Abstract
Perfluorodecanoic acid (PFDA) is widely used in production of many daily necessities based on their surface properties and stability. It was assigned as a Persistent Organic Pollutant in 2009 and became a public concern partly because of its potential for activation of the peroxisome proliferator-activated receptor alpha (PPARα). In this study, wild-type and Ppara-null mice were administered PFDA (80 mg/kg). Blood and liver tissues were collected and subjected to systemic toxicological and mechanistic analysis. UPLC-ESI-QTOFMS-based metabolomics was used to explore the contributing components of the serum metabolome that led to variation between wild-type and Pparα-null mice. Bile acid homeostasis was disrupted, and slight hepatocyte injury in wild-type mice accompanied by adaptive regulation of bile acid synthesis and transport was observed. The serum metabolome in wild-type clustered differently from that in Pparα-null, featured by sharp increases in bile acid components. Differential toxicokinetic tendency was supported by regulation of UDP-glucuronosyltransferases dependent on PPARα, but it did not contribute to the hepatotoxic responses. Increase in Il-10 and activation of the JNK pathway indicated inflammation was induced by disruption of bile acid homeostasis in wild-type mice. Inhibition of p-p65 dependent on PPARα activation by PFDA stopped the inflammatory cascade, as indicated by negative response of Il-6, Tnf-α, and STAT3 signaling. These data suggest disruptive and protective role of PPARα in hepatic responses induced by PFDA.
Keywords: Perfluorodecanoic acid, PPARα, Hepatotoxicity
Introduction
Perfluorodecanoic acid (PFDA) has been used for decades to produce daily necessities, such as food packaging, cookware, carpet, fabric, and firefighting foam (Prevedouros et al. 2006). It is one of perfluorocarboxylic acids that was assigned as Persistent Organic Pollutants by the United Nations Environment Program in 2009 and is found in biological samples from wildlife species and humans (Chang et al. 2008; Houde et al. 2006). PFDA is an activator of the peroxisome proliferator-activated receptor alpha (PPARα) and is capable of triggering oxidative stress (Vanden Heuvel et al. 2006). These responses in liver induced by PFDA are closely related to the uptake and persistence in liver tissues (Lau et al. 2007).
Under the natural exposure, PFDA concentration in human serum in European countries was found to be 0.8 ng/mL in 2013 (Bjerregaard-Olesen et al. 2016). However, no exposure information was found regarding people in developing countries which are supposed to be more polluted than European countries. In Wistar rats, the half-life of PFDA was determined to be 59 days in female and 40 days in male rat (Ohmori et al. 2003). In another study, 0.2 % of the dosed PFDA was eliminated in rat urine within 120 h after intraperitoneal administration. When PFDA 20 mg/kg was intraperitoneally injected, the serum concentration was about 38 μg/mL in male and 46 μg/mL in female mice 5 days after treatment (Kudo et al. 2001). These data indicated a gender-related difference and slow elimination of PFDA. But the contribution of xenobiotic metabolic function to PFDA’s elimination has not been studied, neither the relationship between its toxicokinetics and toxicodynamics.
Besides involvement in metabolism of lipids, glucose, and amino acids, PPARα’s role in mediating bile acid homeostasis was reported (Peters et al. 2005; Zhou et al. 2014). Gemfibrozil, a clinical PPARα activator, increased total bile acid and up-regulated cholesterol 7α-hydroxylase (Cyp7a1) and sterol 12α-hydroxylase (Cyp8b1) mRNA in a dose-dependent manner in wild-type mice but not in Ppara-null mice (Liu et al. 2014a). In a dextran sulfate sodium-induced colitis model, activation of a PPARα-UDP-glucuronosyltransferases (UGTs) signaling pathway accelerated the elimination of intestinal bile acids, resulting in suppression of farnesoid X receptor (FXR)-FGF15 signaling. The subsequent up-regulation of hepatic CYP7A1 (2.5-fold) promoted the de novo bile acid synthesis (Zhou et al. 2014). In Ppara-null mice treated with 1 % cholic acid diet, a marked elevation of taurocholic acid (16-fold) and cholic acid (25-fold) was observed in Pparα-null mice. In contrast, neither metabolic disorder nor the regulation of bile acid synthesis was observed in wild-type mice (Li et al. 2012). Thus, PPARα plays both a positive and a negative role in keeping bile acid homeostasis. PFDA is a PPARα agonist and alters bile acid homeostasis (Johnson and Klaassen 2002), but the role of PPARα in disruption of bile acid homeostasis by PFDA remains unclear.
Hepatic bile acid homeostasis is dependent on a complicated balance among uptake, synthesis, basolateral efflux, and canaliculi export, which are closely regulated by nuclear factors PPARα, FXR, etc (Russell 2003). As a PPARα agonist, PFDA caused a threefold increase in bile acid in mice, and down-regulation of transporters Oatp1a1 (75 %), Oatp1a4 (50 %), and Oatp1b2 (45 %) was observed (Cheng and Klaassen 2008a). But these responses in Pparα-null mice were not reported. In another study, up-regulation of transporters Mrp3 (fourfold) and Mrp4 (31-fold) was observed 2 days after PFDA administration, correlating with the elevated bilirubin and bile acids (Maher et al. 2008). However, the systemic regulation of bile acid metabolism by PFDA and its relationship with bile acid homeostasis was not fully investigated.
UGTs catalyze glucuronidation of xenobiotics as well as metabolism of secondary bile acids and bilirubin. A 4-day treatment of male rats with the PPARα agonist clofibrate produced a modest increases in hepatic Ugt1a1 and Ugt1a3 mRNA levels (Shelby 2006). In transgenic mice carrying the human UGT1 treated with Wy-14,643 for 3 days, induction of UGT1A1, UGT1A3, UGT1A4, and UGT1A6 mRNAs was observed (Senekeo-Effenberger et al. 2007). However, the induction profile of UGTs by PFDA and the role of PPARα were unknown. How induction of CYPs and UGTs by PFDA cooperatively contribute to the toxicokinetic and toxicodynamic endpoints of PFDA was not investigated either.
In the present study, wild-type and Pparα-null mice were administered one dose of PFDA 80 mg/kg in order to determine the role of PPARα in toxicokinetics and toxicodynamics of PFDA. Disruption of bile acid homeostasis by PFDA and protection against inflammation via PPARα activation were demonstrated. These data revealed a dual action of PPARα in hepatotoxicity induced by PFDA.
Materials and methods
Chemicals and reagents
PFDA (98 %), propylene glycol, cholic acid (CA), taurocholic acid (TCA), tauroursodeoxycholic acid (TUDCA), and tauro-α-muricholic acid (TαMCA) were purchased from Sigma-Aldrich (St Louis, MO). TRIzol was obtained from Invitrogen (CA, USA). Alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bile acid (TBA), total bilirubin (TBIL), and direct bilirubin (DBIL) assay kits were from Yonghe Sunshine Technology (Changsha, China). Antibodies against total-JNK (t-JNK) and phospho-JNK (p-JNK), total-ATF2 (t-ATF2) and phospho-ATF2 (p-ATF2), NFκB subunit p65, and active form p-p65 were purchased from Cell Signaling Technology (Danvers, USA). Antibody against GAPDH was acquired from Santa Cruz Biotechnologies (CA, USA). Ultrapure water was freshly prepared using a Milli-Q50 SP Water System (MA, USA). LightCycle 480 SYBR Green I Master Mix was obtained from Roche Diagnostics (Mannheim, Germany). All the other chemicals were of the highest grade available from commercial sources.
