SUMMARY
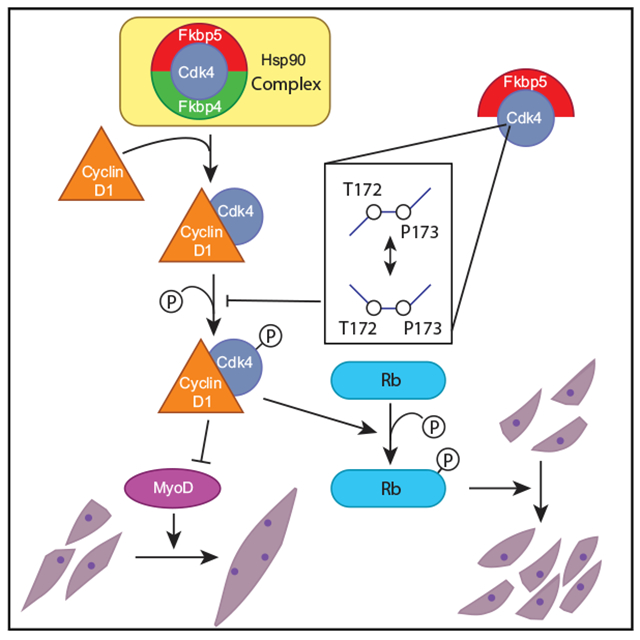
Fkbp5 is a widely expressed peptidyl prolyl isomerase that serves as a molecular chaperone through conformational changes of binding partners. Although it regulates diverse protein functions, little is known about its roles in myogenesis. We found here that Fkbp5 plays critical roles in myoblast differentiation through two mechanisms. First, it sequesters Cdk4 within the Hsp90 storage complex and prevents the formation of the cyclin D1-Cdk4 complex, which is a major inhibitor of differentiation. Second, Fkbp5 promotes cis-trans isomerization of the Thr172-Pro173 peptide bond in Cdk4 and inhibits phosphorylation of Thr172, an essential step for Cdk4 activation. Consistent with these in vitro findings, muscle regeneration is delayed in Fkbp5−/− mice. The related protein Fkbp4 also sequesters Cdk4 within the Hsp90 complex but does not isomerize Cdk4 or induce Thr173 phosphorylation despite its highly similar sequence. This study demonstrates protein isomerization as a critical regulatory mechanism of myogenesis by targeting Cdk4.
Graphical Abstract
INTRODUCTION
Fkbp5 (also called Fkbp51) belongs to the FK506-binding protein (Fkbp) family of molecular chaperones, which contain peptidyl prolyl isomerase (PPIase) domains that accelerate the cis-trans isomerization of a peptide bond preceding a proline (Storer et al., 2011). The two peptide chains flanking a peptide bond can take only two conformations, cis or trans, without any intermediate states because a peptide bond is a partial double bond, restricting free rotation (Hanes, 2015; Schiene-Fischer et al., 2013; Schmidpeter et al., 2015). The trans conformation is generally more stable because steric hindrance between the amino acids on opposite sides of the peptide bond is weaker than in the cis conformation. This makes the cis conformation extremely rare (<0.01%), unless the peptide bond precedes a proline (Weiss et al., 1998). Proline renders the cis conformation more stable and prevalent (around 5%) because it has a cyclic side chain bound to the backbone amide nitrogen, preventing repulsion between the two peptide chains. The higher frequency of the cis conformation leads to increased variation in protein folding patterns, making cis-trans interconversion a rate-limiting step in protein folding that regulates their functions. PPIases accelerate the interconversion by decreasing the energy barrier required for the interconversion, serving as a switch for many protein activities, including transcription, chromatin modification, and signal transduction, as well as pathogenesis of Alzheimer’s disease and cancer (Hanes, 2015; Nigro et al., 2013; Romano et al., 2015; Storer et al., 2011). However, the presence of a PPIase does not necessarily determine the choice between cis and trans conformations; it is also determined by the composition of surrounding amino acids and physiological conditions.
Fkbp5 was discovered as a subunit of the progesterone receptor complex (Smith et al., 1993) and regulates transcriptional activity of several steroid hormone receptors (Makkonen and Palvimo, 2011; Stechschulte and Sanchez, 2011; Storer et al., 2011). For example, activation of androgen receptors by Fkbp5 plays a major role in androgen-mediated proliferation of prostate cancer cells. The interaction between Fkbp5 and glucocorticoid receptors modulates stress-mediated neurological diseases (Hausch, 2015; Storer et al., 2011). In addition, Fkbp5 is involved in steroid-hormone-independent functions, such as isomerization of the tau protein and regulation of the Akt signaling pathway (Cioffi et al., 2011). The immunosuppressants FK506 and rapamycin bind to the PPIase domain of Fkbps, including Fkbp4 and Fkbp5, inhibiting their activity (Hanes, 2015). However, Fkbp1 (also called Fkbp12) is the primary target for FK506-mediated immunosuppression in T cells (Xu et al., 2002). Fkbp4 and Fkbp5 do not seem to play a major role in immunosuppression.
Fkbp5−/− mice are fertile and do not show any major phenotypes other than anti-depressive behavior (O’Leary et al., 2011; Yong et al., 2007). Hypergravity is known to upregulate Fkbp5 expression in the soleus muscle (Shimoide et al., 2016). In addition, overexpressed Fkbp5 in mouse myoblast C2C12 cells induces mRNA encoding sarcomeric myosin heavy chain (MHC) (a marker for differentiating myocytes), which might be relevant to the increased muscle mass due to hypergravity. Other than these findings, little is known about the roles of Fkbp5 in muscle cell proliferation or differentiation. Fkbp4 (also called Fkbp52) is 77% similar to Fkbp5 at the amino acid level (Sivils et al., 2011; Storer et al., 2011) and is also involved in steroid hormone receptor signaling. Fkbp4−/− female mice are sterile due to implantation failure caused by deregulation of the progesterone receptor pathway (Tranguch et al., 2005; Yang et al., 2006). Fkbp4−/− male mice are also sterile with multiple defects in reproductive tissues, such as ambiguous external genitalia and prostate dysgenesis, due to disrupted androgen receptor signaling (Cheung-Flynn et al., 2005). Fkbp4−/−Fkbp5−/− mice are embryonic lethal, indicating that some functions are redundant between the two genes, although details remain unknown (Yong et al., 2007). Muscle phenotypes of Fkbp4−/− and Fkbp4−/−Fkbp5−/− mice have not been characterized.
Myogenesis is tightly coupled with cell cycle regulation (Giacinti and Giordano, 2010; Motohashi and Asakura, 2014; Ruijtenberg and van den Heuvel, 2016). Myogenic stem cells (satellite cells) do not proliferate but will enter the cell cycle when activated by injury or weight bearing. The proliferating cells (myoblasts) will eventually exit from the cell cycle (myocytes) and fuse to form multinucleated cells (myotubes). They will enlarge and mature (myofibers) by incorporating more cells and increasing the cytoplasmic volume per nucleus. The current study illuminates a regulatory mechanism of the cell cycle exit by Fkbp5 through the interaction with Cdk4. Newly translated Cdk4 is first incorporated into a protein complex containing the chaperone Hsp90 and the co-chaperone adaptor Cdc37 in the cytoplasm, where Cdk4 is stabilized in an inactive form (Smith et al., 2015; Stepanova et al., 1996; Vaughan et al., 2008). Activation of Cdk4 and subsequent phosphorylation of Rb play a central role in deciding whether cells proceed through the restriction point in the late G1 phase; cells are then committed to completing the cell cycle independently of mitogens (Asghar et al., 2015; Baker and Reddy, 2012; Bockstaele et al., 2006). The protein level of Cdk4 does not drastically fluctuate during the cell cycle; its activity is controlled by at least three steps of post-translational regulation, although the exact order of events has not been firmly established (Bisteau et al., 2013; Bockstaele et al., 2009; Schachter et al., 2013). First, Cdk4 needs to be released from the Hsp90 complex and bind to cyclin D, whose expression is stimulated by mitogens and increases during the G1 phase. Second, activation of Cdk4 requires phosphorylation of threonine 172 (T172) in the activation loop or T loop by Cdk7 and potentially other Cdk-activating kinases. Finally, Cdk4 requires conformational changes upon binding to its kinase substrates. The cyclin D-Cdk4 complex then enters the nucleus and phosphorylates Rb and weakens the interaction with its binding partner E2F. Further phosphorylation of Rb by cyclin E-Cdk2 releases E2F from the Rb-E2F complex. Released and now active, E2F then binds to its target genes necessary for progression into the S phase. Cyclin D-Cdk4 also inhibits myoblast differentiation by directly interacting with the C terminus of MyoD, preventing its binding to DNA independently of the kinase activity of Cdk4 (Giacinti and Giordano, 2010; Knight and Kothary, 2011). In addition, cyclin D-Cdk4 inhibits the subnuclear localization and interaction of the myogenic transcription factor MEF2 with the coactivator GRIP-1 (Lazaro et al., 2002).
In this work, we compared the roles between Fkbp4 and Fkbp5 in muscle regeneration and myoblast differentiation. This study demonstrated that the PPIase activity of Fkbp5 is a crucial element for the cell cycle exit during myoblast differentiation.
RESULTS
Deregulation of Muscle Regeneration in Fkbp4−/− and Fkbp5−/− Mice
As the first step to understand whether Fkbp4 and Fkbp5 play any roles in muscle regeneration, the tibialis anterior (TA) muscles in wild-type (WT), Fkbp4−/−, and Fkbp5−/− mice were injured by injection of barium chloride. Because the genetic background of Fkbp4−/− (mix of CD-1 and C57BL/6J) and Fkbp5−/− (C57BL/6J) were different, WT of each background was used as control for each strain. Muscle regeneration was followed by hematoxylin and eosin (H&E) staining of histological sections between days 5 and 14 post-injection. Regeneration was quantified by the overall size distribution of regenerating myofibers characterized by the presence of centrally located nuclei (CLN) (Figure 1A). This study indicated that regenerating myofibers between day 5 and 14 as well as myofibers in undamaged mice were smaller in Fkbp4−/− mice than those in WT mice except for day 10 (Figure 1B, left). In contrast, regenerating myofibers in Fkbp5−/− mice were prominently smaller than those in WT mice only on day 10; the difference was subtle at other time points (Figure 1B, right). These results demonstrated that, although TA muscle regeneration was delayed in both knockout (KO) mice, Fkbp5−/− myofibers regenerated more quickly than Fkbp4−/− myofibers.
Figure 1. Regeneration of TA Muscles in Fkbp4−/− and Fkbp5−/− Mice and Differentiation of Their Myoblasts.
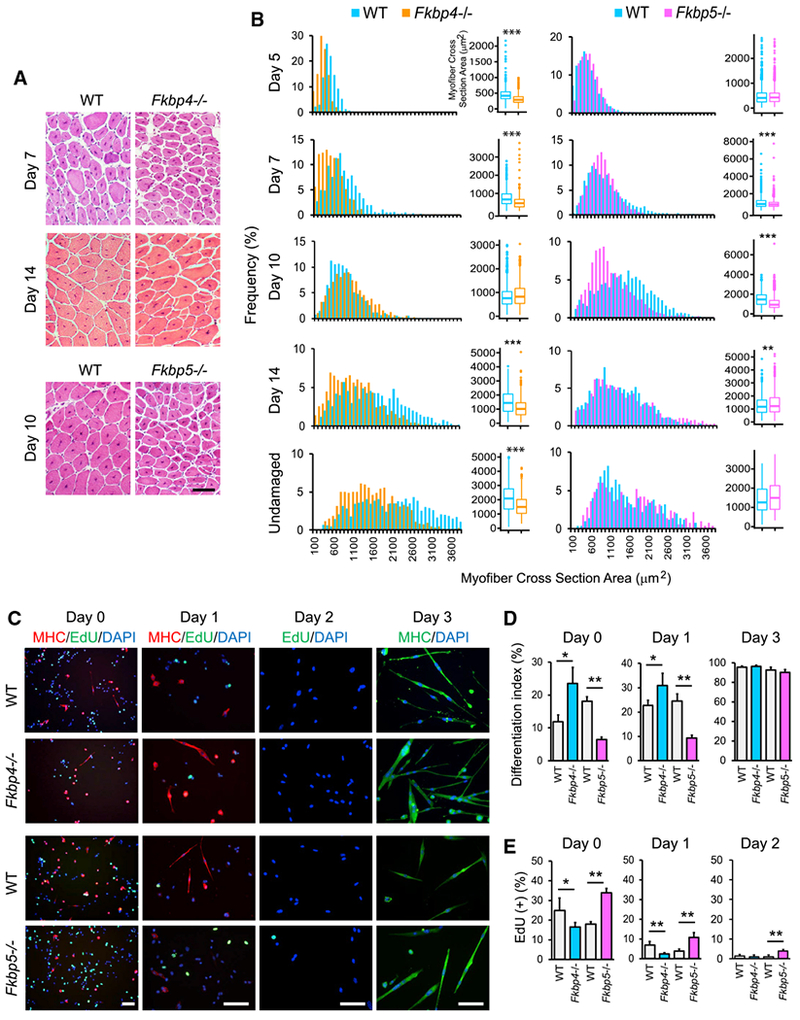
(A) H&E staining of TA muscle sections of KO and WT mice. Scale bar, 100 μm.
