Abstract
Histone deacetylase (HDAC) inhibition has sporadic clinical efficacy in urothelial carcinoma (UC); the genomic basis for clinical response is not known. In two separate phase I clinical trials testing pharmacokinetic aspects of HDAC inhibitors in advanced solid tumors, we identified one patient with advanced UC who had a complete response (CR) to belinostat, and one patient with advanced UC who had a partial response (PR) to panobinostat. The archived tumors of the responders were genomically characterized in comparison to others with UC on the trials. UC cell lines treated with panobinostat and belinostat were studied to elucidate the mechanisms of benefit. Notably, the UC tumors that responded to HDAC inhibition had ARID1A mutations. ARID1A mutations were also noted in the tumors of three patients who had stable disease as their best response to HDAC inhibition. Corroborating the basis of sensitivity, transcriptional profiling of platinum-resistant ARID1A-mutated HT1197 cells treated with panobinostat reveals negative enrichment for both cyto-proliferative (MYC and E2F targets) and DNA repair gene sets, and positive enrichment for TP53 and inflammatory gene sets. Our study identifies ARID1A loss as a basis for clinical response to pan HDAC inhibition and offers avenues for potential rational therapeutic combinations with HDAC inhibitors in advanced UC.
Keywords: HDAC inhibitors, advanced urothelial carcinoma, ARID1A, tumor mutational burden, E2F
Introduction
Bladder cancer, commonly urothelial carcinoma (UC), is the sixth most common cancer in the United States with an estimated 81,190 new cases and 17,240- deaths from it in the year 2018 (1). Advanced UC is an aggressive, fatal disease. First line treatment with cisplatin-based combination chemotherapy affords an objective response rate (ORR) ranging from 46 to 64% and progression-free survival (PFS) of 7.7 to 9.5 months (2–5). In the second line setting, immune checkpoint inhibition with pembrolizumab has an ORR of 21%, compared to single-agent chemotherapy, which has an ORR of 11.4%; with median progression-free survival (PFS) of only 2.1 months and 3.3 months respectively (6). Immune checkpoint inhibition achieves a higher response rate in UC with higher degrees of tumor mutational burden (TMB) (7).
Advanced UC is genomically complex, characterized by multiple overlapping genomic alterations (GA) involving cell cycle regulation, chromatin regulation, and kinase-driven signaling pathways (8, 9). Targeted therapy has demonstrated clinical benefit in some early phase trials, and sporadic exceptional responses have been described (10–12). Genomic characterization of the tumors of patients with an exceptional response can provide the basis for sensitivity, paving the way for biomarker-enriched trials of targeted therapy. For example, one patient in a phase 1 study combining everolimus and pazopanib in solid tumors was noted to benefit with an exceptional complete response at 14 months. Genomic profiling of this patient’s tumor revealed two activating mTOR pathway mutations (11). A phase II trial using everolimus in UC achieved a durable complete response to everolimus in one patient. This patient’s tumor was noted to have TSC1 and NF2 loss of function mutations which were attributed as the basis of the response (12).
Histone deacetylase inhibitors (HDACIs) are a broad and heterogeneous class of anticancer agents that induce differentiation, cell-cycle arrest, and apoptosis in many cancer cell lines (13). HDACIs inhibit tumor growth in animal models and show antitumor activity in patients with advanced breast cancer and sarcoma (14, 15).
Romidepsin, vorinostat and belinostat are approved therapies for certain types of T cell lymphoma, and panobinostat is approved for use in combination with other agents for the treatment of multiple myeloma.
HDACIs when studied in the bladder cancer cell line 5637, result in chromatin modification as well as non-chromatin associated proteomic changes, which lead to inhibition of cell proliferation and the induction of apoptosis through multiple cell death-associated pathways (16). Belinostat has in vivo efficacy in bladder cancer (17). Vorinostat has shown tumor regression in two UC patients in a phase I study (18). Although extensively investigated in a variety of settings, little is known regarding the genomic basis of clinical response to HDAC inhibition in UC. Histone modifying genes are frequently mutated in UC (89%) (8, 9). SWI/SNF alterations are present in 64% of muscle-invasive UC (8). Mutations in genes encoding histone acetyltransferase (HAT) enzymes EP300 and CREBBP, are present in one-third of UC and are implicated in a gene signature that is targetable by HDAC inhibition with mocetinostat (19, 20). A phase II clinical trial using mocetinostat, a specific HDAC inhibitor to class I and II, in patients with previously treated advanced UC harboring HAT (histone acetyltransferase) mutations(21), is complete and the results are unannounced at the time of writing this manuscript..
Two single institution phase I clinical trials testing pharmacokinetic aspects of pan HDACIs, belinostat and panobinostat, enrolled a total of 64 patients with advanced solid tumors (22–24). In these all-comer advanced solid tumor trials, one patient with advanced bladder cancer (or UC), who received belinostat, had a durable complete response (CR) to treatment (22), and another patient with advanced bladder cancer, who received panobinostat, had a partial response (PR) (24).
The objective of our study is to identify the genetic basis of the clinical response to HDAC inhibition in these two trials. We obtained the archived tissue of the two responders, along with all other UC patients who had adequate archived tissue available for genomic analysis, and their recorded best overall response to HDAC inhibition on trial. All advanced UC patients treated on either of the two trials, with tissue and best overall response data, are included this study. We clinically characterized the patients and genomically analyzed their tumors to detect correlative biomarkers. We then performed in vitro studies in bladder cancer cell lines to identify the mechanisms of benefit.
Materials and Methods:
Tissue characterization
The University of Utah Institutional Review Board approved this study. Archived formalin fixed paraffin embedded (FFPE) UC tissue of four patients who received belinostat and three patients who received panobinostat was retrieved. DNA was extracted from 2 × 2 mm FFPE punches. Samples were washed twice with Xylenes (Cardinal Health, Illinois) followed by 200 proof Ethanol (Decon Labs Inc, Pennsylvania) to remove paraffin wax. DNA was isolated using QIAamp DNA FFPE Tissue Kit (Qiagen, Maryland) using the manufacturers recommended protocol and were eluted in 40 μL ATE buffer. DNA samples were quantified using Qubit dsDNA HS Assay Kit using manufacturer’s recommended protocol.
Exome sequencing of the tumor and normal tissue of the patients was performed. Genomic DNA purified from FFPE tissues (approximately 100 ng) was sheared in a 130 μl volume using a Covaris S2 Focused-ultrasonicator with the following settings: Intensity 4.0; Duty Cycle 10%; Cycles per Burst 200; Treatment Time 1:10 minutes. Library construction was performed using the Agilent Technologies SureSelect XT Reagent Kit (cat# G9611A) with eight cycles of PCR. The concentration of the amplified library was measured using a Qubit dsDNA HS Assay Kit (ThermoFisher Scientific cat# Q32851). Amplified libraries (750 ng) were enriched for exonic regions using the Agilent Technologies SureSelect (XT) Human All Exon v6+COSMIC kit (cat# 5190–9307) and PCR amplified (10 cycles) using Agilent Herculase II Fusion DNA polymerase (cat# 600677). Enriched libraries were qualified on an Agilent Technologies 2200 TapeStation using a High Sensitivity D1000 ScreenTape assay (cat# 5067–5584 and 5067–5585) and the molarity of adapter-modified molecules was defined by quantitative PCR using the Kapa Biosystems Kapa Library Quant Kit (cat#KK4824). The molarity of individual libraries was normalized to 5 nM, and equal volumes were pooled in preparation for Illumina sequence analysis.