Animals and treatment
The Pparα-null and wild-type mice on the 129/Sv background were maintained under a standard 12-h light/12-h dark cycle with free access to water and a commercial diet. All mice were acclimated to the environment for 7 days before the experiments. All procedures were performed in accordance with Institute of Laboratory Animal Resource Guidelines, and the protocols were approved by the Medical School of Ningbo University Animal Care and Use Committee.
To design the experiment rightly, pilot tests for dose selection and longitudinal profile of hepatic biochemistry were performed. PFDA was prepared in propylene glycol/H2O (1:1, v/v) before dosing. Firstly, wild-type mice were intraperitoneally injected one dose of PFDA 40 and 80 mg/kg (n = 5), respectively, and TBA level was examined on day 5. In the second pilot test, five wild-type and Pparα–null mice were injected one dose of PFDA 80 mg/kg. Blood was collected by retro-orbital bleeding on days 0, 3, 5, 7, and 10 following PFDA treatment. Serum was collected after centrifugation of the blood samples at 3000×g for 15 min for PFDA exposure and hepatic biochemistry analysis.
For the mechanism of PFDA-induced disruption of bile acid homeostasis, five wild-type and Ppara-null mice were intraperitoneally injected with one dose of PFDA 80 mg/kg. Control groups of both mouse lines were treated with the vehicle only. Five days after dosing, the mice were weighed and then blood was collected. All mice were killed by asphyxiation using carbon dioxide. Liver tissues were harvested and weighed to calculate liver/body weight ratio. Serum was prepared as the above indicated. A section of freshly isolated liver tissues was excised and immediately fixed in 10 % neutral buffered formalin after a brief wash with phosphate-buffered saline. The remaining liver tissues were snap frozen in liquid nitrogen and then stored at −80 °C pending analysis.
Biochemical analysis
The ALT, AST, ALP, TBA, DBIL, and IBIL were assayed using clinical standard methods.
Histopathological assessment
Fixed liver tissues in the above experiments were dehydrated in a serial concentration of alcohol and xylene followed by paraffin embedding. Four-micrometer serial sections were cut and stained with hematoxylin and eosin. Histopathological examination was performed using an Olympus BX41 light microscope.
Serum metabolomic analysis and bile acid identification
Serum samples were diluted using 66.7 % aqueous acetonitrile (1:20) in which bezafibrate (20 μM) was added as an internal standard. Samples were vortexed for 30 s, then centrifuged at 18,000×g for 20 min at 4 °C. An aliquot of 5-μL supernatant was injected into the ultra-performance liquid chromatography-coupled quadrupole time-of-flight mass spectroscopy (UPLC-ESI-QTOFMS, ACQUITY UPLC®, Waters) controlled under conditions as ever reported (Liu et al. 2014a). A reverse-phase 2.1 × 50 mm ACQUITY UPLC® HSS T3 1.8-μm column was used for separation. The gradient of mobile phase started at 100 % A (0.1 % formic acid in water) for 0.5 min, increased to 100 % B (0.1 % formic acid in acetonitrile) over the next 7.5 min, and then returned to 100 % A in last 2 min.
Data were collected in negative mode with full scan (m/z 50–850). Raw data were processed using Marker-Lynx to generate a data matrix consisting of peak areas corresponding to a unique m/z and retention time without normalization. The generated data matrix was imported into SIMCA 13.0.3 (Umetrics, Kinnelon, NJ) to produce score plot using unsupervised principal component analysis (PCA). Loading scatter S-plot was produced by supervised orthogonal projection to latent structures discriminant analysis (OPLS-DA) of PFDA-treated wild-type mice against Ppara-null mice, to explore the contribution of bile acids for the pattern recognition.
Accurate molecular weight of known bile acid components was used to match the data matrix. Four of the contributing items were determined to be CA, TUDCA, Tα/β/ωMCA, and TCA by MS/MS spectra comparison with authentic compound under the same chromatographic conditions as the above. The relative serum levels of these bile acid components were quantified using their peak areas after normalization with internal standard. Their abundance in serum was expressed as fold changes of PFDA-treated mice versus the control groups.
PFDA exposure assessment
Two volumes of acetonitrile with bezafibrate as internal standard were added to serum samples. The mixtures were vortexed for 30 s and centrifuged at 15,000×g for 20 min. An aliquot of 10-μL supernatant was injected to a Shimadzu high-performance liquid chromatography system (Kyoto, Japan) coupled with an API-4000 triple quadrupole mass spectrometer (Toronto, Canada). The ion transition m/z 514.1/469.1 for PFDA was detected in multiple reactions monitoring mode with negative electrospray ionization. A calibration curve with correlation coefficient above 0.99 was constructed for quantification of PFDA.
Quantitative polymerase chain reaction (Q-PCR) analysis
The total RNA from mouse hepatic tissues homogenized in TRIzol reagent was determined by Multiskan GO (ThermoFisher, USA). The reverse transcription system (20 μL) included the following items: 5 × reaction buffer, 10 mM dNTPs, 1 μg oligo dT18, 1 μg random primer, 1 μg total RNA, 200 U M-MLV, and 20 U RNasin. The synthesized cDNA was stored at −20 °C and subjected to analysis within 7 days. The primer sequences, listed in Supplementary Table 1, were extracted from http://mouseprimerdepot.nci.nih.gov/. Each 10-μL PCR system contained total cDNA 1 μL, LightCycle 480 SYBR Green I Master Mix (FastStart Taq DNA Polymerase, reaction buffer, dNTP-mix, SYBR Green I dye, and MgCl2) 5 μL, forward primer 0.2 μL, reverse primer 0.2 μL, and nuclease-free water 3.6 μL. Amplification was performed using reaction cycle at 95 °C 10 s, 55 °C 10 s, and then 72 °C 15 s. The fluorescence signal was detected at the end of each cycle. 18S rRNA was used as an internal control, and melting curve was used to confirm the specificity of the primers.
Western blot analysis
Liver tissues were homogenized by MagNA Lyser (Roche, USA) using RIPA buffer (1:10, g/v) containing 1 % PMSF (Shanghai, China). Tissue debris was removed by centrifugation at 10,000×g and 4 °C for 5 min. The total protein was quantified using a BCA protein assay kit from Beyotime Biotech Co., Ltd (Nantong, China). An equivalent volume of 5 × SDS-PAGE sample loading buffer (Shanghai, China) was added to the tubes which were then boiled for 5 min. The samples were loaded and separated on 10 % SDS–polyacrylamide electrophoresis gels.
