Abstract
Aberrant core fucosylation of proteins has been linked to liver diseases. In this study, we carried out multiple reaction monitoring (MRM) quantification of core fucosylated N-glycopeptides of serum proteins partially deglycosylated by a combination of endoglycosidases (endoF1, endoF2, and endoF3). To minimize variability associated with the preparatory steps, the analysis was performed without enrichment of glycopeptides or fractionation of serum besides the nanoRP chromatography. Specifically, we quantified core fucosylation of 22 N-glycopeptides derived from 17 proteins together with protein abundance of these glycoproteins in a cohort of 45 participants (15 disease-free control, 15 fibrosis and 15 cirrhosis patients) using a multiplex nanoUPLC-MSMRM workflow. We find increased core fucosylation of 5 glycopeptides at the stage of liver fibrosis (i.e., N630 of serotransferrin, N107 of alpha-1-antitrypsin, N253 of plasma protease C1 inhibitor, N397 of ceruloplasmin, and N86 of vitronectin), increase of additional 6 glycopeptides at the stage of cirrhosis (i.e., N138 and N762 of ceruloplasmin, N354 of clusterin, N187 of hemopexin, N71 of immunoglobulin J chain, and N127 of lumican), while the degree of core fucosylation of 10 glycopeptides did not change. Interestingly, although we observe an increase in the core fucosylation at N86 of vitronectin in liver fibrosis, core fucosylation decreases on the N169 glycopeptide of the same protein. Our results demonstrate that the changes in core fucosylation are protein and site specific during the progression of fibrotic liver disease and independent of the changes in the quantity of N-glycoproteins. It is expected that the fully optimized multiplex LC-MS-MRM assay of core fucosylated glycopeptides will be useful for the serologic assessment of the fibrosis of liver.
Biological significance:
We have quantified the difference in core fucosylation among three comparison groups (healthy control, fibrosis and cirrhosis patients) using a sensitive and selective LC-MS-MRM method. Despite an overall increase in core fucosylation of many of the glycoproteins that we examined, core fucosylation changed in a protein- and site-specific manner. Moreover, increased and decreased fucosylation was observed on different N-glycopeptides of the same protein. Altered core fucosylation of N-glycopeptides might be used as an alternative serologic assay for the evaluation of fibrotic liver disease.
Keywords: Core fucosylation, Cirrhosis, Fibrosis, LC-MS-MRM, Serum
1. Introduction
N-glycosylation exerts critical roles in numerous physiological and pathological processes [1]. Core fucosylation is formed by the α−1,6-fucosyltransferase (Fut8) which transfers the fucosyl moiety from GDP-fucose to the innermost GlcNAc residue of N-linked oligosaccharides of glycoprotein [2–4]. Altered core fucosylation (α−1,6) is associated with several diseases [5,6] including gastric tumors [7] and light chain multiple myeloma [8]. An increased degree of core fucosylation has been observed in the progression of lung cancer [9], prostate cancer [10], breast cancer [11], and melanoma [12]. Notably, an overall increased core fucosylation has been associated with hepatocellular carcinoma (HCC) [13–16] and these changes can be observed in serum owing to the homeostatic function of liver secreted glycoproteins [17]. This is already explored in clinical assessment of alpha-fetoprotein L3 (AFP-L3), a core fucosylated form of alpha-fetoprotein (AFP) approved by the US FDA as a biomarker of the risk of HCC [18]. However, AFP-L3 is marginally effective at the early diagnosis of HCC [19] even though hepatocarcinogenesis was significantly suppressed in a Fut8 knock out animal model [20].
Wide distribution of core fucosylated N-glycans in human proteins was documented by improved mass spectrometric methods and it was suggested that specificity of core fucosylation depends on solvent accessibility of the N-glycosylation sites [21]. Due to the complexity of the heterogeneous N-glycoforms and their sub-stoichiometric representation, immunodepletion and further enrichment are often required for the characterization of core fucosylated N-glycoproteins in complex matrices like serum and plasma. High coverage of core fucosylated N-glycopeptides was achieved by combining lectin-based enrichment of core fucosylated proteins/peptides (e.g. Lens culinaris Agglutinin), followed by cleavage of glycans by endoglycosidase F3 (endoF3) and/or fractionation prior to the mass spectrometric analysis [22–28]. However, many of these studies were not conducted in a quantitative manner. For site-specific analysis of core fucosylation, stable isotopic labeling with iTRAQ [26] or TMT [28] was adopted in workflows combining enrichment with tandem mass spectrometry. These workflows were applied to the comparison of core fucosylation in prostate cancer cell lines [28] which identified 973 fucosylated glycopeptides and serum of patients with hepatocellular carcinoma (HCC) [26] which identified 1300 glycopeptides. Comparison of HCC with liver cirrhosis suggested increased fucosylation of 12 glycopeptides in HCC together with 14 glycopeptides increased in cirrhosis [26]. These complex workflows are, however, not directly applicable to clinical samples and a simplified assay comparing intensity of core fucosylated to non fucosylated peptide of isolated ceruloplasmin by nanoLC-LTQMS was published subsequently [23]. In this study, we present a simplified multiplex workflow that combines the endoF digest of glycopeptides with multiple reaction monitoring mass spectrometry (MRMMS), a well-developed targeted quantification workflow [29–36]. The targeted approach is applicable to the glycopeptides simplified by endoF digests because of the specificity and sensitivity of the glyco-peptide transitions in this setting [32,36]. These assays do not require enrichment at the protein or glycopeptide level and allow multiplex quantification of the core fucosylation directly in serum.
We applied the workflow to the analysis of serum samples from a set of control, fibrotic, and cirrhotic patients with the aim to establish a reliable method for the quantification of changes in core fucosylation associated with liver disease progression. Our results demonstrate that this LC-MS-MRM workflow enables simple, sensitive and selective quantification of core fucosylation and reveals protein- and site-specific changes in core fucosylation in the fibrotic liver disease.
2. Materials and methods
2.1. Chemicals and reagents
Ammonium bicarbonate, DL-dithiothreitol (DTT), iodoacetamide (IAA), and endoglycosidases (endo) F1, F2, F3 were supplied by Sigma-Aldrich (St. Louis, MO, USA). Sequencing grade trypsin was purchased from Promega (Madison, WI, USA). 0.1% formic acid in ACN and 0.1% formic acid in H2O were used as supplied from Honeywell (Philadelphia, PA, USA). LC/MS grade trifluoroacetic acid (TFA) and formic acid (FA) were from Thermo Fisher Scientific (Waltham, MA, USA). Water was purified and deionized with a Milli-Q system manufactured by Millipore (Bedford, MA, USA).