(B) Size distribution of regenerating myofibers, which contain centrally located nuclei, and undamaged myofibers stained with H&E. Randomly selected 500 myofibers from two mice each, totaling 1,000 myofibers for each mouse strain, were measured. *p < 0.01, **p < 0.001, and ***p < 0.0001 with Wilcoxon Rank Sum test with Bonferroni adjustment.
(C) Immunostaining of primary myoblasts prepared from WT and KO mice with antibodies against MHC. EdU uptake was also detected. DNA was counter stained with DAPI. Cells were induced to differentiate with 5% horse serum for one and three days. Scale bars, 50 μm.
(D) The differentiation index of primary myoblasts.
(E) EdU uptake of differentiating myoblasts.
*p < 0.05 and **p < 0.01 with Student’s t test in (D) and (E). Data are presented as mean + SD of technical triplicates in (D) and (E).
To understand cell-autonomous phenotypes of myoblasts in each KO mouse, primary myoblasts were prepared and induced to differentiate in vitro. Myoblast differentiation was quantified by the differentiation index, which represented the percentage of nuclei existing in the cells that expressed MHC, and by the expression frequency of myogenin, an early marker for differentiation. Cell cycle exit of myoblasts was assessed by EdU uptake. These indicators showed that Fkbp4−/− and Fkbp5−/− myoblasts demonstrated opposite phenotypes. The differentiation index was higher in Fkbp4−/− myoblasts and lower in Fkbp5−/− myoblasts than WT before differentiation induction (day 0) and in early differentiation with horse serum (day 1; Figures 1C and 1D). However, the index of both genotypes reached higher than 90% by day 3, becoming indistinguishable from WT. Frequency of myogenin expression confirmed the two KO’s opposite effects on myoblast differentiation (Figures S1A–S1C). Consistently, EdU uptake was lower in Fkbp4−/− myoblasts (earlier cell cycle exit) on day 0 and 1 and higher in Fkbp5−/− myoblasts (later cell cycle exit) than WT controls on days 0–2 (Figures 1C and 1E). Furthermore, single myofiber staining revealed that there were less Pax7(−)/MyoD(+) nuclei in Fkbp5−/− myofibers than in WT counterparts 72 hr after isolation of the myofibers (Figures S1D and S1E). Satellite cells were activated as a damage response during the isolation procedure and subsequently differentiated from Pax7(+)/MyoD(−) cells (quiescent satellite cells) to Pax7(+)/MyoD(+) cells (activated satellite cells) and then to Pax7(−)/MyoD(+) cells (differentiating myoblasts). Thus, Fkbp5 KO disrupted differentiation of not only primary myoblasts but also freshly isolated myofiber-associated satellite cells. To characterize more details about the differential functions between Fkbp4 and Fkbp5, we used the mouse myoblast C2C12 cell line as a model in the following sections because of the abundance of the material.
Fkbp4 KD Accelerates and Fkbp5 KD Delays Early Differentiation of Myoblasts
Fkbp4 and Fkbp5 were knocked down (KD) with two independent short hairpin RNAs (shRNAs) to lower than 30% of the control levels in C2C12 cells, and these cells were selected with puromycin (Figure S2A). Although Fkbp4 KD did not decrease EdU uptake in undifferentiated cells unlike in primary myoblast, Fkbp5 KD recapitulated the increased EdU uptake (Figures 2A and S2B). Increased cell proliferation by Fkbp5 KD was also evident with a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay and the S phase frequency with flow cytometry (Figures 2B, 2C, and S2C). In addition, Fkbp5 KD promoted the proliferation of preadipocytes 3T3-L1 and mouse embryonic stem cells ES-E14TG2a and CGR8, indicating that this effect is not limited to muscle cells (Figures S2D and S2E).
Figure 2. Proliferation and Differentiation of Fkbp4 and Fkbp5 KD C2C12 Cells.
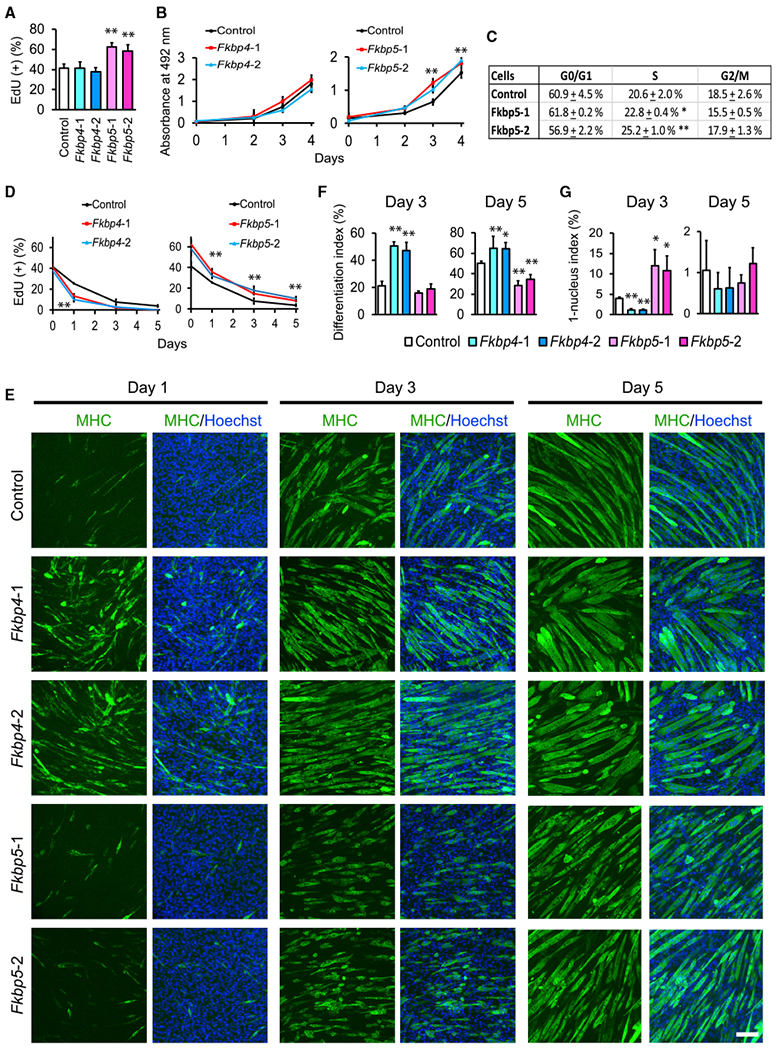
(A) EdU uptake in undifferentiated KD cells. Two shRNA clones were used for each KD. Control shRNA encodes a scrambled non-targeting sequence.
(B) MTS assay for the proliferation of undifferentiated cells after KD. Cell number and absorbance value at 492 nm were proportional in this range.
(C) Flow cytometry analyses of the cell cycle phases in undifferentiated C2C12 cells with Fkbp5 KD.
(D) EdU uptake in KD cells during differentiation.
(E) Immunostaining of KD cells with antibodies against MHC. DNA was counterstained with Hoechst 33342. Cells were induced to differentiate with 5% horse serum. Scale bar, 100 μm.
(F and G) The differentiation index (F) and the 1-nucleus index (G) on day 3 and day 5 based on the observation of 1,000 nuclei.
*p < 0.05 and **p < 0.01 with Student’s t test in comparison to the control cells. Data are presented as mean + or ± SD of technical triplicates.
When differentiation was induced in the KD cells, Fkbp4 and Fkbp5 KD displayed different phenotypes, reproducing the primary myoblast results. First, Fkbp4 KD accelerated but Fkbp5 KD delayed cell cycle exit during differentiation (Figure 2D). In addition, Fkbp4 KD induced more abundant expression of MHC on day 1 and more dense formations of myotubes on day 3 than the control or Fkbp5 KD (Figure 2E). The differentiation index was also higher with Fkbp4 KD on day 3 and 5 than the control (Figure 2F). Furthermore, the 1-nucleus index, defined as the percentage of nuclei in MHC(+) cells that contain only one nucleus, was lower than the control on day 3, indicating more advanced cell fusion (Figure 2G). However, myotubes on day 5 were shorter with Fkbp4 KD than the control (Figure 2E), demonstrating a disruption of further development to fully elongated myotubes. This observation appeared to agree with the small myofibers observed in the Fkbp4−/− TA muscles (Figure 1B). In contrast, Fkbp5 KD myotubes were characterized by shorter myotubes on day 3 and 5, lower differentiation index on day 5, and higher 1-nucleus index on day 3, all of which pointed to delayed differentiation.
These observations were supported by qPCR analysis of differentiation-specific genes, such as myogenin (Myog), myomaker (Mymk), MHC (Myh3), and creatine kinase M-type (Ckm). The transcript levels were higher in Fkbp4 KD cells and lower in Fkbp5 KD cells than the control during differentiation (Figure 3A). Moreover, RNA sequencing (RNA-seq) analysis indicated that more than 1,000 genes were differentially up- or downregulated between Fkbp4 and Fkbp5 KD cells on days 0, 3, and 5 (Figures S3A and S3B). Whereas Fkbp4 KD upregulated muscle-specific genes on day 0 (Figures S3C and S3D), Fkbp5 KD downregulated muscle genes (Figures S4A–S4C). In complementary experiments, retrovirus-mediated overexpression of Fkbp4 did not affect the proliferation, the differentiation index, or expression of differentiation-specific genes compared with the control cells transduced with the empty vector (Figures 3B–3E and S2F). However, Fkbp5 overexpression showed phenotypes opposite to KD: decreased proliferation of undifferentiated cells; increased differentiation index; and increased expression of differentiation-specific genes (Figures 3B–3E). Combining the results of primary myoblasts and C2C12 cells, Fkbp4 inhibits the early phase of myoblast differentiation, whereas Fkbp5 promotes differentiation.
Figure 3. Differentiation of C2C12 Cells after KD or Overexpression of Fkbp4 and Fkbp5.
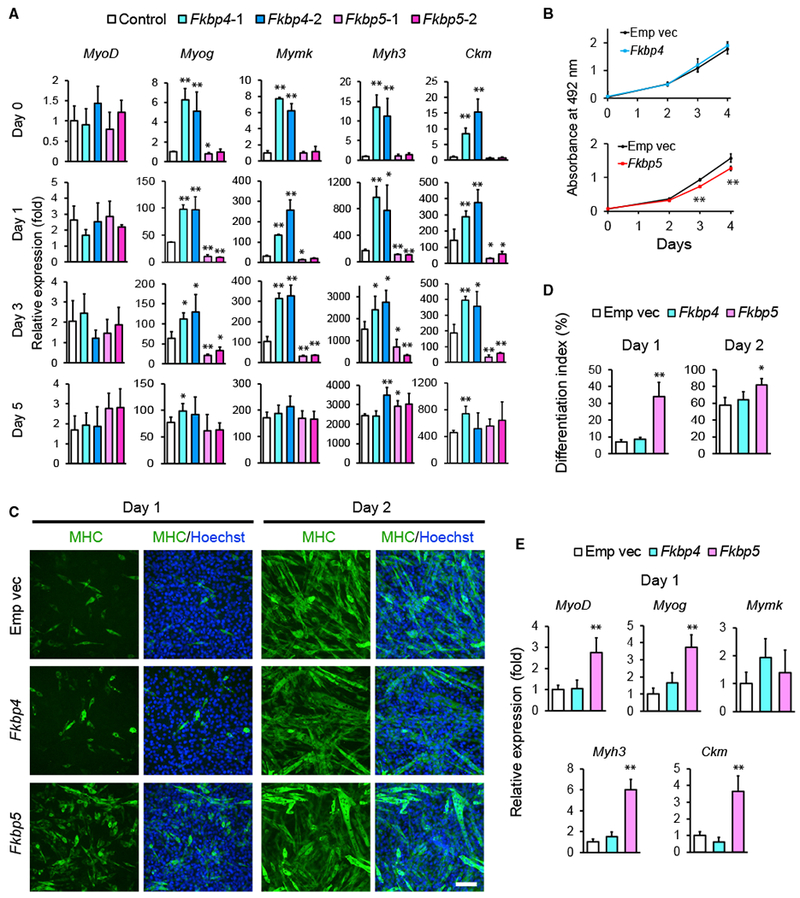
(A) qRT-PCR results comparing the expression levels of indicated muscle genes during differentiation of KD cells. The value obtained with the control on day 0 was defined as 1.0.
(B) MTS assay for the proliferation of undifferentiated cells after overexpression of Fkbp4 and Fkbp5. Empty vector (Emp vec) was transduced as the control.
(C) MHC staining of C2C12 cells overexpressing Fkbp4 and Fkbp5 during differentiation. Scale bar, 100 μm.
(D) The differentiation index of the overexpressing cells on day 1 and day 2 obtained from the observation of 1,000 nuclei.
(E) Relative expression levels of muscle genes determined by qRT-PCR on day 1. The value obtained with the control cells was defined as 1.0.
*p < 0.05 and **p < 0.01 with Student’s t test in comparison to the control cells. Data are presented as mean + or ± SD of technical triplicates.