Exome enriched sequencing libraries (25 pM) were chemically denatured and applied to an Illumina HiSeq 2500 v4 paired end flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq PE Cluster Kit v4-cBot (PE-401–4001). Following transfer of the flowcell to an Illumina HiSeq 2500 instrument (HCS v2.2.38 and RTA v1.18.61), a 125-cycle paired-end sequence run was performed using HiSeq SBS Kit v4 sequencing reagents (FC-401–4003).
Exome Analysis
Fastq sequences were aligned to the B37 genome with BWA (v. 0.7.10) and duplicate pairs marked with samblaster (v. 0.1.22). Alignments were polished using GATK (v. 3.4) IndelRealigner and BaseRecalibrator following recommendations from the Utah Genome Project version 1.3.0 pipeline (http://weatherby.genetics.utah.edu/UGP/wiki). Somatic variants were identified in the tumor sample using the normal unassociated sequence as reference using four different variant callers: SomaticSniper (v. 1.0.4) and MuTect (v. 1.1.7) for SNP detection, and Scalpel (v. 0.5.3) and Strelka (v. 1.0.14) for indel detection. Variants from SomaticSniper were filtered for high quality variants using VarScan (v. 2.4.0) false positive filter. Only variants that passed each caller’s quality filters were retained. Variants from each caller were merged into a single VCF file and annotated using Ensembl Variant Effect Predictor (release 87). Damaging variants were selected based on SIFT and PolyPhen prediction scores and occurrence in protein-coding exons. Copy number analysis was performed using CNVKit (v. 0.8.2).
PRDM1 expression in various cancers was queried from the Human Protein Atlas (http://www.proteinatlas.org/ENSG00000057657-PRDM1/cancer#top).
Cell culture and Cell viabilities
Cell Culture
Human bladder cancer cell lines HT1197, T24 and UMUC3, were obtained from ATCC between 2015 and 2018. Cells were cultured at 37 degrees Celsius and 5% CO2 in appropriate media. All cell lines were obtained from ATCC, and cell line authentication was not felt to be necessary. Early passage cells were frozen down and the frozen stock was thawed every 6 months to replenish the cells for experimentation. All experiments were performed from passages 3 to 20. Testing for mycoplasma was performed periodically and last performedin December 2017 using the Lonza MycoAlert Mycoplasma Detection Kit (# LT07–318).
Cell Viability Screens
Cells were seeded in triplicate in 96 well plates at a density of 1000 cells per well and treated the next day with vehicle (1% DMSO) or increasing doses of drug (inhibitor). Cell viability was assessed using CellTiter-Glo (Promega Corporation, Madison, WI, USA) after 72-hour incubation with the drug. Cell line-specific IC50s were calculated using GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA). The following inhibitors were obtained from Selleck Chemicals, Houston, TX, USA and used in viability screens: panobinostat (LBH589) S1030, belinostat (PXD101) S1085, tubastatin A S8049 (25), cisplatin S1166, mocetinostat (MGCD 0103) S1122.
Transcriptomic Profiling (RNA Sequencing and analysis)
HT1197 cells were seeded in triplicate at a density of 1 × 106 in 10 cm dishes and treated the next day with vehicle (1% DMSO) or 2X cell line IC50 for panobinostat and incubated for 48 hours. RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA, USA). Total RNA samples were hybridized with Ribo-Zero Gold to substantially deplete cytoplasmic and mitochondrial rRNA from the samples. Stranded RNA sequencing libraries were prepared as described using the Illumina TruSeq Stranded Total RNA Kit with Ribo-Zero Gold (RS-122–2301 and RS-122–2302). Purified libraries were qualified on an Agilent Technologies 2200 TapeStation using a D1000 ScreenTape assay (cat# 5067–5582 and 5067–5583). The molarity of adapter-modified molecules was defined by quantitative PCR using the Kapa Biosystems Kapa Library Quant Kit (cat#KK4824). Individual libraries were normalized to 10 nM, and equal volumes were pooled in preparation for Illumina sequence analysis.
RNASeq analysis
Fastq sequences were aligned with Novoalign (v. 2.8) against the hg38 genome with splice junctions. Alignments to splice junctions were converted to genomic coordinates with USeq SamTranscriptomeParser (v.8.8.8). Alignments were counted over annotation using Subread featureCounts (v. 1.5.1) and Ensembl annotation (release 87). Genes with differential expression were identified using DESeq2 (v. 1.14.1). The gene expression data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE10392 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103928).
Cell Cycle Studies
Cells were seeded in triplicate at a density of 1 × 106 in 10 cm dishes and treated the next day with vehicle (DMSO) or 12 nM panobinostat for 24 hours. The cells were collected, fixed in ethanol and stained with Propidium Iodide. Cell Cycle analysis was performed using BD Biosciences LSRFortessa flow cytometer.
Western Blot
P21 expression was assessed by seeding cells in triplicate at a density of 1 × 106 in 10 cm dishes and treating the next day with vehicle (DMSO) or 2X and 4X cell line IC50 panobinostat for 48 hours. To assess gammaH2AX expression, HT1197 cells were seeded in triplicate at a density of 2 × 106 in 10 cm dishes and left to adhere for 48 hours and treated for 1 hour at 1X IC50 treatments of panobinostat or belinostat. Protein was extracted from cell lysates and immunoblotted using the following antibodies: β-Actin (8H10D10) Mouse mAb #3700S, P21 Waf1/Cip1 (12D1) Rabbit mAb #2947S (Cell Signaling Technologies, Boston, MA, USA), gamma H2A.X (phospho S139) Rabbit pAb ab2893 (Abcam, Cambridge, MA, USA).
Quantitative real-time PCR
HT1197 cells were plated in 10 cm dishes at a density of 2 × 106 in 10 cm dishes, allowed to lay down for 48 hours, and with vehicle (DMSO) or 10 nM of panobinostat for 48 hours or 1 μM of belinostat for 24 hours. RNA was extracted using RNeasy kit (Qiagen) and reverse transcribed to cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA was amplified, detected, and quantified with TaqMan Gene Expression Master Mix (Applied Biosystems) using ViiA7 Real-Time PCR System. The following TaqMan gene expression assay primers were used: ACTB (Hs01060665_g1 FAM-MGB), TNF (Hs00174128_m1 FAM-MGB), IFNB1 (Hs01077958_s1 FAM-MGB), IFNA2 (Hs00265051_s1 FAM-MGB).