The samples were transblotted onto PVDF membranes which was blocked with 5 % fat-free milk at 37 °C for at least 2 h. Membranes were incubated overnight with primary antibodies against t-JNK, p-JNK, t-ATF2, p-ATF2, p65, p-p65, and GAPDH. After second antibody incubation for 1 h, the blotted membranes were exposed to ECL substrates (Advansta, USA) and the signals were detected by Tanon 4200SF (Shanghai, China).
Statistical analysis
All data were expressed as the mean ± SD. SPSS 13.0 for Windows was used for the data analysis. The mRNA level and TBA components following treatment were expressed in fold changes versus those in control groups. The results in both wild-type and Pparα-null mice were compared with those in control groups, respectively, using two-tailed Student’s t test compared with their control groups. PFDA exposure and biochemical assessments in pilot tests were statistically analyzed by one-way ANOVA compared with their baseline. Ninety-five percent was set as confidence intervals, and difference was considered significant when the p values were less than 0.05 (p < 0.05).
Results
PFDA disrupted bile acid homeostasis via PPARα
Dose is critically important in toxicological investigation, especially in acute toxicity test. In the pilot tests, TBA increased by sixfold at the dose of 80 mg/kg, but not at 40 mg/kg (Supplementary figure 1a); 80 mg/kg was used for the whole study and the following mechanism investigation. TBA in wild-type mice increased 13-fold 5 days after PFDA treatment, which decreased slowly in the following 5 days compared with the baseline. Increase in ALP became significant (3.0-fold, p < 0.05) on day 5 and was maximal on day 7 (Supplementary figure 1a and c). The ALT increased less than threefold, which maximized on day 5, and returned to baseline on day 10 (Supplementary figure 1d), correlating with the tendency of TBA. In contrast, all these indicators in Pparα–null mice kept unchanged over the 10 days of treatment.
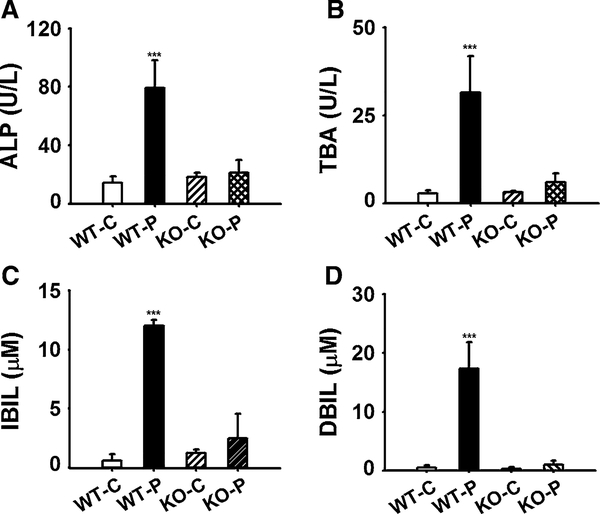
In the experiment continuing for 5 days, ALP and TBA significantly increased (5.6-fold, p < 0.001; 11.7-fold, p < 0.001) in wild-type mice (Fig. 1a, b). IBIL and DBIL are two toxic indicators closely associated with cholestasis. Similar to the ALP and TBA, both IBIL and DBIL were significantly increased (20-fold, p < 0.001; 34.7-fold, p < 0.001) in wild-type mice (Fig. 1c, d). None of the above changes were observed in the Pparα-null mice. These data indicated that PPARα mediates the disruption of bile acid homeostasis induced by PFDA (Fig. 1).
Fig. 1.
Effect of PFDA on serum ALP (a), TBA (b), IBIL (c), DBIL (d) on day 5 after PFDA treatment. Wild-type (WT) and Pparα-null (KO) adult male 129/Sv mice were intraperitoneally injected a single dose of PFDA (80 mg/kg), or the vehicle propylene glycol/water (1/1, v/v). The data were expressed as mean ± SD (n = 5; ***p < 0.001)
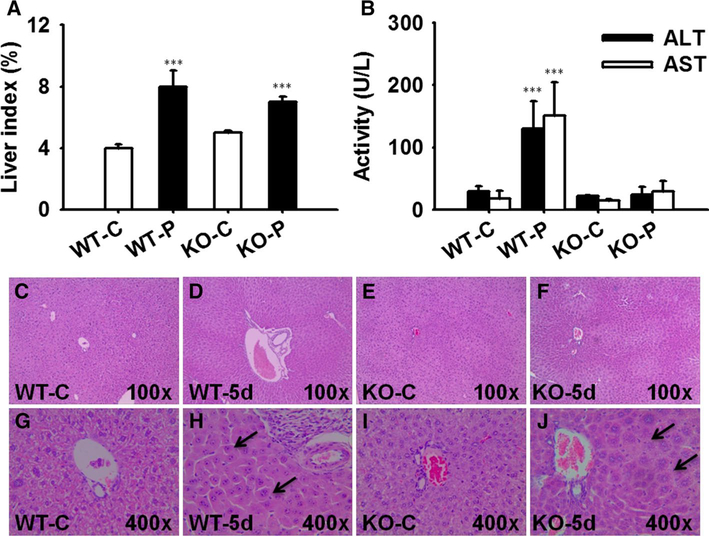
Liver injury biomarkers ALT and AST increased only 4.3- and 7.5-fold in wild-type mice, but not in Pparα-null mice (Fig. 2b), comparable with the response in the pilot test. No overt pathological changes of livers were observed in either mouse line (Fig. 2c–f), because they were not evident enough for observation, considering the slight hepatocyte injury indicated by the above biochemical markers slightly modified. Additionally, both wild-type and Pparα-null mice exhibited elevation of liver/body weight ratio after PFDA administration (Fig. 2a). This could be attributed to hepatocyte proliferation and edema, respectively (Fig. 2h, j).
Fig. 2.
Hepatotoxicity analysis of liver tissues after PFDA treatment. a Increased liver index in mice after PFDA treatment. b Serum ALT and AST activities in mice of four groups. c–j Histopathological staining of representative liver tissues focusing on portal area in wild-type/vehicle, wild-type/PFDA, Pparα-null/vehicle, Pparα-null/PFDA mice. The hepatomegaly (h) in wild-type and edema (j) in Pparα-null mice were marked by arrows (n = 5; ***p < 0.001)
PPARα mediated variation of serum metabolome
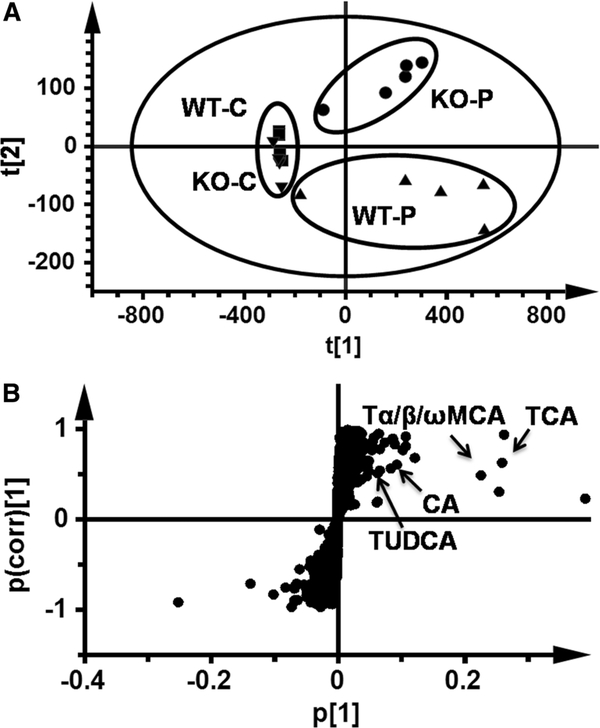
In an unsupervised PCA analysis of the data sets from both control and PFDA-treated wild-type and Pparα-null mice, the control samples of both mouse lines grouped together. Samples of PFDA-treated wild-type and Pparα-null mice clustered away from the control samples but in different directions, indicating different metabolome changes by PFDA treatment between the two mouse lines (Fig. 3a).