Fkbp4 and Fkbp5 Maintain the Hsp90-Cdk4 Complex
To understand how Fkbp4 and Fkbp5 differentially regulate myoblast differentiation, their binding proteins were analyzed. First, C2C12 cells were separately transduced with FLAG-tagged Fkbp4 and Fkbp5 with retrovirus and the transgene-positive cells were selected with puromycin. Immunoprecipitation (IP) was performed with anti-FLAG antibody, and co-precipitated proteins were analyzed with high-resolution peptide tandem mass spectrometry and database searching after proteolytic digestion (Figures S5A–S5C). The co-precipitated proteins included Hsp90α, Hsp90β, Cdk4, and Cdc37, which were known to form a complex with Fkbp4 and Fkbp5 in a breast cancer cell line (Jirawatnotai et al., 2014). Because Cdk4 plays a prominent role in the proliferation and differentiation of myoblasts, we investigated whether Fkbp4 and Fkbp5 differentially regulated the presence of Cdk4 in the complex. First, we verified the co-precipitation between FLAG-Fkbp5 and Cdk4 as well as the reciprocal co-precipitation by western blotting (Figure 4A). Second, we performed IP of endogenous Hsp90α/β to compare co-precipitated proteins between undifferentiated and differentiated (day 5) cells. The amounts of co-precipitated Fkbps and Cdk4 did not display a major difference between the two cell types (Figure 4B). Third, when we repeated IP of endogenous Hsp90α/β with extracts of undifferentiated Fkbp4 and Fkbp5 KD cells, co-IP of Cdk4 was significantly diminished in both KD cells without decreasing the amount of Cdk4 in the input (Figure 4C). Thus, Fkbp4 and Fkbp5 are both necessary to retain Cdk4 within the Hsp90 complex. Finally, IP of cyclin D1 co-precipitated Cdk4 more abundantly from the KD cell extracts than from the control extract, demonstrating that the KD increased the amount of the cyclin D1-Cdk4 complex (Figure 4D). Collectively, KD of Fkbp4 and Fkbp5 similarly released Cdk4 from the Hsp90 complex and promoted the formation of the cyclin D1-Cdk4 complex.
Figure 4. Interacting Protein Partners of Fkbp4 and Fkbp5.
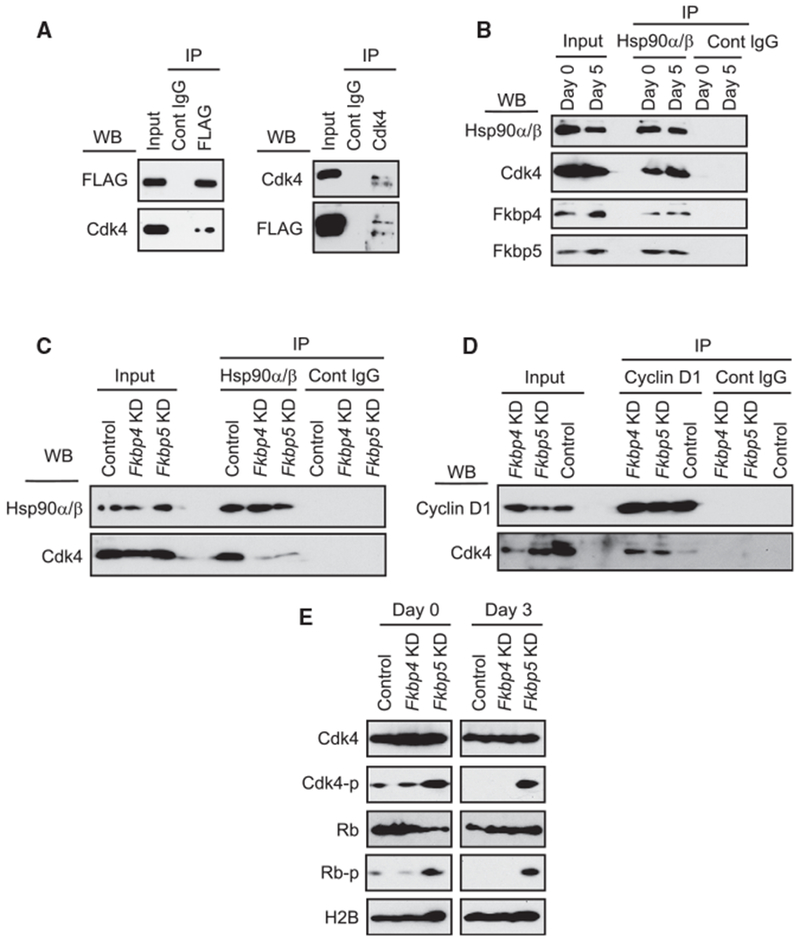
(A) Western blotting of immunoprecipitated proteins. Left images show immunoprecipitation (IP) of 3×FLAG-Fkbp5 with anti-FLAG antibody and co-IP of Cdk4. Right images show IP of Cdk4 and co-IP of 3×FLAG-Fkbp5.
(B) IP of Hsp90α/β and co-IP of Cdk4, Fkbp4, and Fkbp5 from C2C12 cell extracts of differentiation days 0 and 5.
(C) IP of Hsp90α/β and co-IP of Cdk4 from extracts of undifferentiated KD cells.
(D) IP of cyclin D1 and co-IP of Cdk4 from extracts of undifferentiated KD cells.
(E) Detection of phosphorylated Cdk4 and Rb during differentiation of the KD cells. H2B was used as a loading control.
Because there was no clear difference in the above process between the two Fkbps, we investigated whether the Fkbps differentially regulated T172 phosphorylation of Cdk4 (Cdk4-p) as a critical step for Cdk4 activation. Indeed, comparison of the KD cells showed more abundant Cdk4-p by Fkbp5 KD than the control and Fkbp4 KD before (day 0) and on day 2 of differentiation (Figure 4E). Phosphorylation of Rb at S807 and S811, a downstream effect of Cdk4-p, was also increased specifically by Fkbp5 KD. These Rb phosphorylations trigger phosphorylation of additional sites in Rb (Rubin, 2013). The increased Cdk4-p and Rb-pby Fkbp5 KD, but not by Fkbp4 KD, could explain why Fkbp5 KD specifically promoted proliferation and delayed differentiation. Therefore, we further characterized Fkbp5-Cdk4 interactions in the following studies.
Fkbp5 Promotes Isomerization of the T172-P173 Peptide Bond in Cdk4
We hypothesized that Fkbp5 specifically promoted isomerization of the peptide bond between T172 and P173 in Cdk4 as the underlying mechanism for the increased Cdk4-p in Fkbp5 KD cells. This possibility was tested with a well-established in vitro PPIase assay. We synthesized two Cdk4 peptides: Cdk4-P50 (succinyl-GGGLP-FpNA, contains P50) and Cdk4-P173 (succinyl-MALTP-FpNA, contains P173). Phe (F) and a paranitroanaline group (pNA) were linked at the C terminus in each peptide. Chymotrypsin hydrolyzes the bond between F and pNA only when the peptide bond preceding Pro (P) is in the trans conformation (Nelson et al., 2006; Shan et al., 1994). This reaction can be quantified as an increase in the absorbance value at 390 nm caused by the free pNA (Figure 5A). Although the absorbance will also increase without PPIase due to spontaneous cis-trans interconversion, the presence of PPIase will accelerate the increase in the absorbance.
Figure 5. In Vitro Interactions between Fkbp5 and Cdk4.
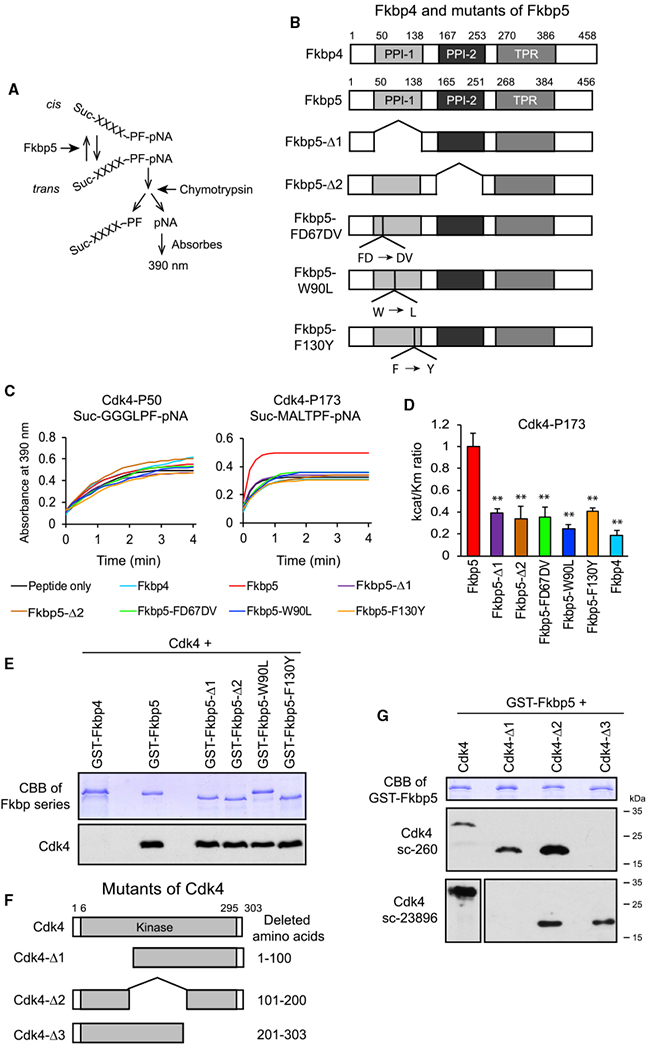
(A) Principle of the in vitro PPIase assay.
(B) Structure of Fkbp5 mutants and Fkbp4. Amino acid numbers are based on UniProt.
(C) PPIase assay comparing the Fkbp proteins using two Cdk4 peptides. Average of technical triplicates is shown.
(D) Catalytic efficiency kcat/KM comparing the Fkbp proteins using the Cdk4-P173 peptide as the substrate. **p < 0.01 with Student’s t test in comparison to Fkbp5. Data are presented as mean + SD of technical triplicates.
(E) Coomassie brilliant blue (CBB) staining of GST-tagged proteins (top) and western blotting of pulled down Cdk4 (bottom). GST-Fkbp5-F130Y ran faster than GST-Fkbp5-W90L for an unknown reason.
(F) Structure of Cdk4 mutants used in the GST-pull-down assay in (G).
(G) CBB staining of GST-Fkbp5 (top) and western blotting of pulled down Cdk4 mutants (middle and bottom). The epitope of the Cdk4 antibody sc-260 is located at the C terminus and that of c-23896 (both from Santa Cruz Biotechnology) is the amino acids 1–20.
Fkbp5 contains a functional PPIase domain (PPI-1) and a nonfunctional pseudo-PPIase domain (PPI-2), in addition to the tetratricopeptide repeat (TPR) domain, which interacts with Hsp90 (Sinars et al., 2003; Stechschulte and Sanchez, 2011; Figure 5B). We prepared bacterial recombinant proteins of five mutant Fkbp5s as well as WT Fkbp4 and Fkbp5 (Figure S6A). Fkbp5-Δ1 and Fkbp5-Δ2 lacked PPI-1 and PPI-2, respectively. Fkbp5-FD67DV lacked PPIase activity due to the replacement of the two amino acids FD with DV at the mouth of the PPIase pocket, potentially causing a major conformational change of the PPIase domain (Barent et al., 1998; Riggs et al., 2007). The single-amino-acid mutations in Fkbp5-W90L and Fkbp5-F130Y were located inside the PPIase pocket (Riggs et al., 2007). Although disruption of the PPIase activity by the last two mutations was originally reported with Fkbp4, these amino acids are conserved between Fkbp4 and Fkbp5. Where as none of the recombinant proteins accelerated the release of pNA from the Cdk4-P50 peptide, only WT Fkbp5 accelerated the release of pNA from the Cdk4-P173 peptide (Figure 5C, red line in the right panel). The promoted isomerization by Fkbp5 was quantified as the higher catalytic efficiency kcat/KM than that of other proteins (Figure 5D). Although Fkbp4 did not show PPIase activity with the two peptides, it was not due to failed preparation of the recombinant protein. Fkbp4 instead showed PPIase activity with a peptide containing P30 (H3P30), but not P16, of histone H3 (Figures S6B and S6C). H3P30 is a known substrate for a yeast Fkbp called Fpr4 (see Discussion).
Next, we studied whether Fkbp5 directly binds to Cdk4 in vitro using a glutathione S-transferase (GST)-pull-down assay with Fkbp5 deletion mutants. Although Fkbp4 did not pull down Cdk4, WT and mutant Fkbp5s pulled it down regardless of PPIase activity. In a reciprocal experiment, the GST-WT Fkbp5 (GST-Fkbp5) was incubated with three deletion mutants of Cdk4 with the removal of around 100 amino acids each (Figure 5F). Western blotting of the pull-down results indicated that each mutant remained bound to Fkbp5, and none of the deleted regions were indispensable for the interaction with Fkbp5 (Figure 5G). The direct interaction between Fkbp5 and Cdk4 and isomerization of Cdk4 by Fkbp5 led us to hypothesize that Fkbp5 inhibits cell proliferation by directly inhibiting Cdk4 through isomerization of the P173 peptide bond. This possibility was tested in the next sections, first focusing on Fkbp5’s PPIase activity and second studying the dependence of the Fkbp5 activity on Cdk4 P173.
