Abstract
Clinical evidence demonstrates that treatment with immune checkpoint inhibitor immunotherapy agents can have considerable benefit across multiple tumours. However, there is a need for the development of predictive biomarkers that identify patients who are most likely to respond to immunotherapy. Comprehensive characterisation of tumours using genomic, transcriptomic, and proteomic approaches continues to lead the way in advancing precision medicine. Genetic correlates of response to therapy have been known for some time, but recent clinical evidence has strengthened the significance of high tumour mutational burden (TMB) as a biomarker of response and hence a rational target for immunotherapy. Concordantly, immune checkpoint inhibitors have changed clinical practice for lung cancer and melanoma, which are tumour types with some of the highest mutational burdens. TMB is an implementable approach for molecular biology and/or pathology laboratories that provides a quantitative measure of the total number of mutations in tumour tissue of patients and can be assessed by whole genome, whole exome, or large targeted gene panel sequencing of biopsied material. Currently, TMB assessment is not standardised across research and clinical studies. As a biomarker that affects treatment decisions, it is essential to unify TMB assessment approaches to allow for reliable, comparable results across studies. When implementing TMB measurement assays, it is important to consider factors that may impact the method workflow, the results of the assay, and the interpretation of the data. Such factors include biopsy sample type, sample quality and quantity, genome coverage, sequencing platform, bioinformatic pipeline, and the definitions of the final threshold that determines high TMB. This review outlines the factors for adoption of TMB measurement into clinical practice, providing an understanding of TMB assay considerations throughout the sample journey, and suggests principles to effectively implement TMB assays in a clinical setting to aid and optimise treatment decisions.
Keywords: Tumor mutational burden, immunotherapy, assay implementation, immune checkpoint inhibitor, next-generation sequencing
Background
Immunotherapy agents have demonstrated significant clinical benefit across multiple tumours; however, as with most therapeutic agents, there is a need for robust biomarkers that predict which patients are most likely to respond to immunotherapy.1 2 Programmed death ligand 1 (PD-L1) expression emerged as a biomarker for immunotherapy,3–5 although challenges with PD-L1 testing exist. PD-L1 immunohistochemistry assays can use different reagents and platforms, and standardisation between them is required.5 There is also uncertainty about whether to assess PD-L1 expression on tumour cells and/or immune cells.6 Furthermore, PD-L1 expression alone is often insufficient for predicting response.7 Despite the challenges, PD-L1 immunohistochemistry assays are approved for companion and complementary testing with immunotherapy.5
Clinical studies to dissect the genetic makeup of tumours revealed that patients with high TMB have increased response rates and improved outcomes to treatment with immunotherapy compared with patients with lower TMB (table 1). This concept was recently validated in a phase III trial with a coprimary endpoint of progression-free survival in patients with high TMB (≥10 mutations/megabase (mut/Mb) measured by the FoundationOne CDx assay8 9).
Table 1.
Summary of clinical evidence demonstrating TMB as a biomarker for response to immunotherapy
| Immunotherapy agent and tumour type | Study/trial* | TMB assay used | Type of benefit |
| Nivolumab | |||
| NSCLC (1 L) | CheckMate 02652 | WES | ORR, PFS |
| NSCLC | Flatiron Health117 | Foundation CGP panel | OS |
| Melanoma (1 L or 2 L) | CheckMate 03822 | WES | ORR, OS, PFS |
| Melanoma | CheckMate 06423 | WES | ORR, OS |
| Bladder | CheckMate 27583 | WES | ORR, OS, PFS |
| GBM | Bouffet et al, 2016118 | WES | DRR |
| Ipilimumab | |||
| Melanoma | Van Allen et al, 2015119 | WES | CBR |
| Snyder et al, 201486 | WES | CBR, OS | |
| Nivolumab and ipilimumab in combination | |||
| NSCLC (1 L) | CheckMate 01235 | WES | ORR, DCB, PFS |
| NSCLC (1 L) | CheckMate 227†8 | FoundationOne CDx | ORR, PFS |
| NSCLC (1 L) | CheckMate 5689 | FoundationOne CDx | ORR |
| SCLC (2 L) | CheckMate 03284 | WES | ORR, OS, PFS |
| Pembrolizumab | |||
| NSCLC (1 L) | KEYNOTE-00136 | WES | ORR, DCB, PFS |
| CRC | Le et al, 201544 | WES | ORR, PFS |
| Multiple solid tumours | KEYNOTE-012/KEYNOTE-028120 121 | WES | ORR |
| Atezolizumab | |||
| NSCLC (2 L) | POPLAR/OAK87 88 | Foundation bTMB | OS, PFS |
| NSCLC (2 L) | POPLAR/FIR/BIRCH85 | FoundationOne | ORR, OS, PFS |
| NSCLC (1 L) | BFAST and B-F1RST122–124 | Foundation bTMB | DOR, ORR, PFS, OS |
| NSCLC | Rizvi et al, 201856 | WES | DCB, ORR, PFS |
| Bladder (1 L or 2 L) | IMvigor 210125 126 | Foundation CGP panel | ORR, OS |
| FoundationOne | ORR | ||
| Bladder (2 L) | IMvigor 211127 | FoundationOne | OS |
| Bladder | Snyder et al, 2017128 | WES | PFS |
| Multiple agents | |||
| NSCLC | Rozenblum et al, 2017129 | FoundationOne and Guardant360 | ORR |
| Melanoma | Johnson et al, 201653 | FoundationOne | ORR, OS, PFS |
| Hugo et al, 201645 | WES | OS | |
| Multiple solid tumours | Goodman et al, 2017130 | FoundationOne | ORR, OS, PFS |
| Yarchoan et al, 201725 | Various (not reported) | ORR | |
| Multiple solid tumours (2 L) | Bonta et al, 2017131 | FoundationOne | ORR |
*Ongoing atezolizumab, durvalumab and avelumab trials have primary completion dates in 2019 and 2020.
†CheckMate 227 has monotherapy and combination therapy arms in the study design.
CBR, clinical benefit rate; CGP, comprehensive genomic profiling; CRC, colorectal cancer; DCB, durable clinical benefit; DOR, duration of response; DRR, durable response rate; GBM, glioblastoma multiforme; NSCLC, non-small cell lung cancer; ORR, objective response rate; OS, overall survival; PFS, progression-free survival; SCLC, small cell lung cancer; TMB, tumour mutational burden; WES, whole exome sequencing.
TMB is defined as the total number of somatic mutations in a defined region of a tumour genome, but the precise definition varies with the sequenced region size and localisation, and the nature of the mutations included.10 11 Testing for genomic alterations, including TMB, is currently performed using next-generation sequencing (NGS). Several NGS approaches exist and the target region ranges from genome-wide analysis (whole genome sequencing (WGS)) to whole exome sequencing (WES, covering the entire coding regions of genes in the genome) and large targeted gene panels (table 1). Many initial clinical studies used WES for TMB assessment, and WES of tumour versus germline DNA is currently considered a reference standard. However, there are many commercial gene panel assays and laboratory-developed tests (LDTs) being used. The US Food and Drug Administration (FDA) recently approved the gene panel assay FoundationOne CDx12 and authorised MSK-IMPACT13 for profiling solid tumours for genomic alterations in a clinical setting, and many other assays are currently available for research use only. As well as testing for actionable mutations, these assays can test for TMB in parallel.14 15 However, they can differ in their methodology (eg, enrichment approach, sequencing system, bioinformatic pipeline), the number of genes and types of mutations included (eg, synonymous and/or nonsynonymous), and the recommendations for interpretation of results. For example, TMB is defined in FoundationOne CDx as the number of somatic, coding, base substitutions (synonymous and nonsynonymous) and short insertions and deletions (indels) per megabase of tumour genome examined.11 14
With a variety of alternative approaches now available, standardisation is required as TMB assays move into clinical practice. It is essential to raise awareness of all assay considerations to ensure that reliable and accurate TMB assessments can be achieved and compared across studies.
