Abstract
Background and aims:
Variation in micro-RNA (miRNA) levels in blood has been associated with alterations of physiological functions of the cardiovascular system. Circulating miRNA have the potential to become reliable biomarkers for risk stratification and early detection of cardiovascular events. Recurrent thrombotic events in patients with established coronary artery disease (CAD) demonstrate the need for personalized approaches to secondary prevention, especially in light of recent novel treatment approaches.
Methods:
In a single center cohort study, whole blood samples were collected from 437 subjects undergoing cardiac catheterization, who were followed for recurrent cardiovascular events during a mean follow up of 1.5 years. We selected a case cohort (n = 22) with recurrent thrombotic events on standard medical therapy (stent thrombosis (n = 6) or spontaneous myocardial infarction (MI) (n = 16)) and a matched cohort with CAD, but uneventful clinical follow up (n = 26), as well as a control group with cardiovascular risk factors, but without angiographic CAD (n = 24). We performed complete miRNA next generation sequencing of RNA extracted from whole blood samples (including leukocytes and platelets).
Results:
A differential pattern of miRNA expression was found among controls, CAD patients with no events, and CAD patients with recurrent events. MiRNA previously associated with MI, CAD, endothelial function, vascular smooth muscle cells, platelets, angiogenesis, heart failure, cardiac hypertrophy, arrhythmia, and stroke were found variably expressed in our case-control cohorts. Seventy miRNA (FDR < 0.05) were linked to the risk of recurrent myocardial infarction and future stent thrombosis, as compared to CAD patients with subsequently uneventful follow up.
Conclusions:
MiRNA next generation sequencing demonstrates altered fingerprint profile of whole blood miRNA expression among subjects with subsequent recurrent thrombotic events on standard medical therapy (‘non-responders’), as compared to subjects with no recurrent cardiovascular events. MiRNA profiling may be useful to identify high risk subjects and provide additional insights into disease mechanisms not currently attenuated with standard medical therapy used in CAD treatment.
Keywords: Coronary artery disease, Myocardial infarction, Stent thrombosis, microRNA
1. Introduction
Coronary artery disease (CAD) remains the predominant cause of increased morbidity and mortality in the elderly American population [1]. Survival of patients has improved after myocardial infarction with rapid use of reperfusion and evolving coronary revascularization technologies during acute coronary syndromes, yet the rate of recurrent myocardial infarction remains ∼30% at 5 years despite optimal medical therapy [1]. Secondary prevention after myocardial infarction routinely consists of dual antiplatelet therapy with aspirin and a P2Y12 receptor inhibitor. While extended dual antiplatelet therapy reduces risk of recurrent events, it is not completely protective from events (5.8% recurrent risk of MI after 12 months in ticagrelor treated arm in PLATO trial), and it is associated with increased risk of bleeding [2]. There is a need for reliable and sensitive biomarkers that would reliably identify patients at increased risk for recurrent myocardial infarction and thrombotic events after initial diagnosis of CAD, to tailor preventive therapy.
MiRNAs are non-coding RNA (18–25 nucleotides) that are usually generated by cleaving of primary mRNA transcripts by ribonuclease III [3–5]. MiRNAs that are synthesized in the nucleus are transported into the cytoplasm to undergo a sequential modifications such as assembly into a dimer, tagging with a ribonuclear protein (RNP) and unwinding by a helicase to become fully functional [6–9].The most commonly function attributed to miRNAs is usually gene repression, but occasionally they can also be associated with gene up-regulation [6–10]. The mechanism of miRNAs induced transcriptional repression involves a wide variety of mechanisms like inhibition of initiation, elongation and degradation of nascent proteins by exolytic pathway [6–9]. MiRNA are believed to be a key system in gene regulation and are likely closely involved in changes related to common cardiovascular pathophysiology, and increasing data on relevance of variable miRNA expression in various presentations of cardiovascular disease and its risk factors has been published [8,11–14].
Most studies using miRNA as biomarkers have focused on circulating miRNA from plasma [15], while fewer studies have employed miRNA derived from fixed whole blood, which includes miRNA derived from platelets and leukocytes [16–18]. In concept, novel miRNA biomarkers may potentially be used to monitor effects of medical therapy on gene expression and posttranslational modifications. We hypothesized that distinctive miRNA patterns would be identifiable and differentiate patient subsets between patients with and without recurrent events.
In this study, we performed miRNA sequencing of whole blood samples collected from a cohort of patients who underwent cardiac catheterization, who were followed prospectively for recurrent clinical events. We hypothesized that miRNA expression patterns may be distinct between patients with coronary artery disease who have uneventful clinical courses after initial diagnosis and management (i.e. ‘responders’ to conventional standard medical therapy), as compared to individuals with recurrent thrombotic coronary events, such as spontaneous myocardial infarction or stent thrombosis (‘non-responders’). We used next generation miRNA sequencing to obtain a complete human miRNA profile from selected subjects, as well as to avoid pre-emptive selection bias of specific target miRNA. Large scale miRNA profiling may help identify novel pharmacological gene targets important in secondary prevention of cardiovascular events in high risk patients with CAD, who are currently not adequately protected with standard medical therapy.
2. Materials and methods
The Krannert cardiac catheterization biobank (GENCATH) study is a prospective biobank study enrolling subjects undergoing cardiac catheterization at Indiana University.