Pro-differentiation Activity of Fkbp5 Requires Its PPIase Activity
To investigate whether Fkbp5 functions as a PPIase while regulating proliferation and differentiation of myoblasts, we attempted to rescue the phenotypes of Fkbp5−/− myoblasts by transducing a series of Fkbp5 mutants lacking the PPIase activity and WT Fkbp4 and Fkbp5 (Figure 6A). EdU uptake was promoted in the myoblasts without transduction (Figures 6B, column no. 2 from the left, 6C, and S6D) and with empty vector (no. 3) compared with WT myoblasts (no. 1) in undifferentiated (day 0) and differentiating cells (day 1). Whereas transduction of Fkbp5 decreased EdU uptake (rescued; no. 4), all the mutants and Fkbp4 failed to decrease the uptake compared with empty vector (nos. 5–10). In particular, the failed rescue by the three PPIase (−) mutants (nos. 7–9) underscores the significance of the PPIase activity of Fkbp5 for the cell cycle exit. The differentiation index also showed that only Fkbp5 (no. 4) rescued the inhibited differentiation in Fkbp5−/− cells on days 0 and 1 (Figures 6D, 6E, and S6E). These results provided evidence that Fkbp5 requires PPIase activity for its pro-differentiation function.
Figure 6. Requirement of the PPIase Activity of Fkbp5 for the Inhibition of Proliferation in Myoblasts.
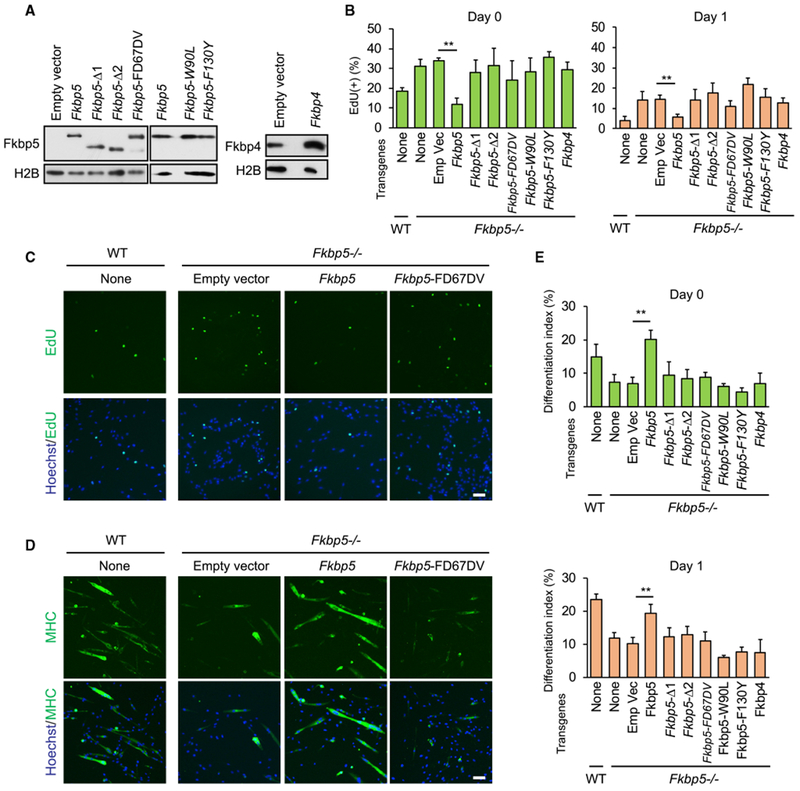
(A) Western blotting of overexpressed Fkbp5 mutants and Fkbp4 in Fkbp5−/− myoblasts.
(B) EdU uptake in WT and Fkbp5−/− myoblasts transduced with the Fkbp genes before (day 0) and during differentiation (day 1).
(C) EdU uptake in WT and Fkbp5−/− myoblasts transduced with the Fkbp5 genes on differentiation day 1.
(D) Immunostaining of MHC in WT and Fkbp5−/− myoblasts transduced with the Fkbp5 genes on differentiation day 1.
(E) The differentiation index in WT and Fkbp5−/− myoblasts transduced with the Fkbp genes before (day 0) and during differentiation (day 1).
**p < 0.01 with Student’s t test in comparison to the empty vector. Data are presented as mean + SD of technical triplicates.
Pro-differentiation Activity of Fkbp5 Is Dependent on Cdk4 P173
We investigated whether Cdk4 is the primary target of the pro-differentiation activity of Fkbp5. If so, Cdk4 KD would cancel the effect of Fkbp5 KD. In the first step, we knocked down Cdk4 alone to understand its effects on the proliferation and differentiation of C2C12 cells. Two independent shRNA clones decreased the Cdk4 mRNA level to less than 30% of the control (Figure S7A). EdU uptake and the differentiation index were not affected by the KD (Figures S7B and S7C). The absence of these phenotypes with Cdk4 KD was consistent with a lack of gross abnormality in Cdk4−/− mouse muscle (Rane et al., 1999; Tsutsui et al., 1999).
In the second step, we made double KD cells of Fkbp5 and Cdk4 and verified efficient KD of each gene (Figure S7D). We then compared EdU uptake and the differentiation index of single- and double-KD cells. Single KD of Fkbp5 (Figures S7E and S7F, column no. 2 from the left) increased EdU uptake and decreased the differentiation index compared with the control (no. 1) as expected. However, an additional KD of Cdk4 erased the effects of Fkbp5 KD (nos. 3–6). Because Cdk4 KD alone did not alter EdU uptake or the differentiation index, this result can be interpreted that Fkbp5 uses Cdk4 as a downstream effector in regulating proliferation and differentiation.
Having determined the essential role of Cdk4 in Fkbp5 activity, we studied whether P173 in Cdk4 is necessary for the activity. The double-KD cells were rescued by transducing retrovirus encoding Cdk4 and Cdk4-P173A. The Cdk4 sequence was mutated (called rCdk4 hereafter) at seven nucleotides within the shRNA target sequence without changing the amino acid sequence to make it resistant to KD. The empty vector was transduced as the control. More than 90% of the transduced cells expressed rCdk4, as shown with immunofluorescence staining, and the expression level was higher than C2C12 cells without any gene manipulation (Figures 7A and S7G). Transduction of rCdk4 (Figure 7B, green column) increased EdU uptake compared with the empty vector (yellow column) as expected, but rCdk4-P173A (pink column) did not cause this effect, indicating that prevention of isomerization of the P173 peptide bond canceled the effect of Fkbp5 KD. Similarly, transduction of rCdk4-P173A erased the effect of Fkbp5 KD in the differentiation index, 1-nucleus index, and expression of three differentiation-specific genes: Myog; Mymk; and Myh3 (Figures 7C–7E). Combined results with all these KDs and rescue experiments identified Cdk4-P173 as the primary target for the PPIase activity of Fkbp5 to promote myoblast differentiation.
Figure 7. Requirement of Cdk4 P173 for the Inhibition of C2C12 Cell Proliferation by Fkbp5.
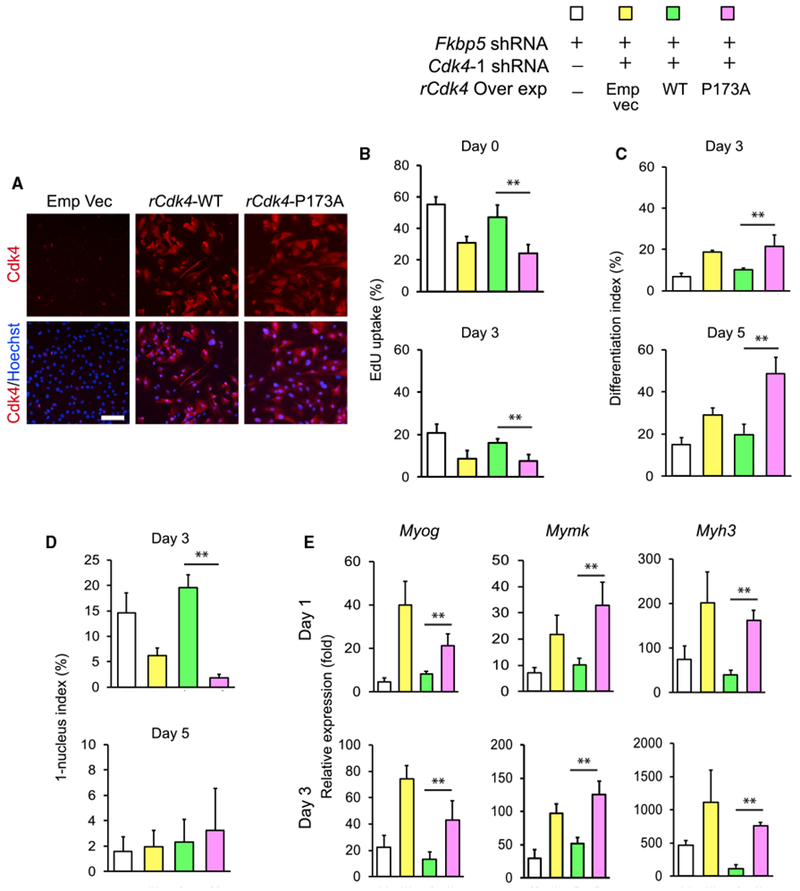
(A) Immunostaining of overexpressed rCdk4-WT and rCdk4-P173A in Cdk4 KD cells. Scale bar, 100 μm.
(B-E) rCdk4-WT and rCdk4-P173A were transduced into C2C12 cells after double KD of Fkbp5 and Cdk4. The cells were used to analyze EdU uptake (B), the differentiation index (C), the 1-nucleus index (D), and qPCR of muscle genes (E). **p < 0.01 with Student’s t test. Data are presented as mean + SD of technical triplicates.
DISCUSSION
This study demonstrates the requirement for an Fkbp family member as a PPIase in myogenesis. The link between Fkbps and muscle proliferation and differentiation has been studied before, in addition to the hypergravity study mentioned earlier. However, they did not evaluate the PPIase activity or identify target molecules of Fkbps. For example, a previous study inhibited Fkbp1 by rapamycin to conclude that Fkbp1 is important for muscle cell proliferation (Jayaraman and Marks, 1993). Rapamycin might have inhibited other target proteins in this work. Indeed, another study found no obvious abnormality in muscle development in skeletal-muscle-specific Fkbp1−/− mice, although disruption of excitation-contraction coupling was observed (Tang et al., 2004). Our study showed the significance of the isomerization of the P173 peptide bond, but it requires further study to determine whether the cis or trans conformation promotes myoblast differentiation. This question may not be solved even if conformation-specific antibody is available because the conformation could be variable depending on whether Cdk4 binds to the Hsp90 complex, cyclin D1, or other partners. This issue and the question of the exact order of Cdk4 isomerization and phosphorylation could be addressed by structural analysis of each protein complex.
The current study also identified two regulatory mechanisms of Cdk4 activity by Fkbp5 in myoblasts: sequestration of Cdk4 in the Hsp90 complex and inhibition of Cdk4 phosphorylation. Through these mechanisms, Fkbp5 inhibits excessive proliferation of undifferentiated myoblasts and promotes differentiation when appropriate external cues are provided. IP results with proliferating myoblast extracts indicated that Fkbp5 is necessary to retain Cdk4 in the Hsp90 complex. The amount of Cdk4 bound to Hsp90 did not increase in differentiating cells, suggesting that the sequestration of Cdk4 to the Hsp90 complex is not a major regulatory process to inactivate Cdk4 during differentiation. Fkbp5 appears to constitutively regulate the level of Cdk4 activity rather than functioning as a switch for the dynamic transition of proliferation to differentiation. As a side note, Fkbp5 KD, but not Fkbp4 KD, substantially decreases the Cdk4 protein in a breast cancer cell line, although the molecular mechanism remains unknown (Jirawatnotai et al., 2014). However, Cdk4 was not downregulated by Fkbp5 KD in C2C12 cells (Figure 4E). The difference between the two results could be due to the difference of the cell types.
In addition, this study illuminates the similarities and differences in the functions of Fkbp4 and Fkbp5 in myoblast proliferation and differentiation. Fkbp4 KD showed two phenotypes, acceleration of early phase, and inhibition of late phase of myoblast differentiation, in contrast to consistent inhibition of differentiation by Fkbp5 KD. Further study of Fkbp4 functions requires the use of cells of these two different stages. The differential functions between the two Fkbps have been extensively studied in the regulation of steroid hormone receptors by competing for receptor binding. Whereas Fkbp4 activates glucocorticoid, progesterone, and androgen receptor, Fkbp5 inhibits steroid hormone receptors in general, except for androgen receptor, which is activated by Fkbp5 (Stechschulte and Sanchez, 2011; Storer et al., 2011). Activation by Fkbp4 is dependent on the PPI-1 domain as an interacting site with the receptors, but catalytic activity is not necessary for activation (Riggs et al., 2007). The opposing roles between the two Fkbps are explained by the structural differences in the TPR domain and the C-terminal sequences (Barent et al., 1998).