2.2. Sample information
Serum samples of 15 disease-free controls were collected at Georgetown University Medical Center under protocols approved by the Institutional Review Board. De-identified serum samples of 30 patients with various degree of liver fibrosis were obtained from the hepatitis C antiviral long-term treatment against cirrhosis trial (HALT-C), as described in our recent study [35]. The HALT-C trial, a prospective randomized controlled trial of 1050 patients, evaluated effect of long-term low-dose peginterferon alpha-2a (IFN) in participants that failed initial anti-HCV therapy with interferon [37,38]. Liver disease status of the HALT-C participants was classified into fibrosis (Ishak score 3–4) or cirrhosis (Ishak score 5–6) based on biopsy-evaluation. All the participants selected for this study were from the control arm of the HALTC trial that did not receive IFN treatment [35]. Method optimization was carried out on a sample pooled from the 45 serum samples of all participants (equal volume of each) and the pooled sample served as a quality control (QC) that was analyzed in parallel with the analyses of the 45 study samples. All the 45 samples were processed in parallel and analyzed individually as described below.
2.3. Sample digestion and deglycosylation
Serum samples (2 μL) were diluted in 140 μL of 50 mM NH4HCO3, 5 mM DTT was added and the reaction was incubated at 60 °C for 1 h. After cooling down to room temperature, 15 mM iodoacetamide was added for 30 min in the dark. Residual iodoacetamide was quenched with another aliquot of 5 mM DTT at room temperature for 20 min. Trypsin was added to the samples (1/40 (w/w)) and incubated at 37 °C overnight. After heating at 95 °C for 10 min, samples were dried down in a vacuum concentrator (Labconco). Deglycosylation with endoF1–F3 was performed according to the manufacturer’s protocol, with slight modifications. Specifically, samples were dissolved in 50 mM sodium acetate (pH 4.5), and then treated with 1 μL of endoF1 (Sigma E9762–1UN), 1 μL of endoF2 (Sigma E0639-.2UN) and 1 μL of endoF3 (Sigma E2264–.2UN) as supplied and 37 μL H2O at 37 °C overnight. After heating at 95 °C for 10 min, the resulting products were dried down and dissolved in 0.1% formic acid for mass spectrometric analysis.
2.4. LC-MS/MS and LC-MS-MRM
To select core fucosylated (HexNAcFuc), non-fucosylated (HexNAc) peptides and unique peptides of N-glycoproteins for the LC-MRM-MS assays, tryptic digests of serum were analyzed on a NanoAcquity UPLC (Waters) coupled with a TripleTOF 6600 mass spectrometer (Sciex). Specifically, 1 μL of each sample prepared above was loaded onto a C18 Trap column (Waters Acquity UPLC Symmetry C18 NanoAcquity 10 K 2G V/M, 100 A, 5 μm, 180 μm × 20 mm) at 15 μL/min for 2 min. Peptides were then separated on an analytical column (Waters Acquity UPLC M-Class, peptide BEH C18 column, 300 A, 1.7 μm, 75 μm × 150 mm) at 40 °C. The flow rate was set at 400 nL/min with a 90-min gradient of buffer A (2% ACN, 0.1% formic acid) and buffer B (0.1% formic acid in ACN) was used for separation: 1% buffer B at 0 min, 5% buffer B at 1 min, 40% buffer B at 60 min, 99% buffer B at 65 min, 99% buffer B at 70 min. The gradient went back to 1% buffer B at 70.1 min and kept for 20 min for column equilibration. Mass spectral acquisition was operated with Analyst TF 1.7.1 software; each cycle consisted of a full scan (m/z 400–1600) and fifty information dependent acquisitions (IDAs) (m/z 100–1600). The dynamic exclusion time was set to 6 s, and 150 counts threshold for two repeated precursors. Collision energy was set automatically according to charge state and m/z of the precursor ion.
For quantification of the glycosylated and unique peptides in serum samples, tryptic digests were analyzed on a NanoAcquity UPLC (Waters) coupled with a QTRAP 6500 mass spectrometer (Sciex). The NanoAcquity UPLC (Waters) and the settings (e.g., gradient and flow rate) were the same as described above. Peptide and glycopeptide quantification on the QTRAP 6500 was carried out in the MRM mode with the ion spray voltage set at 2300 V, curtain gas 11, ion source gas 1 30, and the interface heater temperature 150 °C. The entrance potential (EP) was set as 10 V and collision cell exit potential (CXP) was 13 V. Q1 and Q3 were set at open and unit resolution, respectively. The MRM transitions used for monitoring of modified peptides and unique peptides are listed in Supplementary Tables S1 and S2. Collision energy (CE) for each MRM transition was optimized by a 5 V step optimization followed by a 2 V step fine tuning. Instrument control and data acquisition were performed by Sciex Analyst software (version 1.6.2).
2.5. Data processing and statistical analysis
Raw data files from the TripleTOF 6600 were processed with the Protein Pilot version 5.0 software (Sciex) utilizing the Paragon and Progroup algorithms and the integrated false discovery rate (FDR) analysis function. MS/MS data were searched against the NCBI Homo sapiens of the Uniprot-Sprot database downloaded on June 2, 2015. Search parameters are set as cys alkylation (Iodoacetamide), digestion (Trypsin), instrument (TripleTOF 6600), special factors (HexNAc emphasis and HexNAcFuc emphasis), species (Homo sapiens), ID Focus (Biological modifications), database (uniprot_sprot.fasta), search effort (Thorough), false discovery rate (FDR) analysis (Yes), and user modified parameter files (No). The proteins were inferred based on the ProGroup™ algorithm associated with the ProteinPilot software. The detected protein threshold in the software was set to the value which corresponded to 1% FDR. Peptides were defined as redundant if they had identical cleavage site(s), amino acid sequence, and modification. The mass spectra corresponding to HexNAcFuc/HexNAc-modified peptides were verified by spectral inspection.
LC-MS-MRM data from QTRAP 6500 was analyzed with MultiQuant 3.0 (Sciex) with manual confirmation; transitions with S/N > 10 were adopted for the quantification purposes. Peak intensities were used for peptide and glycopeptide quantification and data normalization. The MRM transition of the highest intensity for each peptide or glycopeptide was selected for quantification, while the remaining transitions were used to assure specificity of the targeted analyses. The core fucosylation level in each peptide was obtained by normalizing the intensity of HexNAcFuc-form peptide to the sum of intensities of both the HexNAcFuc-form and HexNAc-form. Duplicate measurements of each sample were averaged for quantitation.
Statistical analyses were performed with SAS release 9.4 (SAS Institute, Cary, NC). One-way Analysis of Variance (ANOVA) was used to evaluate the difference in core fucosylation among three comparison groups (healthy control, fibrosis and cirrhosis patients). In order to determine which pairs of groups are different after we reject H0 from ANOVA, Bonferroni’s procedure at level 0.05 was performed for multiple testing. Box plots were used to show the distribution of core fucosylation percentage in three groups; all reported p-values are two sided. The same analysis was performed to determine the peak area of unique peptides of core fucosylated N-glycoproteins.