The study protocol was approved by the Indiana University Institutional Review board. All subjects gave written informed consent prior to enrollment.
2.1. Study design
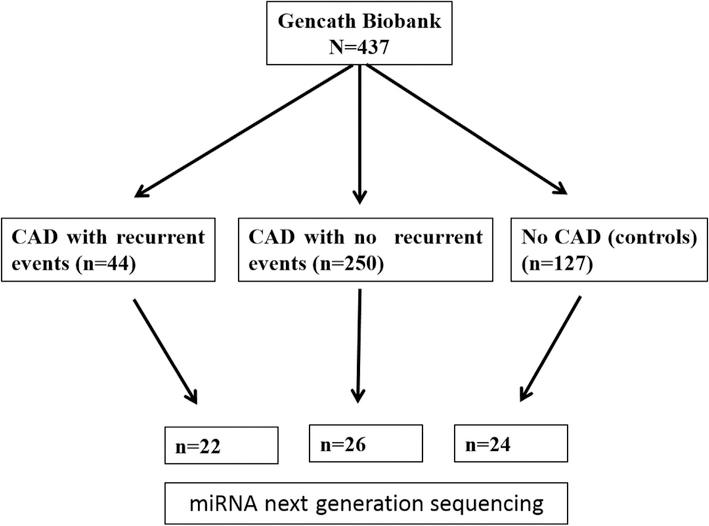
The complete GENCATH biobank cohort includes samples from 437 subjects who underwent cardiac catheterization. Inclusion criteria for the GENCATH biobank were coronary angiography or percutaneous coronary intervention (PCI) completed at one of the participating institutions (Indiana University Health Methodist and Eskenazi Health, Indianapolis, USA) during the index presentation. Exclusion criteria included age < 18 years and inability to provide informed consent. Clinical baseline variables were recorded at time of enrollment, including details of index coronary angiographic findings and index interventions. Subjects were prospectively followed with review of electronic medical records for occurrence of clinical events. Source coronary angiograms were reviewed to ascertain cases of stent thrombosis that occurred during follow up. From the GENCATH cohort we selected 72 subjects for complete miRNA sequencing profiling for use in a matched case-control analysis. The case cohort consisted of subjects with recurrent coronary thrombotic events (n = 22) (defined as either stent thrombosis (n = 6) or spontaneous myocardial infarction (n = 16)) after enrollment in the biobank. The CAD control cohort (n = 26) was a random matched group of patients with CAD and uneventful clinical follow up after enrollment in the biobank. A group of subjects with cardiovascular risk factors, but no diagnosis of heart failure, who underwent cardiac catheterization and had normal coronary arteries on angiogram was included as an additional control group (n = 24). The study design is summarized in Fig. 1.
Fig. 1.
Study flow chart.
2.2. Blood samples
Whole blood was obtained by peripheral venipuncture or from arterial access sheaths and collected in PAXGENE blood RNA vacutainer tubes (Qiagen, MD, USA). Samples were obtained during the index hospitalization prior to cardiac catheterization and frozen at −80 °C until processing. Samples from patients with ST-elevation myocardial infarction on presentation were collected prior to hospital discharge. PAXgene® Blood RNA Tubes were thawed on ice and incubated at room temperature overnight. Total RNA including miRNAs was extracted using PAXgene® Blood miRNA Kit (Qiagen, Germantown, MD; Cat # 763134) according to manufacturer’s instructions for manual purification of RNA from whole blood. Total RNA was quantified using Nanodrop (Thermo Fisher Scientific, Waltham, MA). The RNA quality and the amount of miRNA was measured using the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA).
2.3. MiRNA sequencing
10–20 ng of total RNA was used to create miRNA library. Total RNA samples with < 0.5% miRNA content was enriched for miRNAs. Library was prepared using the small RNA library preparation procedure in the Ion Total RNA-Seq Kit v2 User Guide, Pub. No. 4476286 Rev. E (Life Technologies). If needed, a step of enrichment was conducted, following the small RNA library preparation procedure without smRNA enrichment in the Ion Total RNA-Seq Kit v2 User Guide, Pub. No. 4476286 Rev. E (Life Technologies). The mean RIN for the RNA samples was 8.1 (Min: 6.7, Max: 9.3). Each resulting barcoded library was quantified and its quality accessed, and the libraries pooled in equimolar concentrations. Eight microliters of 100 pM pooled libraries was applied to Ion Sphere Particles (ISP) for template preparation and amplification using Ion OneTouch 2. ISPs were loaded onto the Ion PI™ chip and sequenced on the Ion Proton semiconductor. Each PI chip generated approximately 50 million usable reads of 21–22bp miRNA fragments.
2.4. Bioinformatics analysis
The sequencing data in unmapped bam format generated from ion Proton instrument were first converted to fastq format with the SamToFastq function of Picard (https://broadinstitute.github.io/ picard/). The data were then assessed for quality control with FastQC (v.0.11.4, Babraham Bioinformatics, Cambridge, UK). The Ion Torrent system applies several quality checks to the sequence reads before writing the reads out, this including removal of adapter sequence and low-quality 3’ ends. Therefore, adapter sequence trimming was not included in the analysis steps. The sequencing reads were mapped to the human genome (UCSC hg19) using STAR aligner (v.2.4.2) [19]with the following parameter: -outFilterMultimapNmax 50 –alignIntronMax 1 –outSAMmapqUnique 60”. Reads with mapping quality greater than 10 were assigned to mature miRNAs from mirBase hg19 v20 (www.mirbase.org) with htseq-count (HTSeq-0.6–1) [20]. Differential miRNA expression analysis was performed using edgeR (v.3.12.1) [21,22]. MiRNAs with read count per million (CPM) > 3 in more than 21 of the samples were used for the analysis. False discovery rate (FDR) was computed from p-values using the Benjamini-Hochberg procedure. Pathway analysis of the selected miRNA target genes were carried out with DAVID through the R package RDAVIDWebService [23–25].
