Abstract
Background
Rates of asthma in Jordan have been doubled in the past decade, but this increased prevalence was not met with improved asthma control protocols. The aim of the present study was to assess whether there was any significant association between the level of asthma control and certain single-nucleotide polymorphisms (SNPs) in five genes: (ADRB2; rs1042713 and rs1042714), (CRHR1; rs1876828, rs242939, and rs242941), (STIP1; rs2236647), (ADH5, rs1154400), and (ARG1; rs2781659). These SNPs were selected based on their involvement in enzymes and receptors that are related to asthma pathways and subsequent response to medication and based on a high degree of linkage disequilibrium.
Patients and methods
A cross-sectional genetic association study was conducted from June 2016 to June 2017 in the two major hospitals in Jordan. The present study involved sampling from adult asthmatic patients of Arab descent who were selected from two phenotypic groups, ie, controlled and uncontrolled asthma. The blood samples and medical data were collected from the participants. DNA samples were extracted, quantified, and genotyped according to standard operating procedure. Allelic and haplotypic analyses were performed using the Haploview®.
Results
A total of 245 Arab asthmatic patients were enrolled in this study. Genotyping analysis revealed that the two SNPs (rs1042713 and rs1042714) in ADRB2 gene, along with their related haplotypes, were nominally significantly associated with asthma control in the Jordanian population. The A-allele of rs1042713 and the C-allele of rs1042714 were more common in the uncontrolled asthma group than in the controlled asthma group (P=0.048 and P=0.017, respectively).
Conclusion
This was the first study that identified the nominal significant association between the level of asthma control and genetic variants in ADRB2 gene in Arab population. Further studies in other Arab region with larger sample size are recommended to confirm the relationship.
Keywords: ADRB2, Arab, asthma control, genetic association, Jordan, SNPs
Introduction
Asthma is a chronic airway inflammatory disease that affects ~334 million people of all ages worldwide, and this number is expected to rise by another 100 million by 2025.1,2 This increase in asthma prevalence could be influenced by identification of milder asthma cases.3 Asthma is not a curable disease and hence the ultimate goal of asthma management is to relieve symptoms, prevent the disease progression, and obtain a better quality of life.4 According to asthma management guidelines, there are two main types of asthma medication: controllers and relievers.5–7 A reliever is the rescue inhaler medication that contains short-acting β-2 adrenoceptor agonists (SABAs) as bronchodilators. On the other hand, controller medications include inhaled corticosteroids, leukotriene modifiers, and long-acting β-2 adrenoceptor agonists (LABAs). The cost of several asthma medications comprise a substantial proportion of asthma expenditure and, despite the availability of many therapeutically-effective medications, poor adherence and subsequent poor asthma control are considered serious global health problems.8 Uncontrolled asthmatic patients, in spite of proper asthma management plans, require detailed investigation rather than simply escalating their medications’ regimen. Not only the environmental and pharmacological variables that should be considered in asthma management but also the genetic makeup contributes to the disease progression and response to therapy.9
In the context of asthma, the identification of susceptible genes that are involved in asthma pathogenesis and treatment response is the first step towards developing personalized medicine.10,11 Genome-wide association studies, candidate-gene linkage studies, and positional cloning have already discovered a large number of genetic variants in multiple genes that have been associated with altered therapeutic response to asthma medications and hence affect the level of asthma control.5,10,11 The ADRB2 gene is one of the most widely studied asthma susceptible genes.12,13 It is located on the chromosome 5q31-q32, and nine different polymorphisms have been identified.14,15 Four of the identified polymorphisms are non-synonymous variations that change amino acid sequence (Gly16Arg, Gln27Glu, Val34Met, and Thr164Ile) and have a potential clinical effect on the response to β-2 adrenoceptor agonist (B2A) therapy in asthmatic patients.13,14,16,17 It has been revealed that possession of the Arg allele at position 16 enhanced ADRB2’s downregulation and hence the response to B2A is attenuated in asthmatic patients.17,18
The genetic variant of ADH5 gene was associated with increased risk of asthma and attenuated response to bronchodilator therapy.19 It has been reported that gene–gene interaction between both the ADH5 and ADRB2 genes resulted in a 70% reduction in response to SABA.20 Similarly, ARG1 is a β-agonist response gene that is reported to be significantly associated with response to bronchodilators and inhaled corticosteroids.21,22 CRHR1 is a major regulator of glucocorticoid synthesis and is heavily involved in the inflammatory pathways.5 It is located on chromosome 17q21–22, an area that is linked to asthma in some genome-wide association studies.5,23 The CRHR1 gene was considered as a potential genetic predictor of pulmonary function in asthma patients treated with corticosteroids.23 Likewise, the STIP1 gene is part of the steroid pathway and is composed of 14 exons.24 It has been reported that STIP1 variations might play a role in predicting asthma susceptibility and corticosteroid’s response in patients with reduced lung function.24–26
In Jordan, the prevalence of asthma has been doubled in the last decade, but optimal asthma control has not been achieved.27,28 The present study aimed to investigate the association between the level of asthma control and the incidence of certain genetic variants using a candidate gene approach. Eight single-nucleotide polymorphisms (SNPs) in five candidate genes (ADRB2 [rs1042713 and rs1042714], CRHR1 [rs1876828, rs242939, and rs242941], STIP1 [rs2236647], ADH5 [rs1154400], and ARG1 [rs2781659]) and their common haplotypes were investigated in the Arab-Jordanian ethnic group. This was the first study that examined the relationship between the level of asthma control and these polymorphisms of interest in a unique, carefully phenotyped Jordanian population of Arab descent. Up to the authors’ knowledge, no similar study was conducted in the Middle-Eastern region.