3. Results and discussion
We applied a glycosidase-assisted MRM workflow to the quantification of core fucosylation of serum proteins in fibrotic liver disease. The optimized method allows quantification of the glycosylated peptides without prior fractionation of serum or enrichment of the glycopeptides. In brief, discovery mode proteomics analysis was used to identify core fucosylated proteins and to select tryptic glycopeptides treated with endoglycosidases for the quantification of core fucosylation. This was followed by optimization of the LC-MS-MRM workflow used for multiplex quantification of 17 proteins and core fucosylation of 22 N-glycopeptides in 45 participants at progressing stages of fibrotic liver disease.
3.1. Glycopeptide selection and MRM optimization
In order to analyze core fucosylation, tryptic digests of serum proteins were treated with a combination of endoF1, endoF2, and endoF3. The endoF treatment results in truncated peptide-pairs with HexNAcFuc or HexNAc attached to the asparagine in the NXS/T sequon, as described previously [39–41]. Ratio of these glycoforms serves as a measure of the degree of core fucosylation of the proteins at the specific sites of glycan attachment. For optimal cleavage of the glycans, we optimized the conditions for endoF deglycosylation by testing different endoglycosidases, reaction times, and their combinations. In contrast to previous studies using endoF3 [23–27], we add endoF1 and endoF2 in order to improve the yield of products from different types of glycoforms. EndoF1 cleaves more efficiently oligomannose and hybrid structures, endoF2 prefers biantennary complex-type oligosaccharides, while endoF3 cleaves biantennary and triantennary glycans [39–41].Since the presence of core fucose increases the cleavage performance of endoglycosidases [42,43], we optimized the conditions for the cleavage of the non-fucosylated glycoforms of the peptides which were cleaved with > 90% efficiency for both bi- and tri-antennary structures. Limitation of this method is the known lack of cleavage of tetra-antennary structures by the endoF digests and core fucosylation of these glycoforms is not quantified. However, the tetra-antennary glycoforms generally account for a small percentage of all glycoforms (< 10%) and are not expected to affect substantially the glycopeptides in this study [44,45].
We evaluated 24 glycopeptide pairs (HexNAc and HexNAcFuc forms of the same glycopeptide) identified by LC-MS/MS via data dependent acquisition (Supplementary Tables S1 and S2). Retention time (RT) and fragmentation of each glycopeptide were used to select at least 3 transitions for the quantification of each glycoform with the following criteria: 1) the m/z of product ion usually greater than precursor m/z using singly or doubly charged y ions for the quantification, 2) b ions and transitions close to precursors were avoided to maximize selectivity, and 3) transitions with highest intensity were selected. Of the 24 pairs, 2 were excluded from the final multiplex quantification because one of the glycoforms (HexNAc and HexNAcFuc) was not reliably quantifiable in serum samples of > 20% of the participants; typical reason is the low degree of core fucosylation of the glycopeptide (Supplementary Tables S2).
Simultaneously, we included in the multiplex assays non-glycosylated peptides of the 17 proteins to assess changes in the protein abundance that accompany the changes in core fucosylation of these proteins in liver disease. Uniqueness of each selected peptide for the 17 proteins was confirmed by searching against UniProtKB/Swiss-Prot human protein database and Protein Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Candidate peptides were further filtered using the criteria listed below to reduce potential sources of variability: 1) peptides containing < 6 amino acids (aa) were avoided to ensure uniqueness, 2) large peptides (> 25 aa), potential ragged end peptides, and peptides with known post-translational modifications were excluded, 3) peptides containing missed tryptic cleavage sites (e.g. internal lysine or arginine residues) were avoided. LC-MS-MRM instrument parameters including collision energies (CE) were optimized to yield the highest sensitivity and linearity of response for all transitions. The final multiplex assay consists of the quantification of 22 pairs (HexNAc and HexNAcFuc forms) of glycopeptides and 17 unique peptides for the corresponding glycoproteins (Supplementary Tables S1).
3.2. MRM-based quantification of core fucosylation in liver disease
The optimized workflow was applied to the analysis of 45 serum samples of participants with varying degree of liver fibrosis determined independently by clinical evaluation of liver biopsies. Biopsy is the gold standard for evaluation of fibrosis. The Ishak scores were used in our study to classify participants into fibrosis (n = 15) or cirrhosis (n = 15) groups (cirrhosis is an advanced stage of the fibrotic disease). The disease groups are compared to disease-free participants (n = 15).
To test the reproducibility of the LC-MS-MRM measurement, we included a total of 12 QC runs of a pooled sample. The average RSD in 12 QC runs of the 22 core fucosylated glycopeptides is 9.5% (Supplementary Fig. S1A) and 11.7% for the 17 proteotypic peptides (Supplementary Fig. S1B). For majority (~80%) of all the analytes the RSD was below 15%.
The results of quantification of core fucosylation of the 22 glycopeptides, corresponding to 17 proteins, are presented in Table 1. Approximately half of the glycopeptides (12 of 22) increased significantly in core fucosylation compared to the control group. The increase of core fucosylation of some peptides occurred already at the stage of fibrosis while others increased significantly at the cirrhotic stage (see below). Interestingly, decrease in core fucosylation was observed for several glycopeptides as well, in line with previous observations [26]. We did not observe a correlation between the changes in the quantity of the glycoproteins (Supplementary Tables S3) and their core fucosylation. However, the degree of core fucosylation, and its disease-related change, is clearly protein and site specific.
Table 1.
Quantification of changes in core fucosylation of glycopeptides in liver disease.