3. Results
3.1. Clinical characteristics and endpoints
The demographics and clinical variables of subjects enrolled in the GENCATH biobank cohort are described in Supplemental Table 1. The average age of subjects in the GENCATH biobank cohort is 56 years, with the majority of male gender. The majority of subjects (70.9%) had angiographic coronary artery disease, and the majority presented with acute coronary syndrome. Subjects in the GENCATH cohort were followed for a mean of 1.59 ± 1.1 years. Recurrent myocardial infarction occurred in 10.1%, unplanned PCI in 5.5%, unplanned CABG in 1.8%, stent thrombosis in 1.6%, cardiovascular death in 2.7%, and death in 5% of patients during follow-up.
Demographics and clinical variables of the matched coronary artery case-control cohorts selected from the biobank for miRNA sequencing are listed in Supplemental Table 2.
Subjects with recurrent myocardial infarction or stent thrombosis (recurrent events) were matched with control subjects with coronary artery disease and uneventful follow-up (no recurrent events). No significant difference was observed for age, gender, weight, co-morbidities (diabetes mellitus, hypertension, hyperlipidemia, history of coronary artery bypass grafting, history of PCI, history of MI), risk factors (family history, smoking), P2Y12 antiplatelet therapy, initial presentation, and PCI target vessels (Supplemental Table 2).
Demographics and clinical variables of subjects with coronary artery as compared to control group with angiographically normal coronary arteries is displayed in Supplemental Table 3. Subjects with diagnosis of congestive heart failure (CHF) were excluded from the control cohort, therefore CHF was only found in CAD group. The prevalence of diabetes mellitus, hyperlipidemia, and family history of premature coronary artery disease was higher in the CAD cohort as compared to controls without CAD.
3.2. MiRNA
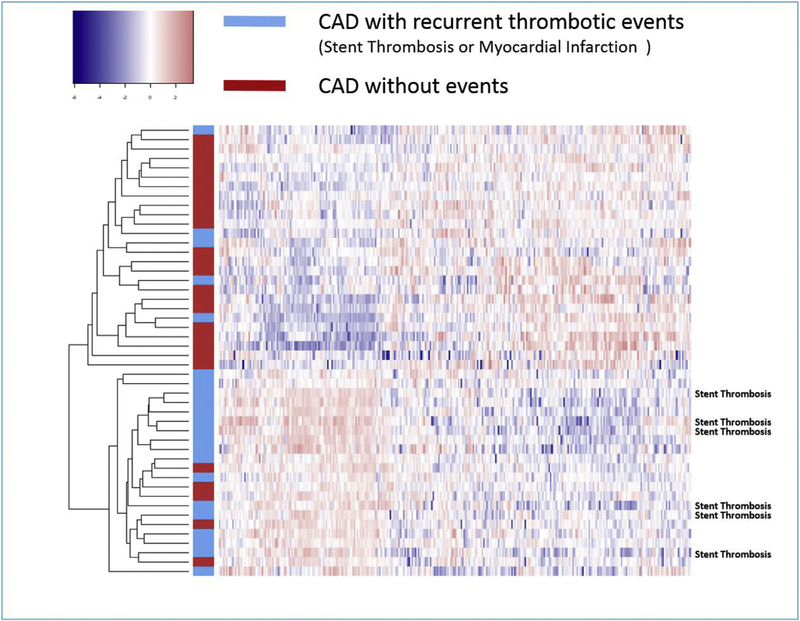
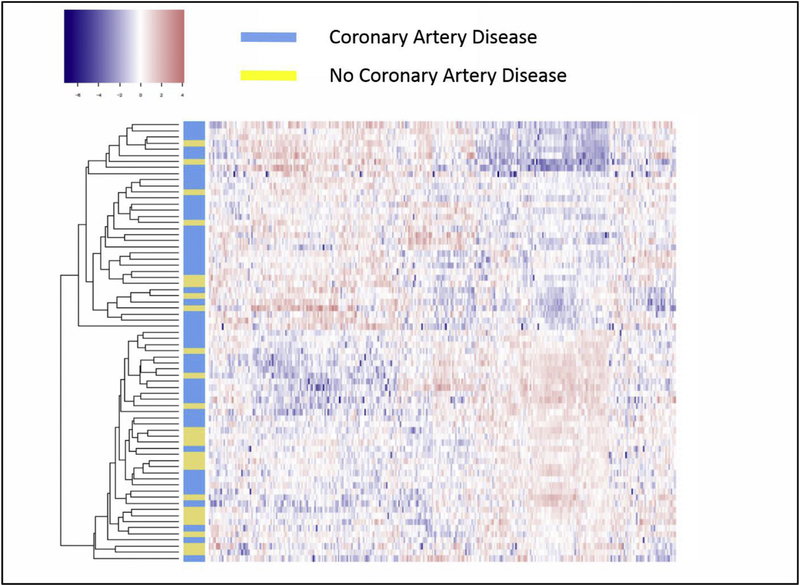
With next generation sequencing, we identified 321 miRNA expressed in controls, CAD patients with recurrent events, and CAD patients with no events. Supervised hierarchical clustering of these 321 miRNA with heat maps demonstrated differential pattern of miRNA expression between CAD patients and controls, as well as among CAD patients with or without recurrent ischemic events (Figs. 2 and 3). The complete list of miRNAs that were upregulated and down-regulated in control, CAD patients with no recurrence and CAD patients with recurrent events are presented in table format in Supplementary Data (Supplementary Table 5 and 6).
