed] [Google Scholar]
- Wang J., Vodovotz Y., Fan L., Li Y., Liu Z., Namas R., Barclay D., Zamora R., Billiar T. R., Wilson M. A., Fan J., Jiang Y. FASEB J. 2015;29:250–262. doi: 10.1096/fj.14-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeke F., Assad A., Findeisen P., Schromm E., Grobholz R., von Gerstenbergk B., Mantovani A., Peri S., Friess H. H., Post S., von Knebel Doeberitz M., Schwarzbach M. H. M. Eur. J. Cancer. 2006;42:2639–2646. doi: 10.1016/j.ejca.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Huang Y.-H., Chou M.-H., Du Y.-Y., Huang C.-C., Wu C.-L., Chen C.-L., Chuang J.-H. Lab. Invest. 2006;87:66–74. doi: 10.1038/labinvest.3700490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated during the current study are available in the ArrayExpress public repository at EMBL-EBI with accession number E-MTAB-6101 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6101).