Combination Studies
Information regarding a clinical trial combining HDACI vorinostat with immune checkpoint inhibitor pembrolizumab was obtained from the clinical trials portal of National Cancer Institute (https://clinicaltrials.gov/ct2/show/NCT02619253).
Results
Patients with objective responses to HDAC inhibition had ARID1A mutations in their tumors
Our study schema is presented in supplemental figure 1. Of a total 17 patients with advanced UC, enrolled in either one of the two trials evaluating belinostat and panobinostat, one patient had a CR, one had a PR, and eight patients had stable disease (SD) (22–24). Three patients had progressive disease (PD), and the remaining five patients were not evaluable for response.
Archived tumor tissue was available in only seven (including the two responders) of the 13 evaluable patients. The seven patients with archived tissue availability were chosen purely by tissue and response availability and tissue testing was performed prospectively from that point onwards. The demographic characteristics, treatment histories and best overall responses in these seven patients are summarized in supplemental table 1. A summary comparing the individual drug characteristics, dosing schedule, maximum drug concentrations and protein binding of each of the two drugs is presented in supplemental table 2. The two HDAC inhibitors are noted to be pan-HDAC inhibitors with activity against Classes I, II and IV HDACs. The maximum drug concentrations (Cmax) achieved are tabulated for the two drugs.
Supplemental figure 2 is a swimmer plot representing the best overall response and duration of benefit with treatment. Patient 10 had a CR and durable clinical benefit (greater than 3.5 years and ongoing at the time of this report). Patient 4 had a PR but had to discontinue treatment due to adverse effects.
Patients 1, 2, 5 and 7 had SD, and patient 3 had PD as the best overall response.
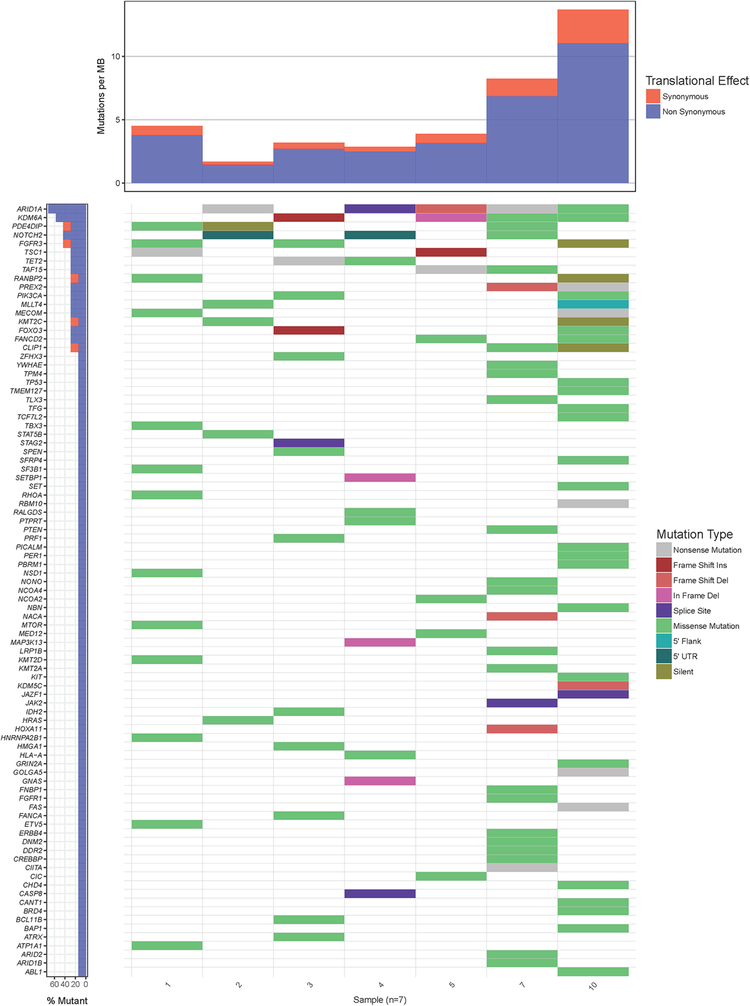
Exome sequencing of tumors revealed multiple genomic alterations (GA), as is typical for UC, depicted in figure 1. The patients with CR and PR (patients 10 and 4, respectively), both had mutations in the ARID1A (AT-Rich Interaction Domain 1A) gene. Patient 10 had the highest TMB of the tumors tested and a FANCD2 (Fanconi Anemia Complementation Group D2) mutation but only at an allele frequency of 23%.
Figure 1.
A waterfall plot of the seven tumors showing cancer causing mutations and tumor mutational burden (TMB). The panel on the left indicates the frequency of genes affected in the 7 tumors. Patients with advanced UC treated with HDACI (belinostat or panobinostat) – referred by numbers indicated at the bottom of the mutation panels. Patient 10 had a complete response to belinostat. Patient 4 had a partial response to panobinostat. Patient 3 had progressive disease. Patients 1, 2, 5 and 7 had stable disease as the best recorded overall response.
Three patients with SD (patients 2, 5 and 7) also had an ARID1A mutation. One patient with PD (patient 3) did not have an ARID1A mutation and had a FANCA (Fanconi Anemia Complementation Group A) mutation (patient 3). Patient 1 had SD for approximately eight months and had mutations in TSC1, mTOR and FGFR3 genes (figure 1).