Patients and methods
Study design
A cross-sectional study was conducted from June 2016 to June 2017 in two major hospitals in North Jordan: King Abdullah University Hospital and Princess Basma Teaching Hospital. The present study used a candidate gene approach and involved adult asthmatic patients who were selected from two major phenotypic groups, ie, controlled and uncontrolled asthma. The level of asthma control was classified according to asthma control test (ACT), a well-known validated questionnaire, as controlled (score of ≥20) or uncontrolled (score of <20).29 The ethical approval for the current study was granted from both human ethics committees at Jordan University of Science and Technology and the Ministry of Health. The study was conducted in accordance with the Declaration of Helsinki.
Patient recruitment
All adult patients visiting outpatient clinics during the study period were invited to participate in the study. Participants were eligible for inclusion in the study if they were of Arab descent and fulfilled the following inclusion criteria: confirmed clinical asthma diagnosis by a physician, managed by a pulmonology specialist for at least 6 months with good clinic attendance records, and adherent to the prescribed medications based on a self-reported medication adherence scale.30 In addition, patients should possess the knowledge of proper inhaler device usage. Written informed consent was obtained from all participants. Patients were excluded if they were suffering from any other respiratory illnesses such as chronic obstructive pulmonary disease, cystic fibrosis, or bronchiectasis. In addition, newly diagnosed patients with asthma, current smokers or former smokers with ≥25 pack-year history of smoking, immunocompromised, or pregnant were excluded from the study. Around 3 mL of blood was withdrawn into EDTA tubes from each patient by a specialist nurse and stored at −20°C until analysis. Patients’ data were collected from both the patients themselves and their hospital records. Unscheduled medical intervention was defined as any hospital admission, emergency department visit, or oral rescue steroid therapy in the previous 12 months.
SNP selection and analysis
In this study, eight SNPs in five genes (two SNPs in ADRB2, one SNP in ADH5, one SNP in ARG1, three SNPs in CRHR1, and one SNP in STIP1) were investigated for their associations with asthma control in patients of Arab descent. These SNPs were selected from the National Center for Biotechnology Information’s database.31 The list of genes as well as their SNPs, reference numbers, and chromosomal positions are summarized in Table 1. These SNPs were selected based on their involvement in enzymes and receptors that are related to asthma pathogenesis and subsequent response to medication and based on the high degree of linkage disequilibrium (such as the two SNPs in ADRB2 and the three SNPs in CRHR1). Previous studies revealed the association of these SNPs with asthma phenotypes such as bronchodilator response and variable response to asthma medications among different ethnic groups.32,33
Table 1.
Candidate genes and their associated SNPs
| Gene | SNP_ID | Major/minor | Chromosome # | SNP location | SNP type |
|---|---|---|---|---|---|
|
| |||||
| ADRB2 | rs1042713 | G/A | 5 | Exon 1 | Gly16Arg |
| rs1042714 | C/G | 5 | Exon 1 | Gln27Glu | |
| ADH5 | rs1154400 | T/C | 4 | Promoter | Synonymous |
| ARG1 | rs2781659 | A/G | 6 | Promoter | Synonymous |
| CRHR1 | rs1876828 | G/A | 17 | Intron 14 | Synonymous |
| rs242939 | A/G | 17 | Intron 4 | Synonymous | |
| rs242941 | G/T | 17 | Intron 3 | Synonymous | |
| STIP1 | rs2236647 | T/C | 11 | Intron 5 | Synonymous |
Abbreviation: SNP, single-nucleotide polymorphism.
The Wizard® Genomic DNA Purification Kit (Promega Corporation, Fitchburg, WI, USA) was used to extract the DNA according to the standard operating procedure. Afterwards, DNA was quantified to determine its purity and concentration using the NanoDrop® 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). As quality control for sample analysis, both blanks and blind duplicate samples per genotyping plate were applied. All samples were sent for genotyping analysis by the Australian Genome Research Facility (Melbourne, VIC Australia) using the Sequenom MassARRAY system (iPLEX Gold; Sequenom, San Diego, CA, USA).