Cdk4−/− mice are viable but show growth retardation and develop diabetes by six weeks after birth due to a reduction in pancreatic b cells (Raneet al., 1999; Tsutsui et al., 1999). Muscle phenotypes have not been reported in these mice. All females are sterile due to diminished progesterone secretion from the corpus luteum in the ovary because of a decreased number of prolactin-secreting cells in the pituitary gland (Moons et al., 2002). Ninety percent of males are also sterile because of hypoplastic seminiferous tubules in the testes. In addition, Cdk4−/− fibroblasts show delayed entrance into the S phase by serum stimulation after quiescence. These observations indicate a cell-type-specific requirement of Cdk4 for cell proliferation. In our result, Cdk4 KD did not affect myoblast proliferation, but it does not necessarily downplay the significance of the Fkbp5-Cdk4 link. Absence of Cdk4 could be compensated by functionally redundant proteins, such as Cdk6 (Malumbres et al., 2004); however, presence of inactive Cdk4 as a dominant negative partner for cyclin D could be detrimental for cell cycle progression.
Our mass spectrometry identified histones H2B and H4 as potential binding partners of Fkbp5 and Fkbp4, respectively. Fkbps have PPIase-dependent and independent functions related to histones. In the PPIase-dependent function, Saccharomyces cerevisiae Fpr4 catalyzes isomerization of the peptide bonds preceding P30 and P38 of histone H3 as demonstrated by the in vitro PPIase assay (Nelson et al., 2006). H3P38 isomerization results in the inhibition of H3K36 methylation, delaying the induction of specific genes in vivo. In the PPIase-independent function, Fpr4 facilitates nucleosome assembly as a histone chaperone in vitro and silences rDNA in vivo without the PPIase domain (Kuzuhara and Horikoshi, 2004; Xiao et al., 2006). However, it is difficult, if not impossible, to study whether Fkbp4 and Fkbp5 function as histone isomerases in mammalian cells. First, antibodies against cis- or trans-specific histones are not commercially available. Additionally, whereas Saccharomyces cerevisiae has only two copies of core histone genes in the haploid genome, the human haploid genome encodes 23 histone H3 open reading frames scattered in eight chromosomes, rendering complete mutagenesis very difficult (Ederveen et al., 2011; Freeman et al., 1992).
There are 16 members of the Fkbp family in the human genome (Hanes, 2015). Systematic analyses of KO and KD phenotypes of these genes would provide unexpected perspectives on how muscle cell differentiation and regeneration are achieved by conformational changes in critical regulators of the processes.
STAR*METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FLAG M2 | MilliporeSigma | Cat# F1804; RRID:AB_262044 |
| MHC | Developmental Studies Hybridoma Bank | Cat# MF 20; RRID: AB_2147781 |
| CD31-PE | eBioscience | Cat# 12-0311; RRID:AB_465633 |
| CD45-PE | eBioscience | Cat# 12-0451; RRID:AB_465669 |
| Sca1-PE | eBioscience | Cat# 12-5981; RRID:AB_466087 |
| Integrin α7-biotin | Miltenyi Biotec | Cat# 130-102-125; RRID:AB_2652471 |
| MyoD | Santa Cruz Biotechnology | Cat# sc-304; RRID:AB_631992_ |
| Myogenin | Developmental Studies Hybridoma Bank | Cat#: F5D; RRID:AB_528355 |
| Pax7 | Developmental Studies Hybridoma Bank | Cat#: PAX7; RRID:AB_528428 |
| Fkbp4 | Bethyl Laboratories | Cat# A301-426A-1; RRID:AB_960992 |
| Fkbp5 | Santa Cruz Biotechnology | Cat# sc-13983; RRID: AB_2278350 |
| Histone H2B | Thermo Fisher Scientific | Cat# MA5-14835; RRID:AB_10982286 |
| Hsp90α/β | Santa Cruz Biotechnology | Cat# sc-13119; RRID: AB_675659 |
| Cdk4 | Santa Cruz Biotechnology | Cat# sc-260; RRID: AB_631219 |
| Phospho-Cdk4-T172 | NeoBioLab | Cat# AP0593; RRID: not found |
| Cyclin D1 | Santa Cruz Biotechnology | Cat# sc-20044; RRID: AB_627346 |
| Rb | Santa Cruz Biotechnology | Cat# sc-74562; RRID: AB_2177334 |
| Phospho-Rb (Ser807/811) | Cell Signaling Technology | Cat# 9308S; RRID: AB_331472 |
| Alexa Fluor 488 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-11029; RRID:AB_2534088 |
| Alexa Fluor 488 donkey anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-21202; RRID:AB_141607 |
| Alexa Fluor 555 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-21429; RRID:AB_2535850 |
| Alexa Fluor 568 donkey anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#: A10042; RRID:AB_2534017 |
| Alexa Fluor 594 donkey anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-21207; RRID:AB_141637 |
| Goat anti-mouse IgG, HRP labeled | Santa Cruz Biotechnology | Cat# sc-2005; RRID:AB_631736 |
| Goat anti-rabbit IgG, HRP labeled | Santa Cruz Biotechnology | Cat# sc-2004; RRID:AB_631746 |
| Bacterial and Virus Strains | ||
| BL21(DE3) | Thermo Fisher Scientific | C600003 |
| Biological Samples | ||
| Collagenase, type 1 | MilliporeSigma | Cat#: C0130 |
| Collagenase, type 2 | Worthington | Cat#: CLS-2 |
| bFGF, human, recombinant | Invitrogen | Cat# PHG0263 |
| Collagen | BD Biosciences | Cat# 354236 |
| Chicken embryo extract | Gemini Bio-products | Cat#: 100-163P |
| Leukemia inhibitory factor | MilliporeSigma | Cat# ESG1106 |
| Normal mouse IgG | Santa Cruz Biotechnology | Cat# sc-2025 |
| Normal rabbit IgG | Santa Cruz Biotechnology | Cat# sc-2027 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Mouse-on-Mouse Blocking Reagent | Vector | Cat# MKB-2213 |
| Isoflurane | Phoenix | N/A |
| Hoechst 33342 | MilliporeSigma | Cat# B2261 |
| DAPI | MilliporeSigma | Cat# 10236276001 |
| Harris Modified Hematoxylin | Thermo Fisher Scientific | Cat# SH26-500D |
| Eosin Y | Thermo Fisher Scientific | Cat# 22-220-104 |
| Permount | Thermo Fisher Scientific | Cat# SP15-100 |
| Anti-PE MicroBeads | Miltenyi Biotec | Cat# 130-048-801 |
| Anti-biotin MicroBeads | Miltenyi Biotec | Cat# 130-090-485 |
| MS columns | Miltenyi Biotec | Cat# 130-042-201 |
| LD columns | Miltenyi Biotec | Cat# 130-042-901 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668019 |
| FuGENE 6 | Promega | Cat# E2691 |
| Polybrene | MilliporeSigma | Cat# H9268 |
| Immobilon P membrane | MilliporeSigma | Cat# IPVH00010 |
| SuperSignal West Dura | Thermo Fisher Scientific | Cat# 34075 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat# 10004D |
| Imperial Protein Stain | Thermo Fisher Scientific | Cat# 24615 |
| Criterion 8-16% Tris-HCl gradient gel | Bio-Rad Laboratories | Cat# 3450038 |
| ReproSil-Pur C18 | Dr. Maisch GmbH | Cat# r13.b9 |
| Glutathione magnetic agarose beads | Thermo Fisher Scientific | Cat# 78601 |
| Ni-NTA agarose | QIAGEN | Cat# 30230 |
| 3X FLAG peptide | MilliporeSigma | Cat# F4799 |
| 2,2,2-Trifluoroethanol | Thermo Fisher Scientific | Cat# AC139750250 |
| α-Chymotrypsin | MilliporeSigma | Cat# C4129 |
| H3-P16, Suc-GGKAPF-pNA | EZBiolab | N/A |
| H3-P30, Suc-RKSAPF-pNA | EZBiolab | N/A |
| Cdk4-P50, Suc-GGGLPF-pNA | EZBiolab | N/A |
| Cdk4-P173, Suc-MALTPF-pNA | EZBiolab | N/A |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 448 imaging kit | Thermo Fisher Scientific | Cat# C10337 |
| Q5 Site-Directed Mutagenesis Kit | New England Biolabs | Cat#: E0552S |
| Quick-RNA Microprep kit | Zymo Research | Cat# R1051 |
| RNeasy Plus Mini kit | QIAGEN | Cat# 74136 |
| GoTaq qPCR Master Mix | Promega | Cat# A6002 |
| CellTiter 96 AQ One Solution Cell Proliferation Assay | Promega | Cat# G3580 |
| Quick-RNA MiniPrep Kit | Zymo Research | Cat# R1055 |
| ProtoScript II Reverse Transcriptase | New England Biolabs | Cat# M0368L |
| Quanti-iT dsDNA Assay Kit | Thermo Fisher Scientific | Cat# P11496 |
| Truseq RNA Sample Preparation Kit | Illumina | Cat# RS-122-2001 |
| Deposited Data | ||
| RNA-seq data | This paper | GEO: GSE109237 |
| Experimental Models: Cell Lines | ||
| C2C12 cells | American Type Culture Collection | Cat# CRL-1772 |
| 3T3-L1 cells | American Type Culture Collection | Cat# CRL-173 |
| ES-E14TG2a cells | American Type Culture Collection | Cat# CRL-1821 |
| CGR8 cells | European Collection of Authenticated Cell Culture | Cat# 07032901 |
| 293FT cells | Thermo Fisher Scientific | Cat# R70007 |
| PLAT-E cells | Toshiyuki Kitamura | N/A |
| BL21(DE3) cells | New England BioLabs | Cat# C2527I |
| Experimental Models: Organisms/Strains | ||
| Fkbp4tm1Dvds/J | Jackson Laboratory | Cat# 017990; RRID:IMSR_JAX:017990 |
| Fkbp5tm1Dvds/J | Jackson Laboratory | Cat# 017989; RRID:IMSR_JAX:017989 |
| Oligonucleotides | ||
| See Table S2 | N/A | |
| Recombinant DNA | ||
| pCMV-VSV-G | Addgene | Cat# 8454 |
| pRSV-Rev | Addgene | Cat# 12253 |
| pMDLg/pRRE | Addgene | Cat# 12251 |
| pMXs-IP | Toshiyuki Kitamura | Cat# |
| pGEX-2T | GE Healthcare | Cat# 28954653 |
| pET-21c(+) | MilliporeSigma | Cat# 69742-3 |
| pMXs-IP-3×FLAG-Fkbp4 | This paper | N/A |
| pMXs-IP-3ൿLAG-Fkbp5 | This paper | N/A |
| pMXs-IP-Fkbp4 | This paper | N/A |
| pMXs-IP-Fkbp5 | This paper | N/A |
| pMXs-IP-Fkbp5-Δ1 | This paper | N/A |
| pMXs-IP-Fkbp5-Δ2 | This paper | N/A |
| pMXs-IP-Fkbp5-FD67DV | This paper | N/A |
| pMXs-IP-Fkbp5-W90L | This paper | N/A |
| pMXs-IP-Fkbp5-F130Y | This paper | N/A |
| pMXs-IP-Cdk4 | This paper | N/A |
| pMXs-IP-Cdk4-P173A | This paper | N/A |
| pET-21c(+)-Cdk4-6×His | This paper | N/A |
| pET-21c(+)-Cdk4-Δ1-6×His | This paper | N/A |
| pET-21c(+)-Cdk4-Δ2-6×His | This paper | N/A |
| pET-21c(+)-Cdk4-Δ3-6×His | This paper | N/A |
| pET-21c(+)-Fkbp4-6×His | This paper | N/A |
| pET-21c(+)-Fkbp5-6×His | This paper | N/A |
| pET-21c(+)-Fkbp5-Δ1-6×His | This paper | N/A |
| pET-21c(+)-Fkbp5-Δ2-6×His | This paper | N/A |
| pET-21c(+)-Fkbp5-FD67DV-6×His | This paper | N/A |
| pET-21c(+)-Fkbp5-W90L-6×His | This paper | N/A |
| pET-21c(+)-Flkbp5-F130Y-6×His | This paper | N/A |
| pGEX-2T-Fkbp4 | This paper | N/A |
| pGEX-2T-Fkbp5 | This paper | N/A |
| pGEX-2T-Fkbp5-Δ1 | This paper | N/A |
| pGEX-2T-Fkbp5-Δ2 | This paper | N/A |
| pGEX-2T-Fkbp5-W90L | This paper | N/A |
| pGEX-2T-Fkbp5-F130Y | This paper | N/A |
| Software and Algorithms | ||
| Metamorph Basic 7.8.6.0 | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| cellSens Entry 1.11 | Olympus | http://www.olympus-lifescience.com/en/software/cellsens/ |
| Photoshop and Illustrator CS6 | Adobe | https://helpx.adobe.com/creative-suite.html |
| ilastik 1.2 | ilastik | http://ilastik.org/index.html |
| ImageJ | RSB, NIH | https://imagej.nih.gov/ij/ |
| FlowJo 7.6.5 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Gopher-pipelines | Publicly available | https://bitbucket.org/jgarbe/gopher-pipelines/overview |
| FastQC version 0.11.5 | Publicly available | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic version 0.33 | Publicly available | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| HISAT2 version 2.0.2 | Publicly available | https://ccb.jhu.edu/software/hisat2/index.shtml |
| Subread version 1.4.6 | Publicly available | http://subread.sourceforge.net/ |
| edgeR | Publicly available | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| PANTHER | Publicly available | http://pantherdb.org/ |
| pheatmap version 1.0.8 | Publicly available | https://cran.r-project.org/web/packages/pheatmap/index.html |
| UniProt | Publicly available | httDs://www.uniDrot.org/uniDrot/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nobuaki Kikyo (kikyo001@umn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Knockout Mice
All protocols were approved by the Institutional Animal Care and Usage Committee of the University of Minnesota (1602-33471A and 1604-33660A). Fkbp4+/− (Fkbp4tm1Dvds/J, stock# 017990) and Fkbp5+/− (Fkbp5tm1Dvds/J, stock # 017989) mice were purchased from Jackson Laboratory. Homozygous mutant, wild-type, and heterozygous mice were obtained by heterozygous breeding and identified by genotyping the Fkbp4 and Fkbp5 alleles according to Jackson Laboratory protocols. Two to four mice of mixed genders at the age of 8–12 weeks old were used in each group unless specified otherwise. Mice were monitored by the Research Animal Resources (RAR) staff of the University of Minnesota in specific pathogen free (SPF) housing. Mice were given standard chow and access to drinking water without restrictions. Mice were euthanized via CO2 inhalation. All methods align with Panel of Euthanasia of the American Veterinary Medical Association recommendations.