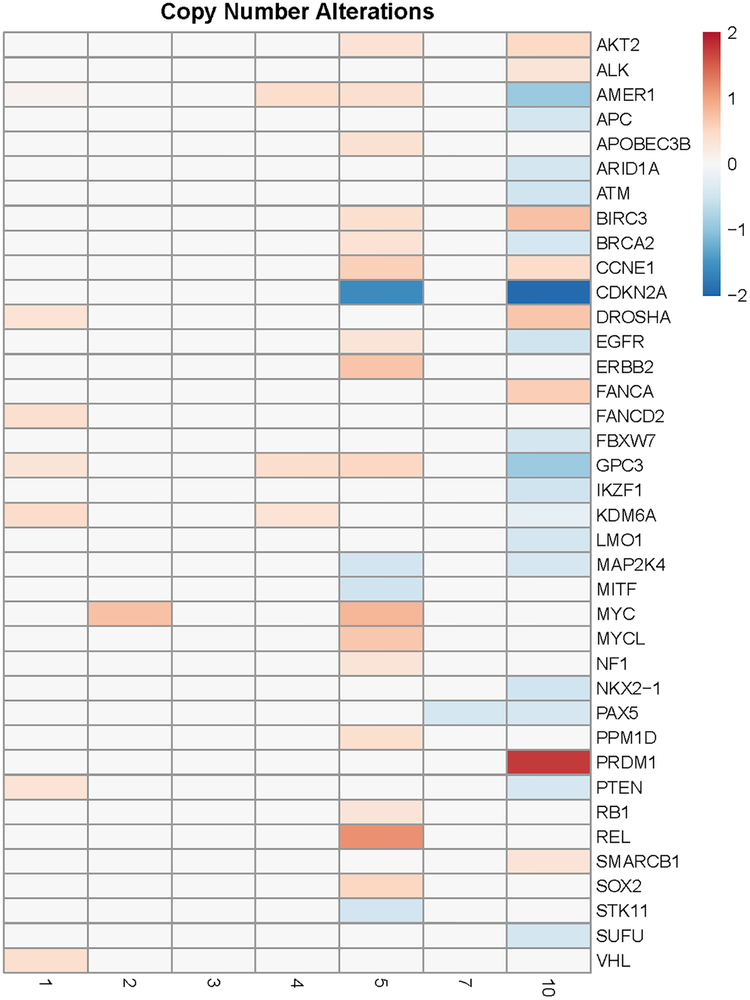
Copy number alterations suggest cell cycle progression, immune escape and DNA repair defects as prominent features of the tumor in the patient who achieved a complete response
Copy number alterations of the tumors are depicted as a tile plot in Figure 2. A log 2 fold (−0.44) copy number deletion of BRCA2 is also noted in Patient 10. A CDKN2A copy number deletion is also noted in the tumor from patient 10. CDKN2A encodes p16 and p14, both of which are tumor suppressors. P16 binds CDK4 and CDK6, which leads to inhibition of cell cycle progression (26). Furthermore, a PRDM1 (Positive Regulatory Domain I-Binding Factor 1) copy number gain is noted. The PRDM1 gene encodes a protein that represses beta-interferon (IFN-β) gene expression by binding to the PRDI (positive regulatory domain I element) of the IFN-β gene promoter.
Figure 2.
A tile plot demonstrating copy number alterations (gain or loss) in the tumors of seven patients with UC treated with HDACI (panobinostat or belinostat) specific to genes implicated in cancer. Patient 4 had PR; Patient 10 had CR; Patient 3 had PD; patients 1, 2, 5 and 7 had SD
Bladder cancer cell lines with diverse genomic alterations and differential sensitivity to cisplatin are very sensitive to HDAC inhibition
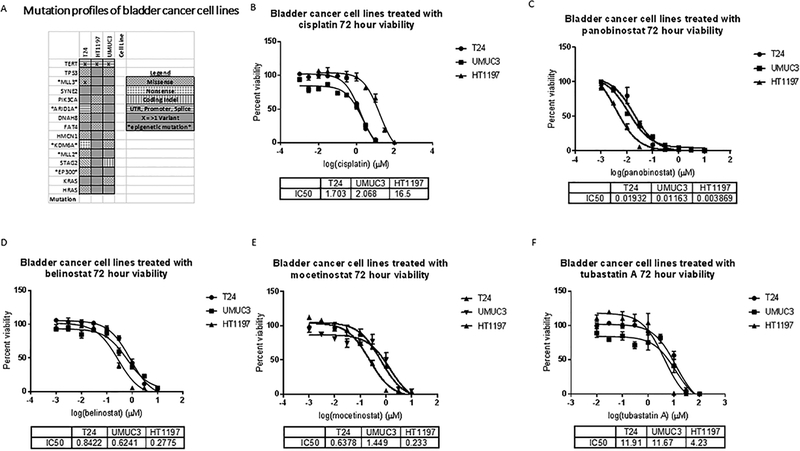
To elucidate the mechanisms of clinical benefit, we evaluated the effect of panobinostat and belinostat on bladder cancer cell lines that have been previously genomically characterized (27). We identified HT1197 as a platinum-resistant UC cell line with an ARID1A mutation and with wildtype (non-mutated) TP53 (figure 3A). T24 is a platinum sensitive ARID1A mutated bladder cancer cell line with several additional genomic alterations including TP53 loss (figure 3A). In addition, we identified UMUC3 as a UC cell line with KDM6A and TP53 mutations (figure 3A) to study alongside the ARID1A mutated lines to isolate the mechanism of targeted benefit in the presence of an ARID1A mutation.
Figure 3.
A. Mutation plots of cell lines adapted from Nickerson, M L et al. “Molecular Analysis of Urothelial Cancer Cell Lines for Modeling Tumor Biology and Drug Response.” Oncogene (2016) (27) under a Creative Commons Attribution- NonCommercial-ShareAlike4.0 International License. Cell viabilities of UMUC3, T24 and HT1197 bladder cancer cell lines treated with increasing doses of B. Cisplatin, C. Panobinostat D. Belinostat, E. Tubastatin, and F. Mocetinostat. Dose response curves showing percent viability normalized to vehicle controls after 72 hours of treatment plotted against the logarithmic drug concentrations. Half-maximal inhibitory concentrations (IC50s) were calculated using GraphPad Prism. The error bars represent biological replicates.
Cell viability assays reveal a differential sensitivity of these cells to cisplatin (figure 3 B). HT1197 is noted to be relatively resistant to cisplatin. All three cell lines were noted to be sensitive to panobinostat and belinostat, in a dose-dependent manner, with IC50s in nanomolar concentrations (figure 3 C, D). Notably, the Cmax achieved for the two drugs (supplemental table 2) are higher than the in vitro IC50 of the two drugs for UC cell lines. When protein binding is taken into account, the free drug Cmax achieved for belinostat clinically is found to be well within the IC50 of all the cell lines studied (~7.5μM). Based on the Cmax values and protein binding ability of panobinostat in patients with normal renal function we calculated 12 nM as the Cmax of unbound drug achieved clinically. Cell cycle studies were performed with this concentration of panobinostat.
A targeted TP53-mediated benefit of HDAC 6 inhibition in ARID1A mutated ovarian carcinoma cells has been reported (28). We proceeded with testing the three cell lines with specific inhibitors of HDAC 6 (tubastatin A), and HDAC classes I and IV (mocetinostat) to identify the HDAC class likely to have contributed to sensitivity HDAC inhibition. We found mocetinostat, causing class I and IV HDAC inhibition, to be growth inhibitory in all tested cell lines (figure 3 E). The specific HDAC6 inhibitor, tubastatin A, was more potent in inhibiting the growth of TP53 intact HT1197 cells (figure 3 F).
HT1197, cell line by virtue of being representative of platinum-resistant UC, with an ARID1A mutation, was chosen to study the transcriptomic changes to explain the mechanism of benefit with HDAC inhibition in ARID1A mutated platinum-resistant UC. In vitro studies were performed with panobinostat as it was found to be the most potent and representative of the hydroxamic class of pan HDAC inhibitors.