Statistical analysis
Demographic and genotypic analyses were performed using the Statistical Package for Social Sciences, version 20 (IBM Corporation, Armonk, NY, USA). The differences in participant responses were analyzed using the chi-squared test, fisher’s exact test, or the independent t-test as appropriate. Deviations from the Hardy–Weinberg Equilibrium (HWE) for all genetic variants were tested using a chi-square goodness of fit test. Allelic and haplotypic analyses were performed using the Haploview® program (version 4.2). Permutation correction (n=1,000) for adjustment of multiple testing for both single-marker analysis and haplotype blocks was conducted in Haploview®. For markers that showed significant association with a single marker level, appropriate codominant genetic model for genotypic analysis was considered. OR and 95% CI adjusted for age and gender were calculated using binary logistic regression. A P-value <0.05 was considered significant for all two-tailed tests in the study.
Results
Demographic and clinical data
A total of 245 Jordanian Arab asthmatic patients were enrolled in the current study. Of the recruited patients, 74 (30%) participants had controlled asthma and 171 (70%) participants had uncontrolled asthma. About three-quarters of the participants in both groups were females, and the mean age was around 30 years. The majority of participants had never smoked and a third of patients in both groups had comorbid diseases. In addition, both groups had comparable of hay fever/eczema, and family history of asthma. Further, 73.7% of the uncontrolled asthma group had at least one unscheduled medical intervention in the last 12 months compared to 44% of the controlled asthma group (P<0.001). Table 2 outlines the demographic and clinical data of the participants.
Table 2.
Demographic and clinical data of the participants
| Characteristicsa | Controlled asthma (n=74) | Uncontrolled asthma (n=171) | P-value |
|---|---|---|---|
|
| |||
| Age at evaluation (years)b | 28.73±16.69 | 30.91±16.34 | 0.655 |
| Gender | 0.581 | ||
| Male | 17 (23) | 45 (26.3) | |
| Female | 57 (77) | 126 (73.7) | |
| Smoking | 1.00 | ||
| Never | 71 (95.9) | 164 (95.9) | |
| Ex-smoker | 3 (4.1) | 7 (4.1) | |
| Level of education | 0.568 | ||
| Basic education | 43 (58.1) | 106 (62) | |
| University degree | 31 (41.9) | 65 (38) | |
| Personal history of hay fever/eczema | 24 (32.4) | 56 (32.7) | 0.961 |
| Family history of asthma | 31 (41.9) | 67 (39.2) | 0.691 |
| Body mass indexb | 29.36±5.3 | 29.48±6.75 | 0.190 |
| Comorbid disease | 24 (32.4) | 65 (38) | 0.404 |
| Unscheduled medical intervention | 33 (44.6) | 126 (73.7) | <0.001 |
| Asthma severity, GINA 2017 | 0.570 | ||
| Mild/moderate (step1–3) | 27 (36.5) | 56 (32.7) | |
| Severe (step 4–5) | 47 (63.5) | 115 (67.3) | |
Notes:
All data expressed as n (%) unless otherwise indicated.
Data described as mean ± SD.
Abbreviation: GINA, Global Initiative for Asthma.
Genotypic and allelic frequencies
In this study, eight SNPs in five genes were investigated. All genetic markers had a 100% genotyping accuracy and a 96.53% successful call rate. All studied SNPs were in HWE (P>0.05) in both the patient and control groups except for the rs2236647 SNP (T/C) in the STIP1 gene and hence it was excluded from the analysis (Table 3).
Table 3.
MAFs and HWE of the included genetic markers
| Gene | SNP_ID | Controlled asthma | Uncontrolled asthma | ||
|---|---|---|---|---|---|
|
| |||||
| MAF | P-value | MAF | P-value | ||
|
| |||||
| ADRB2 | rs1042713 G/A | 0.270 | 0.73 | 0.362 | 0.52 |
| rs1042714 C/G | 0.354 | 0.31 | 0.247 | 0.96 | |
| ADH5 | rs1154400 T/C | 0.326 | 0.37 | 0.247 | 0.96 |
| ARG1 | rs2781659 A/G | 0.425 | 0.58 | 0.404 | 0.14 |
| CRHR1 | rs1876828 G/A | 0.212 | 0.23 | 0.213 | 0.47 |
| rs242939 A/G | 0.110 | 0.88 | 0.135 | 0.06 | |
| rs242941 G/T | 0.356 | 0.52 | 0.357 | 0.23 | |
| STIP1 | rs2236647 C/T | 0.319 | 0.01 | 0.411 | 0.29 |
Abbreviations: HWE, Hardy–Weinberg Equilibrium; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Genetic association between polymorphism of interest and asthma control
The association between the polymorphisms of interest and the level of asthma control was examined in the two groups: controlled and uncontrolled groups. The allelic frequencies (counts) and genotype distributions of all variants in the candidate genes in both study groups are shown in Table 4. The results of allelic analysis indicated no significant association between the polymorphisms of interest and asthma control. Only two SNPs (rs1042713 G/A and rs1042714 C/G) in the ADRB2 gene showed nominal significant association with the risk of uncontrolled asthma (ie, P-value was significant before correcting for multiple testing). The A-allele of rs1042713 and C-allele of rs1042714 were more common in the uncontrolled asthma group than in the controlled asthma group (P=0.048 and P=0.017, respectively). Genotypic analysis using the codominant model for rs1042714 revealed that the risk of having uncontrolled asthma was higher in homozygous (CC) patients (OR =3.93; 95% CI: 1.46–10.58; P=0.007) compared to those who were homozygous for the G-allele.
