Summary
Serum IgA antibodies are readily detected in mice and people, yet the mechanisms underlying the induction of serum IgA and its role in host protection remains uncertain. We report that select commensal bacteria induce several facets of systemic IgA-mediated immunity. Exposing conventional mice to a unique but natural microflora that included several members of the Proteobacteria phylum led to T cell-dependent increases in serum IgA levels and the induction of large numbers of IgA-secreting plasma cells in the bone marrow. The resulting serum IgA bound to a restricted collection of bacterial taxa, and antigen-specific serum IgA antibodies were readily induced after intestinal colonization with the commensal bacterium Helicobacter muridarum. Finally, movement to a Proteobacteria-rich microbiota led to serum IgA-mediated resistance to polymicrobial sepsis. We conclude that commensal microbes overtly influence the serum IgA repertoire, resulting in constitutive protection against bacterial sepsis.
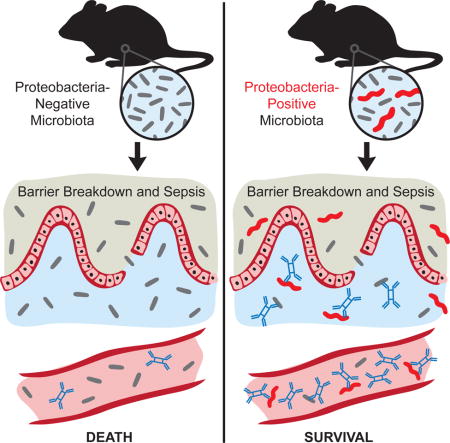
Graphical abstract
In Brief: Wilmore et al. demonstrate a role for serum IgA in protection against polymicrobial sepsis. Induction of protective concentrations of T cell-dependent serum IgA requires colonization of the gut with a complex microbiota that includes bacteria in the phylum Proteobacteria.
Introduction
For many years it was widely accepted that mucosal immune responses are physically and functionally separate from processes regulating systemic immunity. With regard to humoral responses, antigenic invasion of mucosal tissues was shown to result in effective local antibody responses, with little or no contribution to serum antibody concentrations (reviewed in (Macpherson et al., 2008; Tomasi and Bienenstock, 1968)). This viewpoint is supported further by experiments showing that serum IgA antibodies fail to bind to commensal bacterial antigens (Macpherson et al., 2000; Macpherson et al., 2012). Reciprocally, immunization via non-mucosal routes typically results in systemic humoral immunity without inducing protection at mucosal sites. Taken at face value these studies would appear to establish a clear dichotomy between mucosal and systemic responses.
Recent observations however by several groups highlight the capacity of mucosal immune interactions to impact systemic immunity. For instance, colonization of mice with Segmented Filamentous Bacteria (SFB) facilitates the development of autoimmune arthritis associated with the induction of TH17 cells and germinal centers (GCs) (Ivanov et al., 2009; Lecuyer et al., 2014; Talham et al., 1999; Wu et al., 2010; Yang et al., 2014). Additionally, IgA antibodies specific for phosphorylcholine, a common cell wall constituent of many commensal bacteria, are readily observed in sera from common inbred mice (Morahan et al., 1983), and recent work shows that antigen-specific IgA-secreting bone marrow (BM) plasma cells are induced after mucosal immunization with the relatively pro-inflammatory antigen cholera toxin (Lemke et al., 2016). Furthermore, two recent reports revealed that commensal microbes induce serum IgG responses with the capacity to block bacterial infection and modulate mucosal T cell populations (Koch et al., 2016; Zeng et al., 2016). However, whereas it is clear that IgA synthesis plays a critical role in establishing intestinal homeostasis (Fagarasan et al., 2002; Wei et al., 2011), the generation and regulation of serum IgA responses and their role in protective immunity remain largely undefined.
Current models hold that B cells located in the marginal zone (MZ) of the spleen, together with B1 B cells, rapidly generate IgM antibodies against blood-borne bacteria in sepsis and related scenarios (Martin et al., 2001; Pillai et al., 2005). This idea is consistent with the uniquely rapid kinetics with which MZ B cells generate plasma cells in response to toll-like receptor ligands such as lipopolysaccharide (Oliver et al., 1997), and data showing that splenectomy increases susceptibility to bacterial infections including septicemia (Thai et al., 2016). An additional layer of protection may by provided by serum IgG antibodies that result from mucosal B cell responses to the bacterial microbiota in the gut (Koch et al., 2016; Zeng et al., 2016), although the role of such responses in bacterial sepsis has not been tested. Here we report that serum IgA antibodies provide a unique and constitutive protective barrier against polymicrobial sepsis. Our results further show that modulations in the microbial composition of the gut result in heightened serum IgA concentrations that coincide with colonization of the BM by large numbers of IgA-secreting plasma cells and marked changes in the serum IgA repertoire. Such antibodies are induced by a wide variety of bacterial taxa, but are generally enriched with members of the Proteobacteria phylum. Altogether our results demonstrate that commensal microbes can have a substantial impact on the composition of the serum IgA repertoire and the BM plasma cell pool, resulting in protection against widespread and lethal bacterial invasion.
Results
Commensal microbes drive the generation of serum IgA and IgA-secreting BM plasma cells
Recent work indicates that shifts in the GI microbiota can impact the generation of effector and regulatory T cells (Atarashi et al., 2013; Ivanov et al., 2009). Perhaps most notably, the introduction of SFB into mice reared at Jackson Laboratories (JAX-SPF) through co-housing with mice reared at Taconic Farms leads to the induction of TH17 cells in the lamina propria of the small intestine (siLP) (Ivanov et al., 2009; Ivanov et al., 2008). In early experiments, we found that serum IgA concentrations were substantially higher in C57BL/6 (B6) mice reared in our SPF colony (hereafter termed “PENN-SPF” mice) compared to age-matched JAX-SPF B6 mice (Figure 1A). To explore this issue, we cohoused female age-matched B6 JAX-SPF and PENN-SPF adults, a strategy that led to increases in serum IgA levels in mice originally housed under JAX-SPF conditions (Figure 1A). Notably, ELISPOT analyses revealed that PENN-SPF and JAX-SPF mice possess very similar frequencies of IgA-secreting plasma cells in the siLP (Figure 1B), suggesting an alternative source of plasma cells accounted for increased serum IgA levels. Hence, we next quantified IgA-secreting plasma cells in the BM of isolated and cohoused JAX-SPF and PENN-SPF B6 adults. Whereas we detected few if any IgA-secreting plasma cells in JAX-SPF B6 adults, PENN-SPF B6 adults and JAX-SPF B6 adults that had been co-housed with PENN-SPF mice for 10 weeks possessed large numbers of BM IgA-secreting cells (Figure 1C).
Figure 1. Serum IgA concentration and BM IgA+ plasma cell numbers are dependent on housing conditions.