Fig. 2.
Heat map of the expression profiles of the miRNA array analysis of 22 coronary artery disease patients with recurrent coronary events (blue) and 26 patients with no recurrent thrombotic coronary events (red).
Subjects with stent thrombosis during follow up are identified. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Heat map of the expression profiles of the miRNA array analysis of 48 coronary artery disease patients (blue) and 24 controls without coronary disease on angiography (yellow).
Differences in miRNA expression in CAD patients with recurrent thrombotic events vs. CAD patients with no recurrent events. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
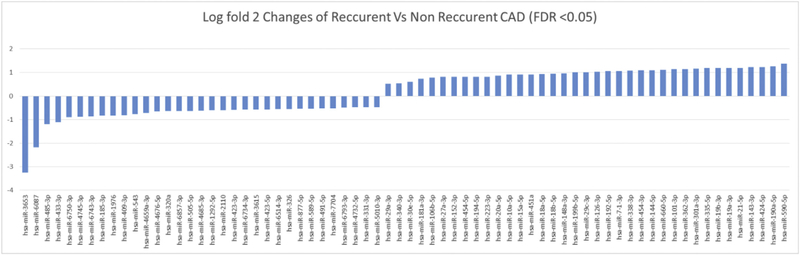
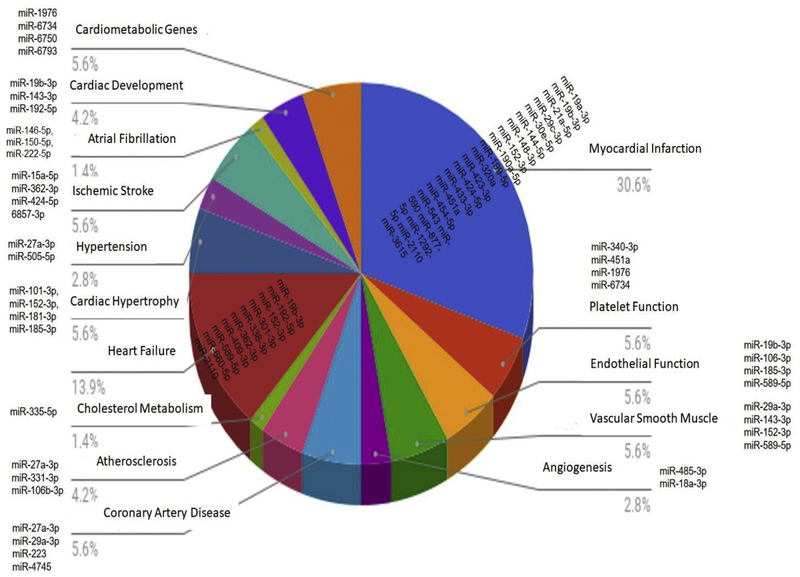
We separated the miRNA on the basis of false discovery rate (FDR) < 0.05 and thus identified 70 miRNA that were significantly differentially expressed between CAD patients with recurrent events as compared to CAD patients with no events. Thirty seven miRNA were upregulated and 33 miRNA were specifically down-regulated in CAD patients who presented with recurrent myocardial infarction or stent thrombosis (recurrent events) as compared to CAD patients with uncomplicated follow-up (no events) (Fig. 4 & Supplemental Fig. 1). A review of the potential functional characteristics of these miRNA revealed that many of the miRNAs that were differentially expressed have been associated with pathophysiologic changes or functions associated with cardiovascular disease state (Supplemental Table 4). Many miRNA (miR-19a-3p, miR-19b-3p, miR-21a-5p, miR-29c-3pmiR-30e-5, miR-144–5p, miR-148–3p, miR-152–3p, miR-190a-5p, miR-199–5p, miR-320a, miR-423–3p, miR-424–5p, miR-433–3p, miR-451a, miR- 454–5p, miR-543, miR-590, miR-877–5p, miR-1292–5p, miR-2110, miR-3615, miR-7704) have been previously associated with coronary artery disease presentations (stable angina, unstable angina, and myocardial infarction). In addition, miRNA that were significantly altered among CAD patients with recurrent events have been previously associated with platelet function (miR-340–3p, miR-451a, miR-1976 and miR- 6734), endothelial function (miR-19b-3p, miR-106–3p, miR-185–3p and miR-589–5p), vascular smooth muscle proliferation (miR-29a-3p, miR- 143–3p, miR-152–3p and miR-589–5p), angiogenesis (miR-485–3p and miR-18a-3p), coronary artery calcification (miR-27a-3p, miR-29a-3p, miR-223 and miR-4745), atherosclerosis (miR-10a-5p, miR-27a-3p, miR-331–3p and miR-106b-3p), atrial fibrillation (miR-409–3p and miR- 7704), stroke (miR-15a-5p, miR-362–3p, miR-424–5p and miR-6857– 3p), heart failure (miR-19b-3p, miR-192–5p, miR-152–3p, miR-301–3p, miR-338–3p, miR-362–3p, miR-409–3p, miR-589–5p, miR-660–5p and miR-2110), cardiac hypertrophy (miR-101–3p,miR-152–3p,miR-181–3p and miR-185–3p) and hypertension (miR-27a-3p and miR-505–5p). For other miRNA, no previous associations with cardiovascular disease states have been published (e.g. miR-3653, miR-6750–3p, miR-6743–3p, miR-1976). In fact the miRNA with the most marked difference in expression between CAD subjects with recurrent events vs those with uneventful follow-up was miR-3653 (log2-fold change = −3.26; FDR = 0.0006), which has not previously been linked to specific disease states. There was only one miRNA significantly different among cohorts with CAD and no recurrent events vs. control group without CAD (miR-618, log2 fold change = 1.29; FDR = 0.03). In contrast, there were 3 miRNA significantly different between CAD subjects with recurrent events vs. controls without CAD (miR-6087: FDR = 2.27*10−6; miR-3653: FDR = 2.62*10−6; miR-551a: FDR = 0.047).