Table 4.
Genotypic distributions and allelic associations of the studied SNPs with asthma control
| Gene | SNP_ID | Genotype distribution and analysis | Allelic distribution and analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Genotype | Controlled asthma, n (%) | Uncontrolled asthma, n (%) | P-value | Allele | Controlled asthma, n (%) | Uncontrolled asthma, n (%) | P-value | Adjusted P-value | ||
|
| ||||||||||
| ADRB2 | rs1042713 | AA | 6 (8.1) | 20 (12) | 0.107 | A | 40 (27) | 121 (36.2) | 0.048 | NS |
| AG | 28 (37.8) | 81 (48.5) | G | 108 (73) | 213 (63.8) | |||||
| GG | 40 (54.1) | 66 (39.5) | ||||||||
| rs1042714 | GG | 11 (15.3) | 10 (6) | 0.041 | G | 51 (35.4) | 82 (24.7) | 0.017 | NS | |
| GC | 29 (40.3) | 62 (37.4) | C | 93 (64.6) | 250 (75.3) | |||||
| CC | 32 (44.4) | 94 (56.6) | ||||||||
| ADH5 | rs1154400 | CC | 6 (8.3) | 16 (9.7) | 0.725 | C | 47 (32.6) | 103 (31.2) | 0.759 | NS |
| CT | 35 (48.6) | 71 (43) | T | 97 (67.4) | 227 (68.8) | |||||
| TT | 31 (43.1) | 78 (47.3) | ||||||||
| ARG1 | rs2781659 | GG | 12 (16.4) | 22 (13.6) | 0.847 | G | 62 (42.5) | 131 (40.4) | 0.678 | NS |
| GA | 38 (52.1) | 87 (53.7) | A | 84 (57.5) | 193 (59.6) | |||||
| AA | 23 (31.5) | 53 (32.7) | ||||||||
| CRHR1 | rs1876828 | AA | 5 (6.8) | 6 (3.6) | 0.361 | A | 31 (21.2) | 71 (21.3) | 0.995 | NS |
| AG | 21 (28.8) | 59 (35.3) | G | 115 (78.8) | 263 (78.7) | |||||
| GG | 47 (64.4) | 102 (61.1) | ||||||||
| rs242939 | GG | 1 (1.4) | 6 (3.5) | 0.833 | G | 16 (11) | 46 (13.4) | 0.449 | NS | |
| GA | 14 (19.2) | 34 (19.9) | A | 130 (89) | 296 (86.6) | |||||
| AA | 58 (79.4) | 131 (76.6) | ||||||||
| rs242941 | TT | 8 (11) | 25 (14.9) | 0.493 | T | 52 (35.6) | 120 (35.7) | 0.983 | NS | |
| TG | 36 (49.3) | 70 (41.7) | G | 94 (64.4) | 216 (64.3) | |||||
| GG | 29 (39.7) | 73 (43.4) | ||||||||
Abbreviations: SNP, single-nucleotide polymorphism; NS, not significant.
Linkage disequilibrium and haplotype analysis
Haplotypic analysis was conducted for SNPs that formed a haplotype block in the groups of uncontrolled vs controlled asthma patients. Haplotype association showed strong linkage disequilibrium (D′=1) between five SNPs in two genes that constructed two blocks as shown in Table 5 and Figure 1. The first block was formed between three selected polymorphisms of CRHR1 in chromosome 17 (rs1876828 G/A, rs242939 A/G, and rs242941 G/T), while the second block was formed between two polymorphisms of ADRB2 in chromosome 5 (rs1042713 G/A and rs1042714 C/G). The GG haplotype in the second block of ADRB2 showed a nominally significant association with uncontrolled asthma (P=0.021). In addition, the AC haplotype in ADRB2 showed a trend toward significant nominal association (P=0.055).
Table 5.
Association between the formed haplotypes and uncontrolled asthma
| Haplotype | Frequency of block | Frequency ratio, % (case:control) | P-value | Adjusted, P-value |
|---|---|---|---|---|
|
Block 1: CRHR1 in chromosome 17 (rs1876828 G/A, rs242939 A/G, and rs242941 G/T) | ||||
| GAG | 0.43 | 42.6:63.6 | 0.84 | NS |
| TAG | 0.23 | 22.6:24.4 | 0.67 | NS |
| GAA | 0.21 | 21.3:21.2 | 0.98 | NS |
| TGG | 0.12 | 13.1:10.7 | 0.46 | NS |
|
Block 2: ADRB2 in chromosome 5 (rs1042713 G/A and rs1042714 C/G) | ||||
| GC | 0.39 | 39.2:37.9 | 0.79 | NS |
| AC | 0.33 | 35.9:27.0 | 0.055 | NS |
| GG | 0.28 | 24.8:35.0 | 0.021 | NS |
Abbreviation: NS, not significant.