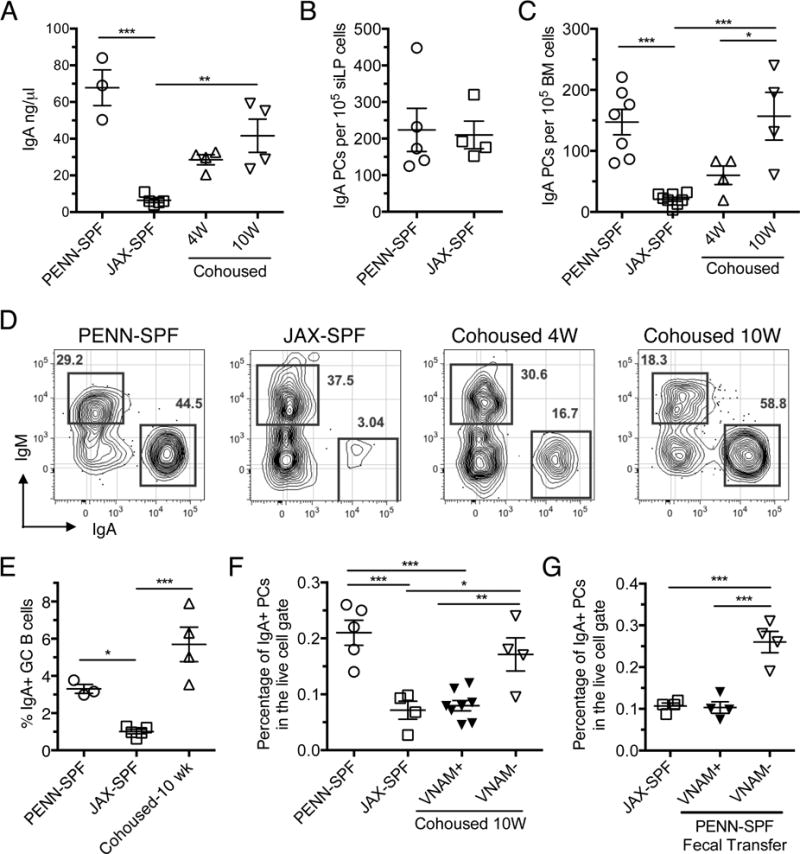
(A) Serum ELISA performed to determine the concentration of IgA from PENN-SPF, JAX-SPF, and cohoused mice, with 3-5 mice per group. (B) Cells from siLP of PENN-SPF or JAX-SPF B6 adults were plated onto ELISpot plates pre-coated with anti-Ig(H+L) and spots were developed using IgA-specific antibodies. (C) BM cells from PENN-SPF, JAX-SPF or JAX-SPF cohoused with PENN-SPF for 4 or 10 weeks were plated onto ELISpot plates as in (B) and detected with IgA-specific antibodies. (D) BM samples from the groups indicated in (C) were stained with the indicated antibodies and 2×106 events collected on an LSR2 flow cytometer. (E) Frequencies of Peyer’s patch GC B cells (CD19+ IgD− CD38− PNA+) in the indicated mice. Means and SEMs are shown for 3-5 mice/grp. (F) Percentages of IgA+ plasma cells within the live cell gate of the BM were determined for mice cohoused with PENN-SPF mice pretreated with VNAM or JAX-SPF, PENN-SPF, and cohoused controls. (G) JAX-SPF mice were gavaged with fecal slurry from PENN-SPF and the treated with VNAM or maintained as controls. Displayed are percentages of IgA+ plasma cells within the live cell gate of the BM. Means and standard error are indicated for at least 3 mice per group. * p <0.05, ** p<0.01, *** p<0.001 determined by one-way ANOVA with Tukey’s posttest or t-test where appropriate. Data are representative of 3 independent experiments.
We also used flow cytometry to confirm and extend these results. Notably, Blanc et al recently showed that viable mouse IgM and IgA-secreting CD138+ BM plasma cells are readily identified via surface IgM and IgA expression (Blanc et al., 2016). With this approach we also detected few IgA-secreting plasma cells in the BM of JAX-SPF mice (Figure 1D). By sharp contrast, the majority of BM plasma cells in PENN-SPF mice were surface IgA+, and these results were recapitulated in JAX-SPF mice after cohousing with PENN-SPF mice for 10 weeks (Figure 1D). Cohousing of JAX-SPF and PENN-SPF adults also led to increased frequencies of IgA+ GC B cells in the former mice (Figure 1E). Notably, these results did not correlate strictly with the induction of TH17 cells in JAX-SPF mice; although as reported the siLP of adults purchased from Taconic Farms contained large numbers of TH17 cells (not shown) (Ivanov et al., 2008), we observed only modest increases in numbers of BM IgA+ plasma cells in mice reared at Taconic Farms (Figure S1). To determine if antibiotic treatment is sufficient to block the increase in IgA+ BM plasma cells seen following cohousing, we placed PENN-SPF mice on an antibiotic cocktail containing Vancomycin, Neomycin, Ampicillin, and Metronidazole (VNAM) for two weeks prior to cohousing. The VNAM treated PENN-SPF mice did not induce significant levels of IgA+ BM plasma cells in JAX-SPF mice (Figure 1F). Additionally, separate cohorts of JAX-SPF mice were given an oral gavage of fecal slurries prepared from PENN-SPF mice, and then half of these mice were placed on VNAM. The VNAM treated JAX-SPF mice that received the fecal transfer had similar levels of IgA+ BM plasma cells as untreated JAX-SPF mice, while fecal transfer alone led to significantly increased levels of IgA+ BM plasma cells (Figure 1G). Together these data suggest that unique bacterial taxa in PENN-SPF mice induce events that enhance serum IgA levels and production of IgA-secreting plasma cells.
Bacterial taxa driving serum IgA
To correlate specific bacterial taxa with systemic IgA responses, we performed 16S ribosomal gene sequencing of stool samples from JAX- and PENN-SPF mice, as well as JAX-SPF mice that were cohoused with PENN-SPF B6 mice for four weeks. Consistent with past work showing that the microbiota between B6 mice housed under disparate conditions is normalized by cohousing within 17 days (Ivanov et al., 2008), within four weeks the intestinal microbiota in JAX-SPF mice was reshaped substantially (Figure 2A). Most notably, members of the Proteobacteria phylum were readily observed in PENN-SPF and cohoused JAX mice only (Figure 2B, 2C). Included were small but significant numbers of Helicobacter sp., shown previously to be highly enriched among IgA-bound GI bacteria (Palm et al., 2014). Furthermore, unlike in JAX-SPF mice, using FISH analyses we easily detected bacteria in the siLP of PENN-SPF and in JAX-SPF mice that were first co-housed with PENN-SPF adults (Figure 2D).
Figure 2. 16S sequencing of JAX-SPF mice demonstrates a lack of commensal bacteria the phylum Proteobacteria.