Preparation of Primary Myoblasts
Mouse muscle mononuclear cells were obtained from hind limbs of 8-12 weeks old male mice as previously described (Motohashi et al., 2014). Briefly, muscles were minced and digested with collagenase type 2 (Worthington, CLS-2) to dissociate muscle cells. Satellite cells were purified with MS columns (Miltenyi Biotec, 130-042-201) and LD columns (Miltenyi Biotec, 130-042-901) by negative selection with antibodies against CD31-PE (eBioscience, 12-0311), CD45-PE (eBioscience, 12-0451), and Sca1-PE (eBioscience, 12-5981), followed by anti-PE MicroBeads (Miltenyi Biotec, 130-048-801). Subsequently, positive selection was applied with an antibody against biotin-conjugated integrin α7-biotin (Miltenyi Biotec, 130-102-125) and anti-biotin MicroBeads (Miltenyi Biotec, 130-090-485). The cells were authenticated by the expression (or the lack thereof) of each surface antigen. Isolated satellite cells were cultured on dishes coated with rat tail collagen (BD Biosciences, 354236) in myoblast growth medium (HAM’s F-10 medium with 20% fetal bovine serum (FBS), 10 ng/ml basic fibroblast growth factor (Thermo Fisher Scientific, PHG0263), penicillin (100 U/ml), and streptomycin (100 mg/ml)) at 37°C with 5% CO2. Low-passage satellite cell-derived primary myoblasts (typically less than eight passages) were used for immunostaining. Differentiation medium (Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 5% horse serum (HS), penicillin, and streptomycin) was used for myogenic differentiation.
Culture of C2C12, CGR8, and ES-E14TG2a cells
Mouse myoblast C2C12 cells were obtained from American Type Culture Collection (ATCC, CRL-1772) and maintained in DMEM with 10% FBS in a 37°C and 5% CO2 incubator. This and other cell lines below were authenticated by each provider. To induce differentiation into myotubes, the cells at 90% confluency (day 0) were rinsed twice with phosphate buffered saline (PBS) before addition of 5% HS in DMEM. Medium was changed every two days.
Mouse embryonic stem cells CGR8 and ES-E14TG2a were purchased from European Collection of Cell Cultures (ECACC, 07032901) and ATCC (CRL-1821), respectively. They were cultured in Glasgow Minimum Essential Medium supplemented with 2 mM glutamine, 0.05 mM 2-mercaptoethanol, 1,000 units/ml leukemia inhibitory factor (MilliporeSigma, ESG1106), and 10% FBS in culture dishes coated with 0.2% gelatin in PBS for 15 min.
METHOD DETAILS
Muscle Injury
Age-matched 8–12-week-old WT, Fkbp4−/−, and Fkbp5−/− mice were anesthetized with 2%–4% isoflurane (Phoenix). Right and left tibialis anterior (TA) muscles were injected with 50 μL 1.2% BaCl2 and 50 μL PBS, respectively. TA muscles of 3–4 mice from each genotype were extracted immediately post-euthanasia at 5, 7, 10, and 14 days post-injury. Muscles were immediately frozen and 10 μm sections were prepared for hematoxylin eosin (H&E) staining.
MTS Cell Proliferation Assay
C2C12, CGR8, and ES-E14TG2a cells were seeded at 1×103 cells/well in triplicate in a 96-well plate. At different days post-seeding, proliferation was measured by adding 20 μL MTS solution (Promega, CellTiter 96 Aqueous One Solution Cell Proliferation Assay, G3581) to 100 mL of culture in each well. Cells were then incubated at 37°C for 2 hr. Absorbance at 492 nm was measured with an LD400 spectrophotometer (Beckman Coulter) and values were calculated by subtracting the absorbance value of a well with the medium only with MTS solution. Average and SD were calculated from technical triplicates.
Gene Knockdown
293FT cells (Thermo Fisher Scientific, R70007) were seeded on day 1 in DMEM with 10% FBS at a concentration of 3×105 cells/well in a 12-well plate. On day 2, cells were transfected with 0.5 μg of a pLKO.1 lentivirus vector encoding an shRNA sequence for the gene of interest (Table S1), together with 0.2 mg each of pCMV-VSV-G (Addgene, 8454), pRSV-Rev (Addgene, 12253), and pMDLg/pRRE (Addgene, 12251) using 2.75 μL Lipofectamine 2000 (Thermo Fisher Scientific, 11668019). Medium was replaced with fresh DMEM with 10% FBS at 5 hr post-transfection. On day 4, target cells were seeded at a density of 1×105 cells/well in 12-well plates. On day 5, medium containing lentivirus from transfected 293FT cells was harvested and the virus was filtered through a 0.45 μm syringe filter and added to the target cells with 0.8 μg/ml polybrene (MilliporeSigma, H9268). Media was replaced with fresh DMEM with 10% FBS on day 6. On day 7, media was replaced with DMEM with 10% FBS containing 1 μg/ml puromycin dihydrochloride (MB Bio, 100552) to select for virus-integrated cells. Puromycin selection continued for 7 days. Proliferative cells were frozen in liquid nitrogen or expanded for further studies.
Gene Subcloning
Following cDNAs were inserted into the pET-21c(+) expression vector (MilliporeSigma, 69742-3) with a 6xHis tag at the carboxyl terminus: Fkbp4, Fkbp5, Fkbp5-Δ1 (deletion of amino acids 50-138), Fkbp5-Δ2 (deletion of amino acids 165-251), Fkbp5-FD67DV (replacement of Phe-Asp with Asp-Val at the amino acids 67 and 68), Fkbp5-W90L (replacement of Trp at the 90th amino acid with Leu), Fkbp5-F130Y (replacement of Phe at the 130th amino acid with Tyr), Cdk4, Cdk4-Δ1 (deletion of amino acids 1-100), Cdk4-Δ2 (deletion of amino acids 101-200), and Cdk4-Δ3 (deletion of amino acids 201–303). Fkbp cDNAs were inserted into the PEGX-2T expression vector (GE Healthcare, 28954653) with a glutathione S-transferase tag at the amino terminus. Fkbp cDNAs were also inserted into the pMXs-IP vector (Kitamura et al., 2003) with and without 3×FLAG (ATGGACTACAAGGACGACGACGACAAGGATTACAAGGATGATGATGATAAGGACTATAAGGACGATGATGACAAA) and a linker (GGCGGCGGAGGCAGC) sequences at the 5′ end.
To prepare shRNA-resistant rCdk4 cDNA, 5′-GCCCTCAAGAGTGTGAGAGTT-3′ in the shRNA target sequence was replaced with 5′-GCaCTgAAaAGcGTcAGgGTa-3′, where lower case letters indicate new nucleotides. Cdk4-P173A mutant cDNA was prepared by replacing CCT (Pro) with gCa (Ala).
Gene Overexpression
PLAT-E cells (Morita et al., 2000) were seeded at a density of 2.5×105 cells/well in a 12-well plate with 10% FBS on day 1. On day 2, cells were transfected with 750 ng of pMXs-IP vector containing the gene of interest (Kitamura et al., 2003) using 2.3 μL FuGENE 6 (Promega, E2691). The culture medium was replaced with fresh DMEM with 10% FBS and target cells were seeded at a concentration of 1×105 cells/well in a 12-well plate on day 3. PLAT-E cell medium containing the virus was harvested on day 4 and filtered through a 0.45 μm syringe filter before transduction into target cells with 0.8 μg/ml polybrene (MilliporeSigma, H9268). Medium was replaced again on day 4 prior to a second transduction on day 5. Starting on day 6, 1 μg/ml puromycin dihydrochloride selection for virus-integrated cells was performed for 5 days. Proliferating cells were frozen in liquid nitrogen or used in experimentation.
Recombinant Cdk4 Protein Series
Recombinant proteins of Cdk4 and its deletion mutants were prepared as described before with some modifications (Gonda et al., 2006). The E. Coli strain BL21(DE3) was transformed with each construct and the protein expression was induced with 1 mM of isopropyl-β-D-thiogalactopyranoside for 3 hr. Cells were harvested and resuspended in Buffer 1 (50 mM sodium phosphate buffer pH 8.0, 300 mM NaCl, 8 M urea, 10 mM imidazole, 0.7 mM 2-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride). The cells were lysed using an Ultrasonic Sonifier 450 (Branson) and 100 μg/ml lysozyme. The lysate obtained from 200 mL culture of E. coli was incubated with 1 mL of Ni-NTA Agarose (QIAGEN, 30230) at 4°Cfor 1 hr with gentle inversion. The mixture was then transferred to a column and washed with Buffer 2 (same as Buffer 1 except for 20 mM imidazole, 6 M urea). The protein was eluted by Elution Buffer (same as Buffer 1 except for 250 mM imidazole, 6 M urea) and collected. Eluted protein (2.5 ml) was renatured by dialysis against a series of buffers at 4°C as follows. First, 500 mL of Buffer 3 (20 mM Tris-HCl pH 8.0,10% glycerol, 10 mM 2-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride) with 2 M NaCl and 1 M urea for 1 hr. Second, 500 mLof Buffer 3 with 2 M NaCl and 0.5 M urea for 1 hr. Third, 500 mL of Buffer 3 with 2 M NaCl for 12 hr. Finally, 500 mL of Buffer 3 with 150 mM NaCl for 2 hr. The dialyzed protein was stored at −80° C.
GST Pull-down Assay
The E. Coli strain BL21(DE3) was transformed with the plasmids encoding Fkbp5 and the mutants. Protein expression was induced with 1 mM of isopropyl-β-D-thiogalactopyranoside for 3 hr. Cells were harvested, lysed in Buffer A (125 mM Tris-HCl pH8.0 and 100 mM NaCl, and 0.5% Tween 20) and 100 μg/ml lysozyme with an Ultrasonic Sonifier 450. Eight hundred microliters of the cell lysate, equivalent to 8 mL of E. coli culture, was incubated with gentle rocking for 1 hr at 4°C with 12.5 μL of Pierce Glutathione Magnetic Agarose (Thermo Fisher Scientific, 78601) previously washed with Buffer A. The beads were then washed three times with Buffer A. Subsequently, 30 μg of the recombinant proteins of Cdk4 and its mutant proteins were separately added to the beads, followed by incubation with gentle rocking for 1 hr at 4°C. The beads were washed three times with Buffer A and bound proteins were eluted with 30 μL of 50 mM reduced glutathione in Buffer A. Eluted proteins were stored at −80°C.
Immunofluorescence Staining of Cells
C2C12 cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 5 min. The cells were stained with an antibody against myosin heavy chain (MHC, Developmental Studies Hybridoma Bank, MF20, 1:200 dilution) and secondary antibody Alexa Fluor 488 anti-mouse IgG (Thermo Fisher Scientific, A21202) for 1 hr each at 25°C. DNA was counter-stained with 5 mg/ml Hoechst 33342 (MilliporeSigma, B2261). Primary myoblasts were fixed with 2% paraformaldehyde for 10 min and blocked with 1% bovine serum albumin in PBS for 30 min. Permeabilized cells were stained with an antibody against MHC and myogenin (Developmental Studies Hybridoma Bank, F5D, 1:50 dilution) and then incubated with secondary antibody Alexa Fluor 488 anti-mouse IgG. DNA was counterstained with 4’,6’-diamidine-2’-phenylindole dihydrochloride (DAPI, MilliporeSigma, 10236276001). Fluorescence images were captured using Metamorph Basic software (Molecular Devices) with LUCPlanFLN 20× or 10× objective lens (Olympus) with 0.45 Ph1 aperture and a C11440-42U digital camera (Hamamatsu) attached to an IX73P2F microscope (Olympus). The images were processed with Adobe Photoshop and Illustrator CS6. Differentiation index was defined as a percentage of nuclei (Hoechst- or DAPI-stained structure) existing within MHC(+) cells among randomly selected 1,000 nuclei in total. 1-nucleus index is a percentage of nuclei that were located in MHC(+) cells containing only one nucleus among randomly selected 1,000 nuclei. To analyze EdU uptake, cells were pulse-labeled with 0.5 μM EdU (5-ethynyl-2′-deoxyuruidine) with a Click-iT EdU Alexa Fluor 448 imaging kit (Invitrogen, C10337) for 30 min. Frequency of EdU uptake was calculated by observing randomly selected 1,000 nuclei stained with Hoechst 33342.