Fig. 3.
Multivariate data analysis of serum metabolome in mice dosed PFDA for 5 days. a Score plot of serum metabolite in PFDA-treated wild-type (WT) and Pparα-null (KO) mice versus their control groups by PCA. b S scatter plot of OPLS-DA recognized serum metabolome in PFDA-treated WT mice versus KO mice, in which contribution of identified bile acids were indicated. Each point in panel a represented an individual mouse serum sample and the points in panel b represented contributing metabolites. The t[1]and t[2] represent principal components 1 and 2, respectively. The p(corr)[1] represents the interclass difference, and p[1] represents the relative abundance of the ions (PCA, principal component analysis; OPLS-DA, orthogonal projection to latent structures discriminant analysis)
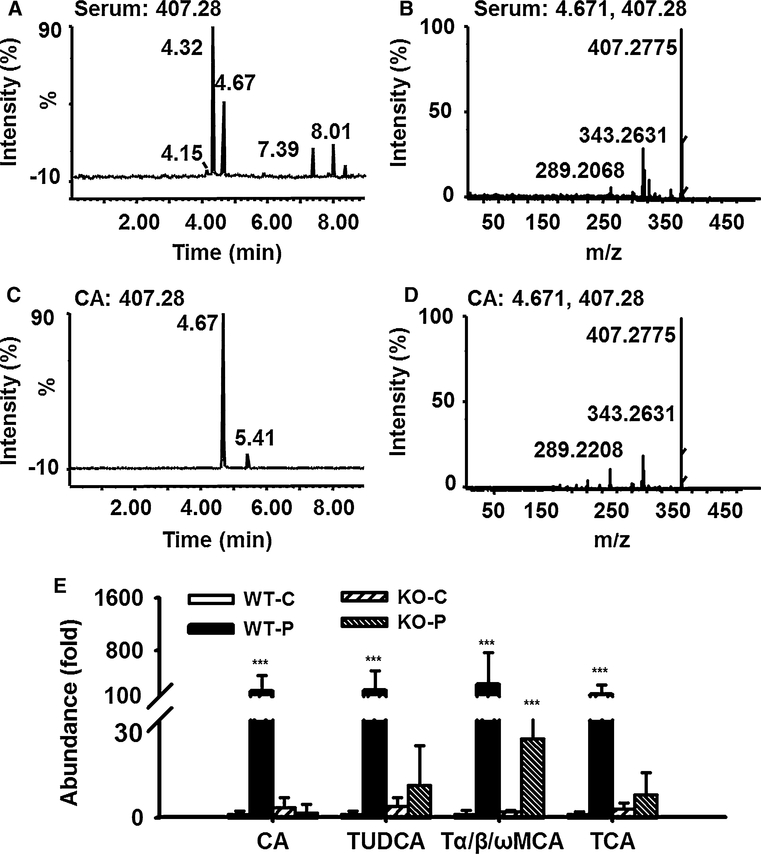
To find the contributing ions for the differences between the two mouse lines, OPLS-DA was used to produce loading scatter S-plots (Fig. 3b) between PFDA-treated wild-type and Pparα-null mice. Three of the contributing ions were CA, TUDCA, and TCA by comparison with authentic standards. A fourth one of them was named Tα/β/ωMCA, because this component determined as TαMCA with RT 3.41 could not be discriminated against TβMCA (RT 3.39) and TωMCA (RT 3.46), respectively. Identification of CA is shown in Fig. 4a–d. Identification of the other three bile acids components are shown in Supplementary Figs. 2, 3 and 4 (comparison between serum component and authentic TαMCA was shown). It was evident that the abundance of the above four bile acids in wild-type mice was 180-, 185-, 280-, and 125-fold higher than those in control group (Fig. 4e). In contrast, these bile acid components were not modified in Pparα-null mice except a 12-fold increase in Tα/β/ωMCA.
Fig. 4.
Exemplary identification of cholic acid (CA) by fragmentation profile and retention time on UPLC-ESI-QTOFMS, and relative abundance of CA, TUDCA, Tα-MCA, and TCA in serum from wild-type (WT), and Pparα-null (KO) mice treated with one dose of PFDA (80 mg/kg). a Total ion count (TIC) chromatogram of MS 407.28 of PFDA-treated WT mouse serum sample when fragmentation was performed. b MS/MS spectra of the peak with retention time 4.67 in WT mouse serum in panel a, c TIC chromatogram of authentic CA when fragmentation was performed at MS 407.28. d MS/MS spectra of the peak with retention time 4.67 in panel c. e The relative abundance CA, TUDCA, Tα/β/ωMCA, and TCA in WT and KO mice on day 5 after PFDA treatment. The data were expressed as mean ± SD (n = 5; ***p < 0.001; TUDCA, tauroursodeoxycholic acid; Tα/β/ωMCA, tauro-α/β/ω-muricholic; TCA, taurocholic acid)
Bile acid metabolism modified by PFDA
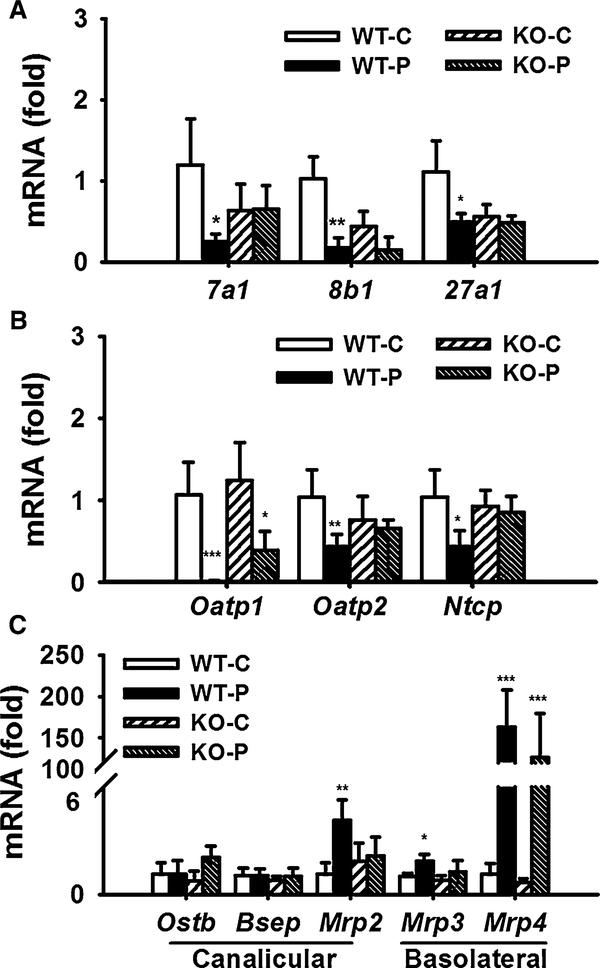
To understand why disruption of bile acid homeostasis was induced by PFDA in wild-type mice, but not in Pparα-null mice, mRNAs encoded by genes involved in bile acid synthesis and transport were analyzed. After PFDA challenge, the Cyp7a1, Cyp8b1, and Cyp27a1 mRNAs were decreased by 73, 71, and 61 %, respectively, in the wild-type mice. In the Pparα-null mice, no changes were observed except for a slight decrease in Cyp8b1 mRNA (Fig. 5a).