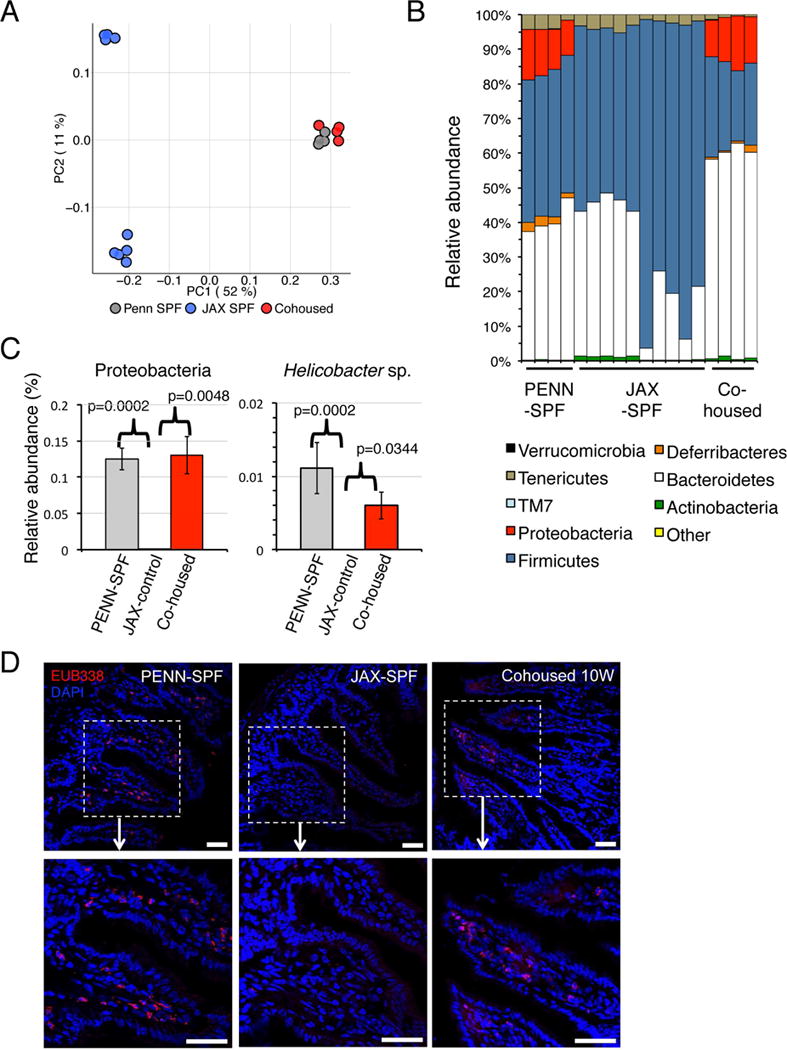
(A-C) Stool samples from JAX-SPF, PENN-SPF, and co-housed mice were subjected to 16S V4 rDNA gene sequencing. (A) Principle coordinate analysis illustrating degree of variance for the indicated mice within each group. (B) Relative abundance of each bacterial phylum for each sample. (C) Comparison of JAX-SPF, PENN-SPF, and co-housed JAX-SPF mice for relative abundance of the phylum Proteobacteria and the genus Helicobacter. (D) Confocal images of FISH analyses performed on sections of the small intestine from JAX-SPF, 10 week cohoused, or PENN-SPF B6 adults. 16S probe EUB338 staining is shown in red, with nuclear (DAPI) in blue. Dashed boxes outlined enlarged areas displayed on the right. Scale bars: 40 μm. Images are representative of 2 different areas of the intestine from 4 mice from each group. P values calculated using one-way ANOVA with Tukey’s posttest. Experiments contain at least 4 mice per group and are representative of 2 independent experiments.
To test directly whether colonization of the gut by a commensal bacterium facilitates the induction of serum IgA and BM IgA-secreting plasma cells, we delivered the Proteobacteria member Helicobacter muridarum to the GI tract of JAX-SPF B6 mice by oral gavage. H. muridarum was selected because it was isolated from healthy rodents and is considered a commensal bacterium (Lee et al., 1992). As shown, 4 weeks after treatment with this strategy we easily detected serum antibodies specific for H. muridarum, but not for the irrelevant skin commensal Staphylococcus epidermidis (Figure 3A, B). Additionally, we readily quantified IgA+ H. muridarum-specific BM and siLP plasma cells in colonized mice, but not in control JAX-SPF mice kept in isolation in our colony (Figure 3C). Therefore, colonization of the GI tract by the commensal bacterium H. muridarum induces the genesis of microbe-specific serum IgA and IgA-secreting BM plasma cells.
Figure 3. Marrow IgA+ plasma cells induced by a commensal bacterium.
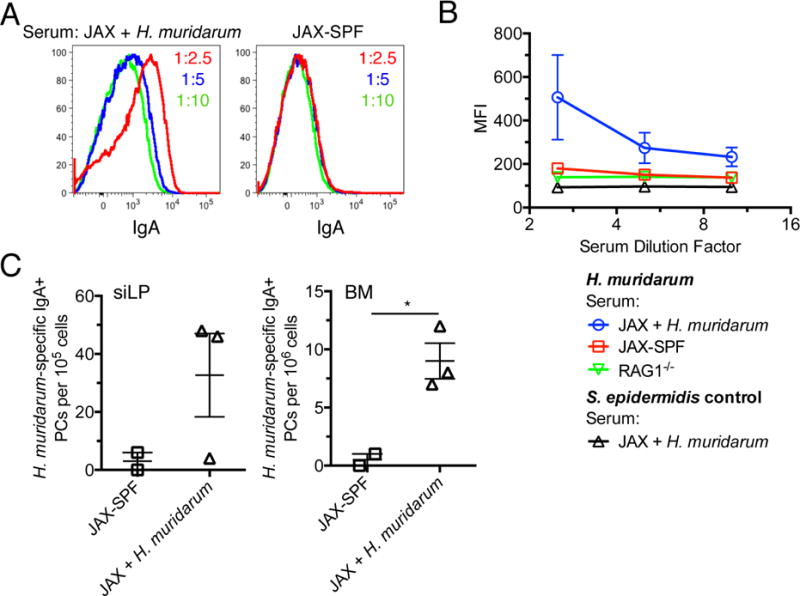
(A) JAX-SPF mice were colonized with Helicobacter muridarum by oral gavage 4 weeks previously; cultured H. muridarum was stained with sera from JAX-SPF controls housed in isolation or JAX-SPF mice colonized with H. muridarum 4 weeks earlier. Bacteria were then stained with PE-anti-IgA and analyzed on an LSR2 flow cytometer. (B) Graphical representation of data presented in (A) with B6.RAG1−/− serum and Staphylococcus epidermidis used as negative controls. S. epidermidis was stained with serum from JAX-SPF H. muridarum mice because it is not found by 16S sequencing in the feces of PENN-SPF or JAX-SPF mice. (C) H. muridarum-specific ELISpot analysis performed using siLP and BM cells from (A). Experiments contain at least 3 H. muridarum treated mice per group and are representative of 2 independent experiments.