Rationale for TMB as a biomarker for response to immunotherapy
Evidence suggests that tumours with higher TMB also carry higher neoantigen loads.16 Tumour-specific somatic mutations can give rise to neoantigens, which are novel protein epitopes specific to tumours that may be presented on the tumour cell surface by major histocompatibility complex molecules.10 17 18 A subset of neoantigens can be recognised as ‘non-self’, leading to T-cell activation and a tumour-targeted immune response.19 20 The clonal nature of tumour evolution implies that T-cell-activating neoantigens will be propagated.21 T-cell receptor sequencing shows that increased clonal neoantigen load may drive clonal T-cell expansion and a sustained antitumor immune response.4 10 22 23
Immune checkpoint molecules negatively regulate T-cell activation, leading to suppression of neoantigen-driven immune responses and allowing tumours to escape immune surveillance.16 Immune checkpoint inhibitors, such as anti-PD-1, anti-PD-L1, and anti-cytotoxic T-lymphocyte antigen 4, restore the antitumour immune response,4 24 and neoantigen-driven activation of the antitumour immune response implies that tumours with high TMB are more likely to respond to immune checkpoint blockade.16 24–26 Moreover, clonal tumour evolution and the effects of increased T-cell activity on other tumour properties may further increase sensitivity to immunotherapy.10 21 27 28
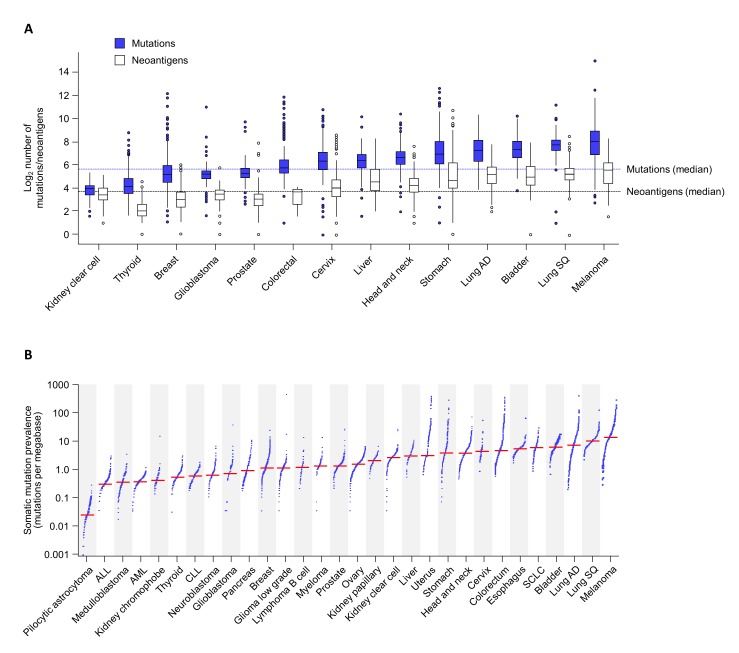
TMB and neoantigen load vary considerably within and across tumour types, with melanomas, lung cancers, and bladder cancers being among those with the highest TMB compared with other tumour types (figure 1).11 29–33 The mutational landscape can also vary across the tumour and evolve during tumour development or following treatment.22 34
Figure 1.
Distribution of TMB and neoantigen load across tumour types. (A) TMB and corresponding predicted neoantigen variation across 14 different tumour types. Data derived from Chen et al.26 (B) TMB variation across 30 different tumour classes. Adapted with permission from Springer: Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500(7463):415–21;30 Copyright 2013. AD, adenocarcinoma; ALL, acute lymphocytic leukaemia; AML, acute myeloid leukaemia; CLL, chronic lymphocytic leukaemia; SCLC, small cell lung cancer; SQ, squamous cell carcinoma; TMB, tumour mutational burden.
Tumours with high TMB may exhibit specific genetic alterations associated with clinical benefit to immunotherapy.35 36 For example, high TMB can be caused by defects in mismatch repair (MMR) genes that are associated with microsatellite instability (MSI).11 16 26 The MMR pathway is responsible for the correction of DNA replication errors; such errors frequently occur in microsatellites (short tandem DNA repeats found across the genome). Mutations in MMR genes can lead to increased mutational incidence within microsatellites; MSI can therefore be a surrogate marker for DNA repair disorders, such as deficient mismatch repair (dMMR).37 Similarly, DNA polymerase epsilon (POLE) is required for high-fidelity DNA replication, ensured by Watson–Crick base pairing and exonuclease (proofreading) activity. Mutations in the exonuclease domain of POLE can cause a hypermutator or ultramutator phenotype with TMB >10 mut/Mb or >100 mut/Mb, respectively.38
While clonal tumour expansion may not increase TMB, POLE mutant, dMMR, and MSI-high tumours have been associated with high TMB. However, the reverse is not always true. For example, melanoma and lung tumours frequently have high TMB but <1% are MSI-high39 40 and POLE deficiency occurs in <3% of lung cancers,41 indicating that other mechanisms, including pathogenic events such as UV exposure or smoking, contribute to increased TMB in these tumour types.11 15 38 42 Testing for driver mutations should therefore occur alongside TMB.35 43 Nevertheless, immunotherapy has demonstrated clinical benefit for patients with dMMR and MSI-high tumours,30 44–48 further supporting that TMB, as a surrogate for genome instability as well as tumour neoantigens, is an appropriate biomarker for response to immunotherapy.
Overview of TMB assays and assay variations
NGS approaches
TMB assessment by NGS can involve WGS, WES, or large targeted gene panels, yet analytical and bioinformatic methods are not standardised across research or clinical studies. Both coding and non-coding sequences are analysed in WGS, but because only coding sequences are relevant for TMB assessment, WES, covering the entire exome, and gene panels, covering selected regions, are each suitable.10 16 Gene panels differ in input sample requirement, gene number or identity, region covered, workflow, and bioinformatic algorithms used (table 2); variation in TMB can arise from one or more of these parameters.11
Table 2.