Fig. 5.
Different regulated expression of genes involved in bile acid metabolism and transport in mouse liver. a mRNA level of Cyp7a1, Cyp8b1, Cyp27a1. b mRNA level of basolateral uptake transporters. c mRNA level of basolateral and canalicular efflux transporters. The mRNA levels were measured by Q-PCR and normalized by 18S rRNA. mRNA levels in vehicle-treated control mice were arbitrarily set as 1, and results were expressed as mean ± SD (n = 5; *p < 0.05; **p < 0.01; ***p < 0.001)
The mRNAs encoding the basolateral transporters, OATP1, OATP2, and NTCP involved in the bile acid uptake were measured. PFDA treatment decreased transcription of Oatp1, Oatp2, and Ntcp mRNAs about 99, 74, and 73 %, respectively, in wild-type mice (Fig. 5b). In the Pparα-null mice, Oatp1 mRNA was only inhibited by 50 %, and no changes in expression of Oatp2 and Ntcp mRNAs were observed.
The basolateral efflux transporters multidrug resistance-related protein 3 (Mrp3) and Mrp4 are involved in export of bile acid into blood. Mrp2, bile salt export pump (BSEP), and organic solute transporter β (OSTB) export bile acids into canaliculi. Mrp4 mRNA was increased in both wild-type and Pparα-null mice (144-fold and 184-fold, p < 0.001). PFDA increased the expression of Mrp2 mRNA by 3.7-fold (p < 0.01) in wild-type, but not in Pparα-null mice. No changes in levels of Mrp3, Bsep, and Ostb mRNA were observed in either mouse line (Fig. 5c).
Exposure of PFDA and toxicokinetic basis
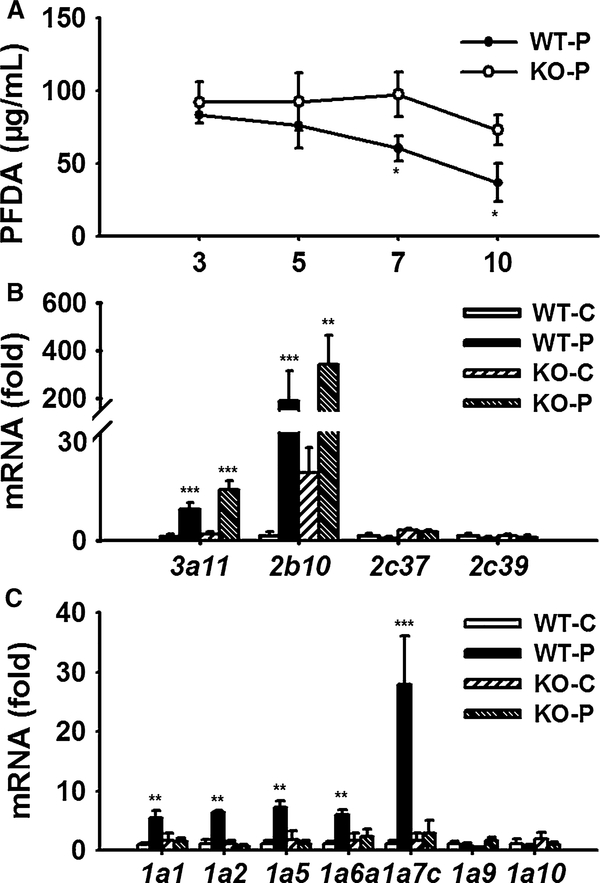
Three days after dosing, the concentration of serum PFDA was about 90 μg/mL, and no difference between wild-type and Pparα-null mice was observed. During the following 7 days, the PFDA concentrations in both wild-type and Pparα-null mice decreased slowly. But the decrease in PFDA in Pparα-null mice was much slower than that in wild-type mice and became significantly different on day 7. By the end of monitoring period, the PFDA concentration in wild-type mice was half of that in Pparα-null mice (Fig. 6a).
Fig. 6.
Toxicokinetic analysis of PFDA and the metabolic basis. a Concentration–time profile of PFDA in mice treated with one dose of PFDA 80 mg/kg within 10 days (n = 3). b, c Hepatic mRNA levels of CYP and UGT isoforms involved in xenobiotic metabolism (n = 5). Date are expressed as mean ± SD (*p < 0.05; **p < 0.01; ***p < 0.001)
Considering the differential PFDA exposure between wild-type and Ppara-null mice, there might be a metabolic basis supporting this finding. As shown in Fig. 6b, expression of Cyp3a11 mRNA was increased by 7.8-fold (p < 0.001) in wild-type mice and by 8.8-fold (p < 0.001) in Pparα-null mice. Cyp2b10 mRNA was also induced in both mouse lines. Although the relative induction was different because of the different control level, the maximal transcription level between two mouse lines was similar (Fig. 6b). Different from the above two P450 s, Cyp3c37 and Cyp2c39 mRNAs were not significantly induced in either mouse line. For the UGT isoforms, Ugt1a1, Ugt1a2, Ugt1a5, Ugt1a6a, and Ugt1a7c mRNA were significantly up-regulated in wild-type mice (5.5-, 5.6-, 6.6-, 5.6-, and 25-fold, respectively, p < 0.001). While none of them were induced in the Pparα-null mice (Fig. 6c), no induction of Ugt1a9 and Ugt1a10 mRNAs was observed.
Up to date, little is known about the metabolism of PFDA. If the above-induced CYPs or UGTs are involved in elimination of PFDA, their regulation in the present study supported the faster decrease in PFDA concentration in wild-type mice than that in Pparα-null mice. However, the exposure level in two mouse lines did not correlate with the hepatotoxic responses. Thus, the differential elimination of PFDA as well as the metabolic basis does not play a key role in the toxicodynamic responses.