Figure 1.
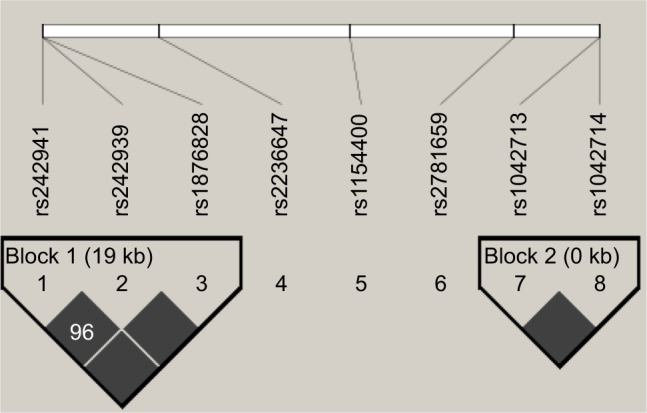
Genetic haplotype blocks: linkage disequilibrium plot of genotyped polymorphisms in CRHR1 (block 1) and ADRB2 (block 2). Boxes without numbers have D′=1.
Discussion
In the present study, analysis of eight genetic markers in five candidate genes (ADRB2, CRHR1, ARG1, ADH5, and STIP1) was carried out in order to investigate their association with uncontrolled asthma in Jordanian Arab population. Due to their immediate onset of action, inhaled SABAs are commonly used agents to induce bronchodilation during asthma exacerbations. On the other hand, LABAs are usually prescribed to maintain the bronchodilation as an add-on therapy to inhaled corticosteroids, in poorly controlled asthmatic patients. However, some patients do not show benefit from using B2A which might lead to further excessive use of SABA inhalers. Their symptoms might deteriorate when they receive such agents,34 and excessive use of SABAs was found to be associated with a noticeably increased risk of asthma related deaths.35 Genetic polymorphisms that could affect pharmacological responses to B2A therapies has been recognized which might lead to future new targeted treatments and help in recognizing uncontrolled-asthma cases.
The Arg16Gly (rs1042713 G/A) polymorphism received the most attention in various clinical studies in relation to its association with poor clinical response to SABAs and LABAs. Both retrospective and prospective data suggested that asthmatic patients who were homozygous for Arg16 might not benefit from regular or as-required administration of SABAs.36,37 Also, research on LABAs demonstrated that patients who were homozygous for Arg16 had more asthma exacerbations36,38 and lower bronchodilator response39 compared to other genotypes at position 16. It has been proposed that genetic variations in the β-2 adrenergic receptors enhanced receptors’ downregulation and, hence, impaired receptors coupling.16,38 Therefore, attenuated response to SABAs and LABAs is expected.16,38 Examining Gln27Glu (rs1042714 C/G) resulted in inconsistent findings with regard to bronchodilator response and lung function.38,40 However, other studies reported no association between these markers and bronchodilator response,41 lung function,42 or difficult asthma.43 The association of asthma control (based on ACT questionnaire) with rs1042713 G/A and rs1042714 C/G in ADRB2 was not investigated before. In the current study, nominal associations were identified between these two SNPs in ADRB2 and uncontrolled asthma. The Arg allele of rs1042713 and the Gln allele of rs1042714 in ADRB2 were more common in the uncontrolled asthma group than in the controlled asthma group. Similar findings were detected in the Spanish population where Arg allele of ADRB2 was linked with an increased risk of having uncontrolled asthma and a reduction in lung function.18 Further, a sub-analysis was conducted to evaluate the potential effect of all studies on SNPs among patients who used small/medium dose of steroid (n=122) vs those who used high dose of steroid (n=102) according to Global Initiative for Asthma 2017.4 The results indicated that the rs1042713 C/G SNP in the ADRB2 gene showed nominal significant associations (P=0.019) with the risk of uncontrolled asthma.
Two non-synonymous SNPs in the ADRB2 gene were not examined for their associations with the asthma control phenotype in the current study. These SNPs were rs1141370 SNP (Val34Met) that was not polymorphic and rs1800888 SNP (Thr164Ile) that was found rare in asthmatic patients in Jordan.25 Moreover, these SNPs were rare in the Caucasian population33 and rs1800888 was not detected in an Omani cohort of Arab descent.44 Previous studies reported the association of other SNPs that were involved in the regulation and signaling of the ADRB2 such as rs1154400 in ADH5 and rs2781659 in ARG1 with the bronchodilator response phenotype.19,21 In the current study, however, neither the allelic nor the genetic analysis showed any significant association of these ADRB2 SNPs with the level of asthma control among the Jordanian Arab population.