| Protein name | Peptidea | Core fucosylation level in percent (mean ± SD) | p value (ANOVA) | ||
|---|---|---|---|---|---|
| Control | Fibrosis | Cirrhosis | |||
| Ig gamma-2 chain C region | EEQF180NSTFR | 91.4 ± 2.75 | 84.9 ± 13.47 | 87.5 ± 3.95 | 0.107 |
| Immuno globulin J chain | E71NISDPTSPLR | 16.8 ± 2.6 | 17.9 ± 4.07 | 20.3 ± 2.34 | 0.013 |
| Ig alpha-2 chain C region | TPLTA205NITK | 36.2 ± 17.03 | 30.2 ± 12.97 | 38 ± 14.7 | 0.340 |
| Hemopexin | SWPAVG187NCSSALR | 9.9 ± 2.41 | 12.8 ± 3.18 | 14 ± 5.24 | 0.016 |
| Complement C3 | TVLTPATNHmG85NVTFTIPANR | 14.3 ± 10.39 | 10.4 ± 5.84 | 11.8 ± 6.6 | 0.430 |
| Alpha-2-HS-glycoprotein | AALAAFNAQN176NGSNFQLEEISR | 15.5 ± 4.57 | 17.7 ± 4.01 | 16.8 ± 5.71 | 0.456 |
| Fibronectin | LDAPTNLQFV1007NETDSTVLVR | 98.6 ± 0.64 | 98.6 ± 0.86 | 98.4 ± 0.67 | 0.671 |
| Complement C1q subcomponent subunit A | NPPMGGNWIFDTVITNQEEPYQ146NHSGR | 45.3 ± 14.86 | 49 ± 11.54 | 49.2 ± 10.63 | 0.624 |
| Serotransferrin | QQQHLFGS630NVTDCSGNFCLFR | 7.7 ± 1.85 | 10.6 ± 2.17 | 11.6 ± 4.69 | 0.004 |
| Plasma protease C1 inhibitor | VLS253NNSDANLELINTWVAK | 34 ± 5.46 | 41.7 ± 5.25 | 43.8 ± 11.84 | 0.005 |
| Pigment epithelium-derived, factor | VTQ285NLTLIEESLTSEFIHDIDR | 79.9 ± 10.68 | 76.9 ± 11.52 | 83.3 ± 6.13 | 0.224 |
| Alpha-2-macroglobulin | GCVLLSYL55NETVTVSASLESVR | 72.3 ± 11.72 | 66.3 ± 11.72 | 66.4 ± 10.88 | 0.271 |
| Clusterin | ML354NTSSLLEQLNEQFNWVSR | 5.5 ± 0.95 | 6.7 ± 1.57 | 8.6 ± 2.4 | 0.000 |
| Alpha-1-antitrypsin | QLAHQS70NSTNIFFSPVSIATAFAMLSLGTK | 27.9 ± 15.05 | 27.3 ± 11.24 | 29.7 ± 11.71 | 0.875 |
| ADTHDEILEGLNF107NLTEIPEAQIHEGFQELLR | 6.9 ± 1.95 | 9.9 ± 2.36 | 11.4 ± 4.48 | 0.001 | |
| Lumican | AFE88NVTDLQWLILDHNLLENSK | 91.8 ± 2.75 | 90.3 ± 10.43 | 94.4 ± 0.89 | 0.234 |
| LHINHN127NLTESVGPLPK | 54.5 ± 3.9 | 58.1 ± 4.84 | 58.9 ± 4.06 | 0.016 | |
| Vitronectin | 169NGSLFAFR | 21.7 ± 7.96 | 14.6 ± 7.92 | 13.8 ± 7.82 | 0.017 |
| N86NATVHEQVGGPSLTSDLQAQSK | 5.6 ± 3.15 | 12.2 ± 6.15 | 12.6 ± 6.2 | 0.001 | |
| Ceruloplasmin | E397NLTAPGSDSAVFFEQGTTR | 9.6 ± 2.34 | 14.4 ± 2.94 | 14.9 ± 4.74 | 0.002 |
| EHEGAIYPD138NTTDFQR | 26 ± 3.86 | 30.3 ± 5.14 | 32.4 ± 8.91 | 0.026 | |
| ELHHLQEQ762NVSNAFLDK | 2.1 ± 0.42 | 2.5 ± 0.38 | 3.2 ± 1.53 | 0.009 | |
Modified glycosylation site is shown in italic.
3.2.1. Increased core fucosylation at the stage of liver fibrosis
Fibrosis of the liver reflects chronic damage associated with the accumulation of extracellular matrix proteins [46,47]. Aberrant glycosylation of these liver proteins has been observed and core fucosylation, in particular, was described in several studies [13–16]. However, quantification of the changes in core fucosylation of the proteins at different stages of the disease progression remains virtually unreported [26]. To the best of our knowledge, multiplex LC-MS-MRM evaluation of core fucosylation of fibrotic liver disease was not previously described. Several studies report increased core fucosylation in the context of hepatocellular carcinoma (HCC), an end-stage complication of the fibrotic liver damage [13–15]. It was, therefore, somewhat surprising that we find the changes in core fucosylation of some proteins already at the stage of liver fibrosis.
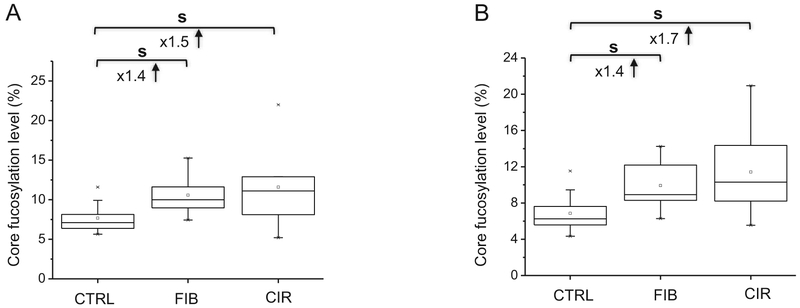
We observe increased core fucosylation of 5 glycopeptides at the stage of liver fibrosis (N630 of serotransferrin, N107 of alpha-1-anti-trypsin, N253 of plasma protease C1 inhibitor, N397 of ceruloplasmin, and N86 of vitronectin). In the case of serotransferrin, we observe approximately 8% core fucosylation of the QQQHLFGS630NVT-DCSGNFCLFR glycopeptide in serum of disease-free controls, in agreement with previous reports [48–50]. However, previous studies compared HCC to disease-free controls [15,45,48] while, in our study, core fucosylation increased to 11% in the fibrotic patients (significant 1.4-fold increase) (Fig. 1A). This elevation occurs therefore at a stage of the disease much earlier than the HCC and could be potentially a risk factor for HCC development. The degree of core fucosylation remains elevated but does not further increase in the cirrhotic patients and we do not see differences in the protein abundance of serotransferrin along the progression of liver damage (Supplementary Tables S3).
Fig. 1.
Increased core fucosylation of QQQHLFGS630NVTDCSGNFCLFR of serotransferrin (A) and ADTHDEILEGLNF107NLTEIPEAQIHEGFQELLR of alpha-1-antitrypsin (B) at the liver fibrosis stage (s, significant at p < 0.05; Ctrl, control; FIB, fibrosis; CIR, cirrhosis).
Similarly, several studies reported core fucosylation of alpha-1-antitrypsin in HCC [15,51–54] but we observe a significant 1.4-fold increase in core fucosylation at the ADTHDEILEGLNF107NLTEIPEAQIHEGFQELLR peptide in fibrosis and further 1.7-fold increase in cirrhosis compared to the disease-free controls (Fig. 1B). The increase occurs at an earlier stage of the disease progression than previously reported [54], further suggesting that it is indispensable to develop and apply an analytical method of more quantitative precision for the earlier monitoring of core fucosylation level for the better evaluation of liver disease progression. Besides the increased core fucosylation at N107, the protein abundance of alpha-1-antitrypsin also increases at the fibrotic stage (Supplementary Tables S3 and Supplementary Fig. S2A).