Inflammation and anti-inflammation mediated by PPARα
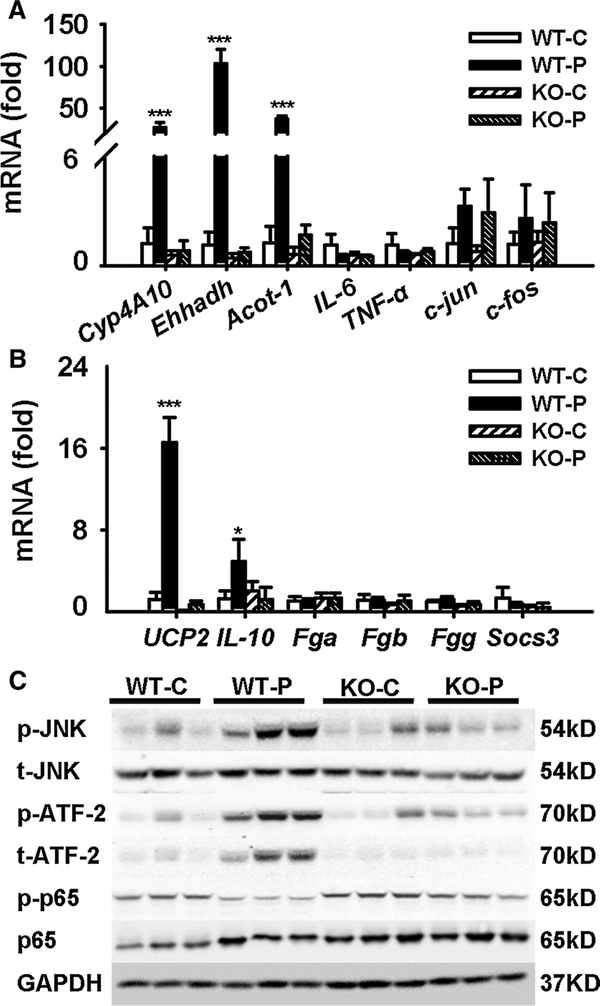
mRNAs for three of the classical PPARα target genes Cyp4A10, Ehhadh, and Acot-1 were increased in wild-type mice, but not in Pparα-null mice, indicating the right response of this transgenic mouse model (Fig. 7a). Transcription of IL-6 and TNF-α were not significantly changed among the four groups (Fig. 7a). The mRNAs encoding c-Jun and c-Fos involved in inflammation were not changed by PFDA in either mouse line. STAT3 is widely involved in hepatotoxicity (Liu et al. 2014b), but in this study, none of the STAT3 target gene mRNAs Fga, Fgb, Fgg, and Socs3 were significantly regulated in either wild-type or Ppara-null mice (Fig. 7b), which supports the negative pathological observation. However, the protective inflammation factor Il-10 was up-regulated by 4.1-fold (p < 0.001) in wild-type mice compared with the control group, and this was not seen in Pparα-null mice (Fig. 7b). These data indicated inflammation occurred in wild-type mice, but not in Pparα-null mice.
Fig. 7.
PFDA induced inflammation and anti-inflammation mediated by PPARα. a, b mRNA level of PPARα target genes, inflammation, apoptosis, necrosis-related factors, and STAT3 target genes. c Western blot of JNK and NFκB signaling in liver extracts. Data were from liver samples collected 5 days after PFDA treatment, and three of them were randomly selected for protein analysis. GAPDH was used as a loading control. The molecular weight was indicated at the left side of each band. Data were expressed as mean ± SD (n = 5; *p < 0.05; ***p < 0.001)
UCP2 was reported to mediate the protection of acetaminophen (APAP)-induced liver injury (Zhou et al. 2014). In this study, PFDA treatment greatly induced Ucp2 mRNA by 14-fold in wild-type mice. But no induction was observed in Pparα-null mice. Thus, the up-regulation of Ucp2 may be involved in the protection of hepatocyte injury by PFDA.
By checking inflammatory pathways, JNK and downstream ATF2 were activated in wild-type mice. Considering the response of IL-10, inflammation occurred in wild-type mice due to disruption of bile acid homeostasis. However, the NFκB pathway was inhibited as indicated by decrease in p-p65 dependent on PPARα activation (Fig. 7c), contributing to the unchanged expression of Il-6, Tnf-α, as well as STAT3 pathway. These results demonstrated that inflammation occurred in PFDA-treated wild-type mice. But its cas-к cade was inhibited, probably by down-regulation of NF B pathway. And these two processes were due to indirect and direct PPARα activation, respectively.
Discussion
PFDA dose in most previous toxicological reports was 40 or 80 mg/kg (Cheng and Klaassen 2008a, b). And PFDA 0 to 80 mg/kg was once used to investigate the dose–response effect for induction of Mrp3 and Mrp4 (Maher et al. 2008). Significant induction of Mrp3 and Mrp4 occurred at 10 and 20 mg/kg. And TBA increased threefold at 80 mg/kg. In the pilot experiment in this study (Supplementary figure 1a), TBA increased only in the 80-mg/kg group, but not in the 40-mg/kg group.
In risk assessment, the acute no-observed-adverse-effect level (NOAEL) in mouse model (if available) should be divided by uncertainty factors (3000–10,000) to calculate human Reference Dose (RfD, a chronic exposure limit for human), to ensure absolute safety. If the exposure by RfD is 0.8 ng/mL or above as reported (Van Rafelghem et al. 1988) and 10,000 is taken as the uncertainty factor, the exposure from NOAEL is 8 μg/mL or above (linear model is supposed to apply). Additionally, when PFDA 20 mg/kg was intraperitoneally injected, the serum concentration was about 38–46 μg/mL (Kudo et al. 2001). In this study, the dose was 80 mg/kg and the serum concentration was about 90 μg/mL. They correlate well with each other. So the doses correlated well with serum exposure levels. Combining the above relationship, the data in this study were of good relevance to risk assessment for human.
In this study, the liver/body weight ratio was increased in both mouse lines after PFDA treatment. This was unexpected because hepatomegaly and hepatocyte hypertrophy induced by PPARα agonists were considered related to a mechanism requiring PPARα. This result accorded with that reported for another perfluoroalkyl carboxylate perfluorobutryate (Foreman et al. 2009). By histopathological examination, slight edema was observed in Pparα-null mice. This probably contributed to the increase in liver weight Pparα-null mice (Fig. 5j). It was clear that bile acid metabolism was modified by PFDA in this study. Although different between two mouse lines, these modifications were expected not to cause bile acid homeostasis disruption. Bile acids are endogenous ligands for FXR, which can feedback to regulate the synthesis (Zhou et al. 2014). An example is down-regulation of Cyp7a1, Cyp8b1, and Cyp27a1 in a mouse model induced by overload of cholic acid challenge (Li et al. 2012). These modifications of bile acid metabolizing enzymes were more likely adaptive or feedback responses to disruption of bile acid homeostasis, serving to protect the liver from potential toxicity of accumulated bile acids (Donner and Keppler 2001; Wagner et al. 2005).