Inhaled corticosteroids are considered the mainstay agents in the treatment of asthma. In the present study, no association was identified between the three studied SNPs in CRHR1 (rs1876828 G/A, rs242939 A/G, and rs242941 G/T) and the asthma control phenotype. This finding is in agreement with Dijkstra et al who showed no association between the CRHR1 polymorphisms and improvement in lung function in response to inhaled corticosteroids therapy.45 In contrast, Tantisira et al reported that the two SNPs (rs242941 and rs1876828) and the common GAT haplotype were linked with significant improvement in lung function.23 However, in the aforementioned study, the 131 SNPs were tested without proper correction for multiple comparisons, which had the potential to generate false positive associations.23 Leukotriene modifiers are the third class of medications that are used for asthma. These agents attenuate asthma inflammation by inhibiting the action of cysteinyl leukotrienes that are one of the main mediators in asthma inflammation. In the present study, the SNPs that are involved in receptors and enzymes of the cysteinyl leukotriene pathway were not examined as only nine patients were using leukotriene modifiers. This could be due to insurance and financial restraints which could limit the number of patients who were placed on these agents. Future studies with a larger number of asthmatic patients who are using this class of medication would highlight any relationship between the leukotriene pathway and the level of asthma control.
The inconsistent findings in the association level in the current study and the previous ones could be explained by the differences in the study design, ethnicity, and number of participants. Further, there are other factors that could explain the aforementioned variations: 1) specific population gene–environment and gene–gene interactions; 2) differences in selected asthma phenotypes and genotypes in addition to the different patterns of linkage disequilibrium across various ethnic groups; and 3) adjustment for multiple testing. It is worth mentioning that the studied SNPs in the current study were corrected for multiple testing to examine genetic associations by performing a permutation analysis on all data sets.
There are few limitations in the current study. Firstly, the limited number of recruited patients in the controlled asthma group restricted the ability to detect significant differences in the outcome of interest. Secondly, replication was not performed for the cohort, but the results could be considered as exploratory findings, stressing on the importance of replication in a larger Arab population of the same phenotype. Finally, other objective methods (lung function test) were not incorporated with the subjective methods (questionnaire) for assessing asthma control due to limited resources. However, in the study, well defined groups based on the presence or absence of asthma control were included. This was confirmed furthermore by unscheduled medical intervention which was significantly higher among uncontrolled asthma group compared to control group. Moreover, ACT is a simple and easy to use tool that can be used in low income and limited health care resources and providers such as Jordan.28
Conclusion
The present study examined, for the first time, the association between different genetic markers and level of asthma control in a genetically isolated adult population of Arab descent. Two SNPs in the ADRB2 (rs1042713 and rs1042714) gene and their related haplotype that were nominally significantly associated with level of asthma control were identified in Jordanian Arab asthmatic patients. At the current stage, additional large-scale studies that recruit participants from different Arab populations in the Middle Eastern region are needed to confirm these associations. Finally, future studies should focus on functional effects and the potential clinical implications of these SNPs to help identify individuals at risk of uncontrolled asthma despite proper management.
Acknowledgments
This study was supported by a grant from the Deanship of Research at Jordan University of Science and Technology, Irbid, Jordan.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Behera D, Sehgal IS. Bronchial asthma–issues for the developing world. Indian J Med Res. 2015;141(4):380–382. doi: 10.4103/0971-5916.159237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Global Asthma Report, Auckland. New Zealand: Global Asthma Network; 2014. [Accessed July 15, 2018]. Available from: http://www.globalasthmanetwork.org/publications/Global_Asthma_Report_2014.pdf. [Google Scholar]
- 3.Chen W, Fitzgerald JM, Lynd LD, Sin DD, Sadatsafavi M. Long-term trajectories of mild asthma in adulthood and risk factors of progression. J Allergy Clin Immunol Pract. 2018;6(6):2024–2032.e5. doi: 10.1016/j.jaip.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 4.Global Initiative for Asthma (GINA) Global strategy for asthma management and prevention (PDF) [Accessed July 15, 2018]. Available from: https://ginasthma.org/2018-gina-report-global-strategy-for-asthma-management-and-prevention/