Gene expression profiling reveals a multifaceted effect of panobinostat on ARID1A mutated bladder cancer cells
HDAC inhibition leads to extensive transcriptional changes in HT1197 cells with the top 50 affected genes depicted in a heat map format (supplemental figure 3). To define the breadth of pathways affected, we performed Gene Set Enrichment Analysis (GSEA) using the Hallmark gene sets. The Hallmark gene sets is a collection of 50 gene sets in the Molecular Signatures Database (MSigDB), which summarize and represent well-defined biological states or processes with minimal overlap or redundancy. The results of the significantly enriched gene sets (FDR <25%) are summarized in table 1. The top 6 affected signatures are presented in supplemental figures 4 A and B. Notably, proliferative genes represented by the MYC Target V1 and V2 gene sets, and cell cycle propagating genes sets associated with E2F and DNA repair gene sets, were the most significantly negatively enriched gene sets with panobinostat treatment. Several more gene sets were significantly positively enriched, many belonging to the pro-inflammatory pathways. The TP53 signature gene set also showed positive enrichment (table 1).
Table 1.
Significantly enriched Hallmark gene sets in HT1197 cells treated with panobinostat for 48 hours
| Downregulated Gene Sets | Gene Set | SIZE | NES | NOM p-val | FDR q-val |
| 1 | HALLMARK_MYC_TARGETS_V2 | 58 | −2.77 | 0 | 0 |
| 2 | HALLMARK_MYC_TARGETS_V1 | 199 | −2.73 | 0 | 0 |
| 3 | HALLMARK_E2F_TARGETS | 199 | −2.58 | 0 | 0 |
| 4 | HALLMARK_G2M_CHECKPOINT | 197 | −1.93 | 0 | 0.002 |
| 5 | HALLMARK_OXIDATIVE_PHOSPHORYLATION | 195 | −1.27 | 0 | 0.134 |
| 6 | HALLMARK_DNA_REPAIR | 141 | −1.25 | 0 | 0.131 |
| Upregulated Gene Sets | Gene Set | SIZE | NES | NOM p-val | FDR q-val |
| 1 | HALLMARK_INTERFERON_ALPHA_RESPONSE | 59 | 1.79 | 0 | 0.002 |
| 2 | HALLMARK_INFLAMMATORY_RESPONSE | 104 | 1.78 | 0 | 0.001 |
| 3 | HALLMARK_COAGULATION | 74 | 1.64 | 0.001 | 0.01 |
| 4 | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 152 | 1.63 | 0 | 0.009 |
| 5 | HALLMARK_ALLOGRAFT_REJECTION | 98 | 1.6 | 0.001 | 0.011 |
| 6 | HALLMARK_IL6_JAK_STAT3_SIGNALING | 48 | 1.59 | 0.005 | 0.011 |
| 7 | HALLMARK_APICAL_JUNCTION | 154 | 1.54 | 0.002 | 0.016 |
| 8 | HALLMARK_INTERFERON_GAMMA_RESPONSE | 118 | 1.54 | 0 | 0.014 |
| 9 | HALLMARK_APOPTOSIS | 127 | 1.52 | 0.001 | 0.016 |
| 10 | HALLMARK_IL2_STAT5_SIGNALING | 144 | 1.5 | 0 | 0.019 |
| 11 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 146 | 1.48 | 0.003 | 0.024 |
| 12 | HALLMARK_PANCREAS_BETA_CELLS | 24 | 1.45 | 0.046 | 0.036 |
| 13 | HALLMARK_XENOBIOTIC_METABOLISM | 135 | 1.43 | 0.005 | 0.043 |
| 14 | HALLMARK_ESTROGEN_RESPONSE_LATE | 146 | 1.42 | 0.004 | 0.04 |
| 15 | HALLMARK_MYOGENESIS | 143 | 1.42 | 0.004 | 0.04 |
| 16 | HALLMARK_KRAS_SIGNALING_UP | 120 | 1.42 | 0.008 | 0.037 |
| 17 | HALLMARK_KRAS_SIGNALING_DN | 98 | 1.4 | 0.012 | 0.046 |
| 18 | HALLMARK_SPERMATOGENESIS | 86 | 1.4 | 0.024 | 0.044 |
| 19 | HALLMARK_ANGIOGENESIS | 26 | 1.4 | 0.07 | 0.043 |
| 20 | HALLMARK_BILE_ACID_METABOLISM | 76 | 1.37 | 0.031 | 0.051 |
| 21 | HALLMARK_HYPOXIA | 167 | 1.36 | 0.006 | 0.057 |
| 22 | HALLMARK_P53_PATHWAY | 169 | 1.35 | 0.013 | 0.063 |
| 23 | HALLMARK_NOTCH_SIGNALING | 30 | 1.32 | 0.111 | 0.078 |
| 24 | HALLMARK_ESTROGEN_RESPONSE_EARLY | 156 | 1.3 | 0.024 | 0.098 |
| 25 | HALLMARK_PEROXISOME | 89 | 1.28 | 0.085 | 0.108 |
Comparison of the expression profile of HT1197 and UMUC3 cell lines revealed a vast difference in the expression profile of the two cell lines. Specifically, E2F targets gene set was significantly more expressed in HT1197 cells compared to UMUC3 cells (Supplemental figure 4 C). Not surprisingly, the TP53 gene set was also significantly more expressed in HT1197 cells (TP53 intact) compared to UMUC3 cells(TP53 mutated).
Panobinostat treatment causes cell cycle arrest in TP53 intact and TP53 mutated bladder cancer cell lines and causes an increase in p21 levels in all three cell lines
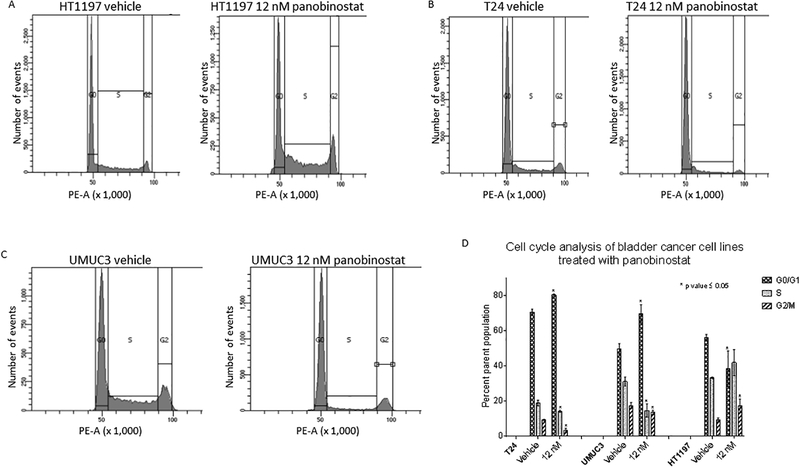
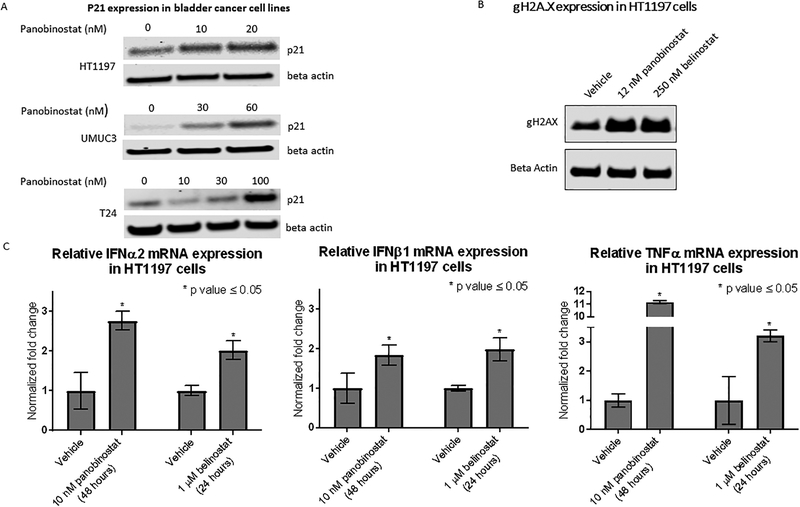
UMUC3 and T24 are TP53 mutated, and HT1197 is TP53 intact (figure 3 A). Cell Cycle studies performed on the three cell lines using panobinostat reveal cell cycle arrest in the G2/M stage in HT1197 cells (figure 4 A) and the G0/G1 stage in T24 and UMUC3 cells (figure 4 B and C). An aggregate of three biological replicates for each cell line shows the difference in cell populations to be statistically significant (figure 4 D). A dose-dependent increase in p21 levels in HT1197, T24 and UMUC3 cells is seen with panobinostat treatment (figure 5 A).