Fig. 4.
Expression pattern of circulating distinct miRNA levels in blood samples of patients undergoing cardiac catheterization.
Next generation miRNA sequence analysis of miRNA of whole blood samples of subjects with coronary disease and recurrent events and subjects with coronary disease and no recurrent events with false discovery rate of < 0.05. The log fold 2 changes of CAD cases with recurrent and non-recurrent events were plotted and shown in the graph.
We classified miRNAs that were upregulated and downregulated in our cohort groups according to previously described functional and clinical associations (Fig. 5).
Fig. 5.
MiRNA differentially expressed in CAD patients with recurrent events vs. no-recurrent events and previously published associations.
3.3. Gene target pathway analysis
In total, 47 miRNA with p < 0.05 have not previously been associated with cardiovascular disease. We identified the gene targets of these 47 non-cardiovascular miRNAs with target scan, with target score less than 80. Furthermore, pathway analysis of the selected miRNA target genes was carried out with DAVID through the R package RDAVIDWebService [23–25]. The methodology and pathway analysis of non-cardiovascular mRNAs identified in this study are included in Supplemental Data section (Supplemental files). The dot plots for the DAVID GO pathway analysis are viewable in the Supplemental Data. They were separated into the three categories of GO analysis (BP, CC and MF). Only pathways with an adjusted p-value < 0.05 were plotted; and only the top 20 were plotted if there are more than 20 enriched pathways for a specific miRNA. No plot was generated if a miRNA has no enriched pathways with an adjusted p-value < 0.05.
4. Discussion
The goal of our study was to examine whole blood next generation miRNA sequencing for use as biomarker fingerprint and in the identification of subjects with coronary artery disease, who are at increased risk for recurrent events with current standard medical therapy. In particular, we focused on patients with established coronary artery disease, who after enrollment in the study, suffered from either coronary stent thrombosis or spontaneous myocardial infarction on standard medical therapy. A random cohort of subjects with coronary artery disease and uneventful follow up served as control, as did a cohort of subjects with normal coronary arteries on angiography. We demonstrated that miRNA sequencing provided a ‘fingerprint’ snapshot that was markedly different in subjects with subsequent recurrent thrombotic events compared to those without events. In particular, subjects with subsequent coronary stent thrombosis exhibited markedly altered miRNA expression as compared to subjects without events (Fig. 2).
There was no significant difference in prevalence of clinical variables in subjects with CAD and recurrent thrombotic events compared to subjects without events. As expected, subjects with angiographically normal coronary arteries in our control cohort were younger, had fewer cardiovascular risk factors, and no congestive heart failure. Whole blood miRNA sequencing also demonstrated altered miRNA expression profiles between coronary artery disease subjects without events and subjects with normal coronary arteries. The large number of miRNA with significantly differential expression among subjects with CAD and uneventful follow up compared to those with recurrent events (70 miRNA with FDR < 0.05) illustrates that although subjects were quite evenly matched based on classic clinical variables used to predict post PCI risk, there appear to be substantial metabolic and epigenetic differences among patient subgroups with increased incidence of recurrent cardiovascular events.
Various miRNAs have been described in the literature as biomarkers in blood plasma for early detection and treatment of stable angina, unstable angina and myocardial infarction. Up-regulation of miR-1, miR-126 and miR-485, and miR-133 in plasma was considered to be an indicator of stable and unstable angina compared to control patients [26–28]. Increases in the plasma levels of miR-208-a/b, miR-499–5p, miR-1 and miR-133 were associated with the onset of MI [27,29,30]. Other miRNA were associated with premature cardiac death or ventricular arrhythmias in MI patients (miR-155, miR-380, miR151–5p) [31,32]. Consistent with several of these findings, we found a differential pattern of expression of miRNAs related to MI disease states that were upregulated and downregulated in patients with recurrent coronary events compared to patients with no events in our cohorts.
Previously described mechanisms linking miRNA function to coronary artery disease phenotypes include downregulation of phospho-AKT/Nitric oxide synthase pathway (miR-206), downregulation of VEGR (miR-361–5p, miR-221, miR-222), promotion of early senescence (miR-217, miR-34), disorganization of cell cycle replication (miR-503, miR-93), or downregulation of monocyte chemoattractant protein-1 (miR-22) [33–37]. However, these specific miRNA were not significantly differentially expressed among cohorts in our study.