In the case of ceruloplasmin, we observe a significant 1.5-fold increase in core fucosylation of E397NLTAPGSDSAVFFEQGTTR at the stage of liver fibrosis; however, core fucosylation of two additional glycopeptides of the same protein increases only in cirrhotic samples (see below). This disease and site-specific change is further pronounced in the case of vitronectin which is also discussed in a later section. The 1.2-fold increase in core fucosylation of the VLS253NNSDANLELINTWVAK glycopeptide of plasma protease C1 inhibitor in liver fibrosis (or other stage of liver disease) is not to our knowledge previously reported (Table 1).
3.2.2. Increased core fucosylation at the stage of liver cirrhosis
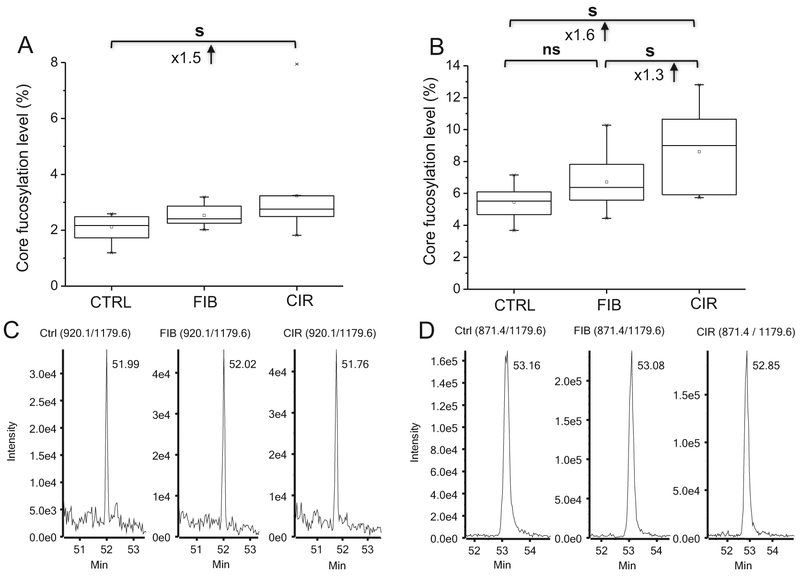
Cirrhosis represents an advanced stage of liver fibrosis and a major risk factor for the development of HCC [55]; several studies of cell and animal models reported that core fucosylation of proteins is a key contributor in the development of HCC [16,20,47]. In our study, two glycopeptides of ceruloplasmin, ELHHLQEQ762NVSNAFLDK (Fig. 2A) and EHEGAIYPD138NTTDFQR (Table 1), have significantly up-regulated core fucosylation at the cirrhosis stage, in contrast to the E397NLTAPGSDSAVFFEQGTTR glycopeptide discussed above. This suggests that site-specific changes in glycosylation should have better prediction accuracy at progressive stages of liver disease progression than the analysis of detached glycans which averages across multiple sites and/or proteins.
Fig. 2.
Increased core fucosylation of ELHHLQEQ762NVSNAFLDK of ceruloplasmin (A) and ML354NTSSLLEQLNEQFNWVSR of clusterin (B) significant at the stage of liver cirrhosis. Representative LC-MRM-MS traces of the transition (920.1/1179.6; [M+3H]3+) of ML354N(HexNAcFuc)TSSLLEQLNEQFNWVSR (C) and the transition (871.4/1179.6; [M+3H]3+) of ML354N(HexNAc)TSSLLEQLNEQFNWVSR (D) of clusterin (s, significant at p < 0.05; ns, non-significant; Ctrl, control; FIB, fibrosis; CIR, cirrhosis).
Clusterin, a well characterized and constitutively secreted extra-cellular chaperone, is also fucosylated [15,26,56,57]. We find a 1.6-fold increase in core fucosylation at ML354NTSSLLEQLNEQFNWVSR of clusterin in liver cirrhosis (Fig. 2B). Fig. 2 shows representative transition of the core fucosylated form (920.1/1179.6; +3 charge; Fig. 2C) and the transition for the HexNAc form (871.4/1179.6; +3 charge; Fig. 2D). The comparison of the retention times on the RP column shows a constant shift of approximately 70 s between the core-fucosylated and non-fucosylated glycoforms of the peptide, which is an expected influence of the addition of core-fucose and thus lower hydrophobicity. Of note, these two glycoforms nearly co-elute for all the other peptides in our study but there is a consistently slightly shorter retention time of the fucosylated glycopeptide (Supplementary Table S1).
The N-glycan profile of hemopexin was suggested as a biomarker for the detection of HCC in patients with cirrhosis [58] and a trend toward increased fucosylation in liver disease was observed in our previously published study [59]. We confirm our observation and show that the core fucosylation of SWPAVG187NCSSALR glycopeptide of hemopexin increases in cirrhosis. And the discrepancy from ref. [58] may be due to differences in the population examined or differences in the methods. At the same time, the observed decrease in the abundance of hemopexin (Supplementary Fig. S2B) shows that the trends in the changes in protein abundance and core fucosylation are independent. Similar trend (increased core fucosylation and decreased protein abundance in cirrhosis) was observed for the E71NISDPTSPLR glycopeptide of the immunoglobulin J chain (Supplementary Table S3 and Fig. S2C). Quantification of the core fucosylation provides therefore independent information and may improve our understanding of the liver disease progression. Our results further demonstrate that the targeted quantification of site-specific core fucosylation is feasible even in samples of unfractionated serum without glycopeptide enrichment and that the LC MS-MRM workflow simplifies the previously reported workflows and will be easier to implement in larger studies.