Single Myofiber Culture and Immunostaining
Single myofibers were isolated by 0.2% collagenase type I digestion (Sigma-Aldrich, C0130) of extensor digitorum longus (EDL) muscles for 90 min at 37°C. Isolated single myofibers were transferred and cultured on 5% HS-coated tissue culture dishes with DMEM supplemented with 10% HS and 2% chicken embryo extract (Gemini Bio-Products, 100-163P)for 72 hr. Single myofibers were fixed with 2% paraformaldehyde in PBS for 20 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with 10% bovine serum albumin in PBS for 30 min. The fibers were stained with primary antibodies (anti-Pax7, Developmental Studies Hybridoma Bank, 1:5 and anti-MyoD, Santa Cruz Biotechnology, sc-304, 1:500) and secondary antibodies (Alexa Fluor 488 donkey anti-mouse IgG (Thermo Fisher Scientific, A21202, 1:500) and Alexa Fluor 568 donkey anti-rabbit IgG (Thermo Fisher Scientific, A10042, 1:500). Nuclei were counterstained with DAPI.
Hematoxylin Eosin Staining
TA muscle sections were fixed with 2% paraformaldehyde for 5 minutes at 25°C priorto staining. The staining procedure was as follows: deionized water for 1 min, Harris Modified Hematoxylin (Thermo Fisher Scientific, SH26-500D) for 2 min, tap water for 1 min, deionized water for 1 min, Eosin-Y (Thermo Fisher Scientific, 22-220-104) for 5 min, 95% ethanol for 30 s, 100% ethanol for 2 min twice, and xylene for 10 min twice. Stained sections were mounted using Permount (Thermo Fisher Scientific, SP15-100). Images were captured with a DP26 camera (Olympus) attached to the microscope above, using the cellSens Entry 1.11 software (Olympus).
Flow Cytometry
Cells were harvested, washed with cold PBS and fixed in ice-cold 70% ethanol for 45 min. Fixed cells were centrifuged at 140 × g for 10 min at 4°C, resuspended in a propidium iodide solution (40 μg/ml propidium iodide and 100 μg/ml RNase I in PBS) at a density of 5×105 cells/ml, and incubated for 30 min at 37°C prior to analysis. The amount of DNA in each cell was quantified with a BD LSRII H4760 (University Flow Cytometry Resource) and the acquired data was analyzed with the FlowJo 7.6.5 software.
Immunoprecipitation
Whole cell extracts were obtained by resuspending 4×106 cells with 300 μL lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM MgCl2, 1% NP40, 0.1 mM phenylmethylsulfonyl fluoride, 2 mM leupeptin, 1.5 mM pepstatin A, and 150 u/ml benzonase). After 20 μL Dynabeads Protein G (Thermo Fisher Scientific, 10004D) were washed twice with 0.1% Tween 20 in PBS, 3 μg of antibody was added and incubated at 25°C for 30 min. The beads were then washed three times with 500 μL of 0.1% Tween 20 in PBS and incubated with the extracts for 20 min at 4°C with gentle rocking. The beads were washed eight times with PBS, 0.1% Tween 20, 0.1 mM phenylmethylsulfonyl fluoride, 2 mM leupeptin, 1.5 mM pepstatin A. The beads were resuspended in 1× Laemmli buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 0.005% bromophenol blue, and 25 mM dithithreitol) and incubated at 95°C for 5 min to elute proteins for SDS-PAGE or mass spectrometry.
Western Blotting
Eluate from immunoprecipitation or whole cell extracts obtained from 2×105 cells were loaded into a 12% SDS-PAGE gel. After completion of electrophoresis, the proteins were transferred to an Immobilon P membrane (EMD Millipore, IPVH00010) at 4°C over-night. The next day, the membrane was blocked with 5% non-fat dry milk (BioRad, 180171A) in PBT (0.2% Tween 20 in PBS) for 1 hr at 25°C. Proteins were then labeled with the primary antibody of interest diluted in 5% milk in PBT at 25°C for 1 hr. After washing with PBT for 5 min three times, the membranes were incubated with secondary antibodies goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology sc-2004) or goat anti-mouse IgG-HRP (Santa Cruz Biotechnology, sc-2005) both diluted at 1:1000 in 5% milk in PBT for 1 hr at 25°C. After washing the membrane with PBT six times, the chemiluminescence signal was detected with a SuperSignal West Dura kit (Thermo Fisher Scientific, 34075) and X-ray films.
Phospho-Cdk4 and phospho-Rb antibodies were used with a second protocol as follows to avoid the sequestration of the antibodies by casein in the milk. The membrane was blocked with 5% non-fat dry milk in TBST (50 mM Tris-HCl pH 7.5, 0.1% Tween 20) for 1 hr at 25°C, followed by wash with TBST for 5 min three times. Primary antibodies were diluted in 5% bovine serum albumin in TBST and three subsequent washes were done with TBST. Secondary antibodies were diluted in 5% milk in TBST and six subsequent washes were done with TBST.
Quantitative RT-PCR
RNA was extracted from cells using a Quick RNA Microprep (Zymo Research, R1051) or RNeasy Plus Mini (QIAGEN, 74136) kit, depending on cell number. RNA quantity and purity were assessed using a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized with ProtoScript II Reverse Transcriptase (New England Biolabs, M0368L). Quantitative PCR (qPCR) was performed with the primers listed in Table S2 and a GoTaq qPCR Master Mix (Promega, A6002) in a Mastercycler realplex2 thermocycler (Eppendorf). PCR conditions were as it follows: an initial denaturation at 95°C for 10 min, 40 cycles of 95°C for 15 s, 30 s at the specific annealing temperature for each set of primers, and 72°C for 30 s, and a melting curve step to check the specificity of the reaction. mRNA expression levels were analyzed by normalizing expression values to glyceraldehyde 3-phosphate dehydrogenase (Gapdh) expression. Mean ± SD of three technical triplicates were calculated.
Peptidyl Prolyl Isomerase Assay
Custom peptides were synthesized at EZBiolab and dissolved at 7.8 mM in 0.47 M LiCl in 2,2,2-trifluoroethanol (Thermo Fisher Scientific, AC139750250). PPIase activity was measured as described before (Nelson et al., 2006; Shan et al., 1994). One hundred and ninety one microliters of PPIase buffer (100 mM Tris-HCl pH 8.0 and 150 mM NaCl), 2 μL of 50 μM recombinant Fkbp5 or its mutants, and 5 μL of 20 μM α-chymotrypsin dissolved in 1 mM HCl and 2 mM CaCl2 were mixed. After an addition of 2 μL of the peptide, absorbance at 390 nm was measured every second for 5 min with a Lambda Bio+ spectrometer (Perkin Elmer). kcat/KM was calculated using nonlinear regression with GraphPad Prism 7.
Mass Spectrometry
This was performed as described previously (Lowe et al., 2018). The proteins obtained with immunoprecipitation were separated with a Criterion 8%–16% Tris-HCl gradient gel (Bio-Rad Laboratories, 3450038). The gel was fixed with 40% ethanol and 10% acetic acid for 30 min, followed by staining with Imperial Protein Stain (Thermo Fisher Scientific, 24615). The stained protein regions were excised and digested with trypsin (Shevchenko et al., 1996). The dried peptide mixtures were solubilized in 2% acetonitrile and 0.1% trifluoroacetic acid in water, cleaned with the Stage Tip protocol (Rappsilber et al., 2003), and dried in vacuo.
Mass spectrometry analysis was performed with approximately 1.5 μg of sample and spectra were acquired on an Orbitrap Fusion (Thermo Fisher Scientific) coupled to an Easy-nLC 1000 (Thermo Fisher Scientific) ultrahigh pressure liquid chromatography pump. Peptides were separated on a 100 μm internal diameter, 20 cm column containing ReproSil-Pur C18 resin (3 μm, 120 Å, Dr. Maisch GmbH, Germany). Liquid chromatography condition was as follows: 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with a gradient consisting of 2 min of 2%–8% B, 40 min of 8%–30% B, 1 min of 30%–90% B, 5 min of 90% B, 1 min of 90%–2% B, and 5 min of 2% B with a flow rate of 330 nl/min.
Mass spectrometric data was acquired in top 8 data dependent mode. The MS1 spectra data was collected at a resolution of 60,000 with an automated gain control (AGC) target of 400,000 and a max injection time of 50 ms. Precursor ions were filtered according to charge state (2-7 z) and an intensity of > 130,000 were selected for MS/MS, in which a dynamic exclusion window of 10 s and ± 10 ppm mass accuracy was employed for previously examined precursor ions. Isolation of MS2 precursor ions was performed using a quadruple mass filter set to 1.6 m/z window and fragmentation of precursor ions was performed using an HCD-normalized collision energy of 35 with first mass set to 100 m/z. The MS/MS scan was acquired with Orbitrap resolution of 15,000 an AGC target of 50,000, and a maximal injection time of 100 ms.
RNA-seq
Total RNA was prepared from KD C2C12 cells before differentiation (day 0) and day 3 and 5 during differentiation. shRNA clone #1 was used for Fkbp4 and Fkbp5 KD. RNA concentration and RNA Integrity Number (RIN) were measured with an Agilent BioAnalyzer 2100. Samples with RIN over 8 were used to create sequencing libraries at University of Minnesota Genomics Center. One microgram of total RNA was used to create each sequencing library using Truseq RNA Sample Preparation Kit (Illumina, RS-122-2001). Briefly, poly-adenylated RNA was first purified using oligo-dT-coated magnetic beads. RNA was then fragmented and reverse-transcribed into cDNA. The cDNA was further fragmented, blunt-ended, and ligated to barcoded adaptors and amplified. Final library size distribution was validated with capillary electrophoresis and quantified with a Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, P11496) and qPCR. Indexed libraries were pooled and size-selected to 320 bp ± 5% with a LabChip XT (PerkinElmer). Libraries were loaded onto a single read flow cell and amplified on a cBot (Illumina) before sequencing using a HiSeq 2500 (Illumina).
Bioinformatics Analysis
On average, 5.96 million reads (5.16 million to 7.31 million) were generated per library. The demultiplexed FASTQ files were analyzed using a customized pipeline (gopher-pipelines;https://bitbucket.org/jgarbe/gopher-pipelines/overview) developed and maintained by the Minnesota Supercomputing Institute. Briefly, FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to check on the sequencing quality of the FASTQ files. Then adapters and low quality reads were trimmed using Trimmomatic v0.33 (http://www.usadellab.org/cms/index.php?page=trimmomatic) (Bolger et al., 2014). An additional quality check with FastQC was performed on the post-trimming sequences to ensure successful adaptor and quality trimming. The remaining sequences were then aligned to GRCm38/mm10 reference genome using HISAT2 v2.0.2 (https://ccb.jhu.edu/software/hisat2/index.shtml) and transcript abundance was counted using subread v1.4.6 (http://subread.sourceforge.net/) (Kim et al., 2015; Liao et al., 2014). Differential gene expression analysis was performed in R v3.4.3 using edgeR package (https://bioconductor.org/packages/release/bioc/html/edgeR.html)(Robinson et al., 2010). Pathway analysis of differentially expressed genes were performed by functionally annotate the genes and perform overrepresentation enrichment test using PANTHER (http://pantherdb.org/) (Mi et al., 2013). Heatmaps were generated using the log transformed counts with pheatmap v1.0.8 (https://cran.r-project.org/web/packages/pheatmap/index.html) packages. Hierarchical clustering was performed using average linkage clustering method with correlation coefficient as similarity metric.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
Images of HE-stained sections were randomly selected for analysis; 1,000 muscle fibers (500 per mouse) from each genotype (WT, Fkbp4−/−, and Fkbp5−/−) at 5, 7, 10, and 14 days post injection were analyzed along with uninjured TA sections. Only regenerating fibers with centrally-located nuclei (CLN) were used. Cross sections were first separated using ilastik 1.2 software (ilastik). Pixel Classification. ImageJ (NIH) was used to make images binary and quantify myofiber cross-section area. Histograms of cross section area were made and are shown in Figure 1.
Statistical Analysis
Student’s t tests were used in the analysis of statistical significance of the difference in the differentiation index, the 1-nucleus index, EdU uptake, and qRT-PCR data. The mean + or ± SD obtained from three experiments was shown in each graph. Wilcoxon Rank Sum test with Bonferroni adjustment was used to determine the statistical significance of the differences in Figure 1B.