- 5.Weiss ST, Litonjua AA, Lange C, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J. 2006;6(5):311–326. doi: 10.1038/sj.tpj.6500387. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Xie X, Song Y, et al. Association of TLR4 (896A/G and 1196C/T) gene polymorphisms with asthma risk: a meta-analysis. Med Sci Monit. 2015;21:3591–3599. doi: 10.12659/MSM.895380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lommatzsch M, Virchow JC. Severe asthma: definition, diagnosis and treatment. Dtsch Arztebl Int. 2014;111(50):847–55. doi: 10.3238/arztebl.2014.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarraf H, Al-Jahdali H, Al Qaseer AH, et al. Asthma control in adults in the Middle East and North Africa: results from the ESMAA study. Respir Med. 2018;138:64–73. doi: 10.1016/j.rmed.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 9.Guilleminault L, Ouksel H, Belleguic C, et al. Personalised medicine in asthma: from curative to preventive medicine. Eur Respir Rev. 2017;26(143):160010. doi: 10.1183/16000617.0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang SQ, Chen XL, Deng JM, et al. Beta-2 adrenergic receptor (ADRB2) gene polymorphisms and the risk of asthma: a meta-analysis of case-control studies. PLoS One. 2014;9(8):e104488. doi: 10.1371/journal.pone.0104488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussein IA, Jaber SH. Genotyping of IL-4 −590 (C>T) gene in Iraqi asthma patients. Disease Markers. 2017;2017(8):1–5. doi: 10.1155/2017/5806236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan I, Ul-Haq Z, Shaheen A, et al. Association of arg16gly and gln27glu, b2-adrenergic receptor gene polymorphism with asthma. A systematic review and meta-analysis of case control studies. J Pak Med Assoc. 2018;68(1):90–97. [PubMed] [Google Scholar]
- 13.Liu ZQ, Jiao Y, Liu CJ, Zhang HC, Hu BR. Association between polymorphisms and haplotypes of the beta-2 adrenergic receptor gene and asthma in a Chinese Han population. Int J Clin Exp Med. 2014;7(10):3812. [PMC free article] [PubMed] [Google Scholar]
- 14.de Paiva AC, Fadel Marson, Ribeiro J, Bertuzzo C. Asthma: Gln27Glu and Arg16Gly polymorphisms of the beta2-adrenergic receptor gene as risk factors. Allergy Asthma Clin Immunol. 2014;10(1):8. doi: 10.1186/1710-1492-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega VE, Hawkins GA, Moore WC. Effect of RareGenetic variants in the β2 adrenergic receptor geneon the risk for exacerbations and symptom control during long-acting beta agonist treatment in a multi-ethnic asthma population. Lancet Respir Med. 2014;2(3):204. doi: 10.1016/S2213-2600(13)70289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor DR, Drazen JM, Herbison GP, Yandava CN, Hancox RJ, Town GI. Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax. 2000;55(9):762–767. doi: 10.1136/thorax.55.9.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jabbal S, Manoharan A, Lipworth J, Anderson W, Short P, Lipworth B. Is Gly16Arg β2 receptor polymorphism related to impulse oscillometry in a real-life asthma clinic setting? Lung. 2016;194(2):267–271. doi: 10.1007/s00408-016-9848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebordosa C, Kogevinas M, Guerra S, et al. ADRB2 Gly16Arg polymorphism, asthma control and lung function decline. Eur Respir J. 2011;38(5):1029–1035. doi: 10.1183/09031936.00146310. [DOI] [PubMed] [Google Scholar]
- 19.Choudhry S, Que LG, Yang Z, et al. GSNO reductase and beta2-adrenergic receptor gene-gene interaction: bronchodilator responsiveness to albuterol. Pharmacogenet Genomics. 2010;20(6):351–358. doi: 10.1097/FPC.0b013e328337f992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore PE, Ryckman KK, Williams SM, Patel N, Summar ML, Sheller JR. Genetic variants of GSNOR and ADRB2 influence response to albuterol in African-American children with severe asthma. Pediatr Pulmonol. 2009;44(7):649–654. doi: 10.1002/ppul.21033. [DOI] [PubMed] [Google Scholar]
- 21.Litonjua AA, Lasky-Su J, Schneiter K, et al. ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med. 2008;178(7):688–694. doi: 10.1164/rccm.200709-1363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vonk JM, Postma DS, Maarsingh H, Bruinenberg M, Koppelman GH, Meurs H. Arginase 1 and arginase 2 variations associate with asthma, asthma severity and beta2 agonist and steroid response. Pharmacogenet Genomics. 2010;20(3):179–186. doi: 10.1097/FPC.0b013e328336c7fd. [DOI] [PubMed] [Google Scholar]
- 23.Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13(13):1353–1359. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 24.Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123(6):1376–1383.e7. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almomani BA, Al-Eitan LN, Samrah SM, Al-Quasmi MN, McKnight AJ. Candidate gene analysis of asthma in a population of Arab descent: a case-control study in Jordan. Per Med. 2017;14(1):51–61. doi: 10.2217/pme-2016-0059. [DOI] [PubMed] [Google Scholar]
- 26.March ME, Sleiman PM, Hakonarson H. Genetic polymorphisms and associated susceptibility to asthma. Int J Gen Med. 2013;6:253. doi: 10.2147/IJGM.S28156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu-Ekteish F, Otoom S, Shehabi I. Prevalence of asthma in Jordan: comparison between Bedouins and urban schoolchildren using the International Study of Asthma and Allergies in Childhood phase III protocol. Allergy Asthma Proc. 2009;30(2):181–185. doi: 10.2500/aap.2009.30.3208. [DOI] [PubMed] [Google Scholar]
- 28.Almomani BA, Al-Sawalha NA, Samrah SM, Gamble JM, Al Momani MA. Asthma insights from Jordan: cross-sectional observational study. J Asthma. 2016;53(4):349–355. doi: 10.3109/02770903.2015.1121493. [DOI] [PubMed] [Google Scholar]
- 29.Nathan RA, Sorkness CA, Kosinski M, et al. Development of the asthma control test: a survey for assessing asthma control. J Allergy Clin Immunol. 2004;113(1):59–65. doi: 10.1016/j.jaci.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Aburuz SM, Bulatova NR, Yousef AM. Validation of a comprehensive classification tool for treatment-related problems. Pharm World Sci. 2006;28(4):222–232. doi: 10.1007/s11096-006-9048-0. [DOI] [PubMed] [Google Scholar]
- 31.National Centre for Biotechnology Information database dbSNP Short Genetic Variations. [Accessed July 2017]. Available from: www.ncbi.nlm.nih.gov/SNP.