A surprising number of miRNA differentially expressed among subjects with subsequent recurrent thrombotic coronary events were previously linked to alterations in endothelial function, platelet function, inflammation, coagulation, or smooth muscle proliferation, which are likely important mediators leading to coronary thrombosis. MiR- 126–3p is highly expressed in endothelial cells and platelets and while its function in the endothelium has been well elucidated, its function in platelets has remained largely unknown. Kaudewitz et al. have demonstrated that mice knockout for miR-126–3p had reduced platelet aggregation. In addition, P2Y12 receptor expression has also been reduced in whole blood of antagomir 126–3p treated mice [38]. In our study, miR-126–3p was significantly upregulated, in patients with recurrent disease vs. controls, as well as in patients with recurrent disease vs. stable CAD. The overexpression of miR-126–3p in platelets of patients with recurrent cardiac events could potentially be linked to either increased tendency towards platelet aggregation or to less response to antiplatelet agents, particularly via the P2Y12 pathway. MiR-223 deficient mice have been shown to form large thrombi and have increased aggregation tendency in response to collagen [38], yet in our study, miR-223 was shown to be upregulated in patients with recurrent events compared to stable disease.
Various miRNA have also been implemented in inflammatory states associated with atherosclerosis leading to cardiac events. MiR-451 and miR-454 were previously reported to be over-expressed in the platelets of patients with CAD [39] and miR-454 has been linked to production of inflammatory cytokines as a part of the thrombus formation cascade. Both of the aforementioned miRNAs are upregulated in our study in the recurrent event group compared to stable disease. Yet miRNA-10p, which has been previously shown to be suppressed in athero-susceptible regions of swine aortas [40] and favored to be a post-translational modulator of the pro inflammatory NF-κB pathway, was up-regulated in our study in patients with recurrent events vs. stable disease. Interestingly though, it was as expected down-regulated in patients with CAD vs. controls. Many inflammatory cytokines have been involved in atherosclerosis including TNF alpha. MiR-19b, which is a part of miR- 17–92 cluster, has been found to be an important modulator of the TNF alpha pathway and its down-regulation in patients with CAD has been shown to increase the inflammatory background in patients with coronary disease [35]. In our study, expectedly, miR-19b was down-regulated in stable CAD compared to controls, yet when looking at recurrent disease vs. stable disease, miR-19b expression is paradoxically upregulated.
MiR-3653 was the most significantly down-regulated miRNA in our dataset in comparison with recurrent events vs. controls. It has numerous predicted targets, but has not been previously linked to cardiovascular disease in the published literature.
Many other miRNA have been previously found differentially expressed in subjects with myocardial infarction, angina, stroke, congestive heart failure, cardiac hypertrophy, and arrhythmias. We have summarized previous reports in the literature of the specific miRNA that were also found to be differently expressed between cohorts in our study in Supplemental Table 4.
The large number of novel miRNA found differentially expressed in sequencing from whole blood samples stabilized in PAXGENE RNA tubes in our cohort, suggests that miRNA derived from leukocytes and platelets may have contributed to differential expressions patterns. This is particularly noteworthy, as most other studies examining the use of miRNA profiling in diagnosis of CAD phenotypes have used plasma samples, which do not contain miRNA derived from leukocytes or platelets, other than the miRNA that were released into plasma in vivo or during sample preparation. The differential expression of miRNA in our study could have been influenced by changes in expression levels at the cellular level or by a modified cellular composition of whole blood. In addition, previous studies using miRNA have focused on the correlation of miRNA with cardiovascular risk factors and disease subtypes, but have not focused on the use of miRNA profiling to identify subjects at risk of subsequent thrombotic events on standard medical therapy. Current evidence based medical therapy for secondary prevention of CAD has substantially lowered event rates with use of dual antiplatelet therapy, high intensity statins, angiotensin converting enzyme inhibitors, beta-blockers, and aldosterone antagonists after MI, however, one year recurrent myocardial infarction event rates in most recent ACS trials remain at ∼5%. This suggests that a substantial subset of patients with CAD are not adequately protected with standard medical therapy, and miRNA profiling of cohorts of subjects with ‘therapy failure’ may potentially lead to novel therapeutic targets in the treatment of cardiovascular disease. We have performed pathway analysis of all novel miRNA not previously associated with cardiovascular disease, and the results are included in the Supplemental Data files.
Future studies are needed to explore the clinical utility and applicability of large datasets derived from miRNA sequencing in care of patients with coronary artery disease, including limitations of applying classic single biomarker models to miRNA ‘fingerprints’.
4.1. Limitations
There are several limitations to our study. The study sample size was relatively small, so we cannot generalize the results of the study, and we did not replicate the findings in a separate cohort. Moreover, the specific source and sequential downstream signaling events that lead to production of these miRNA in patients with recurrent CAD events are unknown. Some of the blood samples, particularly from STEMI patients, were collected after heparin administration, which could have altered some of the miRNA expression. Lastly, we did not investigate temporal changes in miRNA profiles that may occur in individual patients over time in response to medical therapy or disease evolution, and may affect utility in risk prediction.