To gain a more global picture of the commensal bacterial taxa able to induce serum IgA responses, we sought to identify intestinal bacteria bound by serum IgA among complex bacterial intestinal communities. To this end we extended recently described approaches for identifying IgA- or IgG-bound bacteria wherein Ig-bound and unbound bacteria are purified before being subjected to 16S rDNA gene sequencing (Bunker et al., 2015; Koch et al., 2016; Palm et al., 2014). First, we evaluated the degree to which serum IgA from various mice bound to fecal bacteria. To avoid the complication associated with bacterial binding by endogenous IgA, we derived bacterial samples from adult B6.RAG1−/− mice that were first co-housed with B6 mice in our colony. As expected, because cohoused mice are naturally coprophagic (Ebino et al., 1988), 16S data from stool samples indicated similar representation of bacterial taxa in cohoused PENN-SPF B6 and B6.RAG1−/− mice, especially relative to B6 JAX-SPF mice (Figure S2A,B). For flow cytometric analyses of bacterial communities, we used the fluorescent dyes SytoBC and DAPI, which together allow viable bacteria to be identified as SytoBC+ DAPIlow cells (Figure 4A, left-most panel) (Ben-Amor et al., 2005). As shown, whereas sera from JAX-SPF B6 mice stained only small numbers of bacteria, serum from PENN-SPF or previously co-housed JAX-SPF B6 mice stained markedly increased numbers of viable bacteria from B6.RAG1−/− mice (Figure 4A). Of note, the frequency of bacteria bound by serum IgA taken from co-housed JAX-SPF mice was similar to the fraction of bacteria bound by intestinal IgA reported by Palm et al (Palm et al., 2014).
Figure 4. Serum IgA binding to select commensal bacteria including Proteobacteria.
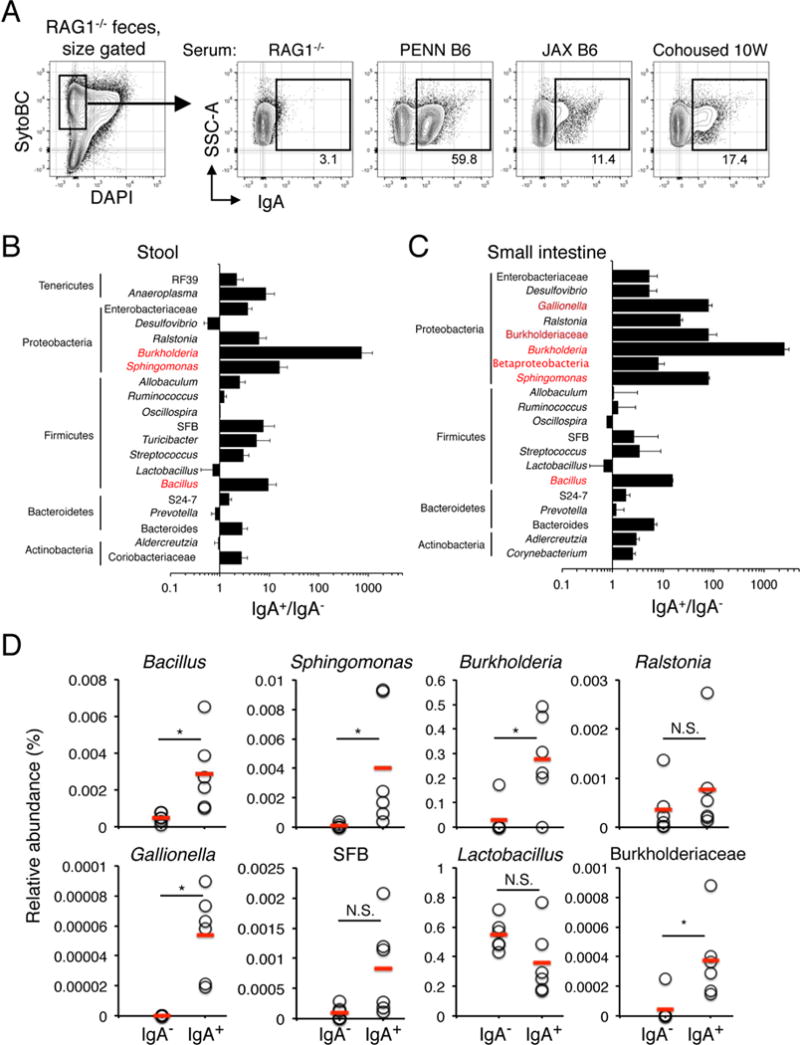
(A) Fecal bacteria from B6.RAG1−/− mice were stained with sera from the indicated mice, and IgA+ bacteria were identified with PE-anti-IgA antibodies by flow cytometry. Bacterial viability was determined by positive staining with SytoBC and minimal DAPI staining, as shown. (B) Fecal bacteria from RAG1−/− mice were stained with sera from PENN-B6 adults and both IgA+ and IgA− fraction sorted twice. DNA from these samples was then subjected to 16S V4 sequencing. The ratio of the relative abundance for each taxon in each fraction (IgA+, IgA−) was used to identify taxa enriched for IgA binding. Bacterial genera that were highly enriched (IgA+/IgA− ≥ 10) are highlighted in red. Genera shown were selected because they possessed ICI scores above or below 3, or because of potential relevance based on previous studies. (C) Small intestine fecal samples from B6.RAG1−/− mice were stained, sorted, and sequenced as in (B). (D) Relative abundance of the indicated taxa for the serum IgA+ and serum IgA− fraction derived from the data in (C). Red lines indicate means for each group. N.S., not significant, *p < 0.05 (Wilcoxon rank-sum).
We purified serum IgA-bound bacteria with two rounds of cell sorting before subjecting DNA from these cells (IgA-coated versus uncoated) to 16S sequencing. We term this strategy “serum IgA-SEQ.” All samples were sorted twice before DNA preparation and PCR by gating on SytoBC+ DAPI− events, however it should be noted that we observed very similar taxa when we analyzed serum IgA bound bacteria in the SytoBC+ DAPI+ fraction (Figure S3A,B). Additionally, we analyzed stool and small intestinal bacteria because recent data indicate that bacteria in the latter site preferentially induce IgA responses (Bunker et al., 2015). Finally, to identify specific highly enriched taxa in serum IgA-bound fractions with the resulting 16S sequence data we exploited the approach of Palm et al. wherein the relative abundance of each taxa in the IgA-bound fraction is divided by its relative abundance in the IgA− fraction. Taxa achieving a score of 10 or more are considered substantially enriched among IgA-bound bacteria (Palm et al., 2014).
16S sequence data for serum IgA+ and IgA− bacteria derived from stool samples or the small intestine revealed that members of the Burkholderia, Spingomonas, and Bacillus genera were each highly enriched among serum IgA-bound bacteria from either source, with enrichment scores for the small intestine of 2546, 80, and 15, respectively (Figure 4B-D). In addition, consistent with the notion that IgA responses target bacteria in the small intestine, we also observed significant enrichment of two additional bacterial taxa among IgA+ bacteria in the small intestine, members of the genus Gallionella, and unidentified members of the Burkholderiaceae family. It should also be noted that SFB, Bacteroides, and Betaproteobacteria were enriched in the IgA-bound fraction in stool and small intestinal samples at levels ranging from 3-9. Finally, IgA-bound bacteria in stool samples from B6 mice were enriched significantly for members of the Burkholderia and Sphingomonas genera, both members of the Proteobacteria phylum (Figure S4). We conclude that select members of the bacterial microbiota induce the production of systemic IgA responses.