Examples of NGS gene panels in development or currently available to assess TMB
| Status | Test name | Number of genes | Coverage (Mb)* | Gene variants | Sample type |
| FDA-approved or authorised diagnostic assays† | MSK-IMPACT15 56 68 | 468 | 1.5 | SNVs, indels, rearrangements/fusions, CNAs, parallel analysis of genomic signatures (eg, TMB and dMMR/MSI) | FFPE |
| Foundation Medicine FoundationOne CDx14 49 | 324 | 0.8 | SNVs, indels, CNAs, select rearrangements, parallel analysis of genomic signatures (eg, TMB and dMMR/MSI) | FFPE | |
| Commercial assays for research use only | Caris Molecular Intelligence132 | 592 | 1.4 | Somatic missense mutations | FFPE |
| Illumina TruSight 500 gene panel133 | 500 | 2.0 | SNVs and indels | FFPE | |
| Thermo Fisher Scientific Oncomine Tumor Mutation Load Assay77 | 409 | 1.7 | SNVs | FFPE | |
| NEO New Oncology NEOplus v2 RUO134 |
>340 | 1.1 | SNVs, indels, fusions, CNAs, parallel analysis of TMB, MSI, and driver mutations | FFPE | |
| Foundation Medicine FoundationOne50 | 315 | 1.1 | SNVs, indels, CNAs, select gene rearrangements, genomic signatures for MSI and TMB | FFPE | |
| Foundation Medicine bTMB assay88 122 | 394 | 1.1 | SNVs | Blood | |
| TruSight Tumor 170135 | 170 | 0.5 | Fusions, splice variants, SNVs, indels, amplifications | FFPE | |
| QIAGEN GeneRead DNAseq Comprehensive Cancer Panel97 | 160 | 0.7 | SNVs, CNAs, indels, and fusions | FFPE | |
| NEO New Oncology NEOplus105 136 | 94 | SNVs, indels, CNAs, rearrangements, and fusions | FFPE |
*Exonic breadth of coverage for the above assays is incomplete because public information may not be available for some assays.
†FoundationOne CDx has FDA premarket approval for mutations associated with several targeted therapies. In addition, FoundationOne CDx can provide tumour mutation profiling to be used by qualified healthcare professionals in accordance with professional guidelines in oncology for patients with solid malignant neoplasms.137 MSK-IMPACT is FDA-authorised to provide information on somatic mutations and MSI. TMB is captured as part of the enhanced report and is considered for investigational use only.13 15
CNA, copy number alteration; dMMR, mismatch repair deficiency; FFPE, formalin-fixed, paraffin-embedded; MSI, microsatellite instability; Mb, megabases; NGS, next-generation sequencing; SNV, single nucleotide variant; TMB, tumour mutational burden.
For most studies to date demonstrating an association between TMB and clinical benefit, TMB was measured by WES. However, it is challenging to routinely implement WES in a clinical setting because this method typically requires complex analysis and a matched normal sample for germline comparisons.11 47 49 50 Interoperability of data by WES is influenced by tumour heterogeneity, artefacts related to tissue preparation of formalin-fixed, paraffin-embedded (FFPE) samples, and discrepancies across assays available from commercial vendors.51
Targeted gene panels have been developed as an alternative method to WES. Whereas WES provides sequence coverage across the entire exome, gene panels are focused on large numbers of cancer-related genes and coupled to bioinformatic algorithms that rapidly report on cancer-targeted genomic alterations, such as copy number alterations (eg, human epidermal growth factor receptor 2 (ERBB2)), gene rearrangements (eg, anaplastic lymphoma kinase (ALK)), MSI, and TMB, along with identification of specific cancer-associated mutations (eg, epidermal growth factor receptor (EGFRT790M)).13 14 Therefore, comprehensive genomic profiling using gene panels can provide targeted information that may not be immediately available in WES data. Three gene panel tests (FoundationOne, FoundationOne CDx, and MSK-IMPACT) have documented good concordance with TMB assessment by WES using both empirical and in silico approaches.15 52–55 Therefore, gene panels potentially provide an effective approach for TMB estimation in clinical practice.11 15 49 50 56
Further concordance studies are needed to bridge TMB data from WES to gene panels, and translation studies aim to create a framework to define common TMB assay parameters and harmonise data between different gene panels. Ongoing studies by the Quality Assurance Initiative Pathology (QuIP) operated by the German Society of Pathology and the Federal Association of German Pathologists, The Friends of Cancer Research, and the International Quality Network for Pathology (IQN Path) together with the European Society of Medical Oncology (ESMO), are essential for the standardisation of TMB assessment.57–60
TMB panel size
The size of the genome area sequenced differs across assays. WES covers the coding regions of all ~22 000 genes (~180 000 exons, equivalent to 30 Mb), making up ~1% of the genome61 and encompassing most of the known disease-causing variants. FoundationOne CDx and MSK-IMPACT assays cover ~0.8 Mb over 324 genes and ~1.5 Mb over 468 genes, respectively.14 62 Some evidence suggests that smaller gene panels may be sufficient for TMB assessment,63 but other evidence indicates that they may only be sufficient to detect hypermutated and ultramutated tumours and that larger gene panels may be more accurate overall.54 64 Tumours with high TMB may be effectively identified by targeted sequencing of several hundred genes, and accuracy may not increase significantly above this threshold.11 36 56 However, variance increases significantly below 0.5 Mb, particularly in samples with low TMB.11 64 For clinical purposes, evidence suggests that gene panels of ≥0.8 Mb are needed.14 64
It is important to consider whether the specific gene sequences in a panel have an impact on the TMB result. Panels targeted to specific cancer-related genes may introduce unwanted bias into the area of genome covered, but if the panel is sufficiently large, then the function of specific genes within it should be negligible and not impact TMB assessment. There are, of course, driver mutations, such as BRCA1, TP53, and POLE, that are associated with large increases in TMB11 15 38 65 and these should be assessed alongside TMB.46 66
Depth of coverage
Alongside sequencing breadth, sequencing depth (the average number of reads that align to a known reference base67) is also variable across NGS assays. The typical sequencing depth for WES is ~100×, and only mutations with allele frequencies of >15% can be detected with confidence.51 The typical sequencing depth for a gene panel is greater at 500×, increasing the likelihood of detecting low-frequency variants at specific loci,68 69 although the minimum depth of coverage required for accurate TMB assessment may be closer to 200×.70 Together, breadth and depth of coverage may impact the sensitivity and specificity of TMB assessment, but strong evidence suggests that gene panels may be adequate to provide a reliable TMB measurement if sequencing quality thresholds are upheld.
Variant calling
A number of calculations and filters are employed downstream of sequencing to include or discard variants in the TMB estimation. The bioinformatic algorithms used are often not publicly reported, but may differ across assays and may impact TMB output.11 Understanding and harmonising these bioinformatic elements are essential.
One important consideration is the types of mutations included in the assessment, such as insertion and deletion alterations (indels), and base substitutions/single nucleotide variants (which can include synonymous and nonsynonymous mutations).11 15 49 50 71 TMB assessment by WES or gene panels commonly includes nonsynonymous somatic tumour mutations, but these may be restricted to missense mutations only. Recent evidence suggests that outcomes following immunotherapy are more strongly associated with TMB that is defined as including nonsynonymous mutations than with TMB that includes both synonymous and nonsynonymous mutations.35 However, some studies using smaller gene panels may also include synonymous mutations to ensure a representative TMB estimate.10 11 15 30 Another consideration is the variant allele frequency (VAF) cut-offs used to ensure that TMB is a reliable estimation of the number of mutations in the tumour, with minimal artefacts from sequencing errors, formalin-induced DNA damage, or sample contamination.72 73 Variant calling may be improved by capturing and sequencing both DNA strands in the target sequence.74 In studies using WES, VAF can vary from 5% to 10%,75 76 FoundationOne CDx and Oncomine assays call all variants with a VAF ≥5%,14 77 and MSK-IMPACT calls those ≥2% for mutation hotspots and ≥5% for nonhotspots.13 TMB reporting adds a further level of complexity when comparing TMB across studies, as most studies report TMB as mut/Mb, whereas others report TMB as total mutations per tumour.