DATA AND SOFTWARE AVAILABILITY
The RNA-seq data have been deposited to Gene Expression Omnibus (GEO) under the accession number of GEO: GSE109237.
Supplementary Material
In Brief.
Ruiz-Estevez et al. show that Fkbp5 inhibits Cdk4 activity by sequestering it in the Hsp90 complex and preventing phosphorylation at T172 through isomerization of P173. Lack of Cdk4 activity inhibits proliferation and promotes myogenesis. This study provides evidence that Fkbp5 functions as an isomerase necessary for myogenesis.
Highlights.
Muscle regeneration is delayed in both Fkbp4 and Fkbp5 knockout mice
Both Fkbps sequester Cdk4 in the Hsp90 complex, preventing cyclin D1 binding
Fkbp5 inhibits Cdk4 by isomerization and decreased phosphorylation
Inhibition of cyclin D1-Cdk4 inhibits proliferation and promotes myogenesis
ACKNOWLEDGMENTS
We thank Matthew Lowe and Michaela Lohman for technical support and Toshio Kitamura for the plasmid pMXs-IP and PLAT-E cells. We acknowledge the Minnesota Supercomputing Institute, University of Minnesota Informatics Institute, and University of Minnesota Genomics Center for RNA-seq, high performance computing resources, and the gopher pipelines. We thank the University Flow Cytometry Resource supported by the NIH (5P30CA077598-20). We recognize the Center for Mass Spectrometry and Proteomics at the University of Minnesota and various supporting agencies, including the National Science Foundation for Major Research Instrumentation grants 9871237 and NSF-DBI-0215759 used to purchase the instruments described in this study. Supporting agencies are listed at the following: http://cbs.umn.edu/msp/about. C.Y. was supported by the Minnesota Stem Cell Institute and the University of Minnesota Informatics Institute Graduate Fellowship (MnDrive). A.A. was supported by the NIH (R01 AR062142 and R21 AR070319). N.K. was supported by the NIH (R01 GM098294, R21 AR066158, R21 HD083648, and R21 CA187232) and Engdahl Family Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.006.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Asghar U, Witkiewicz AK, Turner NC, and Knudsen ES (2015). The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov 14, 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ, and Reddy EP (2012). CDK4: A key player in the cell cycle, development, and cancer. Genes Cancer 3, 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barent RL, Nair SC, Carr DC, Ruan Y, Rimerman RA, Fulton J, Zhang Y, and Smith DF (1998). Analysis of FKBP51/FKBP52 chimeras and mutants for Hsp90 binding and association with progesterone receptor complexes. Mol. Endocrinol 12, 342–354. [DOI] [PubMed] [Google Scholar]
- Bisteau X, Paternot S, Colleoni B, Ecker K, Coulonval K, De Groote P, Declercq W, Hengst L, and Roger PP (2013). CDK4T172 phosphorylation is central in a CDK7-dependent bidirectional CDK4/CDK2 interplay mediated by p21 phosphorylation at the restriction point. PLoS Genet. 9, e1003546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockstaele L, Coulonval K, Kooken H, Paternot S, and Roger PP (2006). Regulation of CDK4. Cell Div. 1, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockstaele L, Bisteau X, Paternot S, and Roger PP (2009). Differential regulation of cyclin-dependent kinase 4 (CDK4) and CDK6, evidence that CDK4 might not be activated by CDK7, and design of a CDK6 activating mutation. Mol. Cell. Biol 29, 4188–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Flynn J, Prapapanich V, Cox MB, Riggs DL, Suarez-Quian C, and Smith DF (2005). Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol. Endocrinol 19, 1654–1666. [DOI] [PubMed] [Google Scholar]
- Cioffi DL, Hubler TR, and Scammell JG (2011). Organization and function of the FKBP52 and FKBP51 genes. Curr. Opin. Pharmacol 11, 308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederveen TH, Mandemaker IK, and Logie C (2011). The human histone H3 complement anno 2011. Biochim. Biophys. Acta 1809, 577–586. [DOI] [PubMed] [Google Scholar]
- Freeman KB, Karns LR, Lutz KA, and Smith MM (1992). Histone H3 transcription in Saccharomyces cerevisiae is controlled by multiple cell cycle activation sites and a constitutive negative regulatory element. Mol. Cell. Biol 12, 5455–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C, and Giordano A (2010). Cell cycle regulation in myogenesis In Cell Cycle Regulation and Differentiation in Cardiovascular and Nerural Systems, Giordano A and Galderisi U, eds. (New York: Springer Science; ), pp. 231–244. [Google Scholar]
- Gonda K, Wudel J, Nelson D, Katoku-Kikyo N, Reed P, Tamada H, and Kikyo N (2006). Requirement of the protein B23 for nucleolar disassembly induced by the FRGY2a family proteins. J. Biol. Chem 281, 8153–8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD (2015). Prolyl isomerases in gene transcription. Biochim. Biophys. Acta 1850, 2017–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausch F (2015). FKBPs and their role in neuronal signaling. Biochim. Biophys. Acta 1850, 2035–2040. [DOI] [PubMed] [Google Scholar]
- Jayaraman T, and Marks AR (1993). Rapamycin-FKBP12 blocks proliferation, induces differentiation, and inhibits cdc2 kinase activity in a myogenic cell line. J. Biol. Chem 268, 25385–25388. [PubMed] [Google Scholar]
- Jirawatnotai S, Sharma S, Michowski W, Suktitipat B, Geng Y, Quackenbush J, Elias JE, Gygi SP, Wang YE, and Sicinski P (2014). The cyclin D1-CDK4 oncogenic interactome enables identification of potential novel oncogenes and clinical prognosis. Cell Cycle 13, 2889–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, and Kumagai H (2003). Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol 31, 1007–1014. [PubMed] [Google Scholar]
- Knight JD, and Kothary R (2011). The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet. Muscle 1, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuhara T, and Horikoshi M (2004). A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol 11, 275–283. [DOI] [PubMed] [Google Scholar]
- Lazaro JB, Bailey PJ, and Lassar AB (2002). Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 16, 1792–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lowe M, Lage J, Paatela E, Munson D, Hostager R, Yuan C, Katoku-Kikyo N, Ruiz-Estevez M, Asakura Y, Staats J, et al. (2018). Cry2 iscritical for circadian regulation of myogenic differentiation by Bclaf1-mediated mRNA stabilization of cyclin D1 and Tmem 176b. Cell Rep. 22, 2118–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkonen H, and Palvimo JJ (2011). Androgen receptor: acting in the three-dimensional chromatin landscape of prostate cancer cells. Horm. Mol. Biol. Clin. Investig 5, 17–26. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Sotillo R, Santamaría D, Galán J, Cerezo A, Ortega S, Dubus P, and Barbacid M (2004). Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118, 493–504. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, and Thomas PD (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons DS, Jirawatnotai S, Parlow AF, Gibori G, Kineman RD, and Kiyokawa H (2002). Pituitary hypoplasia and lactotroph dysfunction in mice deficient for cyclin-dependent kinase-4. Endocrinology 143, 3001–3008. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, and Kitamura T (2000). Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066. [DOI] [PubMed] [Google Scholar]
- Motohashi N, and Asakura A (2014). Muscle satellite cell heterogeneity and self-renewal. Front. Cell Dev. Biol 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N, Asakura Y, and Asakura A (2014). Isolation, culture, and transplantation of muscle satellite cells. J. Vis. Exp (86), 50846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CJ, Santos-Rosa H, and Kouzarides T (2006). Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126, 905–916. [DOI] [PubMed] [Google Scholar]
- Nigro P, Pompilio G, and Capogrossi MC (2013). Cyclophilin A: a key player for human disease. Cell Death Dis. 4, e888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary JC 3rd, Dharia S, Blair LJ, Brady S, Johnson AG, Peters M, Cheung-Flynn J, Cox MB, de Erausquin G, Weeber EJ, et al. (2011). A new anti-depressive strategy for the elderly: ablation of FKBP5/FKBP51. PLoS ONE 6, e24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, and Barbacid M (1999). Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat. Genet 22, 44–52. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, and Mann M (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem 75, 663–670. [DOI] [PubMed] [Google Scholar]
- Riggs DL, Cox MB, Tardif HL, Hessling M, Buchner J, and Smith DF (2007). Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol. Cell. Biol 27, 8658–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano S, D’Angelillo A, and Romano MF (2015). Pleiotropic roles in cancer biology for multifaceted proteins FKBPs. Biochim. Biophys. Acta 1850, 2061–2068. [DOI] [PubMed] [Google Scholar]
- Rubin SM (2013). Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci 38, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S, and van den Heuvel S (2016). Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter MM, Merrick KA, Larochelle S, Hirschi A, Zhang C, Shokat KM, Rubin SM, and Fisher RP (2013).ACdk7-Cdk4 T-loop phosphorylation cascade promotes G1 progression. Mol. Cell 50, 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiene-Fischer C, Aumüller T, and Fischer G (2013). Peptide bond cis/trans isomerases: a biocatalysis perspective of conformational dynamics in proteins. Top. Curr. Chem 328, 35–67. [DOI] [PubMed] [Google Scholar]
- Schmidpeter PA, Koch JR, and Schmid FX (2015). Control of protein function by prolyl isomerization. Biochim. Biophys. Acta 1850, 1973–1982. [DOI] [PubMed] [Google Scholar]
- Shan X, Xue Z, and Melese T (1994). Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J. Cell Biol 126, 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M,Vorm O, and Mann M (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shimoide T, Kawao N, Tamura Y, Morita H, and Kaji H (2016). Novel roles of FKBP5 in muscle alteration induced by gravity change in mice. Biochem. Biophys. Res. Commun 479, 602–606. [DOI] [PubMed] [Google Scholar]
- Sinars CR, Cheung-Flynn J, Rimerman RA, Scammell JG, Smith DF, and Clardy J (2003). Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. USA 100, 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivils JC, Storer CL, Galigniana MD, and Cox MB (2011). Regulation of steroid hormone receptor function by the 52-kDa FK506-binding protein (FKBP52). Curr. Opin. Pharmacol 11, 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Baggenstoss BA, Marion TN, and Rimerman RA (1993). Two FKBP-related proteins are associated with progesterone receptor complexes. J. Biol. Chem 268, 18365–18371. [PubMed] [Google Scholar]
- Smith JR, de Billy E, Hobbs S, Powers M, Prodromou C, Pearl L, Clarke PA, and Workman P (2015). Restricting direct interaction of CDC37 with HSP90 does not compromise chaperoning of client proteins. Oncogene 34, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechschulte LA, and Sanchez ER (2011). FKBP51-a selective modulator of glucocorticoid and androgen sensitivity. Curr. Opin. Pharmacol 11, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova L, Leng X, Parker SB, and Harper JW (1996). Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 10, 1491–1502. [DOI] [PubMed] [Google Scholar]
- Storer CL, Dickey CA, Galigniana MD, Rein T, and Cox MB (2011). FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol. Metab 22, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ingalls CP, Durham WJ, Snider J, Reid MB, Wu G, Matzuk MM, and Hamilton SL (2004). Altered excitation-contraction coupling with skeletal muscle specific FKBP12 deficiency. FASEB J. 18, 1597–1599. [DOI] [PubMed] [Google Scholar]
- Tranguch S, Cheung-Flynn J, Daikoku T, Prapapanich V, Cox MB, Xie H, Wang H, Das SK, Smith DF, and Dey SK (2005). Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc. Natl. Acad. Sci. USA 102, 14326–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Hesabi B, Moons DS, Pandolfi PP, Hansel KS, Koff A, and Kiyokawa H (1999). Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol. Cell. Biol 19, 7011–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Mollapour M, Smith JR, Truman A, Hu B, Good VM, Panaretou B, Neckers L, Clarke PA, Workman P, et al. (2008). Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol. Cell 31, 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MS, Jabs A, and Hilgenfeld R (1998). Peptide bonds revisited. Nat. Struct. Biol 5, 676. [DOI] [PubMed] [Google Scholar]
- Xiao H, Jackson V, and Lei M (2006). The FK506-binding protein, Fpr4, is an acidic histone chaperone. FEBS Lett. 580, 4357–4364. [DOI] [PubMed] [Google Scholar]
- Xu X, Su B, Barndt RJ, Chen H,Xin H,Yan G, Chen L, Cheng D, Heitman J, Zhuang Y, et al. (2002). FKBP12 is the only FK506 binding protein mediating T-cell inhibition by the immunosuppressant FK506. Transplantation 73, 1835–1838. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wolf IM, Chen H, Periyasamy S, Chen Z, Yong W, Shi S, Zhao W, Xu J, Srivastava A, et al. (2006). FK506-binding protein 52 is essential to uterine reproductive physiology controlled by the progesterone receptor A isoform. Mol. Endocrinol 20, 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong W, Yang Z, Periyasamy S, Chen H, Yucel S, Li W, Lin LY, Wolf IM, Cohn MJ, Baskin LS, et al. (2007). Essential role for Co-chaperone Fkbp52 but not Fkbp51 in androgen receptor-mediated signaling and physiology. J. Biol. Chem 282, 5026–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited to Gene Expression Omnibus (GEO) under the accession number of GEO: GSE109237.