3.2.3. Site-specific core fucosylation of serum proteins
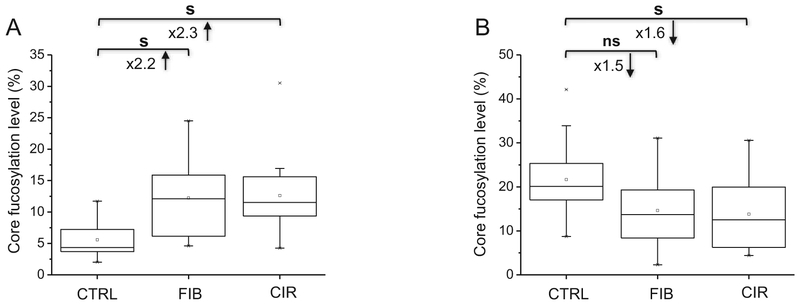
Site-specific changes in core fucosylation are observed for several proteins with more than one core fucosylated glycopeptide. For example, LHINHN127NLTESVGPLPK of lumican has increased core fucosylation in cirrhosis but no change in core fucosylation is observed on the other glycopeptide AFE88NVTDLQWLILDHNLLENSK (Table 1). The degree of core fucosylation of three glycopeptides of ceruloplasmin in disease-free controls ranges from 2% to 26% (Table 1). Although all three sites (N138, N397, N762) of ceruloplasmin increase in core fucosylation but only one of the peptides increases at the stage of liver fibrosis (Table 1). Opposite change in core fucosylation is observed on two glycopeptides of vitronectin (Fig. 3), a candidate biomarker for the detection of HCC in patients with chronic liver disease [60,61]. Specifically, core fucosylation progressively increases in fibrosis and cirrhosis for the N86NATVHEQVGGPS-LTSDLQAQSK glycopeptide (Fig. 3A) but decreases for the 169NGSLFAFR glycopeptide in cirrhosis (Fig. 3B). We fail to detect reliably a third core fucosylation site of vitronectin observed in previous studies of immunodepleted and/or enriched samples of HCC patients [22,26,62,63]. This might be related to higher degree of branching on one of the sites or the presence of bisected glycoforms [64,65] that were reported to down-regulate core fucosylation [66,67]. It could also be related to the selective retention of specific vitronectin glycoforms in the serum of patients during enrichment [62,63]; we do not attempt to further analyze reasons for the observation. Moreover, no change in the abundance of vitronectin is observed (Supplementary Table S3), in agreement with previous studies [63]. In addition, we find a tendency toward a decrease in core fucosylation of GCVLLSYL55NETVTVSASLESVR glycopeptide of alpha-2-macroglobin and EEQF180NSTFR glycopeptide of Ig gamma-2 chain C region in cirrhosis (Supplementary Fig. S3A and C) although a significant increase in the abundance of each protein is observed (Supplementary Fig. S3B and D). Collectively, the observed differences in core fucosylation at specific glycopeptides clearly document the differential regulation of core fucosylation of individual proteins.
Fig. 3.
Site-specific core fucosylation of N86NATVHEQVGGPSLTSDLQAQSK (A) and 169NGSLFAFR (B) of vitronectin (s, significant at p < 0.05; ns, non-significant; Ctrl, control; FIB, fibrosis; CIR, cirrhosis).
4. Conclusion
In this study, we simplified the detection of core fucosylation of N-glycoproteins and demonstrate the feasibility of direct quantification of these glycoforms without any fractionation of serum or enrichment. The multiplex LC-MS-MRM workflow takes advantage of simplification of the N-glycans with endoglycosidases and the advantageous chromatographic/mass spectrometric properties of the truncated glycopeptides. We demonstrate applicability of the method to clinical samples in a study of liver fibrosis intended to improve our understanding of the elevation in core fucosylation of serum proteins at progressive stages of liver disease. The workflow achieves an average RSD of 9.5% and documents significantly elevated core fucosylation of multiple glycoproteins already at the stage of liver fibrosis which suggests involvement of this pathway at the earliest stages of the disease process. We document that the changes in core fucosylation occur in a protein- and site-specific manner; glycopeptides of some proteins even decrease in core fucosylation against the general increasing trend at successive stages of the disease progression. The increased core fucosylation in fibrotic liver disease needs to be taken into account in the studies of HCC. Although we demonstrate that our LC-MS-MRM work-flow is suitable for the quantification of core fucosylated glycopeptides directly in patient serum, the results do not establish clinical utility of such measurements. Further studies are needed to evaluate sensitivity and specificity of the assays for the detection of liver disease at progressing stages of development. In summary, we describe a simplified LC-MS-MRM workflow which has the potential to improve non-invasive serologic monitoring of liver disease by the quantification of site-specific changes in core fucosylation of proteins.
Acknowledgments
This work was supported by National Institutes of Health grants UO1 CA168926, RO1 CA135069 to RG and CCSG Grant P30 CA51008, to Lombardi Comprehensive Cancer Center, supporting the Proteomics and Metabolomics Shared Resource, Georgetown University Medical Center. We thank Dr. Wei Yuan for helpful discussions.
Footnotes
Conflict of interest
The authors have declared no conflict of interest.
Transparency document
The http://dx.doi.org/10.1016/j.jprot.2018.02.003 associated with this article can be found, in the online version.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jprot.2018.02.003.
Supplementary Material
Supplementary Material
References
- [1].Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, et al. , Essentials of Glycobiology, 3rd edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY), 2017. [PubMed] [Google Scholar]
- [2].Wilson JR, Williams D, Schachter H, The control of glycoprotein synthesis: N-acetylglucosamine linkage to a mannose residue as a signal for the attachment of L-fucose to the asparagine-linked N-acetylglucosamine residue of glycopeptide from alpha1-acid glycoprotein, Biochem. Biophys. Res. Commun 72 (1976) 909–916. [DOI] [PubMed] [Google Scholar]
- [3].Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, et al. , Proc. Natl. Acad. Sci. U.S. A 102 (2005) 15791–15996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang Q, Zhang R, Cai H, Wang LX, Revisiting the substrate specificity of mammalian α1,6-fucosyltransferase (FUT8) reveals that it catalyzes core fucosylation of N-glycans lacking α1,3-arm GlcNAc, J. Biol. Chem 292 (2017) 14796–14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Takahashi M, Kuroki Y, Ohtsubo K, Taniguchi N, Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins, Carbohydr. Res 344 (2009) 1387–1390. [DOI] [PubMed] [Google Scholar]
- [6].Miyoshi E, Moriwaki K, Terao N, Tan C-C, Terao M, Nakagawa T, et al. , Fucosylation is a promising target for cancer diagnosis and therapy, Biomol. Ther 2 (2012) 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhao YP, Xu XY, Fang M, Wang H, You Q, Yi CH, et al. , Decreased core-fucosylation contributes to malignancy in gastric cancer, PLoS One 9 (2014) e94536–e94548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen J, Fang M, Zhao Y-P, Yi C-H, Ji J, Cheng C, et al. , Serum N-glycans: a new diagnostic biomarker for light chain multiple myeloma, PLoS One 10 (2015) e0127022–0127036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Geng F, Shi BZ, Yuan YF, Wu XZ, The expression of core fucosylated E-cadherin in cancer cells and lung cancer patients: prognostic implications, Cell Res 14 (2004) 423–433. [DOI] [PubMed] [Google Scholar]