T-cell dependent nature of systemic IgA responses
Recent work suggests that much of the IgA coating of intestinal bacteria occurs via T cell independent mechanisms (Bunker et al., 2015). However, Palm et al reported that IgA responses to colitogenic bacteria are largely T cell dependent (Palm et al., 2014). To address this issue we examined the capacity of serum IgA from PENN-SPF B6.TCRβ−/−δ−/− mice to coat fecal bacteria from co-housed (PENN-SPF B6 plus PENN-SPF B6.RAG1−/−) B6.RAG1−/− mice. Whereas serum from PENN-SPF B6 mice stained substantial numbers of fecal bacteria as expected, serum from B6.TCRβ−/−δ−/−mice did not (Figure 5A). Moreover, very similar to germ-free B6 mice, we were unable to detect BM IgA+ plasma cells in PENN-SPF B6.TCRβ−/−δ−/−adults (Figure 5B). We conclude that systemic IgA responses are largely T cell dependent.
Figure 5. Systemic IgA responses are T cell dependent.
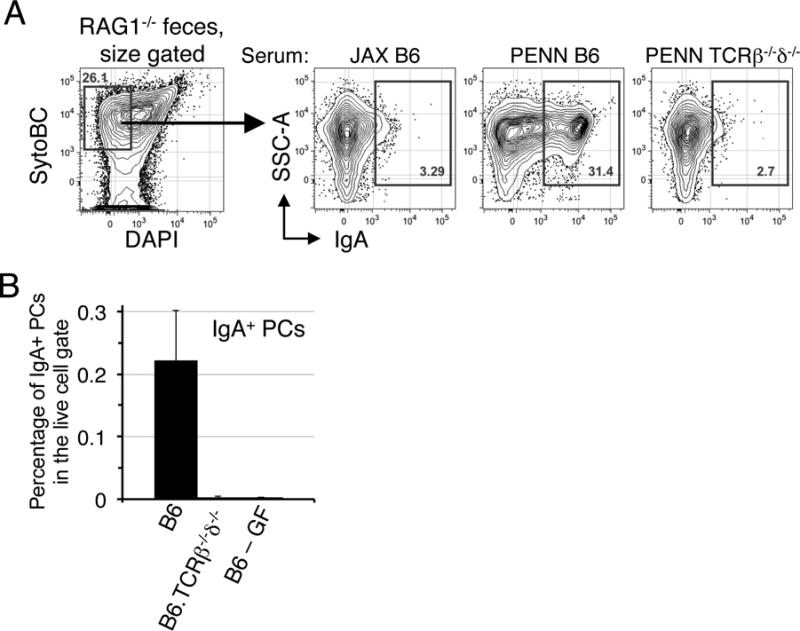
(A) Fecal bacteria from B6.RAG1−/− mice were stained with sera from the indicated mice as in Figure 4A, then IgA-bound bacteria were detected with anti-IgA antibody. Data are representative of 2 independent experiments with 3-4 mice per group. (B) Percentages of IgA+ plasma cells within the live cell gate of the BM were determined by flow cytometry for PENN-SPF B6, B6. TCRβ−/−δ−/−, and germ-free B6 female adults. Data are representative of 2 independent experiments with 4-5 mice per group.
Serum IgA-dependent protection against polymicrobial sepsis
We hypothesized that elevated concentrations of serum IgA, as observed in PENN-SPF mice, would protect against mucosal barrier disruption and the resulting polymicrobial sepsis. Consistent with this idea, a recent report suggests that people with IgA deficiencies are more likely to succumb due to sepsis (Ludvigsson et al., 2016). To test this hypothesis we employed an experimental model of polymicrobial sepsis, namely cecal ligation and puncture (CLP), which results in mortality due to systemic inflammation caused by overwhelming bacteremia (Xiao et al., 2006). Accordingly, we performed CLP on PENN-SPF and JAX-SPF B6 co-housed briefly (1.5 weeks), 4 weeks, or 10 weeks with PENN-SPF B6 mice. The brief 1.5 week cohousing period was insufficient to induce elevations in serum IgA (not shown), but allowed for the transfer of the PENN-SPF microbiota to the JAX-SPF mice. JAX-SPF mice are particularly susceptible to CLP resulting in >85% death, in contrast the PENN-SPF mice that were largely resistant to CLP induced death with <20% succumbing over 14 days (Figure 6A). The cohousing the JAX-SPF for 4 or 10 weeks led to significant levels of protection from death, with the 10-week cohort recapitulating the phenotype of PENN-SPF mice (Figure 6A). To test the relevance of IgA in this context, we next performed CLP on PENN-SPF B6.IgA−/− mice and PENN-SPF B6 mice. Similar to JAX-SPF mice, PENN-SPF B6.IgA−/− mice were susceptible to sepsis-induced mortality (Figure 6B). To determine if serum IgA mediates protection against sepsis, we performed CLP on JAX-SPF mice that subsequently received sera from either PENN-SPF B6 or PENN-SPF B6.IgA−/− mice. All but one animal receiving B6.IgA−/− serum died within 2 days, whereas survival of JAX-SPF mice receiving B6 serum was extended significantly (Figure 6C). We conclude that serum IgA responses provide a constitutive defense mechanism against bacterial sepsis.
Figure 6. Serum-IgA mediated protection from induced sepsis.
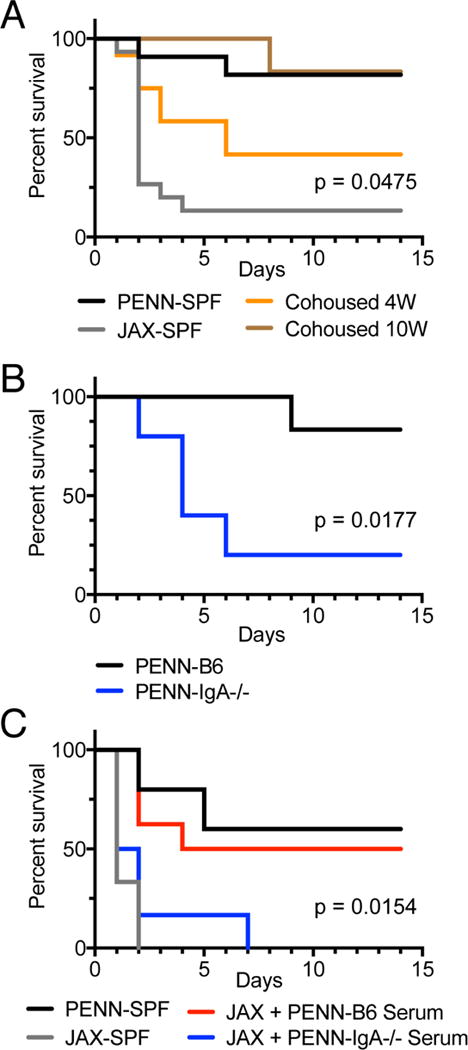
(A) JAX-SPF and PENN-SPF were cohoused for 1.5 (labeled JAX-SPF), 4, or 10 weeks to allow for mice to achieve similar intestinal microflora and in the case of the 4 and 10 week cohoused groups to develop increased serum IgA. Mice were then subjected to CLP surgery with a 1cm ligation followed by 2 punctures with a 21G needle. Survival was monitored daily for 14 days. JAX-SPF n = 15, PENN-SPF n = 11, Cohoused 4W n = 12, and Cohoused 10W n = 6. Reported p value is between JAX-SPF and cohoused 4W. (B) PENN-SPF B6 and PENN-SPF B6.IgA−/− mice were subjected to CLP as in (A). B6 n = 6 and B6.IgA−/− n = 5. (C) CLP was performed on PENN-SPF and JAX-SPF control mice as in (A) while additional JAX-SPF mice received sera from B6 or B6.IgA−/− PENN-SPF mice; 400μl on day zero, and 300μl daily for the next 4 days. Reported p value is between B6 and B6.IgA−/− serum transfer groups. Sample sizes were as follows: Penn-SPF controls, n = 5; JAX-SPF controls, n = 6; JAX-SPF + B6 serum, n = 8; JAX-SPF + B6.IgA−/− serum, n = 6. P values were determined using a log-rank test. Representative of 2 independent experiments.