4.2. Conclusions
In conclusion, our study suggests that whole blood miRNA sequencing may provide an epigenetic, metabolic fingerprint of disease risk in patients with coronary artery disease. In specific, a large number of miRNA were differentially expressed in whole blood in subjects with subsequent coronary thrombotic events compared to subjects with uneventful clinical course on standard therapy. The miRNA found to be differentially expressed among subjects with subsequent events in our study, may potentially serve as indicators for genes involved in disease modulation in subjects with disease refractory to current therapy. Many of the miRNA found to be variably expressed in patients with recurrent MI or stent thrombosis in our study were previously linked to MI, endothelial function, platelet function, coagulation, and inflammation. Further investigation exploring the biologic effects of established key miRNA and novel miRNA variants is needed. Distinct patterns between different groups of patients with coronary artery disease suggest utility in monitoring progression and quiescence of coronary artery disease by measurement of whole blood miRNA profiles, beyond classic biomarkers or ‘simple’ clinical disease classifications.
Supplementary Material
HIGHLIGHTS.
miRNA next generation sequencing performed in CAD patients with and without recurrent events and controls.
Differential miRNA expression pattern between controls, CAD patients with no events, and CAD patients with recurrent events.
Seventy miRNA (FDR < 0.05) were linked with risk of recurrent myocardial infarction and future stent thrombosis.
MiRNA profiling may identify high risk subjects and provide insights into CAD disease mechanisms.
Acknowledgements
MiRNA sequencing was performed by Dr. Howard Edenberg at the Center for Medical Genomics and statistical analysis by the Center for Computational Biology and Bioinformatics at Indiana University School of Medicine. We would like to thank research nurses and coordinators involved in enrollment for the Gencath biobank project (Toni Lathrop, Andrea Schaffter, Elise Hannemann, Roxanne Kovacs, Keith Wright). SK is supported by NIH/NIGMS (T32GM008425). The study was supported by the Indiana University – Indiana University Health strategic research initiative, and by the Charles Fisch Cardiovascular Research Award endowed by Dr Suzanne B. Knoebel of the Krannert Institute of Cardiology.
Footnotes
Conflict of interest
The authors declared they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
References
- [1].Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB, Heart disease and stroke Statistics-2016 update: a report from the American heart association, Circulation 133 (2016) e38–360. [DOI] [PubMed] [Google Scholar]
- [2].Alfredsson J, Neely B, Neely ML, Bhatt DL, Goodman SG, Tricoci P, Mahaffey KW, Cornel JH, White HD, Fox KA, Prabhakaran D, Winters KJ, Armstrong PW, Ohman EM, Roe MT, Predicting the risk of bleeding during dual antiplatelet therapy after acute coronary syndromes, Heart 103 (2017) 1168–1176. [DOI] [PubMed] [Google Scholar]
- [3].Gulyaeva LF, Kushlinskiy NE, Regulatory mechanisms of microRNA expression, J. Transl. Med 14 (2016) 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ha M, Kim VN, Regulation of microRNA biogenesis, Nat. Rev. Mol. Cell Biol 15 (2014) 509–524. [DOI] [PubMed] [Google Scholar]
- [5].Scalbert E, Bril A, Implication of microRNAs in the cardiovascular system, Curr. Opin. Pharmacol 8 (2008) 181–188. [DOI] [PubMed] [Google Scholar]
- [6].Ali SS, Kala C, Abid M, Ahmad N, Sharma US, Khan NA, Pathological microRNAs in acute cardiovascular diseases and microRNA therapeutics, J. Acute Dis 5 (2016) 9–15. [Google Scholar]
- [7].Catalucci D, Latronico MV, Condorelli G, MicroRNAs control gene expression: importance for cardiac development and pathophysiology, Ann. N. Y. Acad. Sci 1123 (2008) 20–29. [DOI] [PubMed] [Google Scholar]
- [8].Condorelli G, Latronico MV, Cavarretta E, microRNAs in cardiovascular diseases: current knowledge and the road ahead, J. Am. Coll. Cardiol 63 (2014) 2177–2187. [DOI] [PubMed] [Google Scholar]
- [9].Ono K, Kuwabara Y, Han J, MicroRNAs and cardiovascular diseases, FEBS J 278 (2011) 1619–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jonas S, Izaurralde E, Towards a molecular understanding of microRNA-mediated gene silencing, Nat. Rev. Genet 16 (2015) 421–433. [DOI] [PubMed] [Google Scholar]
- [11].Romaine SP, Tomaszewski M, Condorelli G, Samani NJ, MicroRNAs in cardiovascular disease: an introduction for clinicians, Heart 101 (2015) 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].P G, MicroRNAs in cardiovascular disease, Curr. Opin. Cardiol 31 (2016) 249–254. [DOI] [PubMed] [Google Scholar]
- [13].Barwari TJA, Mayr M, MicroRNAs in cardiovascular disease, J. Am. Coll. Cardiol 68 (2016) 2577–2584. [DOI] [PubMed] [Google Scholar]
- [14].Small EMFR, Olson EN, MicroRNAs add a new dimension to cardiovascular disease, Circulation 121 (2010) 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jakob P, Kacprowski T, Briand-Schumacher S, Heg D, Klingenberg R, Stähli BE, Jaguszewski M, Rodondi N, Nanchen D, Räber L, Vogt P, Mach F, Windecker S, Völker U, Matter CM, Lüscher TF, Landmesser U, Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction, Eur. Heart J 38 (2017) 511–515. [DOI] [PubMed] [Google Scholar]