- [10].Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM, Core fucosylation and α2–3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia, Glycobiology 21 (2011) 195–205. [DOI] [PubMed] [Google Scholar]
- [11].Tu CF, Wu MY, Lin Y-C, Kannagi R, Yang R-B, FUT8 promotes breast cancer cell invasiveness by remodeling TGF-β receptor core fucosylation, Breast Cancer Res 19 (2017) 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, et al. , A systems biology approach identifies FUT8 as a driver of melanoma metastasis, Cancer Cell 31 (2017) 804–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hutchinson WL, Du MQ, Johnson PJ, Williams R, Fucosyltransferases: differential plasma and tissue alterations in hepatocellular carcinoma and cirrhosis, Hepatology 13 (1991) 683–688. [PubMed] [Google Scholar]
- [14].Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, et al. , Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, et al. , Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma, J. Proteome Res (2) (2006) 308–315. [DOI] [PubMed] [Google Scholar]
- [16].Zhou Y, Fukuda T, Hang Q, Hou S, Isaji T, Kameyama A, Gu J, Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation, Sci. Rep 7 (2017) 11563–11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lee SJ, Evers S, Roeder D, Parlow AF, Risteli J, Risteli L, et al. , Mannose receptormediated regulation of serum glycoprotein homeostasis, Science 295 (2002) 1898–1901. [DOI] [PubMed] [Google Scholar]
- [18].Shimizu K, Katoh H, Yamashita F, Tanaka M, Tanikawa K, Taketa K, et al. , Comparison of carbohydrate structures of serum alpha-fetoprotein by sequential glycosidase digestion and lectin affinity electrophoresis, Clin. Chim. Acta 254 (1996) 23–40. [DOI] [PubMed] [Google Scholar]
- [19].Gupta S, Bent S, Kohlwes J, Test characteristics of a-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis, Ann. Intern. Med 139 (2003) 46–50. [DOI] [PubMed] [Google Scholar]
- [20].Wang Y, Fukuda T, Isaji T, Lu J, Im S, Hang Q, et al. , Loss of α1,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways, FASEB J 29 (2015) 3217–3227. [DOI] [PubMed] [Google Scholar]
- [21].Thaysen-Andersen M, Packer NH, Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching, Glycobiology 22 (2012) 1440–1452. [DOI] [PubMed] [Google Scholar]
- [22].Jia W, Lu Z, Fu Y, Wang HP, Wang LH, Chi H, et al. , A strategy for precise and large scale identification of core fucosylated glycoproteins, Mol. Cell. Proteomics 8 (2009) 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yin H, Lin Z, Nie S, Wu J, Tan Z, Zhu J, et al. , Mass-selected site-specific core-fucosylation of ceruloplasmin in alcohol-related hepatocellular carcinoma, J. Proteome Res 13 (2014) 2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cao Q, Zhao X, Zhao Q, Lv X, Ma C, Li X, et al. , Strategy integrating stepped fragmentation and glycan diagnostic ion-based spectrum refinement for the identification of core fucosylated glycoproteome using mass spectrometry, Anal. Chem 86 (2014) 6804–6811. [DOI] [PubMed] [Google Scholar]
- [25].Ma C, Zhang Q, Qu J, Zhao X, Li X, Liu Y, et al. , A precise approach in large scale core-fucosylated glycoprotein identification with low- and high-normalized collision energy, J. Proteome 114 (2015) 61–70. [DOI] [PubMed] [Google Scholar]
- [26].Yin H, Tan Z, Wu J, Zhu J, Shedden KA, Marrero J, et al. , Mass-selected site-specific core-fucosylation of serum proteins in hepatocellular carcinoma, J. Proteome Res 14 (2015) 4876–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ma C, Qu J, Li X, Zhao X, Li L, Xiao C, et al. , Improvement of core-fucosylated glycoproteome coverage via alternating HCD and ETD fragmentation, J. Proteome 146 (2016) 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou J, Yang W, Hu Y, Höti N, Liu Y, Shah P, et al. , Site-specific fucosylation analysis identifying glycoproteins associated with aggressive prostate cancer cell lines using tandem affinity enrichments of intact glycopeptides followed by mass spectrometry, Anal. Chem 89 (2017) 7623–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, et al. , High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites, Mol. Cell. Proteomics 6 (2007) 1809–1817. [DOI] [PubMed] [Google Scholar]
- [30].Lsmeier AJ, Paesold-Burda P, Hennet T, N-glycosylation site occupancy in serum glycoproteins using multiple reaction monitoring liquid chromatography-mass spectrometry, Mol. Cell. Proteomics 6 (2007) 2132–2138. [DOI] [PubMed] [Google Scholar]
- [31].Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R, Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MSMRM) analysis of site-specific glycoforms of haptoglobin in liver disease, Mol. Cell. Proteomics 12 (2013) 1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goldman R, Sanda M, Targeted methods for quantitative analysis of protein glycosylation, Proteomics Clin. Appl 9 (2015) 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hong Q, Ruhaak LR, Stroble C, Parker E, Huang J, Maverakis E, Lebrilla CB, A method for comprehensive glycosite-mapping and direct quantitation of serum glycoproteins, J. Proteome Res 14 (2015) 5179–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yuan W, Sanda M, Wu J, Koomen J, Goldman R, Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LC-MS-MRM in liver disease, J. Proteome 116( (2015) 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sanda M, Benicky J, Wu J, Wang Y, Makambi K, Ahn J, Smith CI, Zhao P, Zhang L, Goldman R, Increased sialylation of site specific O-glycoforms of hemopexin in liver disease, Clin. Proteomics 13 (2016) 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhao Y, Jia W, Wang J, Ying W, Zhang Y, Qian X, Fragmentation and site-specific quantification of core fucosylated glycoprotein by multiple reaction monitoring mass spectrometry, Anal. Chem 83 (2011) 8802–8809. [DOI] [PubMed] [Google Scholar]
- [37].Di Bisceglie AM, Shiffman ML, Everson GT, Lindsay KL, Everhart JE, Wright EC, et al. , Prolonged therapy of advanced chronic hepatitis C with low dose peginterferon, N. Engl. J. Med 359 (2008) 2429–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lok AS, Everhart JE, Chung RT, Kim HY, Everson GT, Hoefs JC, et al. , Evolution of hepatic steatosis in patients with advanced hepatitis C: results from the hepatitis C antiviral long-term treatment against cirrhosis (HALTC) trial, Hepatology 49 (2009) 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Trimble RB, Tarentino AL, Identification of distinct endoglycosidase (Endo) activities in Flavobacterium meningosepticum: Endo F1, Endo Fa, and Endo F3, J. Biol. Chem 266 (1991) 1646–1651. [PubMed] [Google Scholar]