Discussion
Our work demonstrates that a variety of commensal bacterial taxa elicit serum IgA responses, resulting in protective activity against polymicrobial sepsis and a marked remodeling of the BM plasma cell pool. In contrast to conventional models that predict a critical role for MZ and B1 B cells in generating IgM antibodies to blood-borne bacteria (Martin et al., 2001; Pillai et al., 2005), we demonstrate that pre-existing serum IgA antibodies play a unique role in the context of bacterial sepsis, especially when the microbes in question originate from the gut microbiota. Moreover, marginal zone and B1 B cells require stimulation via the B cell receptor or pattern recognition receptors to secrete large quantities of immunoglobulin (Fairfax et al., 2007; Oliver et al., 1997). By comparison, IgA+ plasma cells actively secrete antibody without receptor engagement, thus providing a constitutive humoral shield against systemic bacterial invasion. Notably, both pools of IgA-secreting cells consist mainly of long-lived cells (Bemark et al., 2016)(our unpublished results), and therefore provide a mechanism for the continuous production of serum antibodies with anti-microbial activity. These ideas therefore build on recent evidence that serum IgG antibodies specific for intestinal microbes form a barrier of protection against systemic invasion by commensal bacteria (Koch et al., 2016; Zeng et al., 2016). Therefore, we propose that serum IgA and IgG antibodies may play similar roles to the protective role proposed for natural IgM antibodies (Ochsenbein et al., 1999), with the IgA component providing a noninflammatory mechanism for keeping invading bacteria in check.
Our results also shed light on the role of systemic IgA responses for enteric pathogens. Systemic antibody responses may be of little advantage for certain enteric pathogens. However, in scenarios associated with breakdown of intestinal barrier function, certain enteric pathogens readily spread to distant tissues to cause or exacerbate disease. One classic example is poliovirus (Blondel et al., 2005). Even rotavirus, thought originally to remain localized to the intestine, was shown recently to spread to extra-intestinal tissues (Blutt and Conner, 2007). Thus, heightened serum IgA responses, induced perhaps through appropriate adjuvants, could increase availability of neutralizing antibodies throughout the body, and consequently limit the spread of infection for a variety of enteric pathogens.
Past work indicates that recently generated mucosal plasma cells readily travel through the efferent lymph and into the blood before eventually returning to mucosal sites (Gowans and Knight, 1964). Consistent with this idea, more recent work indicates that mucosal B cells are induced to express gut homing receptors by specialized dendritic cells (Mora and von Andrian, 2008). Our data reveal another layer of complexity to these processes, and suggest that certain consortia of commensal microbes push the boundaries between the mucosal and systemic immune systems, increasing the production of mucosal IgA-secreting plasma cells and the infiltration of many of these cells into the BM. Notably, colitogenic bacteria characterized by a propensity to invade the inner GI mucus layer are preferentially coated with IgA (Palm et al., 2014). Such microbes include members of the Helicobacter genus such as H. muridarum, shown here to induce the generation of IgA+ BM plasma cells.
A recent study highlights the strong propensity of Helicobacter species to induce mucosal T cell differentiation (Chai et al., 2017). Additionally, Atarashi et al. showed that SFB, Citrobacter rodentium, and Escherichia coli are especially adept at attaching to intestinal epithelial cells, which in turn enhances the local generation of TH17 cells (Atarashi et al., 2015). Given that mucosal TH17 cells can support the induction of large numbers of IgA+ GCs (Hirota et al., 2013), it follows that select members of the GI microbial community possess an intrinsic propensity to invade the inner mucus later and drive the generation of TH17-supported IgA responses, which in turn lead to increases in the differentiation of IgA+ plasma cells and serum IgA titers. This idea is consistent with our data demonstrating that bacterial microbes are readily and uniquely detected in the siLP of PENN-SPF mice. Additional work is ongoing to identify these microbes and characterize their capacity to generate local and systemic IgA responses. It is tempting to speculate that these microbes will include members of the Burkholderia, Sphingomonas, and Bacillus genera, identified by our serum-IgA seq experiments.
Several studies show that certain intestinal bacterial taxa influence T cell mediated processes beyond local mucosal tissues (Klaasen et al., 1993; Wu et al., 2010). In this regard, serum IgA can increase with age or exposure of germ-free mice to commensal bacteria. However, these results have been attributed to increased numbers of IgA+ siLP plasma cells (Kamata et al., 2000). By contrast our results with conventional SPF mice show that changes in the commensal microbiota lead to increases in serum IgA without altering frequencies of siLP IgA-secreting cells, suggesting that the bulk of the increased serum IgA is due to increased production of IgA+ BM plasma cells.
Moving forward, how B-lineage cells regulate intestinal homeostasis as microbial communities evolve throughout life while also providing protection for dangerous enteric pathogens remains unclear. Defining the cell-cell interactions and molecular mechanisms through which effective and lasting IgA responses to enteric microbes are generated is essential for establishing the fundamental mechanisms underlying intestinal homeostasis, and for improving approaches to treat and avoid inflammatory bowel disease and intestinal barrier disruption. We suggest that defining the mechanisms whereby intestinal homeostasis is maintained will require elucidation of the roles played by long-lived IgA-secreting plasma cells in this process including a better understanding of how and why certain commensal microbes promote the induction of systemic IgA responses.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Allman (dallman@pennmedicine.upenn.edu).
Experimental Model and Subject Details
Vertebrate Animals
B6.Blimp1+/GFP (Kallies et al., 2004), B6.RAG1−/−, B6.TCRβ−/−δ−/−, B6.IgA−/− and C57BL/6J (B6) mice were bred and maintained at the University of Pennsylvania. B6 mice termed ‘JAX-SPF’ were from Jackson Laboratories and housed in our SPF facility; these cages were kept in isolation and changed separately from other mice. Mice were all female to facilitate cohousing and aged 8-20 weeks at the time of euthanasia. All experiments were performed in accordance with the Office of Regulatory Affairs Institutional Animal Care and Use Committee.
Bacterial strains
Helicobacter muridarum (ATCC 49282) was cultured on blood agar plates (BD BBL Prepared Plated Media R01202). Isolates were incubated for 48-72 hours at 37° C under microaerophilic conditions (Pack-Micro, Mitsubishi Gas Chemical America Inc R681005). Pure cultures were harvested and suspended in PBS before centrifugation and measurement of bacterial density by weight. Mice were given an oral gavage of 5 mg of H. muridarum re-suspended in PBS.