CYPs and UGTs are widely involved in the elimination of many xenobiotics, as well as endogenous compounds including bile acids and bilirubin. Induction of Cyp2b10, Cyp3a11, and Ugt mRNAs by PFDA was ever reported (Arand et al. 1991; Cheng and Klaassen 2008b). Differential induction of Cyp2b10 in wild-type (9.5-fold) and Pparα-null mice (29-fold) by PFDA 80 mg/kg, and slight induction of Cyp3a11 (2.1-fold) was reported. Activation of CAR by PFDA was involved in Cyp2b10 induction (Cheng and Klaassen 2008b). In this study, induction fold of Cyp3a11 and Cyp2b10 in both mouse lines was much higher than the reference data. But neither of their induction was dependent on PPARα, which accorded with previous study.
However, the induction profile of UGT isoforms, the role of PPARα, and the contribution of their induction to bile acid homeostasis disruption were not reported. In this study, similar induction of Cyp3a11 and Cyp2b mRNAs was observed in both mouse lines, but the induction of five Ugt1a isoforms was observed only in wild-type mice. These modifications probably contributed to the faster decrease in PFDA exposure in wild-type mice. However, bile acid homeostasis disruption developed only in wild-type mice whose PFDA exposure was lower than that in Pparα-null mice. Thus, the modified toxicokinetics were not critical factor for the bile acid homeostasis disruption in wild-type mice, suggesting toxicodynamics mediated by PPARα was more important in hepatic toxicity.
In cholestasis induced by LCA, the liver injury is caused by an overload of bile acids (Woolbright et al. 2014; Yu et al. 2015). Super increase in ALT and severe necrosis in the liver tissues were reported when the ALP increased by twofold (Woolbright et al. 2014). This was different from serious hepatic injury induced by overload of TBA (more than eightfold) in cholestatic models where the ALP increased by 2.5-fold where liver injury was cleared observed (Tanaka et al. 2009; Zeng et al. 2016). In this study, in the wild-type mice treated with PFDA, serum TBA increased 11.7-fold and ALP increased 5.6-fold. Mild hepatocyte injury was determined based on the slight increase in ALT and AST, although the pathological changes are not evident (Fig. 5). Comparing with typical cholestatic models where cholestasis is often accompanied by super increase in biomarkers and serious pathological modifications (Yu et al. 2015; Zeng et al. 2016), the protective effect mediated by PPARα was concluded in this study.
The increase in protective Il-10 transcription and activation of JNK and downstream ATF2 indicated the occurrence of inflammation in wild-type mice, which accorded with increase in ALT and AST (4.3-fold, 9.2-fold). These results were quite similar to perfluorobutryate which causes focal necrosis with inflammatory cell infiltrate only in wild-type mice, but diminished in Pparα-null mice (Foreman et al. 2009). Considering the disruption of bile acid homeostasis, this inflammation was a reaction to bile acid load, not a direct PPARα-dependent reaction.
An anti-inflammatory role of PPARα was ever reported. PPARα agonist fibrates and Wy-14,643 inhibited p65-mediated activation of the proinflammatory Il-1β by inducing the inhibitory IкBα in a PPARα-dependent manner (Delerive et al. 2000). In human endothelial cells, fenofibrate and Wy-14,643 reduced vascular cell adhesion molecule 1 promoter activity by inhibiting NFκB translocation (Marx et al. 1999). In this study, although JNK and ATF2 in wild-type mice were activated, the activation of NFκB was inhibited, supporting the inflammation process in wild-type mice supposed above. The inhibition of NFκB signaling was PPARα dependent, contributed to anti-inflammation, and corroborated the negative response of Tnf-α, Il-6, and STAT3 pathway (Fig. 7a).
The induction of Ucp2 and inhibition of c-Jun and c-Fos was proven to be dependent on PPARα activation (Patterson et al. 2012). Similarly, in mice treated with PFDA, Ucp2 mRNA in wild-type mice was induced by PFDA treatment (14.0-fold), and this was not seen in Pparα-null mice. The expression of c-Jun and c-Fos mRNAs was not induced in either mouse line. These results were in accordance with the protective role of UCP2 mediated by PPARα (Patterson et al. 2012).Thus, inhibited hepatic injury under the PFDA-disrupted bile acid homeostasis involved the induction of Ucp2.
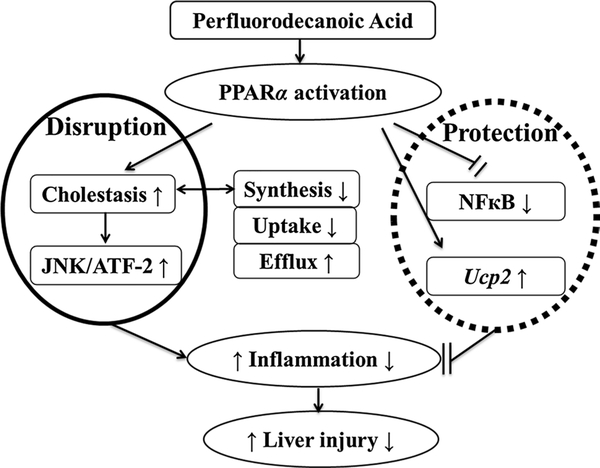
Taken together, no hepatic injury was induced by PFDA-disrupted bile acid homeostasis which was dependent on PPARα. Induction UGTs contributed to differential PFDA toxicokinetics between wild-type and Pparα-null mice, but did not support hepatotoxicity. TBA load led to inflammation involving activation of JNK pathway. However, the inflammatory response was inhibited by PPARα-dependent inhibition of NFκB signaling (Fig. 8). These data suggested both disruptive and protective role of PPARα in hepatotoxicity induced by PFDA.
Fig. 8.
Proposed mechanism underlying the dual action of PPARα in hepatotoxicity induced by perfluorodecanoic acid. The responses in wild-type (WT) mice were initiated by cholestasis mediated PPARα. JNK pathway was involved in the inflammation driven by overload of total bile acid. No liver injury was induced because the toxicity cascade was inhibited by anti-inflammation and induction of Ucp2 also mediated PPARα directly. The data in Pparα-null (KO) mice were negative because the initiating cholestasis was not induced, indicating the role of PPARα in the dual action stated
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China [Grant 81273582, 81302848], Public Projects of Zhejiang Province (2015C37110, 2015R405055), K.C.Wong Magna Fund in Ningbo University and the National Cancer Institute Intramural Research Program.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that there is no conflict of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s00204–016-1779–7) contains supplementary material, which is available to authorized users.