- [40].Freeze HH, Kranz C, Endoglycosidase and glycoamidase release of N-linked gly-cans, Curr. Protoc. Mol. Biol 89 (2010) 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee JE, Fusco ML, Saphire EO, An efficient platform for screening expression and crystallization of glycoproteins produced in human cells, Nat. Protoc 4 (2009) 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Plummer TH Jr., A.W. Phelan, A.L. Tarentino, Porcine fibrinogen glycopeptides: substrates for detecting endo-beta-N-acetylglucosaminidases F2 and F3(1), Anal. Biochem 235 (1996) 98–101. [DOI] [PubMed] [Google Scholar]
- [43].Huang W, Li J, Wang LX, Unusual transglycosylation activity of Flavobacterium meningosepticum endoglycosidases enables convergent chemoenzymatic synthesis of core fucosylated complex N-glycopeptides, Chembiochem 12 (2011) 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Clerc F, Reiding KR, Jansen BC, Kammeijer GS, Bondt A, Wuhrer M, Human plasma protein N-glycosylation, Glycoconj. J 33 (2016) 309–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ji ES, Hwang H, Park GW, Lee JY, Lee HK, Choi NY, et al. , Analysis of fucosylation in liver-secreted N-glycoproteins from human hepatocellular carcinoma plasma using liquid chromatography with tandem mass spectrometry, Anal. Bioanal. Chem 408 (2016) 7761–7774. [DOI] [PubMed] [Google Scholar]
- [46].Bataller R, Brenner DA, Liver fibrosis J Clin. Invest 115 (2005) 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Blomme B, Van Steenkiste C, Callewaert N, Van Vlierberghe H, Alteration of protein glycosylation in liver diseases, J. Hepatol 50 (2009) 592–603. [DOI] [PubMed] [Google Scholar]
- [48].Yamashita K, Koide N, Endo T, Iwaki Y, Kobata A, Altered glycosylation of serum transferrin of patients with hepatocellular carcinoma, J. Biol. Chem 264 (1989) 2415–2423. [PubMed] [Google Scholar]
- [49].Satomi Y, Shimonishi Y, Hase T, Takao T, Site-specific carbohydrate profiling of human transferrin by nano-flow liquid chromatography/electrospray ionization mass spectrometry, Rapid Commun. Mass Spectrom 18 (2004) 2983–2988. [DOI] [PubMed] [Google Scholar]
- [50].Nagae M, Morita-Matsumoto K, Arai S, Wada I, Matsumoto Y, Saito K, et al. , Structural change of N-glycan exposes hydrophobic surface of human transferrin, Glycobiology 24 (2014) 693–702. [DOI] [PubMed] [Google Scholar]
- [51].Mills K, Mills PB, Clayton PT, Johnson AW, Whitehouse DB, Winchester BG, Identification of alpha(1)-antitrypsin variants in plasma with the use of proteomic technology, Clin. Chem 47 (2001) 2012–2022. [PubMed] [Google Scholar]
- [52].Kolarich D, Weber A, Turecek PL, Schwarz H-P, Altmann F, Comprehensive glyco-proteomic analysis of human α1-antitrypsin and its charge isoforms, Proteomics 6 (2006) 3369–3380. [DOI] [PubMed] [Google Scholar]
- [53].Ruhaak LR, Koeleman CA, Uh H-W, Stam JC, van Heemst D, Maier AB, et al. , Targeted biomarker discovery by high throughput glycosylation profiling of human plasma alpha1-antitrypsin and immunoglobulin A, PLoS One 8 (2013) e73082–e73093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, et al. , Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma, PLoS One 5 (2010) e12419–12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fattovich G, Stroffolini T, Zagni I, Donato F, Hepatocellular carcinoma in cirrhosis: incidence and risk factors, Gastroenterology 127 (2004) S35–50. [DOI] [PubMed] [Google Scholar]
- [56].Kapron JT, Hilliard GM, Lakins JN, Tenniswood MP, West KA, Carr SA, et al. , Identification and characterization of glycosylation sites in human serum clusterin, Protein Sci 6 (1997) 2120–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gbormittah FO, Bones J, Hincapie M, Tousi F, Hancock WS, Iliopoulos O, Clusterin glycopeptide variant characterization reveals significant site-specific glycan changes in the plasma of clear cell renal cell carcinoma, J. Proteome Res 14 (2015) 2425–2436. [DOI] [PubMed] [Google Scholar]
- [58].Debruyne EN, Vanderschaeghe D, Van Vlierberghe H, Vanhecke A, Callewaert N, Delanghe JR, Diagnostic value of the hemopexin N-glycan profile in hepatocellular carcinoma patients, Clin. Chem 56 (2010) 823–831. [DOI] [PubMed] [Google Scholar]
- [59].Benicky J, Sanda M, Pompach P, Wu J, Goldman R, Quantification of fucosylated hemopexin and complement factor H in plasma of patients with liver disease, Anal. Chem 86 (2014) 10716–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee HJ, Na K, Choi EY, Kim KS, Kim H, et al. , Simple method for quantitative analysis of N-linked glycoproteins in hepatocellualr carcinoma specimens, J. Proteome Res 9 (2010) 308–318. [DOI] [PubMed] [Google Scholar]
- [61].Lee JY, Kim JY, Park GW, Cheon MH, Kwon KH, et al. , Targeted mass spectrometric approach for biomarker discovery and validation with nonglycosylated tryptic peptides from N-linked glycoproteins in human plasma, Mol. Cell. Proteomics 10 (2011) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lee HJ, Cha HJ, Lim JS, Lee SH, Song SY, Kim H, et al. , Abundance-ratio-based semiquantitative analysis of site-specific N-linked glycopeptides present in the plasma of hepatocellular carcinoma patients, J. Proteome Res 13 (2014) 2328–2338. [DOI] [PubMed] [Google Scholar]
- [63].Hwang H, Lee JY, Lee HK, Park GW, Jeong HK, Moon MH, et al. , In-depth analysis of site-specific N-glycosylation in vitronectin from human plasma by tandem mass spectrometry with immunoprecipitation, Anal. Bioanal. Chem 406 (2014) 7999–8011. [DOI] [PubMed] [Google Scholar]
- [64].Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, et al. , N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus, Hepatology 46 (2007) 1426–1435. [DOI] [PubMed] [Google Scholar]
- [65].Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, et al. , Detection of hepatocellular carcinoma using glycomic analysis, Clin. Cancer Res 15 (2009) 1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kurimoto A, Kitazume S, Kizuka Y, Nakajima K, Oka R, Fujinawa R, et al. , The absence of core fucose up-regulates GnT-III and Wnt target genes: a possible mechanism for an adaptive response in terms of glycan function, J. Biol. Chem 289 (2014) 11704–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nishima W, Miyashita N, Yamaguchi Y, Sugita Y, Re S, Effect of bisecting GlcNAc and core fucosylation on conformational properties of biantennary complex-type NGlycans in solution, J. Phys. Chem. B 116 (2012) 8504–8512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.