Method Details
Bacterial flow cytometry and sorting
For analyses and purification of serum IgA-bound bacteria samples were prepared and stained according to Palm et al (Palm et al., 2014). Bacteria were harvested by physical disruption of stool samples followed by light centrifugation to pellet debris. The resulting bacteria were stained with PE-anti-mouse IgA antibodies at a concentration of 1-12.5. Samples were then washed twice then stained with SytoBC and DAPI to identify viable bacteria (Ben-Amor et al., 2005). For serum IgA, samples were stained first with 50 μl of a 1:2 serum dilution for 30 minutes on ice, then washed twice before adding PE-anti-IgA antibodies. Bacteria were run on LSRII or FACSAria cytometers equipped with green (532nm) lasers. All purified bacteria were sorted twice as SytoBC+ DAPIlow IgA+/− cells.
Tissue preparation
Spleen and Peyer’s patch cells were isolated by mechanical disruption of the tissue between frosted glass slides and filtration through 66 μm nitex mesh. BM cells were isolated by flushing bones using a 23G needle and syringe with FACs buffer (PBS, 0.5% BSA, 1mM EDTA). siLP preparations were made using a modified protocol from Hall et al. (Hall et al., 2011). Small intestines were harvested and Peyer’s patches, fat, and intestinal contents were removed. The cleaned small intestine was cut into ~2cm pieces before incubating at 37° C on a shaker for 20 minutes in RPMI 1640 with 20mM HEPES, 5mM EDTA, 1mM DTT, Penicillin/Streptomycin, and 5% FBS. The small intestine pieces are then filtered over a large pore strainer and washed 2 times in RPMI with Pen/Strep, 2mM EDTA, and 20 mM HEPES. The pieces were minced and put into RPMI with Pen/Strep, 0.1 mg/mL Liberase TL (Roche), 0.05% DNase I (Sigma D5025), and 20 mM HEPES for 30 minutes at 37° C while shaking. The Liberase reaction is then quenched with cold RPMI with 10% FBS, 0.05% DNAse I, Pen/Strep, 20 mM HEPES and the small intestine is filtered through 70 μm Corning cell strainers (Corning 352350). Before running siLP cells through the cell sorter or flow cytometer and before plating on ELISpot plates they are filtered again with 66 μm nitex.
Flow cytometry
Mouse cells were stained for flow cytometric analysis as previously described (Wilmore et al., 2017). Briefly, cell suspensions were stained with the Zombie Aqua Fixable Viability kit (BioLegend) according to the manufacturer’s instructions. Then, cells were stained with a cocktail of optimal concentrations of antibodies in 100 μl on ice for 30min., washed twice, and when needed stained with fluorescently labeled streptavidin at a predetermined concentration. The following antibodies were used for staining and analysis: PE-anti-CD138 (BD; 281-2), APC-Cy5.5-anti-CD19 (ThermoFisher; RM7719), PerCP-eFluor710-anti-IgM (eBio; II/41), PE-Cy7-anti-CD4 (eBio; GK1.5), -CD8α (eBio; 53-6.7), -Gr1 (eBio; RB6-8C5), -F4/80 (eBio; BM8), -TER-119 (BioLegend), AF700-anti-CD38 (eBio; HIT2), APC-anti-B220 (Tonbo; RA3-6B2), FITC-PNA (Sigma), APC-Cy7-anti-IgD (BioLegend; 11-26c.2a), Biotin-anti-IgA (BioLegend; RMA-1), BrilliantViolet(BV)605-anti-Thy1.2 (BioLegend; 53-2.1), and BV785-anti-CD19 (BioLegend; 6D5). To detect biotin-conjugated antibodies we used streptavidin-BV421 (BioLegend).
ELISA/ELISpot assays
ELISA and ELISpot assays were performed according to standard protocols. Briefly, plates (ThermoFisher 442404) were coated with anti-mouse Ig(H+L) (Southern Biotech 1010-01) in a sodium carbonate/bicarbonate solution pH 9.6. Helicobacter ELISpot plates were coated with 200ng/well of heat-killed H. muridarum. Serum was serially diluted and detected using an HRP conjugated anti-mouse IgA antibody (Sigma A4789) and BD OptEIA TMB substrate (BD 555214). ELISpot plates (EMD Millipore MSIPS4W10) were coated the same as ELISA plates, but detection was performed with anti- IgM, IgA, IgG, κ, and λ antibodies conjugated to biotin (Southern Biotech). Biotinylated antibodies were revealed with streptavidin-alkaline phosphatase (Sigma E2636) and developed with BCIP/NBT (Sigma B1911). ELISpot plates were imaged and spots were counted using a CTL Immunospot Analyzer (Cellular Technologies Limited).
16S sequencing
All sequencing analysis of the 16S V4 region was performed according to the SOP detailed by Kozich et al. (Kozich et al., 2013). Specifically, DNA from fecal bacteria was isolated using the DNeasy PowerSoil Kit (Qiagen). To isolate DNA from sorted bacteria we first used the MasterPure Yeast DNA Purification kit combined with a 10-minute high-speed vortex in 0.5mm glass bead tubes (Mo Bio). Then, the DNA extraction was completed with the PureLink Genomic DNA Mini Kit (ThermoFisher). PCR was performed with barcoded 16S V4 oligonucleotides using AccuPrime Pfx SuperMix (ThermoFisher), PCR products were then normalized using the SequalPrep Normalization Kit (ThermoFisher), then pooled and sequenced on a MiSeq (Illumina) using a MiSeq Reagent Kit v2 500 cycle (Illumina) with 5% PhiX (Illumina).
Fluorescence In Situ Hybridization
4 mm-long pieces of the small intestine were cryoprotected in OCT compound (Fisher Health Care). 7 μm-thick transversal tissue slices were sectioned with a cryostat. FISH was performed following the protocol by Lowry et al. (Lowrey et al., 2015). Briefly, slides were fixed in 3.7% formaldehyde at RT and then incubated overnight at 4°C in 70% ETOH. Hybridization was performed overnight at 37°C in 10% formamide and 1X SSC (Saline Sodium Citrate) using 2 μg/mL of the probe EUB338 labeled with Alexa Fluor 594 (IDT). The EUB338 probe targets the 16S rRNA of ~90% of all bacteria found in the gut (Amann et al., 1990). Afterwards slides were washed thoroughly and mounted in ProLong Diamond Antifade with Dapi (Molecular Probes). All solutions were prepared with DEPC treated water. Images were obtained in a Zeiss LSM710 confocal microscope and analyzed using Volocity 6.3 (Perkin Elmer) software.
Quantification and Statistical Analysis
Statistical analysis
Data presented were analyzed with Prism v6 software (GraphPad) for statistical analysis. Data shown are presented as mean ± SEM and * denotes a significant p value of < 0.05. The p values were determined by a two-tailed unpaired student’s t-test with Welch’s correction or one-way ANOVA with Tukey’s posttest where appropriate. Differences in survival were analyzed using a log-rank test.