Somatic variants
It is important to distinguish between tumour-specific acquired (somatic) mutations and those that are present in normal tissue (also known as germline mutations). For WES and MSK-IMPACT, paired sample mutation calling is typically performed on tumour samples and matched germline control (blood) samples.11 13 15 Using control samples for assessment of germline variants may not be feasible, practical, or even legal, because germline testing may have further-reaching implications outside cancer diagnosis.78–80 Somatic mutations can be inferred in tumours without matched controls in silico from databases of known polymorphisms or variant data in tissue from different patients.11 14 54 81 Some assays filter potential germline mutations using a somatic-germline zygosity algorithm, which identifies the origin of the mutation by leveraging allele frequencies and genome-wide copy number.11 14 82 Although these approaches simplify the TMB assessment workflow, they may result in false positives and higher TMB, particularly in patients with ethnic backgrounds that are less well represented in reference databases.11 54 These effects may be negligible for TMB assessment by WES or large gene panels, but increased in smaller panels.54
Defining TMB thresholds
TMB thresholds were initially defined retrospectively in exploratory analyses of immunotherapy efficacy, with patients being grouped by TMB tertile or quartile35 52 83–85 or by numeric cut-offs, such as 100 or 178 mutations per exome22 36 86 or 9–20 mut/Mb.11 87 88 To date, the TMB threshold of ≥10 mut/Mb, measured by FoundationOne CDx (equivalent to ~200 mutations by WES) in first-line patients with non-small cell lung cancer (NSCLC), is the only cut-off established for enhanced response to immunotherapy in one trial and clinically validated in a separate study using a preplanned approach.8 9 55
Identifying a single, fixed TMB threshold that could be applicable across different tumours may be difficult, because the median number of somatic mutations differs across tumour types (figure 1).26 30 In silico analyses suggested that clinical response in NSCLC and melanoma may increase and then plateau at ~260 nonsynonymous mutations (measured by WES),31 but this prediction would need stringent validation to determine which types of mutations contributed to this number and how to translate this value into other patient groups. Further studies evaluating a clinically validated TMB cut-off will be instrumental in providing an agreed TMB cut-off within particular tumour types. Increased use of specific approved commercial assays will also assist in cross-trial comparisons.
Considerations for sample preparation and TMB assays
Samples used for TMB assessment may be cytology or liquid biopsies, or FFPE tumour tissue. Although some evidence exists for the feasibility of using cytology and fine-needle aspiration for analysis by NGS,89 90 these samples can yield variable amounts of DNA that may not always be sufficient for in-depth molecular analysis.91 92 FFPE samples are the most commonly available patient samples for clinical testing. FFPE slides and extracted DNA are reliable for TMB assessment for 6 months and likely beyond.14 However, FFPE samples present a risk for use with high-throughput genomic assays, because preanalytical factors common to NGS testing can affect the sample integrity or amount of DNA that can be extracted.72 73 93 94 These factors include clinical sample availability; sample collection, preparation, and fixation (optimally 24 hours in neutral buffered formalin for surgical specimens and 12 hours for biopsies95 96); neoplastic cell content in the sample; batch processing; and ensuring sufficient DNA quality and yield for assessment (10–100 ng for gene panels and around 250 ng for WES).14 62 97–100 Insufficient DNA is a common cause of sample attrition in clinical trials,52 101 so assays that require less DNA may be advantageous.
In addition to variability in sample handling, technical processes and working practices employed by clinical laboratories can vary extensively.102 Specifically, centralised and decentralised laboratories differ in infrastructure, equipment, and gene panels used, and different platforms may vary in bioinformatic workflow, data integration, and/or how the data are reported.47 103 104
TMB from liquid biopsies
Interest in isolating cell-free DNA (cfDNA) from liquid biopsies for assessment of circulating tumour DNA (ctDNA) is increasing because of the less invasive nature of sample collection from patients and greater opportunity for obtaining frequent samples. TMB measurements from cfDNA may have advantages over tissue biopsy in the follow-up of early responses to immunotherapy due to the ease and speed of obtaining them.105 TMB assays that use cfDNA have been developed and validated, with ongoing efforts for harmonisation with TMB data from tissue samples.87 106–109 Data for TMB concordance between tissue and cfDNA by gene panel assays are conflicting, with reports of both good concordance110 and a lack of concordance.111 Some studies have demonstrated that the sensitivity of detecting mutations from cfDNA is decreased compared with solid tumour samples for both WES and gene panels, although the correlation between TMB assessed from tissue and cfDNA is greater by WES than a panel.75 105 107 Liquid biopsies may yield a variable mix of normal and tumour DNA, and the tumour DNA could reflect heterogeneity across different tumour sites. Thus, variable data exist for concordance between solid and liquid samples, and there is a need for further studies. Nevertheless, future clinical use of TMB assessment from liquid biopsies remains feasible.
Turnaround time
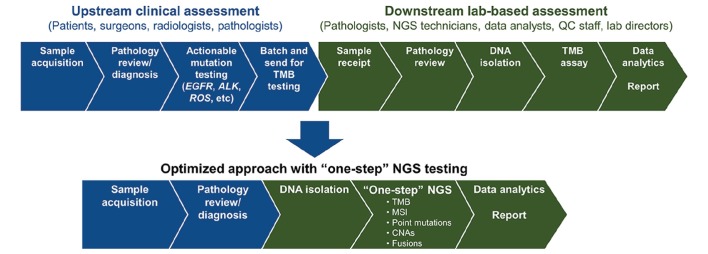
Alongside technical specifications of the tests, other regulatory, practical, and logistic factors are important, such as assay turnaround time, runtime, and cost. Turnaround time is impacted by the time required for steps in the workflow and, therefore, the availability of key stakeholders to carry out their role in the generation of a reliable TMB result (figure 2). Given the comprehensive nature of the assay, it is difficult to balance turnaround time with cost, and microcosting analyses are urgently needed.112 The common turnaround time for TMB assessment by commercial assays is approximately 2 weeks, including 4–5 days of library preparation time.14 69 Moving forward, there is a need to establish acceptable TMB assay turnaround times, especially if the test is applied in the first-line cancer setting. A test for both actionable mutations and TMB is preferable for first-line patients. Moreover, TMB is a surrogate marker for neoantigen load, which may impact immune response and response to therapy, and studies monitoring TMB suggest that follow-up testing could detect residual disease or recurrence.21 22 113
Figure 2.
Biopsy sample workflow for TMB testing. A proposed, optimised workflow is shown to streamline diagnostic testing for TMB alongside other genomic markers. ALK, anaplastic lymphoma kinase; CNA, copy number alteration; EGFR, epidermal growth factor receptor; MSI, microsatellite instability; NGS, next-generation sequencing; QC, quality control; ROS, ROS proto-oncogene 1, receptor tyrosine kinase; TMB, tumour mutational burden.