Figure 4.
Cell cycle changes by flow cytometry using 12 nM panobinostat in A. HT1197, B. UMUC3, and C. T24 vs vehicle treatment for 24 hours. D. Changes in percent population of cells quantified by graph pad prizm. The change in the cell population distribution was replicated for a total of three times. An unpaired student t-test to compare the percent population with and without treatment showed the changes in G0/G1 and G2/M populations to be statistically significant. The error bars represent biological replicates.
Figure 5.
A. Western blot to assess p21 levels in bladder cancer cell lines treated with panobinostat at 2 and 4 times the IC50 dose for 48 hours. This was repeated for a total of three times B. Western blot showing accumulation of gH2Ax marks in HT1197 upon treatment with panobinostat and belinostat. This was repeated for a total of three times. C. RT PCR to assess expression of TNFα, IFNβ1 and IFNα2 in HT1197 cells treated with panobinostat and belinostat. Results from three biological replicates are represented with error bars and the change in expression is tested for significance by the unpaired t-test (Graphpad Prism).
The pathways affected by HDAC inhibition inform choices about rational therapeutic combinations
Noting the negative enrichment for DNA repair pathway gene set, we immunoblotted for gamma H2AX (a byproduct of double-stranded DNA breaks) and observed increased density upon treatment with panobinostat and belinostat (figure 5 B). Given positive enrichment for multiple inflammatory gene sets on GSEA, we evaluated the expression of TNFα, IFNβ1 and IFNα2 in HT1197 cells treated with panobinostat and belinostat by RT-PCR. An increase in mRNA levels of three pro-inflammatory cytokines occurs with both of these drugs (figure 5 C).
Discussion
In two separate early phase advanced solid tumor trials using two different pan HDACIs of the hydroxamic acid group, two responders were identified (supplemental figure 1). Both responders had platinum-resistant disease. ARID1A mutation emerged as the common biomarker in these two responders (figure1). Besides the responders, three patients studied (patients 2, 5 and 7) had ARID1A mutations (figure 1) and SD with PFS ranging from 3.5 to 6 months (supplemental figure 2).
ARID1A encodes a protein component of the ATP dependent chromatin remodeling SWI/SNF complex, influencing nucleosome-DNA topology and exerting transcriptional control. ARID1A acts as a tumor suppressor in a ‘gatekeeper’ capacity by repressing E2F-mediated cell cycle progression and, myc-mediated cell proliferation, bringing about p21 induction (29). Additionally, ARID1A regulates p21 in a TP53-dependent fashion. Supporting our findings on the tumor tissue genome, HT1197 and T24, both ARID1A mutated UC cell lines (figure 3 A), when treated with panobinostat, are exquisitely sensitive, with IC50 in nanomole concentrations (figure 3C). The gene expression profile changes seen in HT1197 with panobinostat show that MYC, TP53 and E2F target gene sets (table 1, supplemental figure 4 A, B) are primarily affected, a benefit apropos to ARID1A mutated states. In summary, the multiple roles of ARID1A as a tumor suppressor coincide exactly with the multiple antitumor effects of pan HDAC inhibition. Our cell line expression studies comparing ARID1A mutated HT1197 cells and ARID1A wildtype UMUC3 cells reveal a more significant presence of the E2F target signature in ARID1A mutated cells, underpinning the attribution of “targeted” clinical benefit (supplemental figure 4C). In summary, the oncogenic pathways affected by ARID1A loss are largely targeted by pan HDAC inhibition, and this may partly explain the targeted clinical benefit seen in ARID1A mutated bladder cancer.
ARID1A loss affects the ability of the SWI/SNF chromatin remodeling complex to perform its DNA repair functions, making the cell dependent on alternative DNA repair pathways (30). As noted in table 1, treatment with panobinostat results in significant downregulation of DNA repair gene sets in HT1197 cells. Synthetic lethality of DNA repair downregulation in the presence of a DNA repair defect is a possible mechanism of clinical benefit with HDAC inhibition seen in tumors with ARID1A loss. We see an accumulation of gH2AX marks representing DNA damage in HT1197 cells upon treatment with panobinostat and belinostat (figure 5 B).
ARID1A is frequently mutated across human cancers, most prominently in ovarian clear cell carcinomas (range 46 to 57%) (31), and is associated with progressively aggressive forms of UC (32). TCGA data on muscle-invasive bladder cancer quantitates ARID1A loss at 25 % of these tumors (33). Comprehensive genomic profiling of advanced UC tissue has revealed a similar prevalence of ARID1A mutations (26%). In a separate study evaluating the prevalence of ARID1A mutation in various stages and grades of UC, ARID1A mutations were identified in 23% of high-grade non-muscle invasive bladder tumors and associated with a higher risk of recurrence after BCG treatment (34). ARID1A loss tumors are scattered across the various expression clusters of UC. Multiple additional genomic alterations result in a varied group of tumors (8).