References
- Arand M, Coughtrie MW, Burchell B, Oesch F, Robertson LW (1991) Selective induction of bilirubin UDP-glucuronosyl-transferase by perfluorodecanoic acid. Chem Biol Interact 77(1):97–105 [DOI] [PubMed] [Google Scholar]
- Bjerregaard-Olesen C, Bach CC, Long M et al. (2016) Time trends of perfluorinated alkyl acids in serum from Danish pregnant women 2008–2013. Environ Int 91:14–21. doi: 10.1016/j.envint.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Chang SC, Das K, Ehresman DJ et al. (2008) Comparative pharmacokinetics of perfluorobutyrate in rats, mice, monkeys, and humans and relevance to human exposure via drinking water. Toxicol Sci 104(1):40–53 [DOI] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD (2008a) Critical role of PPAR- in perfluorooctanoic acid- and perfluorodecanoic acid-induced downregulation of oatp uptake transporters in mouse livers. Toxicol Sci 106(1):37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD (2008b) Perfluorocarboxylic acids induce cytochrome P450 enzymes in mouse liver through activation of PPAR-alpha and CAR transcription factors. Toxicol Sci 106(1):29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive P, Gervois P, Fruchart JC, Staels B (2000) Induction of IkappaBalpha expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-alpha activators. J Biol Chem 275(47):36703–36707 [DOI] [PubMed] [Google Scholar]
- Donner MG, Keppler D (2001) Up-regulation of basolateral multidrug resistance protein 3 (Mrp3) in cholestatic rat liver. Hepatology 34(2):351–359 [DOI] [PubMed] [Google Scholar]
- Foreman JE, Chang SC, Ehresman DJ et al. (2009) Differential hepatic effects of perfluorobutyrate mediated by mouse and human PPAR-alpha. Toxicol Sci 110(1):204–211 [DOI] [PubMed] [Google Scholar]
- Houde M, Martin JW, Letcher RJ, Solomon KR, Muir DC (2006) Biological monitoring of polyfluoroalkyl substances: a review. Environ Sci Technol 40(11):3463–3473 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Klaassen CD (2002) Regulation of rat multidrug resistance protein 2 by classes of prototypical microsomal enzyme inducers that activate distinct transcription pathways. Toxicol Sci 67(2):182–189 [DOI] [PubMed] [Google Scholar]
- Kudo N, Suzuki E, Katakura M, Ohmori K, Noshiro R, Kawashima Y (2001) Comparison of the elimination between perfluorinated fatty acids with different carbon chain length in rats. Chem Biol Interact 134(2):203–216 [DOI] [PubMed] [Google Scholar]
- Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A, Seed J (2007) Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicol Sci 99(2):366–394 [DOI] [PubMed] [Google Scholar]
- Li F, Patterson AD, Krausz KW, Tanaka N, Gonzalez FJ (2012) Metabolomics reveals an essential role for peroxisome proliferator-activated receptor in bile acid homeostasis. J Lipid Res 53(8):1625–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Krausz KW, Fang Z-Z, Brocker C, Qu A, Gonzalez FJ (2014a) Gemfibrozil disrupts lysophosphatidylcholine and bile acid homeostasis via PPARα and its relevance to hepatotoxicity. Arch Toxicol 88(4):983–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Tanaka N, Sun L et al. (2014b) Saikosaponin d protects against acetaminophen-induced hepatotoxicity by inhibiting NF-kappaB and STAT3 signaling. Chem Biol Interact 223C:80–86. doi: 10.1016/j.cbi.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JM, Aleksunes LM, Dieter MZ et al. (2008) Nrf2- and PPAR alpha-mediated regulation of hepatic Mrp transporters after exposure to perfluorooctanoic acid and perfluorodecanoic acid. Toxicol Sci 106(2):319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx N, Sukhova GK, Collins T, Libby P, Plutzky J (1999) PPARalpha activators inhibit cytokine-induced vascular cell adhesion molecule-1 expression in human endothelial cells. Circulation 99(24):3125–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori K, Kudo N, Katayama K, Kawashima Y (2003) Comparison of the toxicokinetics between perfluorocarboxylic acids with different carbon chain length. Toxicology 184(2–3):135–140 [DOI] [PubMed] [Google Scholar]
- Patterson AD, Shah YM, Matsubara T, Krausz KW, Gonzalez FJ (2012) Peroxisome proliferator-activated receptor alpha induction of uncoupling protein 2 protects against acetaminophen-induced liver toxicity. Hepatology 56(1):281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Cheung C, Gonzalez FJ (2005) Peroxisome proliferator-activated receptor-alpha and liver cancer: where do we stand? J Mol Med (Berl) 83(10):774–785 [DOI] [PubMed] [Google Scholar]
- Prevedouros K, Cousins IT, Buck RC, Korzeniowski SH (2006) Sources, fate and transport of perfluorocarboxylates. Environ Sci Technol 40(1):32–44 [DOI] [PubMed] [Google Scholar]
- Russell DW (2003) The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712 [DOI] [PubMed] [Google Scholar]
- Senekeo-Effenberger K, Chen S, Brace-Sinnokrak E et al. (2007) Expression of the human UGT1 locus in transgenic mice by 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid (WY-14643) and implications on drug metabolism through peroxisome proliferator-activated receptor alpha activation. Drug Metab Dispos Biol Fate Chem 35(3):419–427 [DOI] [PubMed] [Google Scholar]
- Shelby MK (2006) Induction of rat UDP-glucuronosyltransferases in liver and duodenum by microsomal enzyme inducers that activate various transcriptional pathways. Drug Metab Dispos 34(10):1772–1778 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Cui YJ, Klaassen CD (2009) ANIT-induced intrahepatic cholestasis alters hepatobiliary transporter expression via Nrf2-dependent and independent signaling. Toxicol Sci 108(2):247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rafelghem MJ, Vanden Heuvel JP, Menahan LA, Peterson RE (1988) Perfluorodecanoic acid and lipid metabolism in the rat. Lipids 23(7):671–678 [DOI] [PubMed] [Google Scholar]
- Vanden Heuvel JP, Thompson JT, Frame SR, Gillies PJ (2006) Differential activation of nuclear receptors by perfluorinated fatty acid analogs and natural fatty acids: a comparison of human, mouse, and rat peroxisome proliferator-activated receptor-alpha, -beta, and -gamma, liver X receptor-beta, and retinoid X receptor-alpha. Toxicol Sci 92(2):476–489 [DOI] [PubMed] [Google Scholar]
- Wagner M, Halilbasic E, Marschall H-U et al. (2005) CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology 42(2):420–430 [DOI] [PubMed] [Google Scholar]
- Woolbright BL, Li F, Xie Y et al. (2014) Lithocholic acid feeding results in direct hepato-toxicity independent of neutrophil function in mice. Toxicol Lett 228(1):56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SJ, Bae S, Kang JS et al. (2015) Hepatoprotective effect of vitamin C on lithocholic acid-induced cholestatic liver injury in Gulo(−/−) mice. Eur J Pharmacol 762:247–255. doi: 10.1016/j.ejphar.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Zeng H, Li D, Qin X et al. (2016) Hepatoprotective effects of schisandra sphenanthera extract against lithocholic acid-induced cholestasis in male mice are associated with activation of the pregnane X receptor pathway and promotion of liver regeneration. Drug Metab Dispos Biol Fate Chem 44(3):337–342 [DOI] [PubMed] [Google Scholar]
- Zhou X, Cao L, Jiang C et al. (2014) PPARalpha-UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nat Commun 5:4573. doi: 10.1038/ncomms5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.