Quality assurance
Multidisciplinary teams are involved in oncology sampling and diagnostics, from sample collection by the surgeon through to TMB assay result interpretation (figure 2). There is a need for all to be coordinated and well informed on the necessary approaches to achieve a reliable assay result, and molecular tumour boards have been set up to ensure global harmonisation in tumour-sequencing practices.114 Recent NGS guidelines from pathologists emphasise the role of the laboratory director in using an error-based approach that can identify potential sources of errors that may occur throughout the analytical process.72
The robustness of a TMB assessment method, reflected by its approval status (FDA, CLIA, CE-IVD approved, or for research use only), is likely to influence the choice of assay used in clinical practice. Many steps within the TMB assessment pathway can vary and their impact should be assessed. For example, changing an extraction protocol may pose a minor risk if quality control is in place to verify sample purity and concentration; however, changes to which NGS platform is used are likely to require a new validation process.72 Appropriately trained personnel at each step of the workflow to carry out quality-control processes will minimise variance and impact.72
As TMB becomes more established, an increase in the number of commercial or LDT panels is anticipated. Recommendations for how to implement TMB assays will be developed further with input from the international harmonisation efforts described above. It is therefore important to understand how LDTs can augment TMB testing. The identification of reference standards or external quality evaluation is a way to compare LDTs with other methods. It is likely that the clinical evidence for robust TMB assessment will be limited to a few key assays, with a need for calibration and concordance studies to facilitate the transition from clinical trials to the real-world setting.
Challenges and solutions to adopting TMB assays in clinical practice
Because data have demonstrated that high TMB is a predictive marker for clinical benefit from immunotherapy, the ability to classify TMB as high or low is essential for predicting response. Defining cut-offs for TMB may directly lead to inclusion or exclusion of some patients from treatment. However, most published clinical studies of TMB to date have been exploratory, using a number of different methods, and it is challenging to compare data across studies. The availability of preanalytic, sequencing, and bioinformatic protocols will help to harmonise TMB estimation with clinical outcomes.
Lessons can also be learnt from experiences with PD-L1 expression testing. The need for sample quality control, concordance studies across PD-L1 testing platforms to determine the interchangeability of tests, and appropriate training for key personnel to interpret the data are all applicable for any biomarker. Even though TMB outputs are based on computational assessment, there is still a need for education and training on how these values are derived.
With PD-L1 expression and TMB as independent and potentially complementary predictive markers for response to immunotherapy,52 questions turn to the possible need for coordinated multiparameter or composite testing to continue to advance the field. Interplay between TMB, PD-L1, tumour-infiltrating lymphocytes, and cytolytic activity is emerging from comprehensive immunogenomic analyses,26 raising the possibility of composite biomarker testing within the tumour microenvironment.
Conclusions
Validated predictive biomarkers should accurately predict patient responses to immunotherapy. The use of TMB assessment has been actively addressed by ongoing studies, including those clinically validating TMB as a biomarker for response to immunotherapy. Using valuable lessons learnt from PD-L1 testing, including the need for concordance studies across testing platforms and the need for trained personnel to assess the assay results, the development of TMB as a clinical diagnostic tool is being defined.
As the field evolves, solutions emerge to ensure that reliable TMB assessments are carried out in clinical practice. However, definitive recommendations may not be possible until more data are available from prospective clinical trials. From the knowledge gained so far, key parameters can be highlighted throughout the TMB testing workflow, including collection of samples, methods, and analysis (table 3). These include current suggestions to use solid tissue samples, to use gene panels with a coverage >0.8 Mb (preferably >1 Mb), to ensure sufficient information is given when reporting TMB values (including number of genes, capture region, germline subtraction, types of mutations called, variant calling threshold, and sequence depth72 103), to participate in external quality-control schemes when available and to ensure trained stakeholders are available for efficient workflow and assay interpretation.
Table 3.
Key parameters for the harmonisation of TMB analysis and workflow
| Parameter | Principles |
| Sample |
|
| Methodology |
|
| Platforms |
|
| Germline |
|
| Algorithm |
|
| Cut-off |
|
| Concordance |
|
| Reporting |
|
BRAF, B-Raf proto-oncogene, serine/threonine kinase; EGFR, epidermal growth factor receptor; FFPE, formalin-fixed, paraffin-embedded; FNA, fine-needle aspiration; KRAS, KRAS proto-oncogene, GTPase; mut/Mb, mutations per megabase; NGS, next-generation sequencing; TMB, tumour mutational burden; WES, whole exome sequencing.
It will be necessary to accumulate larger datasets, set minimal tissue sample purity thresholds, and standardise sample processing and robust bioinformatic pipelines such that TMB can be calculated in a reliable manner.1 11 99 115 116 Further analyses will also be required to understand how biomarkers, such as TMB, PD-L1, and other genetic/immune markers, interact.
Despite method variability, scoring the number of somatic mutations in tumour DNA is relatively simple, and exhaustive characterisation of tumours will increase understanding of their characteristics. As momentum for TMB as a biomarker of immunotherapy response increases and standardised approaches begin to emerge to allow for clinical implementation, it is likely that TMB assessment, with or without additional biomarkers, will be at the forefront of precision medicine in the foreseeable future.
Footnotes
Contributors: All authors contributed to writing the manuscript and approved the final version.
Funding: Medical writing support and editorial assistance were provided by Stuart Rulten, PhD, and Jay Rathi, MA, of Spark Medica Inc, funded by Bristol-Myers Squibb, according to Good Publication Practice guidelines.