In ovarian and endometrial cancer in which ARID1A and TP53 mutations are mutually exclusive (35), ARID1A loss has been shown to inactivate the apoptosis-promoting function of TP53 by up-regulating HDAC6. Hence there is a case for specific HDAC 6 inhibition as a therapeutic strategy in ARID1A mutated ovarian cancers (28). In contrast, TP53 mutations are present in about half of muscle-invasive UC and can overlap with ARID1A loss. T24 cells are ARID1A mutated but have multiple other genomic alterations including TP53. The IC50 for panobinostat is also higher, likely due to lack of functional TP53 (figure 3A). Cell viabilities reveal cytotoxicity with specific inhibitors of HDAC 6 and Class I/ IV, but maximum cytotoxicity is seen with pan HDAC inhibition (figure 3 C, D, E, F). The cell cycle is arrested with 12nM of panobinostat in both T24 and HT1197 cells but is arrested in G0/G1 in T24 and G2M in HT1197 cells (figure 4 A–D). P21 induction is noted in TP53 wildtype and TP53 mutated bladder cancer cells treated with panobinostat (figure 5 A). In conclusion, pan HDAC inhibition rather than class-specific HDAC inhibition is more likely to be clinically effective in UC given the presence of overlapping ARID1A and TP53 mutations.
With regard to additional GA noted in patient 10, CDKN2A loss is present in 47% of UC (33). A CDKN2A copy number deletion suggests cell cycle checkpoint inhibition as a mechanism of tumor growth. A BRCA 2 copy number (homozygous) deletion in the tumor (figure 2) suggests an acquired tumor DNA repair defect which may have contributed to the TMB and immunogenicity with the creation of neoantigens. PRDM1 amplification suggests immune escape as a possible tumor survival pathway. Overexpression of PRDM1 is typically noted in head and neck, lung and skin tumors, as well as melanoma. PRDM1 overexpression has been shown in some UC cases as well (36). It is possible that PRDM1 gene amplification may play a role in immune evasion by tumors by suppressing the expression of IFN-β. Inflammatory gene sets are significantly upregulated upon treatment of HT1197 cells with panobinostat (table 1). Specifically, IFN-β expression increases with panobinostat and belinostat in HT1197 cells (figure 5 C). The durable benefit with belinostat to patient 10 with a tumor with moderate TMB indicates an immune-mediated benefit as well. Thus, characterization of this patient’s tumor corroborates several additional mechanisms of HDAC inhibition efficacy in the clinical setting. When patient 10 is compared to patient 4 (who had a PR on panobinostat), ARID1A mutation stands out as the common genomic alteration in the two responders.
While ARID1A loss appears to be predictive of clinical benefit (CR, PR, SD) from HDAC inhibition, it’s possible that the benefit from HDAC inhibition is not exclusive to this particular GA. Patient 1 had GA involving TSC1, mTOR and FGFR3 genes (figure 1) and eight months of stable disease on HDAC inhibition (supplemental figure 2). The strong cytotoxicity and p21 induction in ARID1A intact UMUC3 cells indicate in vitro efficacy of panobinostat in a variety of mutational landscapes. On comparing the expression profile of HT1197 and UMUC3 cells, we found the E2F target gene set is more strongly expressed in HT1197 cells (supplemental figure 4C). Downregulation of E2F target genes are a significant signature effect of HDACIs (table 1) and can account for the targeted benefit seen in ARID1A mutated UC.
Historically, HDACIs have shown promising preclinical efficacy in solid tumors but mostly negative clinical trials for efficacy (37). Complex effects of HDAC inhibitors, with no targeting specificity apparent in preclinical studies, underlines the importance of sequencing the tumor genome of the rare responders. Our study indicates clinical evidence of response (CR and PR) specifically in patients with ARID1A gene mutations (patients 4 and 10), using two different pan HDACIs (figure 1) with different Cmax and potency (supplemental table 2). Our findings strongly indicate a rather specific targeted benefit with HDACIs in the setting of ARID1A loss.
In the two clinical trials analyzed here, of the patients who derived clinical benefit, some discontinued treatment due to side effects despite PR or stable disease (supplemental figure 2). Rational therapeutic combination therapies can improve the response rate at lower doses of HDAC inhibition and expand therapeutic benefit to UC with other genomic alterations.
Our findings of complete and durable benefit of HDAC inhibition corresponding with the degree of TMB (figure1), HDACIs resulting in upregulation of inflammatory gene sets (table 1) and increased expression of inflammatory cytokines (figure 5 C), theorize a possible role of HDAC inhibition in enhancing immunogenicity and thus enhancing responses of UC to immune checkpoint inhibition. A clinical trial combining vorinostat and pembrolizumab in UC is currently underway and is likely to shed light on the efficacy of the combination. Based on the observation of DNA repair inhibition by HDACIs, combinations with DNA damaging agents or with DNA repair inhibitors may be explored. A limitation of our study is the reliance upon availability of archived tissue; hence, tumor tissue could be obtained for seven out of 12 evaluable patients only. There may be other factors such as DNA methylation that may confer sensitivity to HDAC inhibition in UC, which we have not explored in our study, as our patient cohort was limited.
Our study has, for the first time, identified ARID1A gene mutation as a biomarker for clinical response and benefit from HDAC inhibition in patients with advanced UC. ARID1A is one of the most prominent chromatin remodeling genomic alterations seen in UC and is present in approximately 25% of UC. The genomic complexity of UC with multiple overlapping genomic alterations, limits the efficacy of targeting a single mutation. Our study provides grounds for biomarker enrichment and combination strategies in future clinical trials of HDACIs in UC.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Neeraj Agarwal for providing samples and clinical data for this project and supporting this project.
The authors would also like to thank the patients and their families who contributed to the research.
Research Support
The work described in the manuscript was supported by the faculty development funds from the Huntsman Cancer Institute/Huntsman Cancer Foundation and Department of Medicine at University of Utah and a grant from the Genitourinary Malignancy Disease Oriented Team (GUMDOT) at Huntsman Cancer Institute.
The project described was also supported by the High-Throughput Genomics and Bioinformatic Analysis core at Huntsman Cancer Institute supported by Award Number P30CA042014 from the National Cancer Institute. This work was supported by the University of Utah Flow Cytometry Facility in addition to the National Cancer Institute through Award Number 5P30CA042014–24. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Conflict of Interest
The authors report no conflict of interest with regard to the work presented in this manuscript.