- 32.Isidoro-García M, Sánchez-Martín A, García-Sánchez A, Sanz C, García-Berrocal B, Dávila I. Pharmacogenetics and the treatment of asthma. Pharmacogenomics. 2017;18(13):1271–1280. doi: 10.2217/pgs-2017-0024. [DOI] [PubMed] [Google Scholar]
- 33.Portelli M, Sayers I. Genetic basis for personalized medicine in asthma. Expert Rev Respir Med. 2012;6(2):223–236. doi: 10.1586/ers.12.9. [DOI] [PubMed] [Google Scholar]
- 34.Taylor DR, Hall IP. ADRB2 polymorphisms and β2 agonists. Lancet. 2007;370(9605):2075–2076. doi: 10.1016/S0140-6736(07)61876-5. [DOI] [PubMed] [Google Scholar]
- 35.Lanes SF, García Rodríguez LA, Huerta C. Respiratory medications and risk of asthma death. Thorax. 2002;57(8):683–686. doi: 10.1136/thorax.57.8.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basu K, Palmer CNA, Tavendale R, Lipworth BJ, Mukhopadhyay S. Adrenergic β2-receptor genotype predisposes to exacerbations in steroid-treated asthmatic patients taking frequent albuterol or salmeterol. J Allergy Clin Immunol. 2009;124(6):1188–1194.e3. doi: 10.1016/j.jaci.2009.07.043. [DOI] [PubMed] [Google Scholar]
- 37.Israel E, Chinchilli VM, Ford JG, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364(9444):1505–1512. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 38.Palmer CNA, Lipworth BJ, Lee S, Ismail T, Macgregor DF, Mukhopadhyay S. Arginine-16 2 adrenoceptor genotype predisposes to exacerbations in young asthmatics taking regular salmeterol. Thorax. 2006;61(11):940–944. doi: 10.1136/thx.2006.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wechsler ME, Lehman E, Lazarus SC, et al. beta-Adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006;173(5):519–526. doi: 10.1164/rccm.200509-1519OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giubergia V, Gravina LP, Castaños C, Chertkoff L, Grenoville M. Influence of beta2-adrenoceptor polymorphisms on the response to chronic use of albuterol in asthmatic children. Pediatr Pulmonol. 2008;43(5):421–425. doi: 10.1002/ppul.20759. [DOI] [PubMed] [Google Scholar]
- 41.Bleecker ER, Yancey SW, Baitinger LA, et al. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J Allergy Clin Immunol. 2006;118(4):809–816. doi: 10.1016/j.jaci.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 42.Wechsler ME, Kunselman SJ, Chinchilli VM, et al. Effect of β2-adrenergic receptor polymorphism on response to longacting β2 agonist in asthma (LARGE trial): a genotype-stratified, randomised, placebo-controlled, crossover trial. Lancet. 2009;374(9703):1754–1764. doi: 10.1016/S0140-6736(09)61492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almomani B, Hawwa AF, Millership JS, et al. Can certain genotypes predispose to poor asthma control in children? A pharmacogenetic study of 9 candidate genes in children with difficult asthma. PLoS One. 2013;8(4):e60592. doi: 10.1371/journal.pone.0060592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Balushi K, Zadjali F, Al-Sinani S, Al-Zadjali AM, Bayoumi R. Frequencies of the Arg16Gly, Gln27Glu and Thr164Ile Adrenoceptor β2 Polymorphisms among Omanis. Sultan Qaboos Univ Med J. 2015;15(4):e486–e490. doi: 10.18295/squmj.2015.15.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dijkstra A, Koppelman GH, Vonk JM, Bruinenberg M, Schouten JP, Postma DS. Pharmacogenomics and outcome of asthma: no clinical application for long-term steroid effects by CRHR1 polymorphisms. J Allergy Clin Immunol. 2008;121(6):1510–1513. doi: 10.1016/j.jaci.2008.04.015. [DOI] [PubMed] [Google Scholar]