Competing interests: ER reports personal fees and non-financial support from AstraZeneca and Bristol-Myers Squibb. JL reports grants from Agilent Technologies and Roche Tissue Diagnostics, and personal fees from AstraZeneca, Bristol-Myers Squibb, Genentech, Merck, Pfizer, and Roche Tissue Diagnostics. FL-R reports personal fees from AstraZeneca, Bristol-Myers Squibb, Life Technologies, Merck, Pfizer, and Roche. FP-L reports grants and personal fees from AstraZeneca, Bristol-Myers Squibb, Merck, and Roche, and grants from NanoString. NN reports grants and personal fees from AstraZeneca, Bristol-Myers Squibb, Qiagen, Roche, and Thermo Fisher Scientific, and grants and non-financial support from Merck. SM-B reports grants and personal fees from AstraZeneca and Novartis, and personal fees from Bristol-Myers Squibb and Roche. RB declares no conflicts of interest.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 2016;17:e542–e551. 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehnert JM, Monjazeb AM, Beerthuijzen JMT, et al. The challenge for development of valuable immuno-oncology biomarkers. Clin Cancer Res 2017;23:4970–9. 10.1158/1078-0432.CCR-16-3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hersom M, Jørgensen JT. Companion and complementary diagnostics-focus on PD-L1 expression assays for PD-1/PD-L1 checkpoint inhibitors in non-small cell lung cancer. Ther Drug Monit 2018;40:9–16. 10.1097/FTD.0000000000000460 [DOI] [PubMed] [Google Scholar]
- 4.Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol 2016;27:1492–504. 10.1093/annonc/mdw217 [DOI] [PubMed] [Google Scholar]
- 5.Büttner R, Gosney JR, Skov BG, et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non-small-cell lung cancer. J Clin Oncol 2017;35:3867–76. 10.1200/JCO.2017.74.7642 [DOI] [PubMed] [Google Scholar]
- 6.Rimm DL, Han G, Taube JM, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol 2017;3:1051–8. 10.1001/jamaoncol.2017.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voong KR, Feliciano J, Becker D, et al. Beyond PD-L1 testing-emerging biomarkers for immunotherapy in non-small cell lung cancer. Ann Transl Med 2017;5:376 10.21037/atm.2017.06.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093–104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramalingam S, Hellmann MD, Awad M. Tumor mutational burden (TMB) as a biomarker for clinical benefit from dual immune checkpoint blockade with nivolumab (nivo) + ipilimumab (ipi) in first-line (1L) non-small cell lung cancer (NSCLC): identification of TMB cutoff from CheckMate 568. Presented at the AACR Annual Meeting, Chicago, IL, 2018. [Google Scholar]
- 10.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 11.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FDA FDA announces approval, CMS proposes coverage of first breakthrough-designated test to detect extensive number of cancer biomarkers. 2017. Available from:https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm587273.htm
- 13.FDA FDA unveils a streamlined path for the authorization of tumor profiling tests alongside its latest product action. 2018. Available from:https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm585347.htm
- 14.FDA FoundationOne CDx: Summary of Safety and Effectiveness Data (SSED). 2018. Available from:https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019B.pdf
- 15.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabanon RM, Pedrero M, Lefebvre C, et al. Mutational landscape and sensitivity to immune checkpoint blockers. Clin Cancer Res 2016;22:4309–21. 10.1158/1078-0432.CCR-16-0903 [DOI] [PubMed] [Google Scholar]
- 17.Gilboa E. The makings of a tumor rejection antigen. Immunity 1999;11:263–70. 10.1016/S1074-7613(00)80101-6 [DOI] [PubMed] [Google Scholar]
- 18.Łuksza M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017;551:517–20. 10.1038/nature24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geyer RJ, Tobet R, Berlin RD, et al. Immune response to mutant neo-antigens: Cancer's lessons for aging. Oncoimmunology 2013;2:e26382 10.4161/onci.26382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. Embo J 2013;32:194–203. 10.1038/emboj.2012.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017;171:934–49. 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber J, Horak C, Hodi FS, et al. Baseline tumor T cell receptor (TcR) sequencing analysis and neo antigen load is associated with benefit in melanoma patients receiving sequential nivolumab and ipilimumab. Ann Oncol 2016;27:1047O 10.1093/annonc/mdw378.0127029704 [DOI] [Google Scholar]
- 24.Liontos M, Anastasiou I, Bamias A, et al. DNA damage, tumor mutational load and their impact on immune responses against cancer. Ann Transl Med 2016;4:264 10.21037/atm.2016.07.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YP, Zhang Y, Lv JW, et al. Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: Immunotherapeutic implications. Theranostics 2017;7:3585–94. 10.7150/thno.21471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulie PG, Van den Eynde BJ, van der Bruggen P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135–46. 10.1038/nrc3670 [DOI] [PubMed] [Google Scholar]
- 29.Grivas PD, Melas M, Papavassiliou AG. The biological complexity of urothelial carcinoma: Insights into carcinogenesis, targets and biomarkers of response to therapeutic approaches. Semin Cancer Biol 2015;35:125–32. 10.1016/j.semcancer.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colli LM, Machiela MJ, Myers TA, et al. Burden of nonsynonymous mutations among TCGA cancers and candidate immune checkpoint inhibitor responses. Cancer Res 2016;76:3767–72. 10.1158/0008-5472.CAN-16-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109–21. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 35.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018;33:843–52. 10.1016/j.ccell.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buecher B, Cacheux W, Rouleau E, et al. Role of microsatellite instability in the management of colorectal cancers. Dig Liver Dis 2013;45:441–9. 10.1016/j.dld.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 38.Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell 2017;171:1042–56. 10.1016/j.cell.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 2017;2017:1–15. 10.1200/PO.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takamochi K, Takahashi F, Suehara Y, et al. DNA mismatch repair deficiency in surgically resected lung adenocarcinoma: Microsatellite instability analysis using the Promega panel. Lung Cancer 2017;110:26–31. 10.1016/j.lungcan.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 41.Song Z, Cheng G, Xu C, et al. Clinicopathological characteristics of POLE mutation in patients with non-small-cell lung cancer. Lung Cancer 2018;118:57–61. 10.1016/j.lungcan.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 42.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 2018;8:822–835. 10.1158/2159-8290.CD-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801–6. 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viale G, Trapani D, Curigliano G. Mismatch repair deficiency as a predictive biomarker for immunotherapy efficacy. Biomed Res Int 2017;2017:4719194 10.1155/2017/4719194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuchs CS, Doi T, Jang RW, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol 2018;4:e180013 10.1001/jamaoncol.2018.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campesato LF, Barroso-Sousa R, Jimenez L, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 2015;6:34221–7. 10.18632/oncotarget.5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu P, Pang L, Arreaza G, et al. Data interoperability of whole exome sequencing (WES) based mutational burden estimates from different laboratories. Int J Mol Sci 2016;17:651 10.3390/ijms17050651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017;376:2415–26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res 2016;4:959–67. 10.1158/2326-6066.CIR-16-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garofalo A, Sholl L, Reardon B, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med 2016;8:79 10.1186/s13073-016-0333-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szustakowski JD, Green G, Geese WJ, et al. Evaluation of tumor mutation burden as a biomarker for immune checkpoint inhibitor efficacy: a calibration study of whole exome sequencing with FoundationOne®. Presented at the AACR Annual Meeting, Chicago, IL, 2018. [Google Scholar]