References
- 1.Cancer Facts & Figures 2018 [Internet]. American Cancer Society, Inc; 2018. [Google Scholar]
- 2.von der Maase H, Hansen SW, Roberts JT, Dogliotti L, Oliver T, Moore MJ, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol. 2000;18(17):3068–77. [DOI] [PubMed] [Google Scholar]
- 3.Sternberg CN, de Mulder PH, Schornagel JH, Theodore C, Fossa SD, van Oosterom AT, et al. Randomized phase III trial of high-dose-intensity methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC) chemotherapy and recombinant human granulocyte colony-stimulating factor versus classic MVAC in advanced urothelial tract tumors: European Organization for Research and Treatment of Cancer Protocol no. 30924. J Clin Oncol. 2001;19(10):2638–46. [DOI] [PubMed] [Google Scholar]
- 4.Sternberg CN, de Mulder P, Schornagel JH, Theodore C, Fossa SD, van Oosterom AT, et al. Seven year update of an EORTC phase III trial of high-dose intensity M-VAC chemotherapy and G-CSF versus classic M-VAC in advanced urothelial tract tumours. Eur J Cancer. 2006;42(1):50–4. [DOI] [PubMed] [Google Scholar]
- 5.von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, et al. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23(21):4602–8. [DOI] [PubMed] [Google Scholar]
- 6.Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee J-L, Fong L, et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. New England Journal of Medicine. 2017;376(11):1015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross JS, Wang K, Khaira D, Ali SM, Fisher HAG, Mian B, et al. Comprehensive genomic profiling of 295 cases of clinically advanced urothelial carcinoma of the urinary bladder reveals a high frequency of clinically relevant genomic alterations. Cancer. 2016;122(5):702–11. [DOI] [PubMed] [Google Scholar]
- 10.Nogova L, Sequist LV, Garcia JMP, Andre F, Delord J-P, Hidalgo M, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1–3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. Journal of Clinical Oncology. 2017;35(2):157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagle N, Grabiner BC, Van Allen EM, Hodis E, Jacobus S, Supko JG, et al. Activating mTOR Mutations in a Patient with an Extraordinary Response on a Phase I Trial of Everolimus and Pazopanib. Cancer Discovery. 2014;4(5):546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome Sequencing Identifies a Basis for Everolimus Sensitivity. Science (New York, NY). 2012;338(6104):221-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ververis K, Hiong A, Karagiannis TC, Licciardi PV. Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents. Biologics : Targets & Therapy. 2013;7:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapani D, Esposito A, Criscitiello C, Mazzarella L, Locatelli M, Minchella I, et al. Entinostat for the treatment of breast cancer. Expert Opin Investig Drugs. 2017:1–7. [DOI] [PubMed] [Google Scholar]
- 15.Tang F, Choy E, Tu C, Hornicek F, Duan Z. Therapeutic applications of histone deacetylase inhibitors in sarcoma. Cancer Treat Rev. 2017;59:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li QQ, Hao J-J, Zhang Z, Hsu I, Liu YI, Tao Z, et al. Histone deacetylase inhibitor-induced cell death in bladder cancer is associated with chromatin modification and modifying protein expression: A proteomic approach. International Journal of Oncology. 2016;48(6):2591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckley MT, Yoon J, Yee H, Chiriboga L, Liebes L, Ara G, et al. The histone deacetylase inhibitor belinostat (PXD101) suppresses bladder cancer cell growth in vitro and in vivo. Journal of Translational Medicine. 2007;5(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly WK, Richon VM, O’Connor O, Curley T, MacGregor-Curtelli B, Tong W, et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin Cancer Res. 2003;9(10 Pt 1):3578–88. [PubMed] [Google Scholar]
- 19.Duex JE, Swain KE, Dancik GM, Paucek RD, Owens C, Churchill MEA, et al. Functional Impact of Chromatin Remodeling Gene Mutations and Predictive Signature for Therapeutic Response in Bladder Cancer. Molecular Cancer Research. 2018;16(1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou N, Moradei O, Raeppel S, Leit S, Frechette S, Gaudette F, et al. Discovery of N-(2-aminophenyl)-4-[(4-pyridin-3-ylpyrimidin-2-ylamino)methyl]benzamide (MGCD0103), an orally active histone deacetylase inhibitor. J Med Chem. 2008;51(14):4072–5. [DOI] [PubMed] [Google Scholar]
- 21.Noah M Hahn JP, Richard Martin Bambury, Sumanta Kumar Pal, Lowell L. Hart, Petros Grivas, Matthew I. Milowsky, Ajjai Shivaram Alva, Guru Sonpavde, Amir Mortazavi, Joaquim Bellmunt, Elizabeth A. Guancial, Sumati Gupta, Richard C. Chao, Mary A. Collier, James Christensen, Isan Chen, Jonathan E. Rosenberg. A phase 2 study of the histone deacetylase (HDAC) inhibitor mocetinostat in patients with urothelial carcinoma (UC) and inactivating alterations of acetyltransferase genes. J Clin Oncol 33, 2015. (suppl; abstr TPS4575). 2015. [Google Scholar]
- 22.Agarwal N, McPherson JP, Bailey H, Gupta S, Werner TL, Reddy G, et al. A phase I clinical trial of the effect of belinostat on the pharmacokinetics and pharmacodynamics of warfarin. Cancer Chemother Pharmacol. 2016;77(2):299–308. [DOI] [PubMed] [Google Scholar]
- 23.Bailey H, McPherson JP, Bailey EB, Werner TL, Gupta S, Batten J, et al. A phase I study to determine the pharmacokinetics and urinary excretion of belinostat and metabolites in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2016/October/28 ed2016. p. 1059–71. [DOI] [PubMed] [Google Scholar]
- 24.Sharma S, Witteveen PO, Lolkema MP, Hess D, Gelderblom H, Hussain SA, et al. A phase I, open-label, multicenter study to evaluate the pharmacokinetics and safety of oral panobinostat in patients with advanced solid tumors and varying degrees of renal function. Cancer Chemother Pharmacol. 2015;75(1):87–95. [DOI] [PubMed] [Google Scholar]
- 25.Leonhardt M, Sellmer A, Krämer OH, Dove S, Elz S, Kraus B, et al. Design and biological evaluation of tetrahydro-β-carboline derivatives as highly potent histone deacetylase 6 (HDAC6) inhibitors. European Journal of Medicinal Chemistry. 2018;152:329–57. [DOI] [PubMed] [Google Scholar]
- 26.Canepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, et al. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59(7):419–26. [DOI] [PubMed] [Google Scholar]
- 27.Nickerson ML, Witte N, Im KM, Turan S, Owens C, Misner K, et al. Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitler BG, Wu S, Park PH, Hai Y, Aird KM, Wang Y, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu R-C, Wang T-L, Shih I-M. The emerging roles of ARID1A in tumor suppression. Cancer Biology & Therapy. 2014;15(6):655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discovery. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shain AH, Pollack JR. The Spectrum of SWI/SNF Mutations, Ubiquitous in Human Cancers. PLoS ONE. 2013;8(1):e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balbas-Martinez C, Rodriguez-Pinilla M, Casanova A, Dominguez O, Pisano DG, Gomez G, et al. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8(5):e62483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. Next-generation Sequencing of Nonmuscle Invasive Bladder Cancer Reveals Potential Biomarkers and Rational Therapeutic Targets. European urology.72(6):952–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan B, Wang T-L, Shih I-M. ARID1A, a Factor That Promotes Formation of SWI/SNF-Mediated Chromatin Remodeling, Is a Tumor Suppressor in Gynecologic Cancers. Cancer Research. 2011;71(21):6718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 37.Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: a review of clinical trials. Clinical Epigenetics. 2015;7:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.