16S Sequencing analysis
Sequencing data was analyzed using the QIIME 1.8.0 software package (Caporaso et al., 2010). A total of 2,973,731 sequences were analyzed. Operational taxonomic unit (OTU) selection was performed using CD-HIT (97% identity threshold) (Li and Godzik, 2006). The RDP Classifier was used to assign taxonomy within QIIME at a 0.8 confidence threshold (Wang et al., 2007). Chimeric sequences were removed with ChimeraSlayer and rarefaction analysis was performed on a subsampled set of 15000 genes. PCoA plots were generated using unweighted UniFrac distances based on OTU tables generated in QIIME.
Supplementary Material
Highlights.
Serum IgA concentrations depend on the composition of the gut microbiota
Cohousing transfers Proteobacteria to naïve mice resulting in increased serum IgA
T-cell dependent serum IgA is induced in response to select bacteria in the gut
Serum IgA protects against lethal sepsis following intestinal barrier disruption
Acknowledgments
We gratefully thank Drs. Yasmine Belkaid, David Artis, and Gregory Sonnenberg for helpful discussions, Dr. Margaret Conner for providing IgA−/− mice, and Kelly Owens for artistic input. We also thank the UPenn Flow Cytometry and Cell Sorting facility, the Human Immunology Core facility, as well as the UPenn Molecular Profiling core. This work was supported by National Institutes of Health (NIH) grants R01-AI097590 and R01-AI097590 to D. Allman, an F32-AI114089 to J. Wilmore, and a pilot grant from the PennCHOP Microbiome Program. D. Jones and B. Gaudette were supported by NIH training grant T32CA009140.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
JRW and DA designed experiments and wrote the manuscript; BTG performed bioinformatics analyses; JRW, TH, DDJ, and CAG performed experiments; DGA performed all imaging experiments; SDC cultured and provided anaerobic bacteria; AM and DB analyzed 16S data.
Declaration of Interests
The authors declare no competing interests.
References
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Hazanov H, Stromberg A, Komban R, Holmqvist J, Koster S, Mattsson J, Sikora P, Mehr R, Lycke NY. Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nat Commun. 2016;7:12698. doi: 10.1038/ncomms12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Amor K, Heilig H, Smidt H, Vaughan EE, Abee T, de Vos WM. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl Environ Microbiol. 2005;71:4679–4689. doi: 10.1128/AEM.71.8.4679-4689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc P, Moro-Sibilot L, Barthly L, Jagot F, This S, de Bernard S, Buffat L, Dussurgey S, Colisson R, Hobeika E, et al. Mature IgM-expressing plasma cells sense antigen and develop competence for cytokine production upon antigenic challenge. Nat Commun. 2016;7:13600. doi: 10.1038/ncomms13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel B, Colbere-Garapin F, Couderc T, Wirotius A, Guivel-Benhassine F. Poliovirus, pathogenesis of poliomyelitis, and apoptosis. Curr Top Microbiol Immunol. 2005;289:25–56. doi: 10.1007/3-540-27320-4_2. [DOI] [PubMed] [Google Scholar]
- Blutt SE, Conner ME. Rotavirus: to the gut and beyond! Curr Opin Gastroenterol. 2007;23:39–43. doi: 10.1097/MOG.0b013e328011829d. [DOI] [PubMed] [Google Scholar]
- Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z, Perry JSA, Knoop KA, Tanoue T, Narushima S, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebino KY, Yoshinaga K, Saito TR, Takahashi KW. A simple method for prevention of coprophagy in the mouse. Lab Anim. 1988;22:1–4. doi: 10.1258/002367788780746548. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- Fairfax KA, Corcoran LM, Pridans C, Huntington ND, Kallies A, Nutt SL, Tarlinton DM. Different kinetics of blimp-1 induction in B cell subsets revealed by reporter gene. J Immunol. 2007;178:4104–4111. doi: 10.4049/jimmunol.178.7.4104. [DOI] [PubMed] [Google Scholar]
- Gowans JL, Knight EJ. The Route of Re-Circulation of Lymphocytes in the Rat. Proc R Soc Lond B Biol Sci. 1964;159:257–282. doi: 10.1098/rspb.1964.0001. [DOI] [PubMed] [Google Scholar]
- Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34:435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, Nutt SL. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T, Nogaki F, Fagarasan S, Sakiyama T, Kobayashi I, Miyawaki S, Ikuta K, Muso E, Yoshida H, Sasayama S, et al. Increased frequency of surface IgA-positive plasma cells in the intestinal lamina propria and decreased IgA excretion in hyper IgA (HIGA) mice, a murine model of IgA nephropathy with hyperserum IgA. J Immunol. 2000;165:1387–1394. doi: 10.4049/jimmunol.165.3.1387. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beynen AC. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, Barton GM. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Lee A, Phillips MW, O’Rourke JL, Paster BJ, Dewhirst FE, Fraser GJ, Fox JG, Sly LI, Romaniuk PJ, Trust TJ, et al. Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int J Syst Bacteriol. 1992;42:27–36. doi: 10.1099/00207713-42-1-27. [DOI] [PubMed] [Google Scholar]
- Lemke A, Kraft M, Roth K, Riedel R, Lammerding D, Hauser AE. Long-lived plasma cells are generated in mucosal immune responses and contribute to the bone marrow plasma cell pool in mice. Mucosal immunology. 2016;9:83–97. doi: 10.1038/mi.2015.38. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lowrey L, Woodhams DC, Tacchi L, Salinas I. Topographical Mapping of the Rainbow Trout (Oncorhynchus mykiss) Microbiome Reveals a Diverse Bacterial Community with Antifungal Properties in the Skin. Appl Environ Microbiol. 2015;81:6915–6925. doi: 10.1128/AEM.01826-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2012;245:132–146. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal immunology. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity. 2001;14:617–629. doi: 10.1016/s1074-7613(01)00129-7. [DOI] [PubMed] [Google Scholar]
- Mora JR, von Andrian UH. Differentiation and homing of IgA-secreting cells. Mucosal immunology. 2008;1:96–109. doi: 10.1038/mi.2007.14. [DOI] [PubMed] [Google Scholar]
- Morahan G, Berek C, Miller JF. An idiotypic determinant formed by both immunoglobulin constant and variable regions. Nature. 1983;301:720–722. doi: 10.1038/301720a0. [DOI] [PubMed] [Google Scholar]
- Ochsenbein AF, Fehr T, Lutz C, Suter M, Brombacher F, Hengartner H, Zinkernagel RM. Control of early viral and bacterial distribution and disease by natural antibodies. Science. 1999;286:2156–2159. doi: 10.1126/science.286.5447.2156. [DOI] [PubMed] [Google Scholar]
- Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27:2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annual review of immunology. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai LH, Mahevas M, Roudot-Thoraval F, Limal N, Languille L, Dumas G, Khellaf M, Bierling P, Michel M, Godeau B. Long-term complications of splenectomy in adult immune thrombocytopenia. Medicine (Baltimore) 2016;95:e5098. doi: 10.1097/MD.0000000000005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi TB, Jr, Bienenstock J. Secretory immunoglobulins. Adv Immunol. 1968;9:1–96. doi: 10.1016/s0065-2776(08)60441-1. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Wilmore JR, Jones DD, Allman D. Protocol for improved resolution of plasma cell subpopulations by flow cytometry. Eur J Immunol. 2017;47:1386–1388. doi: 10.1002/eji.201746944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