- 56.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36:633–41. 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tkach Tuzman K Shared burden: how tests developers are seeking harmony for tumor mutational burden assays. 2018. Available from:https://www.biocentury.com/bc-innovations/tools-techniques/2018-03-15/how-test-developers-are-seeking-harmony-tumor-mutational-
- 58.Deans ZC, Costa JL, Cree I, et al. Integration of next-generation sequencing in clinical diagnostic molecular pathology laboratories for analysis of solid tumours; an expert opinion on behalf of IQN Path ASBL. Virchows Arch 2017;470:5–20. 10.1007/s00428-016-2025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Krieken H, Deans S, Hall JA, et al. Quality to rely on: meeting report of the 5th Meeting of External Quality Assessment, Naples 2016. ESMO Open 2016;1:e000114 10.1136/esmoopen-2016-000114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.IQN Path International quality network for pathology. Annual Report. 2017. Available from:http://www.iqnpath.org/wp-content/uploads/2018/04/IQNPath_AnnualReport2017-26032018.pdf
- 61.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272–6. 10.1038/nature08250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.FDA Evaluation of Automatic Class III Designation for MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets): decision summary. 2018. Available from:https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN170058.pdf
- 63.Yourstone S, Mola N, Weigman VJJ. Effects of sequencing parameters and panel size on mutational burden calculations. Presented at the AACR Annual Meeting, Chicago, IL, 2018. [Google Scholar]
- 64.Baras AS, Stricker T. Characterization of total mutational burden in the GENIE cohort: Small and large panels can provide TMB information but to varying degrees. Cancer Res 2017;77:LB-105 10.1158/1538-7445.AM2017-LB-105 [DOI] [Google Scholar]
- 65.Spigel DR, Schrock AB, Fabrizio D, et al. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol 2016;34:9017 10.1200/JCO.2016.34.15_suppl.9017 [DOI] [Google Scholar]
- 66.Chaudhuri A, Chabon JJ, Lovejoy AF, et al. Analysis of circulating tumor DNA in localized lung cancer for detection of molecular residual disease and personalization of adjuvant strategies. J Clin Oncol 2017;35:8519 10.1200/JCO.2017.35.15_suppl.8519 [DOI] [Google Scholar]
- 67.Sims D, Sudbery I, Ilott NE, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 2014;15:121–32. 10.1038/nrg3642 [DOI] [PubMed] [Google Scholar]
- 68.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feliubadaló L, Tonda R, Gausachs M, et al. Benchmarking of whole exome sequencing and ad hoc designed panels for genetic testing of hereditary cancer. Sci Rep 2017;7 10.1038/srep37984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee C, Bae JS, Ryu GH, et al. A method to evaluate the quality of clinical gene-panel sequencing data for single-nucleotide variant detection. J Mol Diagn 2017;19:651–8. 10.1016/j.jmoldx.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 71.Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn 2013;15:607–22. 10.1016/j.jmoldx.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 72.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 2017;19:341–65. 10.1016/j.jmoldx.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 2002;161:1961–71. 10.1016/S0002-9440(10)64472-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moens LN, Falk-Sörqvist E, Ljungström V, et al. HaloPlex targeted resequencing for mutation detection in clinical formalin-fixed, paraffin-embedded tumor samples. J Mol Diagn 2015;17:729–39. 10.1016/j.jmoldx.2015.06.009 [DOI] [PubMed] [Google Scholar]
- 75.Koeppel F, Blanchard S, Jovelet C, et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS One 2017;12:e0188174 10.1371/journal.pone.0188174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lauss M, Donia M, Harbst K, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun 2017;8:8 10.1038/s41467-017-01460-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.ThermoFisher ThermoFisher Oncomine™ tumor mutation load assay user guide. 2018. Available from:https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0017042_TumorMutLoad_UG.pdf
- 78.Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med 2016;13:3–11. 10.20892/j.issn.2095-3941.2016.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology Policy Statement Update: genetic and genomic testing for cancer susceptibility. J Clin Oncol 2015;33:3660–7. 10.1200/JCO.2015.63.0996 [DOI] [PubMed] [Google Scholar]
- 80.Robson ME, Storm CD, Weitzel J, et al. American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol 2010;28:893–901. 10.1200/JCO.2009.27.0660 [DOI] [PubMed] [Google Scholar]
- 81.Tian Y, Jones W, Mola N. TMB calling in absence of matched normal: learning germline behavior. Presented at the AACR Annual Meeting, Chicago, IL, 2018. [Google Scholar]
- 82.Sun JX, He Y, Sanford E, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol 2018;14:e1005965 10.1371/journal.pcbi.1005965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galsky MD, Saci A, Szabo PM, et al. Impact of tumor mutation burden on nivolumab efficacy in second-line urothelial carcinoma patients: Exploratory analysis of the phase II CheckMate 275 study. Ann Oncol 2017;28:v295–v329. 10.1093/annonc/mdx371.003 [DOI] [Google Scholar]
- 84.Antonia S, Callahan MK, Awad MM, et al. Impact of tumor mutation burden on the efficacy of nivolumab or nivolumab + ipilimumab in small cell lung cancer: an exploratory analysis of CheckMate 032. Presented at the IASLC 18th World Conference on Lung Cancer, Yokohama, Japan, 2017. [Google Scholar]
- 85.Kowanetz M, Zou W, Shames D, et al. OA20.01 Tumor Mutation Burden (TMB) is Associated with Improved Efficacy of Atezolizumab in 1L and 2L+ NSCLC Patients. J Thorac Oncol 2017;12:S321–S322. 10.1016/j.jtho.2016.11.343 [DOI] [Google Scholar]
- 86.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fabrizio D, Lieber D, Malboeuf C, et al. A blood-based next-generation sequencing assay to determine tumor mutational burden (bTMB) is associated with benefit to an anti-PD-L1 inhibitor, atezolizumab. Presented at the AACR Annual Meeting, Chicago, IL, 2018. [Google Scholar]
- 88.Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441–8. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- 89.Stoy SP, Segal JP, Mueller J, et al. Feasibility of endobronchial ultrasound-guided transbronchial needle aspiration cytology specimens for next generation sequencing in non-small-cell lung cancer. Clin Lung Cancer 2018;19:230–8. 10.1016/j.cllc.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 90.DiBardino DM, Rawson DW, Saqi A, et al. Next-generation sequencing of non-small cell lung cancer using a customized, targeted sequencing panel: Emphasis on small biopsy and cytology. Cytojournal 2017;14:7 10.4103/1742-6413.202602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roy-Chowdhuri S, Chow CW, Kane MK, et al. Optimizing the DNA yield for molecular analysis from cytologic preparations. Cancer Cytopathol 2016;124:254–60. 10.1002/cncy.21664 [DOI] [PubMed] [Google Scholar]
- 92.Chen H, Luthra R, Goswami RS, et al. Analysis of pre-analytic factors affecting the success of clinical next-generation sequencing of solid organ malignancies. Cancers 2015;7:1699–715. 10.3390/cancers7030859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang P, Lehmann BD, Shyr Y, et al. The utilization of formalin fixed-paraffin-embedded specimens in high throughput genomic studies. In Int J Genomics. Hindawi Publishing Corporation 2017;1926304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grizzle WE. Special symposium: fixation and tissue processing models. Biotech Histochem 2009;84:185–93. 10.3109/10520290903039052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Einaga N, Yoshida A, Noda H, et al. Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation. PLoS One 2017;12:e0176280 10.1371/journal.pone.0176280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Howat WJ, Wilson BA. Tissue fixation and the effect of molecular fixatives on downstream staining procedures. Methods 2014;70:12–19. 10.1016/j.ymeth.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.QIAGEN GeneRead DNAseq targeted panels V2. 2018. Available from:https://www.qiagen.com/us/shop/sample-technologies/dna/genomic-dna/generead-dnaseq-gene-panels-v2/?catno=NGHS-501X