- [16].Freedman JE, Ercan B, Morin KM, Liu CT, Tamer L, Ayaz L, Kanadasi M, Cicek D, Seyhan AI, Akilli RE, Camci C, Cengiz B, Oztuzcu S, Tanriverdi K, The distribution of circulating microRNA and their relation to coronary disease, F1000Res 1 (2012) 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang J, Pei Y, Zhong Y, Jiang S, Shao J, Gong J, Altered serum MicroRNAs as novel diagnostic biomarkers for atypical coronary artery disease, PloS One 9 (2014) e107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xu ZWQ, Pan J, Sheng X, Hou D, Chong H, Wei Z, Zheng S, Xue Y, Zhou Q, Cao H, Zhang CY, Wang D, Jiang X, Characterization of serum miRNAs as molecular biomarkers for acute Stanford type A aortic dissection diagnosis, Sci. Rep 7 (2017) 13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29 (2013) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Anders SPP, Huber W, HTSeq–a Python framework to work with high-throughput sequencing data, Bioinformatics 31 (2015) 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26 (2010) 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McCarthy DJ, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation, Nucleic Acids Res 40 (2012) 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang DWSB, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources, Nature Protoc 4 (2009) 44–57. [DOI] [PubMed] [Google Scholar]
- [24].Huang DWSB, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res 37 (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fresno CFE, RDAVIDWebService: a versatile R interface to DAVID, Bioinformatics 29 (2013) 2810–2811. [DOI] [PubMed] [Google Scholar]
- [26].D’Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, Rubino M, Marenzi G, Colombo GI, Achilli F, Maggiolini S, Capogrossi MC, Pompilio G, Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina, PloS One 8 (2013) e80345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Economou EK, Oikonomou E, Siasos G, Papageorgiou N, Tsalamandris S, Mourouzis K, Papaioanou S, Tousoulis D, The role of microRNAs in coronary artery disease: from pathophysiology to diagnosis and treatment, Atherosclerosis 241 (2015) 624–633. [DOI] [PubMed] [Google Scholar]
- [28].Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, Chen C, Wang DW, Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis, J. Transl. Med 11 (2013) 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fichtlscherer S, Zeiher AM, Dimmeler S, Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler. Thromb. Vasc. Biol 31 (2011) 2383–2390. [DOI] [PubMed] [Google Scholar]
- [30].Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, Kempf T, Wollert KC, Thum T, Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome, J. Mol. Cell. Cardiol 51 (2011) 872–875. [DOI] [PubMed] [Google Scholar]
- [31].Matsumoto S, Sakata Y, Nakatani D, Suna S, Mizuno H, Shimizu M, Usami M, Sasaki T, Sato H, Kawahara Y, Hamasaki T, Nanto S, Hori M, Komuro I, A subset of circulating microRNAs are predictive for cardiac death after discharge for acute myocardial infarction, Biochem. Biophys. Res. Commun 427 (2012) 280–284. [DOI] [PubMed] [Google Scholar]
- [32].Zhang Y, Wang R, Du W, Wang S, Yang L, Pan Z, Li X, Xiong X, He H, Shi Y, Liu X, Yu S, Bi Z, Lu Y, Shan H, Downregulation of miR-151–5p contributes to increased susceptibility to arrhythmogenesis during myocardial infarction with estrogen deprivation, PloS One 8 (2013) e72985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Caporali A, Emanueli C, MicroRNA regulation in angiogenesis, Vasc. Pharmacol 55 (2011) 79–86. [DOI] [PubMed] [Google Scholar]
- [34].Chen B, Luo L, Zhu W, Wei X, Li S, Huang Y, Liu M, Lin X, miR-22 contributes to the pathogenesis of patients with coronary artery disease by targeting MCP-1: an observational study, Medicine (Baltim.) 95 (2016) e4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tang Y, Zhang YC, Chen Y, Xiang Y, Shen CX, Li YG, The role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery disease, Sci. Rep 5 (2015) 15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang HW, Lo HH, Chiu YL, Chang SJ, Huang PH, Liao KH, Tasi CF, Wu CH, Tsai TN, Cheng CC, Cheng SM, Dysregulated miR-361–5p/VEGF axis in the plasma and endothelial progenitor cells of patients with coronary artery disease, PloS One 9 (2014) e98070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang Q, Kandic I, Kutryk MJ, Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease, Biochem. Biophys. Res. Commun 405 (2011) 42–46. [DOI] [PubMed] [Google Scholar]
- [38].Kaudewitz D, Skroblin P, Bender LH, Barwari T, Willeit P, Pechlaner R, Sunderland NP, Willeit K, Morton AC, Armstrong PC, Chan MV, Lu R, Yin X, Gracio F, Dudek K, Langley SR, Zampetaki A, de Rinaldis E, Ye S, Warner TD, Saxena A, Kiechl S, Storey RF, Mayr M, Association of MicroRNAs and YRNAs with platelet function, Circ. Res 118 (2016) 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen F, Zhao X, Peng J, Bo L, Fan B, Ma D, Integrated microRNA-mRNA analysis of coronary artery disease, Mol. Biol. Rep 41 (2014) 5505–5511. [DOI] [PubMed] [Google Scholar]
- [40].Fang Y, Shi C, Manduchi E, Civelek M, Davies PF, MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.