- 98.Kircher M, Kelso J. High-throughput DNA sequencing--concepts and limitations. Bioessays 2010;32:524–36. 10.1002/bies.200900181 [DOI] [PubMed] [Google Scholar]
- 99.Roh W, Chen PL, Reuben A, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med 2017;9:eaah3560 10.1126/scitranslmed.aah3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cree IA, Deans Z, Ligtenberg MJ, et al. Guidance for laboratories performing molecular pathology for cancer patients. J Clin Pathol 2014;67:923–31. 10.1136/jclinpath-2014-202404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Al-Kateb H, Nguyen TT, Steger-May K, et al. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS). Mol Oncol 2015;9:1737–43. 10.1016/j.molonc.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pant S, Weiner R, Marton MJ. Navigating the rapids: the development of regulated next-generation sequencing-based clinical trial assays and companion diagnostics. Front Oncol 2014;4:78 10.3389/fonc.2014.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roy S, Coldren C, Karunamurthy A, et al. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn 2018;20:4–27. 10.1016/j.jmoldx.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 104.Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med 2016;14:168 10.1186/s12916-016-0705-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Müller JN, Falk M, Talwar J, et al. Concordance between comprehensive cancer genome profiling in plasma and tumor specimens. J Thorac Oncol 2017;12:1503–11. 10.1016/j.jtho.2017.07.014 [DOI] [PubMed] [Google Scholar]
- 106.Sholl LM. Next-generation sequencing from liquid biopsies in lung cancer patients: advances in comprehensive biomarker testing. J Thorac Oncol 2017;12:1464–6. 10.1016/j.jtho.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 107.Khagi Y, Goodman AM, Daniels GA, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res 2017;23:5729–36. 10.1158/1078-0432.CCR-17-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 109.Sato KA, Hachiya T, Iwaya T, et al. Individualized mutation detection in circulating tumor DNA for monitoring colorectal tumor burden using a cancer-associated gene sequencing panel. PLoS One 2016;11:e0146275 10.1371/journal.pone.0146275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mehrotra M, Singh RR, Loghavi S, et al. Detection of somatic mutations in cell-free DNA in plasma and correlation with overall survival in patients with solid tumors. Oncotarget 2018;9:10259–71. 10.18632/oncotarget.21982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Davis AA, Chae YK, Agte S, et al. Comparison of tumor mutational burden (TMB) across tumor tissue and circulating tumor DNA (ctDNA). J Clin Oncol 2017;35:e23028 10.1200/JCO.2017.35.15_suppl.e23028 [DOI] [Google Scholar]
- 112.Sabatini LM, Mathews C, Ptak D, et al. Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the association for molecular pathology. J Mol Diagn 2016;18:319–28. 10.1016/j.jmoldx.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van der Velden DL, van Herpen CML, van Laarhoven HWM, et al. Molecular tumor boards: current practice and future needs. Ann Oncol 2017;28:3070–5. 10.1093/annonc/mdx528 [DOI] [PubMed] [Google Scholar]
- 115.Jamieson NB, Maker AV. Gene-expression profiling to predict responsiveness to immunotherapy. Cancer Gene Ther 2017;24:134–40. 10.1038/cgt.2016.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mola N, Schu M, Stiegelmeyer S, et al. Tumor mutational burden: guidelines for derivation and robustness of measurement. Presented at the AACR Annual Meeting, Chicago, IL, 2018. [Google Scholar]
- 117.Singal G, Miller PG, Agarwala V, et al. Analyzing biomarkers of cancer immunotherapy (CIT) response using a real-world clinico-genomic database. Ann Oncol 2017;28:v403–v427. 10.1093/annonc/mdx376.005 [DOI] [Google Scholar]
- 118.Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol 2016;34:2206–11. 10.1200/JCO.2016.66.6552 [DOI] [PubMed] [Google Scholar]
- 119.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seiwert TY, Cristescu R, Mogg R, et al. Genomic biomarkers in relation to PD-1 checkpoint blockade response. J Clin Oncol 2018;36:25 10.1200/JCO.2018.36.5_suppl.2529035642 [DOI] [Google Scholar]
- 122.Fabrizio D, Malboeuf C, Lieber D, et al. Analytic validation of a next generation sequencing assay to identify tumor mutational burden from blood (bTMB) to support investigation of an anti-PD-L1 agent, atezolizumab, in a first line non-small cell lung cancer trial (BFAST). Ann Oncol 2017;28:v22–v24. 10.1093/annonc/mdx363.018 [DOI] [Google Scholar]
- 123.Velcheti V, Kim ES, Mekhail T, et al. Prospective clinical evaluation of blood-based tumor mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC): Interim B-F1RST results. J Clin Oncol 2018;36:12001 10.1200/JCO.2018.36.15_suppl.12001 [DOI] [Google Scholar]
- 124.Mok TSK, Gadgeel S, Kim ES, et al. Blood first line ready screening trial (B-F1RST) and blood first assay screening trial (BFAST) enable clinical development of novel blood-based biomarker assays for tumor mutational burden (TMB) and somatic mutations in 1L advanced or metastatic NSCLC. Ann Oncol 2017;28:v460–v496. 10.1093/annonc/mdx380.084 [DOI] [Google Scholar]
- 125.Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 2017;389:67–76. 10.1016/S0140-6736(16)32455-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909–20. 10.1016/S0140-6736(16)00561-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Powles T, Loriot Y, Ravaud A, et al. Atezolizumab (atezo) vs. chemotherapy (chemo) in platinum-treated locally advanced or metastatic urothelial carcinoma (mUC): Immune biomarkers, tumor mutational burden (TMB), and clinical outcomes from the phase III IMvigor211 study. J Clin Oncol 2018;36(6_suppl):409 10.1200/JCO.2018.36.6_suppl.409 [DOI] [Google Scholar]
- 128.Snyder A, Nathanson T, Funt SA, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med 2017;14:e1002309 10.1371/journal.pmed.1002309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rozenblum AB, Ilouze M, Dudnik E, et al. Clinical impact of hybrid capture-based next-generation sequencing on changes in treatment decisions in lung cancer. J Thorac Oncol 2017;12:258–68. 10.1016/j.jtho.2016.10.021 [DOI] [PubMed] [Google Scholar]
- 130.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598–608. 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bonta I, Isac JF, Meiri E, et al. Correlation between tumor mutation burden and response to immunotherapy. J Clin Oncol 2017;35(15_suppl):e14579 10.1200/JCO.2017.35.15_suppl.e14579 [DOI] [Google Scholar]
- 132.Hussain MA, Heath EI, Somer BG, et al. Spectrum of tumor mutational load (TML) in genitourinary cancers (GU CA). J Clin Oncol 2017;35(15_suppl):4535 10.1200/JCO.2017.35.15_suppl.4535 [DOI] [Google Scholar]
- 133.So AS, Kaplan S, Zhang S, et al. Accurate measurement of tumor mutation burden through tumor-only sequencing using a 500-gene panel. Presented at the AACR Annual Meeting, Chicago, IL, 2018 10.1158/1538-7445.AM2018-435 [DOI] [Google Scholar]
- 134.New Oncology , 2018. Assays. Available: http://www.newoncology.com/en/assays
- 135.Trusight , Tumor 170. 2018. Available: https://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/trusight-tumor-170-data-sheet-1170-2016-017.pdf
- 136.Menon R, Mariotti E, Mueller J, et al. PUB053 panel based hybrid capture sequencing assay to correlate mutational load with response to immunotherapy. J Thorac Oncol 2017;12:S1478–S1479. 10.1016/j.jtho.2016.11.2023 [DOI] [Google Scholar]
- 137.FDA Approval of FoundationOne CDx - P170019. 2017. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019a.pdf